Abstract
Loss of function in the kinase IRAK-4 or the adapter MyD88 in humans interrupts a pathway critical for pathogen sensing and ignition of inflammation. Yet patients with loss of function mutations are surprisingly only susceptible to a limited range of pathogens. We employed a systems approach to investigate transcriptome responses following in vitro exposure of patients’ blood to Toll-like receptor and interleukin-1 receptor agonists, and whole pathogens. Responses to purified agonists were globally abolished but variable residual responses were present following exposure to whole pathogens. Further dissection of the latter responses identified a narrow repertoire of immune transcriptional programs affected by loss of MyD88 or IRAK-4 function. This work introduces the use of a systems approach for the global assessment of innate immune responses, and the characterization of human primary immunodeficiencies.
Keywords: Toll-like receptors, Toll-like-IL-1 receptors, blood profiling, primary immunodeficiencies, MyD88, IRAK-4, human, microarray, innate immunity
Studies of children with recurrent invasive pneumococcal infection have led to the discovery of two primary immunodeficiencies (PIDs), caused by autosomal recessive mutations in IRAK4 and MYD88, which impair Toll-like and Interleukin 1 family Receptors (TIR) signaling1, 2. Toll-like receptors (TLRs) act as sensors of the innate immune system by recognizing specific components conserved among a variety of microorganisms that invade the host. The ligation of TLRs by their agonists induces an inflammatory response to control the infection3. In humans 10 functional members of the TLR family have been identified (TLR1-10). MyD88 is a key downstream adapter for most interleukin-1 receptors (IL-1Rs) and all TLRs except for TLR3 and partially TLR4; IRAK-4 is selectively recruited to TLRs and IL-1Rs by MyD884–6. Thus, loss of IRAK-4 or MyD88 function interrupts a pathway critical for pathogen sensing (TLR) and inflammation (IL-1R), known as TIR deficiency7. Indeed, animal models indicate that deficiency in either one of the two molecules results in heightened susceptibility to a wide range of pathogens8. Yet surprisingly MyD88- and IRAK-4-deficient patients display a narrow and transient infectious phenotype mostly limited to Gram-positive bacteria, Streptococcus pneumoniae and Staphylococcus aureus, and to Gram-negative bacteria, Pseudomonas aeruginosa, in particular, and mostly from birth until adolescence when the condition improves8–10.
The contrast between a broad and profound immunological phenotype and a narrow and transient infectious phenotype in these patients is intriguing. When assessing immune competency of patients with PIDs the choice of the panel of analytes used as readout is crucial. Indeed, a normal response does not exclude the possibility of a defect affecting a pathway that is not covered by the panel. Conversely, a defective response detected using a given panel does not exclude the possibility of a conserved, potentially redundant, response being present. Adopting systems-scale profiling approaches as the readout can address such limitations. A commonly used systems approach is ex-vivo blood transcriptome profiling. This approach has led to the identification of novel therapeutic targets and the development of biomarker signatures11–15. Subsequently the same approach was adopted to investigate responses to vaccines16, 17.
We have developed here an unbiased approach to evaluate the transcriptional profiles of blood cells from MyD88- and IRAK-4-deficient patients in response to a broad array of TLR- and IL-1R-agonists as well as whole pathogens (bacteria, virus, and fungus). Our hypothesis was that MyD88- and IRAK-4-dependent and independent immunological mechanisms of recognition of pathogens would be revealed in vitro, which could contribute to understanding the in vivo protection against most microbes in these patients. This approach allowed the assessment of the range of responses that can be elicited in the absence of functional IRAK-4 or MyD88 and provided insights into essential mechanisms for the maintenance of immunity to pathogens.
RESULTS
In vitro transcriptome responses to purified TIR agonists
Before evaluating the impact of MyD88 and IRAK-4 deficiencies on innate immune function we set out to establish in a set of control subjects the baseline response elicited by the engagement of purified TIR agonists We measured transcriptional responses to TIR agonists in 14 control subjects on a genome-wide scale. Whole blood was stimulated in vitro for 2 hours with an array of agonists spanning all TLRs (PAM3 (TLR1/2), PAM2 (TLR2/6), Poly (I:C) (TLR3), LPS (TLR4), Flagellin (TLR5), 3M2 (TLR7), 3M13 (TLR8), R848 (TLR7/8), CpG-D19, CpG-C (TLR9), and IL-1Rs (IL-1β, IL-18, IL-33) along with two positive controls, tumor necrosis factor (TNF) and phorbol ester (PMA) plus ionomycin. Transcripts displaying consistent differences in stimulated expression levels across healthy control subjects were selected as described earlier (in 2 and online Methods). This filter identified sets of transcripts responding to each stimulation. They included a number of known chemokines, cytokines, co-stimulatory molecules, antibacterial peptides and transcription regulators involved in the TIR signaling pathway3, 5. Pathway analysis confirmed that immune cell trafficking and inflammatory response genes were significantly over-represented across these gene lists (p<0. 0001) (Ingenuity pathway analysis software, Ingenuity Systems, www.ingenuity.com), as well as type I interferon (IFN) signaling for LPS, Poly (I:C), 3M13, 3M2 and R848 stimulations (p<0. 001). The magnitude of transcriptional changes varied for each stimulus: LPS, R848 and TNF induced stronger responses (713, 535 and 550 probes, respectively) and PAM3, PAM2, 3M13, IL-1β and IL-18 lower responses (101, 183, 80, 99 and 119 probes, respectively). Several transcripts were found to be overlapping among stimuli. For instance, 70 annotated probes (59 genes) were induced by all TLR agonists except Poly (I:C) (non-specific TLR3 ligand for which signaling is MyD88-independent) (Supplementary Fig. 1). CpG-A (D19), CPG-C and IL-33 were weak inducers at this early time-point and these conditions were not included in further analyses. This first step established the in vitro blood transcriptional response to TIR agonists in healthy individuals.
Characterizing responses in patients with TIR deficiency
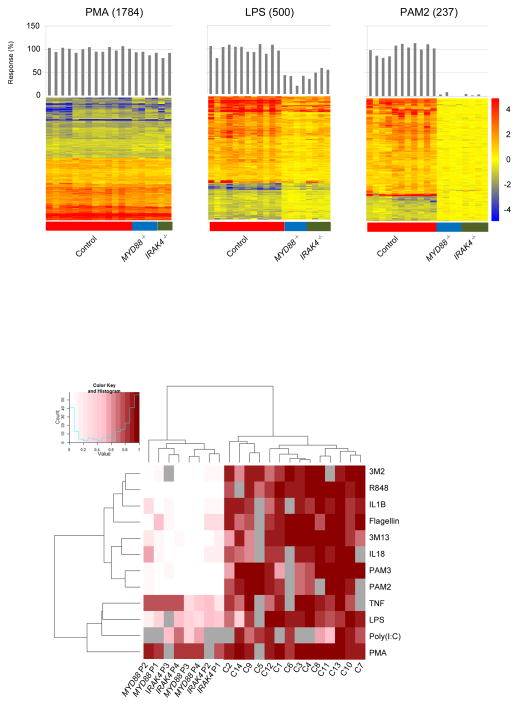
Next we assessed the ability of IRAK-4- and MyD88-deficient patients to respond following engagement of TIRs by purified agonists. Our initial cohort included 4 patients with IRAK-4 deficiency and 4 patients with MyD88 deficiency (patients P1 to P4, Table 1). All IRAK4 and MYD88 mutations were loss-of-function1, 2, 9. All patients were asymptomatic and without any active infectious process at the time of sample collection. They were aged 1–18 years. The responses to PMA + ionomycin (positive control) and Poly (I:C) (non-specific TLR3 ligand for which signaling is MyD88-independent), were conserved -- 92. 4% of transcripts responsive in healthy controls were also found in patients. The response to LPS (partially MyD88-dependent) was reduced but not abolished (40. 2% of healthy response). However, a dramatic drop in the number of responsive transcripts was observed in both IRAK-4- and MyD88-deficient patients to PAM3 (TLR1/2 agonist; 8. 8% of healthy response), PAM2 (TLR2/6 agonist; 6% of healthy response), Flagellin (TLR5 agonist; 19. 7% of healthy response), 3M13 (TLR7 agonist;13. 8% of healthy response), 3M2 (TLR8 agonist; 16. 3% of healthy response), R848 (TLR7/8 agonist; 3. 9% of healthy response), IL-1β (IL-1R agonist; 19. 5% of healthy response) and IL-18 (IL-18R agonist; 23. 4% of healthy response). The extent of the defects for each given stimulus was consistent across patients both quantitatively and qualitatively (Fig. 1a and Supplementary Fig. 2). When all stimuli were considered together, clustering according to levels of responsiveness across subjects resulted in a clear separation between MyD88-dependent and MyD88-independent signals (Fig. 1b). Conversely, clustering subjects across all stimuli resulted as expected in a clear separation between patients and controls. No differences in responsiveness were observed between MyD88- and IRAK-4-deficient patients. The residual levels of response (10–20%) observed for IL-1β and Flagellin could be explained because the agonists used were E. coli derived, and, despite adding polymyxin B to the culture, minimal levels of LPS may be present, and thus, activation might have occurred through TLR4 engagement. Indeed, this residual response was not observed (<10%) when the agonists used were not produced in bacteria (PAM2, PAM3, R848). In summary, whole genome transcriptional responses to MyD88-dependent TIR agonists were nearly abolished in patients compared to controls. This finding unequivocally demonstrates that the loss of signaling downstream of TIRs resulting from MyD88 and IRAK-4 deficiency remains uncompensated in these patients.
Table 1.
Patient information.
| Batch 1 | Batch 2 | Batch 3 | |
|---|---|---|---|
| IRAK4−/− |
P11, 9 male, age
12yo; Mutation: 1188+520A>G/1189- 1G>T; Splice mutation;
on IVIG P2 1, 9 male, age 12yo; Mutation: E402X/E402X; Nonsense mutation; on Cotrimoxazole P3 1, 9 male, age 18yo; Mutation: Q293X/BAC210N13del; Nonsense mutation/large deletion; on Cotrimoxazole P4 9 female, age 14yo; Mutation:Q293X/Q293X; Nonsense mutation; on Azythromycin and IVIG |
|

|
| MYD88−/− |

|

|

|
| Controls | 14 (6 for bacterial stimulation) | 5 | 8 (2 young children) |
| Stimulations | Toll-IL1R (PAM3, PAM2, PolyIC, Flagellin, LPS, R848, CpGs, IL1R, IL18R) + TNFa + PMAiono + 4 HK bacteria1 for IRAK-4P3, P4 and MyD88 P3, P4. | Toll-R (PAM2, LPS) + TNFa + PMAiono + 4HK bacteria1 all patients | Toll-R (PAM2, PAM3, PolyIC, R848) + PMAiono + 4HK bacteria1 + BCG + HSV + HK. CA all patients |
siblings. In blue, patients reanalyzed in different batches to include new pathogen stimulations. IVIG: intravenous immunoglobulin.
4 HK bacteria include 3 strains of heat killed S. pneumoniae and S. aureus.
Figure 1.
Blood transcriptional responses following in vitro exposure to purified TLR or IL-1R agonists. (a) Changes in expression levels of transcripts responsive to TIR agonists are represented on a heatmap. Blood from healthy controls or patients was stimulated in vitro for 2 hours with TLR agonists and cytokines (batch 1); responsive transcripts were arranged by rows via hierarchical clustering (1784, 500 and 237 for PMA + ionomycin, LPS and PAM2 stimulations, respectively), and individual subjects by columns from left to right: healthy controls (n=14), MYD88−/− patients (n=4), IRAK4−/− patients (n=4). Changes versus the non-stimulated condition are represented by a color scale: red = up-regulated; blue = down-regulated; yellow = no change. Bar graphs represent overall individual levels of responsiveness relative to the average of controls: the number of responsive probes in a given subject/average of differentially expressed probes in healthy controls x100. Responses to 8 additional TIR agonists and TNF are presented similarly in Supplementary Fig 2a. (b). The overall responsiveness of individual subjects along the horizontal axis relative to the average of controls is shown across all TIR ligand stimulations on a heatmap. Subjects and stimulations were grouped by hierarchical clustering.
Responses to whole bacteria are only marginally impaired
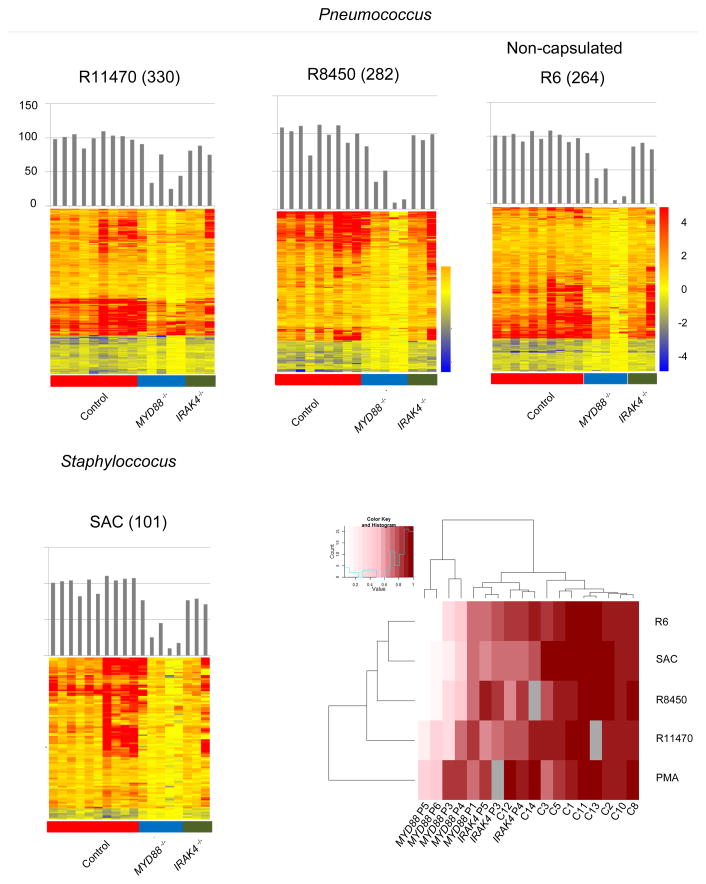
To use a stimulus more akinto that experienced by patients during infection, we exposed samples from 5 MyD88- and 3 IRAK-4-deficient patients with whole heat-killed bacteria (Table 1). The bacteria chosen in this assay were those IRAK-4- and MyD88-deficient patients are most susceptible to:S. pneumoniae and S. aureus9. For S. pneumoniae, 3 different strains were used, including a less virulent non-encapsulated R6 strain. In controls, the magnitude of transcriptional change varied according to the bacteria: S. aureus (SAC) induced the lowest responses (354 transcripts), compared to S. pneumoniae (1553, 865 and 1439 transcripts for R11470, R8450 and R6 strains, respectively).
Patients displayed substantial levels of transcriptional activity in response to heat-killed bacteria (Fig. 2a, 2b). The response in MyD88-deficient patients was consistently lower than in IRAK-4-deficient patients (R11470: 63% versus 84% for MyD88- and IRAK-4-deficient patients respectively; R8450: 45% versus 99%; R6: 46% versus 89%; SAC: 47% versus 75%). This difference was not observed earlier when using purified TIR agonists. Thus the evaluation of transcriptional response to bacterial pathogens shows that, as opposed to what was observed using purified agonists to TIRs, patients with MyD88 or IRAK-4 deficiency maintain the capacity to respond to S. pneumoniae and S. aureus at the transcriptional level (>50% on average; range 27% to 102% of healthy responses) through MyD88- and IRAK-4-independent mechanisms, highlighting the redundancy of the microbial sensing system in blood leukocytes. Furthermore, our findings suggest that patients with MyD88 deficiency could differ from those with IRAK-4 deficiency in their ability to respond to Gram-positive bacteria (50% versus 87% of transcriptional change compared to healthy controls respectively, on average for all 4 bacterial stimulations).
Figure 2.
Blood transcriptional responses following in vitro exposure to whole bacteria. (a). Responsive transcripts in controls and patients are represented on a heatmap for individual bacterial stimulations. Blood from healthy controls or patients (batch 1 and 2) was stimulated in vitro for 2 hours with three strains of heat-killed S. pneumoniae (R11470, R8450, R6) as well as S. aureus (SAC); responsive transcripts were arranged by rows via hierarchical clustering, (330, 282, 264 and 101 for R11470, R8450, R6 and SAC stimulations, respectively) and individual subjects by columns from left to right: healthy controls, MyD88-deficient patients, IRAK-4-deficient patients. Changes versus the non-stimulated condition are represented by a color scale: red = up-regulated; blue = down-regulated; yellow = no change. Bar graphs represent overall individual levels of responsiveness relative to the average of controls: the number of responsive probes in a given subject/average of differentially expressed probes in healthy controls x100. (b) The overall responsiveness of individual subjects along the horizontal axis relative to the average of controls is shown across all whole bacteria stimulations on a heatmap. Subjects and stimulations were grouped by hierarchical clustering.
Modular repertoire mapping of TIR transcriptome responses
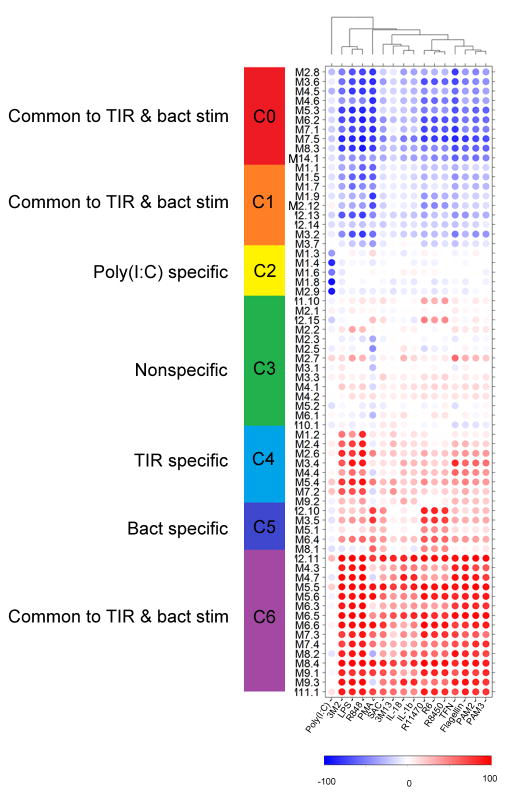
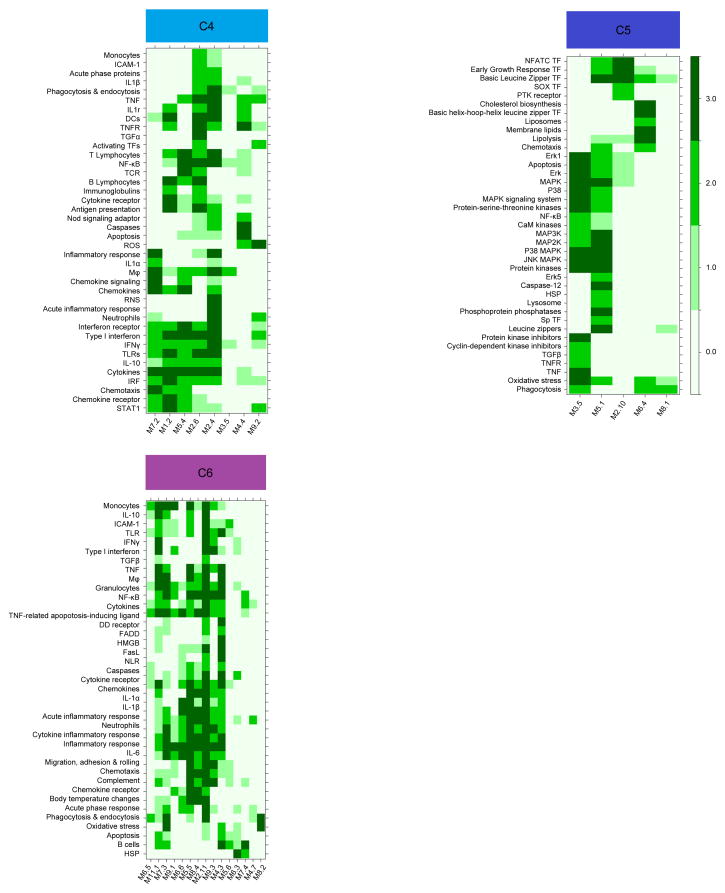
We next sought to characterize the transcriptional programs affected by loss of MyD88- and IRAK-4-dependent signaling. We employed a data mining strategy that consists in mapping relationships among group of genes based on similarities in expression patterns across a wide range of conditions. The approach devised for the construction of such modular transcriptional repertoires has been described in detail elsewhere18 For this study a large gene co-clustering network was constructed using responses to each stimulus across all subjects as input datasets. Network analysis identified co-clustered gene sets – also referred to as transcriptional modules. The resulting collection of modules served as a framework for downstream data analysis and interpretation. This data-driven process identified a repertoire consisting of 66 modules comprised of 1, 088 transcripts. Functional annotations were obtained for each module (Supplementary Table 1). In addition, modules were broadly categorized based on their patterns of response in control subjects to the different TIR agonists and bacteria (Fig. 3). Thus all 66 modules were grouped into 7 clusters (C0 to C6). Clusters 0, 1, and 6 consist of modules responsive to both TIR and bacteria. Cluster C6 is uniformly up-regulated upon these stimulations and is mainly composed of inflammation-related modules, including cytokines (IL-1β, IL-6, IL-8), chemokines, NF-κB, acute-phase response elements and neutrophil function and phagocytosis (Fig. 4, Table 2). Cluster C4 is composed of modules responsive to TIR agonists. For instance, M5. 4 and M7. 2 were preferentially induced by LPS (TLR4), 3M2 or R848 (TLR8) and, to a lesser extent, Poly (I:C), and correspond to an IFN-related inflammatory response that is not induced by Gram-positive bacteria. Bacterial stimulations triggered a set of modules that were not induced by pathogen-associated molecular pattern (PAMP) stimulation of individual TLRs, and thus could be considered whole bacteria-specific (Cluster C5). The annotation of C5 suggests a role in cell signaling (Fig. 4, Table 2). This indicates that in blood cultures exposed to whole bacteria, receptors other than TIR are engaged (possibly ITAM-DAP12-associated receptors19 such as CD300, integrins, Fc Receptors (FcRs) C-type lectins and complement), and pathways or transcription factors other than NF-κB triggered (e. g. MAP Kinases, Protein kinase C, phosphatidylinositol, basic-leucine zipper and NFAT transcription factors).
Figure 3.
Modular transcriptome repertoire of whole blood responses to TIRs agonists and Gram-positive bacteria stimulations. Whole genome transcriptional responses to TIRs agonists and whole Gram-positive bacteria (columns) measured in healthy control subjects were mapped against a modular repertoire composed of 66 sets of co-clustered transcripts (rows). Spots represent the percent of module probes that are up (red) or down (blue) regulated in a sample. The average “modular activity” of healthy control subjects is shown. Hierarchical clustering identified seven clusters (C0 to C6) of modules with related expression pattern across conditions. C0, C1, and C6 contain modules induced by both TIRs and bacteria stimulations, whereas C4 and C5 consist of modules uniquely activated by TLRs or bacteria respectively. Cluster C3 contained modules that did not show a clear pattern of induction by either TIR or bacterial stimulations.
Figure 4.
Literature profiles of module clusters (C4, C5, C6). Accumenta Literature Lab™ was used to obtain literature abstract terms association scores for the modules in clusters C4, C5 and C6. Term associations suggest that C4 and C6 are functionally related to inflammation and that C5 is functionally related to cell signaling. Association scores values ranging from 0 to 3 are shown on the heatmaps, with 0 indicating that the term is not associated with the module (white); and 3 indicating that the term is highly associated with the module (dark green). The intensity of the color is proportional to the score. Modules and terms are ordered by hierarchical clustering based on similarities in patterns of term association.
Table 2.
Functional annotation, and specificity, for each module cluster.
| Module Cluster | Specificity | Transcriptional activity upon stimulation | Functionality |
|---|---|---|---|
| C0 | Common to TIR and bacterial stimulations | downregulation | Cell differentiation, proliferation, adhesion, and metabolism |
| C1 | Common to TIR and bacterial stimulations | Cell signaling, ubiquitination, cell movement, type I IFN, and inflammation | |
| C2 | Poly (I:C) specific | Erythrocytes, hemoglobin, and platelets | |
| C3 | up and downregulation | Inflammation, phagocytosis, apoptosis | |
| C4 | TIR specific | upregulation | Type I and II IFN, Inflammation, caspases, and apoptosis |
| C5 | Bacterial specific | Cell signaling | |
| C6 | Common to TIR and bacterial stimulations | Inflammation |
Impact of TIR deficiencies on the transcriptome repertoire
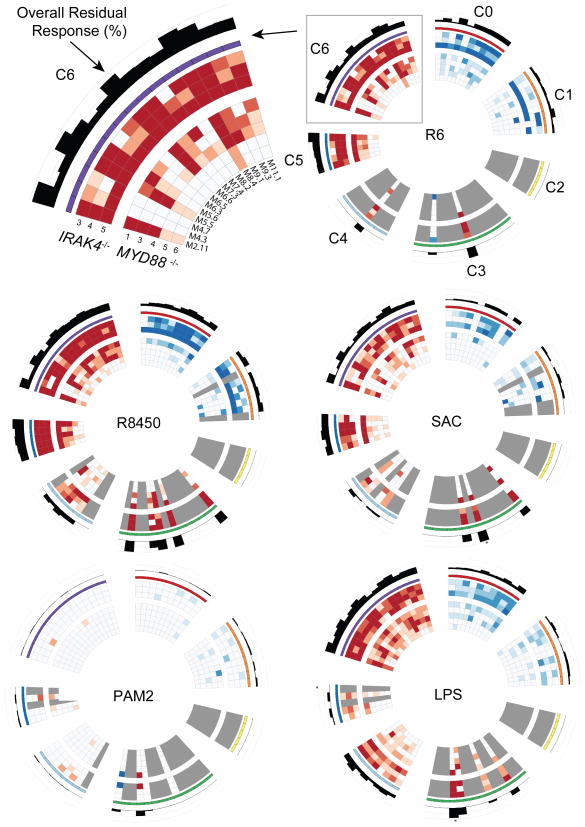
We next used this modular repertoire as a framework for dissecting the transcriptional programs impacted by MyD88 and IRAK-4 deficiencies. Circular heatmaps20 were employed to represent residual patient responses for each TIR stimulation across the 7 module clusters, where each circle represents a patient and each spoke represents a module (Fig. 5, Supplementary Fig. 3). As was expected based on our earlier findings, patients were unable to mount a response to MyD88-dependent purified TIR agonists for any of the modules described above, including those found in clusters C4 and C6, which are associated with inflammation (residual responses to PAM2 Fig. 5, bottom left; all others Supplementary Fig. 3). Residual responses were observed to LPS stimulations (partially MyD88-dependent) with C6 modules (Inflammation) displaying overall well-conserved responses, in contrast with C4 modules (Interferon), which displayed poor residual responses (Fig 5, bottom right). Next, IRAK-4- and MyD88-deficient patients’ responses to heat-killed bacteria were analyzed in a similar fashion. The overall responses to all 4 heat-killed bacteria were relatively well preserved (Fig 5). The most preserved modular responses to Gram-positive bacteria in both MyD88- and IRAK-4-deficient patients were: 1) M2. 11, which contains IL1A, IL1B, TNF, CCL20, CCL3L1, CCL4L1 and CXCL1; 2) M5. 5, which contains IL8, IRAK2, CCL3L3, CXCL2, IL1RN, PLAU and PTGS2; 3) M6. 6, which contains CCRL2, CYP4B1, NRLP3, OSM, PTGS2, TAGAP; and 4) M8. 4, which contains CCL3, CCL3L1, CCL4L2, NFKBIA, NLRP3 and PLAUR. These 4 modules, all from cluster C6, displayed high levels of up-regulation for all donors (>90% for controls and IRAK-4-deficient patients, and >50% for MyD88-deficient patients, Fig. 6). This indicates that patients were able to induce a pro-inflammatory program in response to S. pneumoniae and S. aureus activation, probably through the participation of other sensors, such as NLRP3-activators in the form of inflammasome components, which are present in 2 of the 4 most preserved modules.
Figure 5.
Modular transcriptome repertoire mapping of patient residual responsiveness to TIRs agonists and whole bacteria. For visualization purposes, circular heatmaps were generated to represent the transcriptional modular activity of IRAK-4- (outer rings) and MyD88- (inner rings) deficient patients with respect to healthy controls in response to HK bacteria stimulations and TIRs agonists (batch 1 and 2). The values plotted represent residual responsiveness relative to the average of healthy subjects on a color scale ranging from −1 (saturated blue, intact module down-regulation) to +1 (saturated red, intact module up-regulation). Values close to zero are shown in white or very pale color. The cases where a value was missing are represented by a gray color. The histogram values shown on the hub are the absolute value of the average across patients of the normalized module scores (calculated above). Values shown range between 0 and 1, where cases with values >1 have an asterisk above the bar. Residual responses to 10 additional TIRs agonists and TNF are presented similarly in Supplementary Fig. 3.
Figure 6.
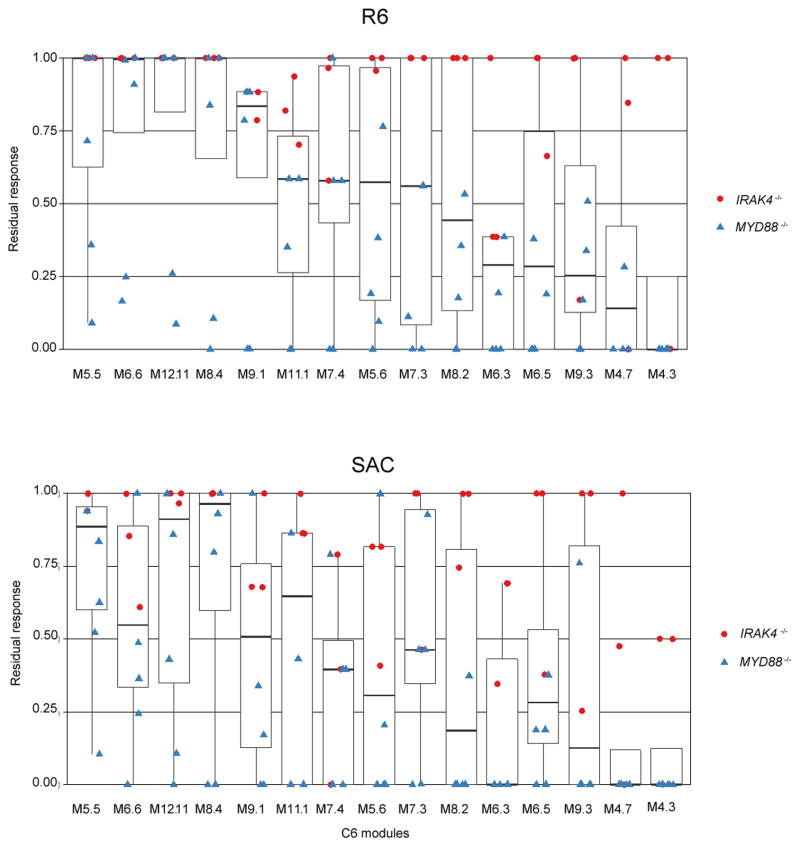
Residual responsiveness following in vitro exposure to whole bacteria for C6 modules. Box plot displays residual responsiveness of individual patients relative to the average of healthy subjects following 2 hours in vitro exposure to heat-killed S. pneumoniae (R6 strain) and S. aureus (SAC). Red dots = IRAK4−/− patients; Blue triangles = MYD88−/− patients (batch 1 and 2). Modules represented here belong to cluster C6. Supplementary Fig. 4 shows residual responses to S. pneumoniae R11470 and R8450 strains.
Three modules display defective responsiveness to bacteria
Further examination of modular patterns of responsiveness to whole bacteria conversely revealed impairment of specific transcriptional programs. Indeed, residual levels of responsiveness among modules constituting cluster C6 diverged markedly (Fig. 5 top left corner, Fig. 6). This heterogeneity was observed both across modules and bacterial species or strains (Fig. 6, and Supplementary Fig. 4). However, three modules, M4. 3, M4. 7, M6. 3, did present consistently low levels of residual responsiveness across these conditions. Of the three modules, responsiveness was most consistently impaired for M4. 3. Module M4. 3 is related to NF-κB activation (contains NFKB1, NFKB2, IRAK3), regulation (SRC, TNIP1), apoptosis (BIRC3, CFLAR, DENND5A-activator of Rab39- IER5), IL-1β and the inflammasome (CARD16 and P2RX7). Responses to S. pneumoniae R6 strain were abolished in all but one IRAK-4-deficient patient for M4. 3 (Fig. 6a), who however, displayed a blunted response when exposed to S. aureus (Fig. 6b), thus suggesting species- or strain-specific fluctuations in pathogen susceptibility (responses to S. pneumoniae strains R8450 and R11470 are shown in Supplementary Fig. 4). Overall responses were also similarly blunted for module M6. 3, which includes TNFAIP8, IRG1 and molecules related to cell metabolism (ACSL1, PDE4B, RIN2). Of interest, IRG1 enhances macrophage bactericidal activity21 and ACSL1 has recently been found to play a role as an inflammatory mediator in LPS-stimulated macrophages22. M4. 7 is another module displaying overall decreased responsiveness. Notably, mutation of NBN (nibrin), which is one of the 9 genes constituting module M4. 7, causes Nijmegen breakage syndrome, a DNA repair PID characterized by combined cellular and humoral immunodeficiency with severe recurrent sinopulmonary infections causing significant mortality in this patient population23. A single nucleotide polymorphism for NFKBIE, another gene belonging to this same module, has been associated with susceptibility to invasive pneumococcal diseases24. Finally, a third gene from M4. 7, CLIC4, has been found to play an important role in mediating innate resistance to bacterial infection in an animal model25. These module-specific defects were observed again in a third set of samples (batch 3, Table 1), including blood drawn from 2 repeat patients (IRAK4−/−P5 and MYD88−/− P4) and 2 new patients (IRAK4−/−P6 and MYD88−/−P7). This new set of patients showed the same degree of preserved transcriptional activity in response to heat-killed bacteria when compared with previous set of patients (Supplementary Fig. 5), and when dissecting this response, again, all showed low residual responsiveness for M4. 3, M4. 7 and M6. 3 (Supplementary Fig. 6).
Residual responses to viral, fungal and bacterial pathogens
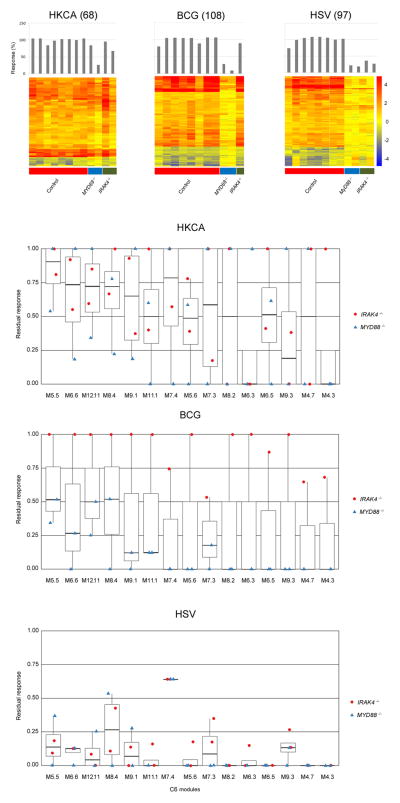
Mice lacking IRAK-4 or MyD88 are susceptible to a wide range of pathogens, which is in striking contrast with the narrow range of susceptibility observed in humans with inborn errors in IRAK-4 or MyD888. Thus we subsequently investigated patterns of transcriptional response in these patients upon whole blood stimulation with pathogens deficient mice but not humans a resusceptible to9. Responses to Heat-killed Candida albicans (HK. CA), Bacillus Calmette-Guérin (BCG), and herpes simplex virus 1 (HSV) were measured in 2 MyD88- and 2 IRAK-4-deficient patients, as well as 8 control subjects (batch 3, Table 1). Patients displayed substantial levels of transcriptional activity in response to HK. CA with near normal residual responsiveness observed in 3 of the 4 patients (Fig. 7a, left panel). In response to BCG stimulation (Fig. 7a, middle panel), MyD88-deficient patients showed markedly deficient responses (<30% globally) that affected all the inflammatory modules, while an IRAK-4 deficient patient had preserved responsiveness both globally (90%) and at the module level (Fig. 7b, middle panel) (data could not be obtained for the second IRAK4 patient). These observations could not be attributed to prior BCG vaccination status (MyD88 P1: no vaccination, MyD88 P2: vaccination, IRAK-4 P2: vaccination). Finally, both MyD88- and IRAK-4-deficient patients showed globally defective transcriptional responses to whole blood stimulation with HSV (<20% responses) (Fig. 7a, right panel). At the module level one single inflammatory module, M7. 4, was selectively preserved upon HSV stimulation (Fig. 7b, right panel). This module contains molecules such as NFIL3 (E4BP4), related to macrophage, dendritic cell and NK differentiation and function, that are required for antiviral immunity26, and MYC and REL, related to NF-κB signaling. This finding suggests that while TLR signaling to HSV is for the most part abolished, alternative pathways involving NF-κB signaling may be preserved and contribute to maintain resistance to this pathogen in MyD88 and IRAK-4 deficient patients.
Figure 7.
Blood transcriptional responses following in vitro exposure to Candida, BCG and HSV. (a). Responsive transcripts in 8 controls and four patients (2 MYD88−/− and 2 IRAK4−/−) are represented on a heatmap for individual pathogen stimulations. Blood from healthy controls and patients (batch 3) was stimulated in vitro for 2 hours with heat-killed Candida albicans (HKCA), Bacillus Calmette-Guérin (BCG) and herpes simplex virus (HSV); responsive transcripts were arranged by rows via hierarchical clustering, (68, 108 and 97 for HKCA, BCG and HSV stimulations, respectively) and individual subjects by columns from left to right: healthy controls, MyD88-deficient patients, IRAK-4-deficient patients. Changes versus the non-stimulated condition are represented by a color scale: red = upregulated; blue = downregulated; yellow = no change. Bar graphs represent overall individual levels of responsiveness relative to the average of controls: the number of responsive probes in a given subject/average of differentially expressed probes in healthy controls x100. (b) Box plot displays residual responsiveness of individual patients (2IRAK4−/− and 2 MYD88−/−) relative to the average of healthy subjects following 2 hours in vitro exposure to HKCA, BCG and HSV. Red dots = IRAK4−/− patients; Blue triangles = MYD88−/− patients. Modules represented in the figure belong to the cluster C6.
In summary, the use of a systems approach revealed a multifaceted pattern of innate immune responsiveness in patients with MyD88 or IRAK-4 deficiencies. Our results demonstrate a profound defect in the ability of blood leukocytes from MyD88- and IRAK-4-deficient patients to respond to pathogen- and host-derived TIR agonists. Responses to specific TIR agonists were completely abrogated, while responses to whole organisms ranged from being relatively normal (HK. CA) to dramatically reduced (HSV). Differences in levels of responsiveness between MyD88- and IRAK-4- deficient subjects were also observed for several conditions. Transcriptional programs elicited by pyogenic bacteria to which both groups are clinically susceptible were not associated with a dramatic reduction in overall responsiveness, but rather impairment of specific inflammatory transcriptional programs.
DISCUSSION
We have devised a systems approach for the assessment of TIR and antibacterial immunity inpatients with increased susceptibility to infection. The distinct benefit of using a genome-wide approach is that it provides an unbiased means to interrogate responses to innate immune stimuli as opposed to more traditional approaches that require relying on a priori knowledge to select a small panel of analytes as readout1, 2, 27. Here we assessed the global impact of defects in IRAK-4 and MyD88 on innate immune transcriptional programs. Patients’ responses to TLR2 agonists were less than 10% of healthy subjects. TLR2 is known to be crucial for Gram-positive bacteria recognition (S. pneumoniae28 and S. aureus29), which are pathogens to which IRAK-4- and MyD88-deficient patients are most susceptible.
We have confirmed the close dependence between IRAK-4 and MyD88 molecules in the signaling pathway downstream of TIR, concordant with the functional structure they form known as the Myddosome6. However, stimulation with whole pathogens, as opposed to purified TIR agonists, revealed differences in IRAK-4 and MyD88 deficient patient’s ability to mount transcriptional responses. This might be explained considering that MyD88 protein is involved in Ras/MAPK signaling pathway through direct interaction with ERK without IRAK participation30. Nevertheless, these differences do not translate into cytokine production, which has been shown equally impaired 1, 2, 27, nor in the infectious clinical phenotype, which is indistinguishable8–10.
Despite the profound defect in the common TIR signaling pathway, IRAK-4- and MyD88-deficient patients were able to up-regulate major inflammatory modules (modules M2. 11, M5. 5, M8. 4, M6. 6) when their blood was exposed to S. pneumoniae and S. aureus. This indicates that a MyD88-independent, but NF-κB- and MAPK-dependent pathway would be responsible for this proinflammatory response since the same inflammatory program is observed with specific TIR stimulation28, 31. S. aureus and S. pneumoniae can be recognized by NOD232 and TNFR133, both of which are expressed in leukocytes, activate NF-κB and MAPKs, initiating the same inflammatory processes as TLRs34, and they do not signal via MyD88. There is consistent evidence that both TLRs and NOD2 receptors synergize to induce combined responses35. The lack of synergism with TLR2 might explain the suboptimal response observed (lack of up-regulation of M4. 3, M4. 7, M6. 3). Those results are consistent with previously reported data obtained at the protein level: TNFα secretion in whole blood from IRAK-4 patients was undetectable 24h after TLR stimulation, but detectable upon heat-killed Staphylococcus stimulation, although to a lower extent compared to healthy controls1. The residual induction of cytokines via MyD88/IRAK-4-independent signaling pathways may account for both the resistance to other infections and the fever/inflammation that can be observed at late stages of infection.
These results raise an important question: can the susceptibility to bacteria in TIR deficiencies be ascribed to this partially defective inflammatory response, or other mechanisms contribute to the phenotype? Our assay only evaluated responses in whole blood, and not in mucosae or skin36. IL-1Ris crucial to maintain protective immunity against invasive staphylococcal skin infection37. Indeed neutrophils play a key role in bacterial clearance of epithelial sites38 and IL-1R-mediated signaling by resident skin cells would be essential for adequate neutrophil recruitment to the site of infection. We previously published that skin-derived fibroblasts from both IRAK-4- and MyD88-deficient patients showed no response to IL-1R activation2. Also, IL-1R has proven crucial in other S. aureus infections39, 40. It might be that IRAK-4- and MyD88-deficient patients are able to generate an initial proinflammatory response upon initial encounter with Gram-positive bacteria in whole blood, as we show, but presumably at epithelial sites as well (NOD2 expression has been described in keratinocytes and the lung41; TNFR1 is widely distributed on the airway epithelium33). Nevertheless, defective IL1-R signaling could result in an impaired systemic amplification of this initial response, ultimately increasing the risk of not only epithelial infections, but also bacteremia starting from skin and mucosa, which is characteristic of these patients. This would also explain why IRAK-4-and MyD88-deficient patients are clinical phenocopies despite different degrees of response to whole bacteria in blood, since they display similar levels of defect to IL-1β stimulation.
MyD88-deficient patients showed marked impairment in transcriptional responses to whole blood activation with BCG, despite prior BCG vaccination. These results are consistent with the strong TLR2 agonist activity to BCG in mice42. However, MyD88-deficient patients are not susceptible to mycobacteria nor to, BCG 9. In humans resistance to BCG is known to be dependent on IFN-γ and its production is likely to be preserved through MyD88-and IRAK-4-independent pathways, 43. Similarly, while the abnormal responses to HSV observed in both in MyD88- and IRAK-4-deficient patients attest to the role of TLR2 and TLR9 in the initiation of immune responses to HSV44, it does not result in increased susceptibility to this pathogen. Earlier work from our group demonstrated that HSV control in the central nervous system is exquisitely dependent on a functional TLR3-pathway in neurons and oligodendrocytes but not in blood leukocytes45.
Technologies available for profiling transcript abundance on a genome-wide scale have become robust and cost-effective46. Its application in whole blood has been used extensively for the investigation of disease pathogenesis, identification of biomarkers, and assessment of responses to vaccines16, 17. However, molecular phenotypes may not always be apparent in ex vivo blood profiled at the steady state, as is the case in patients with MyD88 and IRAK-4 deficiencies (data not shown). We show here that using global profiling as readout in an in vitro functional assay can produce comprehensive innate immune phenotypes in patients with PID. Possible clinical applications for a targeted assay derived from the modular transcriptional framework constructed in this study can be foreseen beyond PID, in clinical settings where defects in innate immunity are suspected, such as recurrent pyogenic infections47 or aging48, and also for the prediction of responses to vaccines16, 17. The choice of conditions for the in vitro stimulations is obviously critical; here we selected a panel tailored for inborn errors in TIR pathway. This can be adapted depending on the population being screened.
In conclusion, this work demonstrates the use of systems approaches as a robust means for the global assessment of innate immune competence. Applied to the study of patients with inborn defects of TIR signaling, this strategy revealed that patients with MyD88 and IRAK-4 deficiencies suffer from a profound loss of responsiveness to soluble TIR agonists and that their ability to respond to whole bacteria is selectively impacted. Such findings are consistent with the delayed clinical and biological signs of inflammation observed in those patients during the course of infection1, 2, 9, 29. More broadly by lever aging high throughput profiling technology together with an effective analytic framework, this work also opens new avenues for the widespread use of systems approaches as a global readout in functional immunological assays.
Online METHODS
1. Patient information and sample collection
A total of 40 blood samples were collected from three groups of subjects: 7 patients with complete MyD88 deficiency, 6 patients with complete IRAK-4 deficiency, and 27 healthy donors (Table 1). IRAK4 mutations from P1, P2, and P3 are null (no IRAK-4 protein detected27). Mutations from IRAK-4- P4, P5 and P69, and MyD88- patients 1 to 7 are loss-of-function. MyD88- P1 and P2 have normal levels of non-functional MyD88 protein; MyD88- P3-P7 and IRAK-4- P4 and P5 have small amounts of non-functional protein2. Subjects were recruited in four different sets: the first set included 4 healthy controls, MyD88- P1 and P2, IRAK-4- P1 and P2. The second set included 10 different healthy controls, MyD88-P3 and P4 and IRAK-4-P3 and P4. First and second set (batch 1) were analyzed to evaluate patients’ responses to 13 different TIR agonists (10 TLR agonists, 3 IL1-R agonists. A third set (batch 2) was included afterwards to increase the number of MyD88- and IRAK-4-deficient patients for bacterial stimulations (3 strains of heat killed S. pneumoniae and S. aureus); batch 2 included 5 new healthy controls, MyD88- P5 and P6, new samples from MyD88-P1, and IRAK-4-P5. Four patients from batch 1 and four from batch 2 were analyzed to evaluate TIR patients’ responses to bacteria. The fourth set (batch 3) was included to test for alternative pathogens stimulations (1 new IRAK-4-P6, 1 new MyD88-P7 patient, new samples from IRAK-4-P5 and MyD88-P4, and 8 new healthy controls). Data on patient antimicrobial prophylaxis at the time of blood drawn is detailed in Table 1.
Healthy individuals were considered so based on past medical history (no recurrent infections) and current health status. All healthy controls were adult subjects except for batch 3, which included 2 healthy young children age 3 and 7 years-old. Indeed, Toll-like receptor function is mainly age dependent for the newborn49, and there was an evident limitation in obtaining the required high volume of blood from healthy children to use as controls. Blood samples were obtained at Necker Hospital, Paris, France, with approval of the local Research Ethics Committee. All participants aged 18 or their legal tutors gave written informed consent.
2. Blood culture
Peripheral blood was drawn into sodium heparin vacuum tubes on clinic site (Necker Hospital, Paris). Immediately, 500μl whole blood (WB) was diluted with equal amount of RPMI before adding different stimulus, each experimental condition was performed in replicates. Diluted WB was activated for 2 hours with 10μg/ml of polymyxin B to clear LPS contamination plus a wide range of agonists Pam3CSK4 (TLR1/2; 100 ng/ml, InvivoGen®); Pam2CSK4 (TLR2/6; 100 ng/ml, InvivoGen®); poly(I:C) (TLR3; 25 μg/ml, InvivoGen®); LPS (TLR4; 100ng/ml, Sigma®); Flagellin (TLR5; 100 ng/ml, InvivoGen®); 3M-13 (TLR7; 3μg/ml, 3M pharmaceuticals®); 3M-2 (TLR8; 3μg/ml, 3M pharmaceuticals®); R848 (TLR7/8; 3μg/ml, InvivoGen®); CpG-D19 and CpG-C (TLR9; 3μg/ml, from collaborator);IL1B (IL-1R; 20ng/ml, R&D systems®); IL-18 (IL-1R; 50ng/ml, R&D systems®); IL-33 (IL-1R; 50ng/ml, R&D systems®); TNF (TNFR; 20ng/ml, R&D systems®); PMA (25 ng/ml, Sigma®) + Ionomycin (1μg/ml, Sigma®); 3 heat-killed pneumococcal strains (R6, 5.106 particle/ml; R8450 108 particle/ml; R11470, 5.106 particle/ml, heat-killed at 65°C for 15 min, from collaborator), and heat-killed Staphylococcus aureus (107 particle/ml, InvivoGen®); HSV-1, strain KOS-1 (MOI 1:1); live BCG (M. bovis BCG, Pasteurj sub-strain at an MOI of 20 BCG/leukocytes); HK Candida albicans (1.106 particules/ml, InvivoGen®), or left unstimulated for 2h. After stimulation, WB was lysed with Tempus solution (from Tempus tubes, Applied Biosystems) to stabilize the RNA at 1:3 ratio after stimulation. The lysates were kept at −80°C until mRNA extraction.
3. RNA extraction and processing for microarray analysis
Total RNA was isolated from WB lysate using Tempus MagMAX-96 Blood RNA isolation kit (Applied Biosystems/Ambion). RNA quality and quantity were assessed using Agilent 2100 Bioanalyzer (Agilent Technologies) and NanoDrop 1000 (NanoDrop Products, Thermo Fisher Scientific). Samples with RNA integrity numbers values >6 were retained for further processing. Globin mRNA was depleted using the GLOBIN clear™ (Applied Biosystems/Ambion). Globin-reduced RNA was amplified and labeled using the Illumina Total Prep-96 RNA Amplification Kit (Applied Biosystems/Ambion). This process was performed in 3 different batches (batch 1, 2, and 3, Table 1). Biotin-labeled cRNA was hybridized overnight to Human HT-12 V4 BeadChip arrays (IIlumina), which contains 47,231 probes, and scanned on an Illumina BeadStation 500 (batch 1), iScan (batch 2), or HiScan (batch 3) to generate signal intensity values.
4. Microarray data analysis
To reduce the potential batch effect between the sets, the background subtracted raw signal values, extracted with Illumina Beadstudio (version 2), were processed using ComBat50. The data was then quantile normalized and the minimum intensity was set to 10. These data are available at GEO Series accession number GSE25742. Only the probes called present in at least 10% of the samples (p<0.01) were retained for downstream analysis (n=19,152). Transcripts differentially regulated upon stimulation were defined based on a minimum 1.5-fold change (up- or down-regulation) and a minimum absolute raw intensity difference of 150 with respect to the respective unstimulated sample. Probes passing these filters in at least 75% of healthy control samples were considered substantially affected by stimulation, and were used in Fig. 1, 2,5, 7, Supplementary Fig.2, 3, 5online). No explicit statistical test of differential expression was performed. The “percent of healthy response” quantifies the number of probes changed in the patients compared to the healthy controls. It is calculated as the number of stimulation-affected probes divided by the average number of stimulation-affected probes in the healthy control samples for the same stimulus. Red/white plots (Fig. 1b, 2b) represent the normalized probe counts (number of probes passing the cutoffs for samples divided by number of probes passing the cut-offs for 75% of controls) for each subject-stimulation pair.
5. Module construction
Clustering of probes into co-expressed modules
We generated sets of co-expressed probes as described previously21, with variations as follows. The data used for clustering includes 10 healthy control and 4 patient samples (2 MyD88- and 2 IRAK-4-deficient) for 19 stimulations (all TLR + IL-1R, TNF and 4 bacteria), available as GEO Series GSE25742. The raw expression data was background-subtracted and average-normalized before clustering. Only probes having at least a 1.5-fold change and ±100 difference compared to their matched non-stimulated sample for all stimulations were clustered. To improve clustering efficiency, the expression values of the selected probes were first converted into trinary values (e.g. −1, 0, 1) as follows: for cases where the absolute difference is <100 or the absolute value of the log2 fold change was < 0.585, then the signal was considered to be unchanged, and was set to 0. Otherwise, the signal was set to 1 if it is greater than the baseline and −1 if it was smaller than the baseline. Next, within the samples for each stimulus, a group reduction was performed such that for each probe, each group of samples (healthy, MyD88-/-, IRAK-4-/-) was replaced by a single value that indicated whether the trinary signal was consistent within 75% of the samples of the group. This yielded three trinary values (one per group) per probe. We clustered the group-reduced trinary probe values separately for each stimulus by grouping probes with the same pattern of values, creating as many clusters as necessary for all probes to be included in a cluster. The clusters were then used as input for our module extraction algorithm. The extraction required that the maximal clique used as the seed of a module contained at least ten probes. We did not use paraclique to expand the module. When completed, fourteen rounds of modules were produced, the number corresponding to the number of input clustered datasets. A total of 66 modules were obtained composed of 1088 transcripts.
Analysis of module-level data
Module activity was calculated as the differences between the percent of up-regulated and down-regulated probes (Fig. 3). To compare the module response of patient samples to healthy controls, we calculated a “residual response” value which is the per-patient module score divided by the mean of the healthy controls module score for each stimulus. Module scores for any stimuli with a healthy control absolute value mean score < 10% were considered not detected, so residual responses were not computed for these modules (shown in gray in (Fig. 5, Supplementary Fig.3 online). For cases where healthy controls’ mean scores and an individual patient’s scores were of opposite signs, the residual response was set to zero. Circular heat map plots were generated using Circos20.
Functional annotation of modules
Acumenta Literature Lab™ was used to associate each probe within a particular module to terms in PubMed abstracts. Association scores reflecting the strength of the associations were used in heat maps and to calculate “cumulative LitLab Scores”. The terms that showed a strong association with a module were used to create the functional annotation. Gene networks for each module were created, using “direct interactions” in GeneGo MetaCore™ (version 6.10). Modules were annotated using the network processes that were indicated for each module. The final annotations are a summary of Acumenta Literature Lab™ and GeneGo MetaCore™. In the cases where no terms had a strong association to a module and the gene networks were inconclusive, the modules were left un-annotated (14 out of 66 modules) (Table 2, Supplementary Table 1 online). The association scores obtained from Acumenta Literature Lab™ were plotted in heat maps (Fig. 4).
Supplementary Material
Acknowledgments
We thank Drs L. Marodi, J. C. Rodriguez Gallego, G. Davies, Y. Camcioglu, A. Bousfiha, J. Vasconcelos and J. I Aróstegui for sending blood from the patients with IRAK-4 and MyD88 deficiencies. We thank Benjamin Lemoine and Phuong Nguyen for sample processing. We thank Emmanuelle Varon for providing bacterial strains and Maya Chrabieh for technical help. We also thank the patients and their families for their invaluable help. The present work was supported by grants from NIH (1R21AI085523-01, U01AI082110, U19-AI089987, U19-AI08998, U19-AI057234, R01ARO50770-02) and the Dana Foundation. Laia Alsina was supported from 2008 to 2010 by a grant from La Fundació La Caixa, and since 2012 by the Instituto de Salud Carlos III, grant PFIS12/01990 co-financed by the European Regional Development Fund (ERDF).
Abbreviations
- PIDs
Primary Immunoeficiencies
- TLRs
Toll-like Receptors
- IL-1Rs
interleukin-1 receptors
- TIR
TLR/Interleukin-1 receptors
- PAMPs
Pathogen Associated Molecular Patterns
- SAC
Staphylococcus aureus
- S. pneumoniae
Streptococcus pneumoniae
- ITAM
Immunoreceptor Tyrosin-based Activation Motif
- FCRs
Fc Receptors
- IFN
Interferon
- HK
Heat-killed
- WB
whole blood
- BCG
Bacillus Calmette-Guérin
- HSV
herpes simplex virus
- CA
Candida albicans
Footnotes
AUTHOR CONTRIBUTION
L. A was responsible for acquisition of data, analysis and interpretation of data, and drafting of manuscript. E. I, MC. A, and K K. D were responsible for analysis and interpretation of data, and drafting of manuscript. KK. D is responsible for Circos figures. Z. J contributed to acquisition of data. C. P sent patients’ blood. P. G, H. B, A. P, C. P contributed to sample collection and whole blood stimulation. A. P, C. P contributed to critical revision of the manuscript. N. B generated the modular analysis. H. Q, R. B, and, A. I contributed to statistical analysis of microarray data. E. A contributed to sample collection and data acquisition. L. Abel, V. P, JL. C, and D. C. are responsible for the study conception and design, and critical revision of the manuscript.
DATABASE ACCESSION NUMBER: GEO Series accession number GSE25742.
References
- 1.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 2.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 5.Lee CC, Avalos AM, Ploegh HL. Accessory molecules for Toll-like receptors and their function. Nat Rev Immunol. 12:168–179. doi: 10.1038/nri3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in Host Defense: Natural Insights from Evolutionary, Epidemiological, and Clinical Genetics. Annual Review of Immunology. 2011;29:447–491. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 8.von Bernuth H, Picard C, Puel A, Casanova JL. Experimental and natural infections in MyD88- and IRAK-4-deficient mice and humans. Eur J Immunol. 2012;42:3126–3135. doi: 10.1002/eji.201242683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picard C, et al. Clinical features and outcomes of patients with IRAK-4 and MyD88 deficiency. Medicine. 2010;89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picard C, Casanova JL, Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IkappaBalpha deficiency. Clin Microbiol Rev. 2011;24:490–497. doi: 10.1128/CMR.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allantaz F, Chaussabel D, Banchereau J, Pascual V. Microarray-based identification of novel biomarkers in IL-1-mediated diseases. Curr Opin Immunol. 2007;19:623–632. doi: 10.1016/j.coi.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–571. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramilo O, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banchereau R, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 7:e34390. doi: 10.1371/journal.pone.0034390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trautmann L, Sekaly RP. Solving vaccine mysteries: a systems biology perspective. Nat Immunol. 2011;12:729–731. doi: 10.1038/ni.2078. [DOI] [PubMed] [Google Scholar]
- 17.Obermoser G, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14:271–80. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivashkiv LB. Cross-regulation of signaling by ITAM-associated receptors. Nat Immunol. 2009;10:340–347. doi: 10.1038/ni.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall CJ, et al. Immunoresponsive Gene 1 Augments Bactericidal Activity of Macrophage-Lineage Cells by Regulating beta-Oxidation-Dependent Mitochondrial ROS Production. Cell Metab. 2013;18:265–278. doi: 10.1016/j.cmet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Rubinow KB, et al. Acyl-CoA synthetase 1 is induced by Gram-negative bacteria and lipopolysaccharide and is required for phospholipid turnover in stimulated macrophages. J Biol Chem. 2013;288:9957–9970. doi: 10.1074/jbc.M113.458372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chrzanowska KH, Gregorek H, Dembowska-Baginska B, Kalina MA, Digweed M. Nijmegen breakage syndrome (NBS) Orphanet J Rare Dis. 2012;7:13. doi: 10.1186/1750-1172-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman SJ, et al. IkappaB genetic polymorphisms and invasive pneumococcal disease. Am J Respir Crit Care Med. 2007;176:181–187. doi: 10.1164/rccm.200702-169OC. [DOI] [PubMed] [Google Scholar]
- 25.He G, et al. Role of CLIC4 in the host innate responses to bacterial lipopolysaccharide. Eur J Immunol. 2011;41:1221–1230. doi: 10.1002/eji.201041266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Male V, Nisoli I, Gascoyne DM, Brady HJ. E4BP4: an unexpected player in the immune response. Trends Immunol. 2012;33:98–102. doi: 10.1016/j.it.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Ku CL, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204:2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppe U, Suttorp N, Opitz B. Recognition of Streptococcus pneumoniae by the innate immune system. Cell Microbiol. 2012;14:460–466. doi: 10.1111/j.1462-5822.2011.01746.x. [DOI] [PubMed] [Google Scholar]
- 29.Mullaly SC, Kubes P. The role of TLR2 in vivo following challenge with Staphylococcus aureus and prototypic ligands. J Immunol. 2006;177:8154–8163. doi: 10.4049/jimmunol.177.11.8154. [DOI] [PubMed] [Google Scholar]
- 30.Coste I, et al. Dual function of MyD88 in RAS signaling and inflammation, leading to mouse and human cell transformation. J Clin Invest. 2010;120:3663–3667. doi: 10.1172/JCI42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nau GJ, Schlesinger A, Richmond JF, Young RA. Cumulative Toll-like receptor activation in human macrophages treated with whole bacteria. J Immunol. 2003;170:5203–5209. doi: 10.4049/jimmunol.170.10.5203. [DOI] [PubMed] [Google Scholar]
- 32.Moreira LO, et al. The TLR2-MyD88-NOD2-RIPK2 signalling axis regulates a balanced pro-inflammatory and IL-10-mediated anti-inflammatory cytokine response to Gram-positive cell walls. Cell Microbiol. 2008;10:2067–2077. doi: 10.1111/j.1462-5822.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez MI, et al. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med. 2004;10:842–848. doi: 10.1038/nm1079. [DOI] [PubMed] [Google Scholar]
- 34.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tada H, Aiba S, Shibata K, Ohteki T, Takada H. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun. 2005;73:7967–7976. doi: 10.1128/IAI.73.12.7967-7976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zola TA, Lysenko ES, Weiser JN. Mucosal clearance of capsule-expressing bacteria requires both TLR and nucleotide-binding oligomerization domain 1 signaling. J Immunol. 2008;181:7909–7916. doi: 10.4049/jimmunol.181.11.7909. [DOI] [PubMed] [Google Scholar]
- 37.Miller LS, et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity. 2006;24:79–91. doi: 10.1016/j.immuni.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Molne L, Verdrengh M, Tarkowski A. Role of neutrophil leukocytes in cutaneous infection caused by Staphylococcus aureus. Infect Immun. 2000;68:6162–6167. doi: 10.1128/iai.68.11.6162-6167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hultgren OH, Svensson L, Tarkowski A. Critical role of signaling through IL-1 receptor for development of arthritis and sepsis during Staphylococcus aureus infection. J Immunol. 2002;168:5207–5212. doi: 10.4049/jimmunol.168.10.5207. [DOI] [PubMed] [Google Scholar]
- 40.Kielian T, Bearden ED, Baldwin AC, Esen N. IL-1 and TNF-alpha play a pivotal role in the host immune response in a mouse model of Staphylococcus aureus-induced experimental brain abscess. J Neuropathol Exp Neurol. 2004;63:381–396. doi: 10.1093/jnen/63.4.381. [DOI] [PubMed] [Google Scholar]
- 41.Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce antimicrobial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–3111. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Nicolle DM, et al. Chronic pneumonia despite adaptive immune response to Mycobacterium bovis BCG in MyD88-deficient mice. Lab Invest. 2004;84:1305–1321. doi: 10.1038/labinvest.3700149. [DOI] [PubMed] [Google Scholar]
- 43.Zhang SY, et al. Inborn errors of interferon (IFN) -mediated immunity in humans: insights into the respective roles of IFN–alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol Rev. 2008;226:29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 44.Ahmad R, El Bassam S, Cordeiro P, Menezes J. Requirement of TLR2-mediated signaling for the induction of IL-15 gene expression in human monocytic cells by HSV-1. Blood. 2008;112:2360–2368. doi: 10.1182/blood-2008-02-137711. [DOI] [PubMed] [Google Scholar]
- 45.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 47.Picard C, Puel A, Bustamante J, Ku CL, Casanova JL. Primary immunodeficiencies associated with pneumococcal disease. Curr Opin Allergy Clin Immunol. 2003;3:451–459. doi: 10.1097/00130832-200312000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Caruso C, et al. Mechanisms of immunosenescence. Immun Ageing. 2009;6:10. doi: 10.1186/1742-4933-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levy O, et al. Selective impairment of TLR-mediated innate immunity in human newborns: neonatal blood plasma reduces monocyte TNF-alpha induction by bacterial lipopeptides, lipopolysaccharide, and imiquimod, but preserves the response to R-848. J Immunol. 2004;173:4627–4634. doi: 10.4049/jimmunol.173.7.4627. [DOI] [PubMed] [Google Scholar]
- 50.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.