Abstract
In higher eukaryotes, tRNAs with the same anticodon are encoded by multiple nuclear genes and little is known about how mutations in these genes affect translation and cellular homeostasis. Similarly, the surveillance systems that respond to such defects in higher eukaryotes are not clear. Here, we discover that loss of GTPBP2, a novel binding partner of the ribosome recycling protein Pelota, in mice with a mutation in a tRNA gene that is specifically expressed in the central nervous system causes ribosome stalling and widespread neurodegeneration. Our results not only define GTPBP2 as a ribosome rescue factor, but also unmask the disease potential of mutations in nuclear-encoded tRNA genes.
In higher eukaryotes, the nuclear genome typically contains several hundred tRNA genes, which fall into isoacceptor groups, each representing an anticodon (1). Relative to the total number of tRNA genes, the number of isodecoders, i.e. tRNA molecules with the same anticodon but differences in the tRNA body, increases dramatically with organismal complexity leading to speculation that isodecoders might not be fully redundant with one another (2). Overexpression of reporter constructs with rare codons that are decoded by correspondingly low-abundance tRNAs in bacteria and yeast, or mutations in single-copy mitochondrial tRNA genes, may result in stalled elongation complexes (3–5). However, the consequences of mutations in multicopy, nuclear-encoded tRNA isodecoder genes or in the surveillance systems that eliminate the effect of such tRNA mutations are not known in higher eukaryotes.
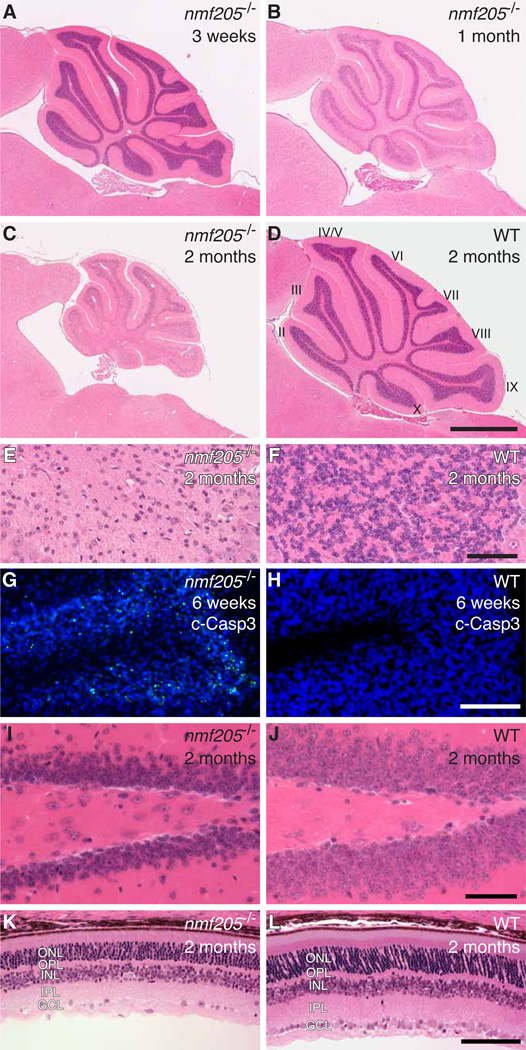
The nmf205 mutation was identified in an ENU-mutagenesis screen of C57BL/6J (B6J) mice for neurological phenotypes (6). B6J-nmf205−/− mice were indistinguishable from wild-type mice at 3 weeks of age, but showed clear truncal ataxia at 6 weeks (movie S1). Mice died at 8 – 9 weeks with severe locomotor deficits. Progressive apoptosis of neurons in the inner granule layer (IGL) in the mutant cerebellum was initially observed between 3 and 4 weeks of age (Fig. 1A to H). Apoptosis of mutant granule cells in the dentate gyrus, CA2 pyramidal neurons, and layer IV cortical neurons occurred between 5 and 8 weeks of age (Fig. 1I and J, and fig. S1A to H). Further, many neurons in the retina, including photoreceptors and amacrine, horizontal, and ganglion cells degenerated during this time (Fig. 1K and L, and fig. S1I to T). Histological analysis of other organs did not reveal obvious pathology, nor was neurodegeneration observed in mice heterozygous for this mutation.
Fig. 1. Progressive neurodegeneration in the nmf205−/− mice.
(A to F) Hematoxylin and eosin (H and E) stained sagittal sections of nmf205−/− and wild-type (WT; +/+) cerebella. (E and F) Higher-magnification images of cerebellar lobule IX from (C and D). (G and H) Cerebellar sections were immunostained with antibodies to cleaved caspase-3 (c-Casp3; green) and counterstained with Hoechst 33342. (I to L) H and E-stained sagittal sections of the dentate gyrus (I and J) and retina (K and L). Scale bars, 1 mm (D), 50 µm (F and J), and 100 µm (H and L). ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
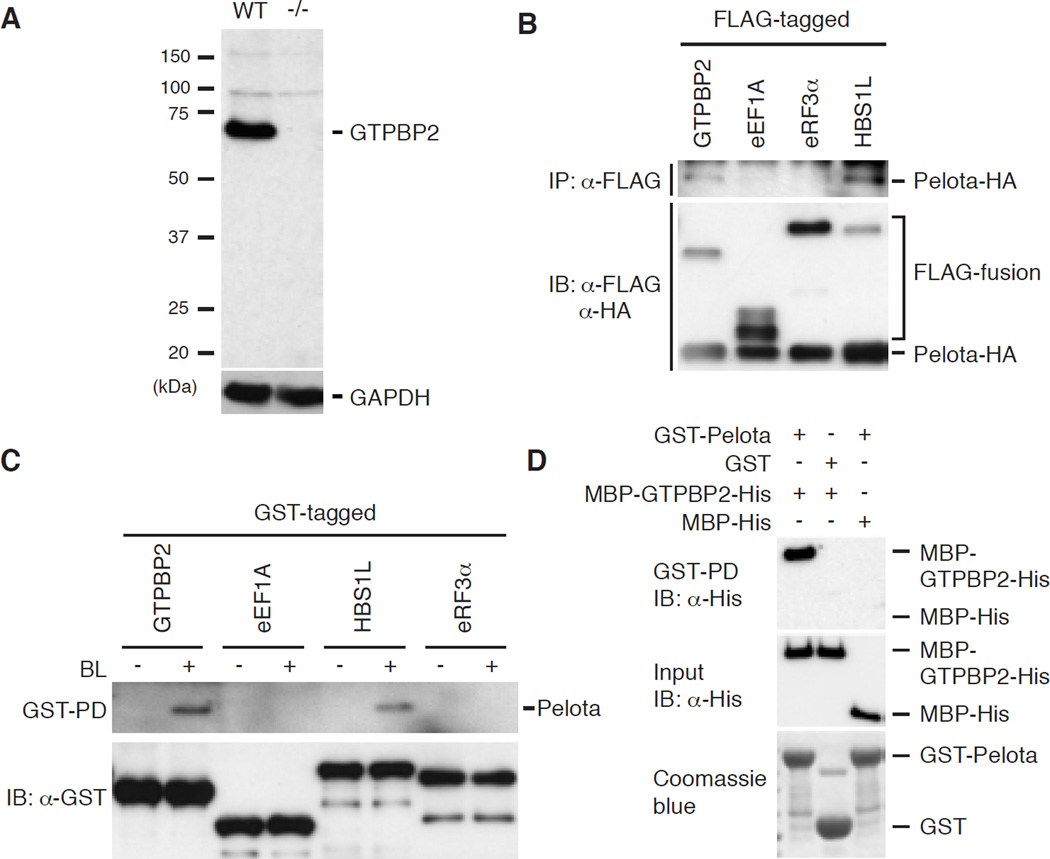
We identified the nmf205 mutation as a point mutation in the consensus splice donor site of intron 6 of Gtpbp2, which results in mis-spliced mRNAs with premature stop codons (fig. S2). Accordingly, western blot analysis of mutant cerebellar extracts failed to detect GTPBP2 (Fig. 2A). Complementation tests using nmf205 mice and mice with a targeted deletion of Gtpbp2 confirmed that loss of Gtpbp2 results in neurodegeneration (fig. S3).
Fig. 2. The nmf205 mutation disrupts the Pelota-interacting protein, GTPBP2.
(A) Western blot analysis of GTPBP2 in wild-type (WT, +/+) and nmf205−/− (−/−) cerebellar extracts using antibodies to the N-terminal of GTPBP2. GAPDH was used as a loading control. (B) Co-immunoprecipitation (IP) from HEK293T cells co-transfected with FLAG-fusion proteins as indicated, and Pelota-HA. Input levels were determined by immunoblotting (IB). (C) GST-pull down of brain lysate (BL). The pulldown eluate and GST-fused baits were immunoblotted as indicated. (D) Bacterially expressed MBP-GTPBP2-His or MBP-His were purified and were incubated with GST or GST-Pelota. GST-pull down products (top panel) and input (middle and bottom panels) were immunoblotted with anti-His antibody (top two panels) or visualized by Coomassie blue (bottom panel).
GTPBP2 shares domain homology with a translational GTPase family that is characterized by the no-go and non-stop decay/ribosome recycling protein Hbs1, and the eukaryotic release factor eRF3, which bind Dom34 and eRF1, respectively (fig. S4A) (7–9). Although no interaction was observed between GTPBP2 and eRF1 in co-immunoprecipitation assays, Pelota (the mammalian Dom34 homolog) was immunoprecipitated by GTPBP2 (Fig. 2B). GST-GTPBP2 also pulled down Pelota from brain extracts demonstrating that GTPBP2 can interact with endogenous Pelota (Fig. 2C). Affinity capture of bacterially expressed GTPBP2 by Pelota demonstrated that these proteins directly interact (Fig. 2D).
Analysis of mice from our mapping cross and B6J.BALBChr1 congenic mice revealed that homo-or heterozygosity for a BALB/cJ-derived locus (Mod205) on distal Chromosome 1 suppressed neurodegeneration in nmf205−/− and Gtpbp2−/− mice (fig. S5). Mutant mice carrying this BALB locus routinely survived to 18 months or longer. Further analysis of multiple other inbred strains including C57BL/6NJ (B6N) suggested that neurodegeneration in B6J-nmf205−/− mice is likely due to an epistatic mutation in the B6J strain (table S1).
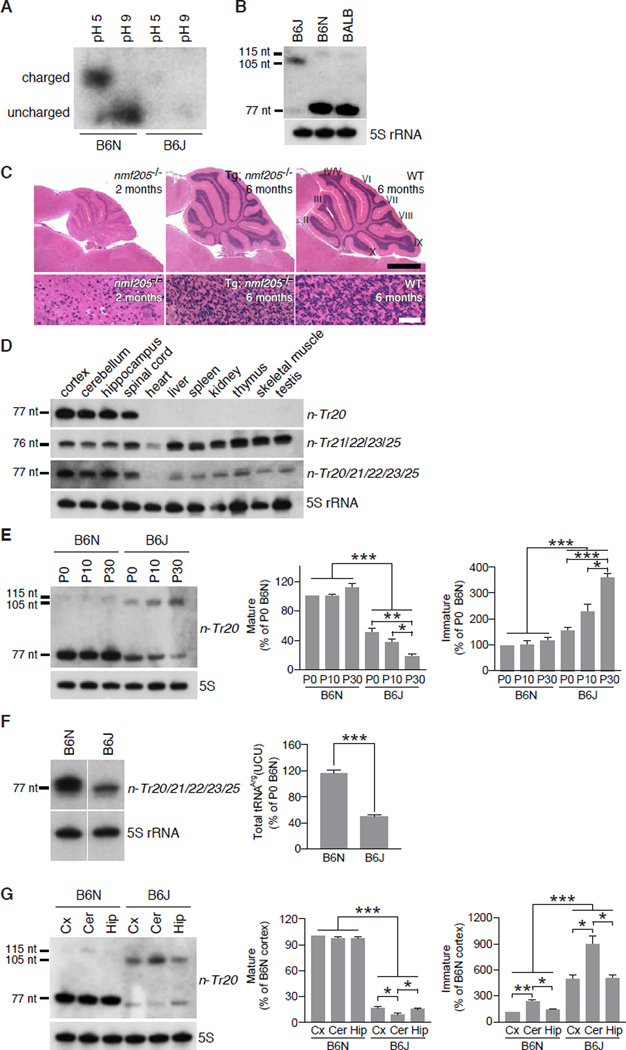
One SNP in the Mod205 region, rs46447118, was determined to be a T in the B6J genome but a C in all other tested strains (fig. S6A). This SNP resides at nucleotide 50 in the stem of the T-loop of n-Tr20, one of 5 isodecoders of the nuclear-encoded tRNAArgUCU family (fig. S6B and C). Orthologs of n-Tr20 are widely found in both vertebrates and invertebrates (fig. S6D). We assayed n-Tr20 aminoacylation and found that the majority of this tRNA was charged in the B6N brain, but very low levels were observed in B6J (Fig. 3A). Mutations in the T-stem of tRNAs have been shown to affect pre-tRNA processing and function (10, 11). In agreement, a 105-nt band was detected in the B6J brain, which was confirmed to be the pre-tRNA retaining the leader and trailer sequences (Fig. 3B, and fig. S7A). In wild-type brains the pre-tRNA is 115-nt, suggesting the C-to-T mutation changes the length of the primary transcript. Examination of n-Tr20 processing in reciprocal congenic strains confirmed that this mutation underlies the observed maturation defect (fig. S7B).
Fig. 3. Mutation of the CNS-specific tRNAArg gene n-Tr20 underlies nmf205-mediated neurodegeneration.
(A) Acylated (pH 5) and deacylated (pH 9) n-Tr20 detected by northern blot analysis of acid PAGE gels. (B) Northern blot analysis of n-Tr20 in brain RNA from P30 mice. (C) H and E-stained sagittal sections of cerebella from 2-month-old B6J-nmf205−/− (nmf205−/−), 6-month-old Tg(n-Tr20B6N)609; B6J-nmf205−/−and 6-month-old wild-type (WT; B6J) mice. Higher magnification images of lobule IX from are shown in the lower panel. Scale bars: 1 mm (C, top panel) and 50 µm (bottom panel). (D) Northern blot analysis of n-Tr20, its isodecoders, and all tRNAArgUCU genes. 5S rRNA was used as loading control. (E) Northern blot analysis of n-Tr20 in P0, P10, and P30 B6N and B6J brain RNA. Mature and immature n-Tr20 transcripts were quantified relative to the P0 B6N brain. (F) Northern blot analysis of n-Tr20 in the P30 B6N and B6J cortex (Cx), cerebellum (Cer), and hippocampus (Hip). Mature and immature n-Tr20 was quantified relative to the B6N cortex. (G) Northern blot analysis of B6N and B6J P30 brain RNA using pooled probes to n-Tr20/21/22/23/25 tRNAs. Bands were quantitated relative to the P0 B6N brain. Error bars indicate SEMs. All data are representative of independent experiments with ≥ 3 mice. *p < 0.05, **p < 0.005 and ***p < 0.0005 (two-tailed Student's t test).
To confirm that loss of mature n-Tr20 underlies neurodegeneration in B6J-nmf205−/− mice (which are mutant for both Gtpbp2 and n-Tr20), B6J mice transgenically expressing wild-type n-Tr20 and harboring the nmf205 mutation (Tg; nmf205−/−) were examined (fig. S8A and B). At 6-months of age, neuron death was greatly attenuated in the brain and retina (Fig. 3C).
Although Gtpbp2 is widely expressed (fig. S4B) (12, 13), pathology in mice lacking this gene is restricted to the CNS. In contrast to other members of the tRNAArgUCU family, expression of both mouse and human n-Tr20 was surprisingly confined to the CNS (Fig. 3D, and fig. S8, C and D). In addition, overall expression of the tRNAArgUCU isodecoder family was higher in the CNS than other tissues (Fig. 3D). Compared to age-matched B6N brains, which show steady postnatal expression, levels of processed n-Tr20 in the B6J brain fell from 50% of B6N levels at P0 to 19% by P30 (Fig. 3E) and a concomitant increase in immature n-Tr20 was also observed. Although B6J brains have a slight increase in expression of the other members of the tRNAArgUCU family, a dramatic reduction in the B6J total tRNAArgUCU pool was observed demonstrating that n-Tr20 normally comprises approximately 60% of the expression of this isodecoder family (Fig. 3F, and fig. S9). Spatial differences in processing of mutant n-Tr20 were also observed within the B6J brain with significantly lower levels of processed and higher levels of unprocessed n-Tr20 in the cerebellum compared to the cortex and hippocampus (Fig. 3G). Together, these data define a CNS-specific tRNA in which levels of mature transcript correlate with the onset and severity of cell death in Gtpbp2-deficient mice.
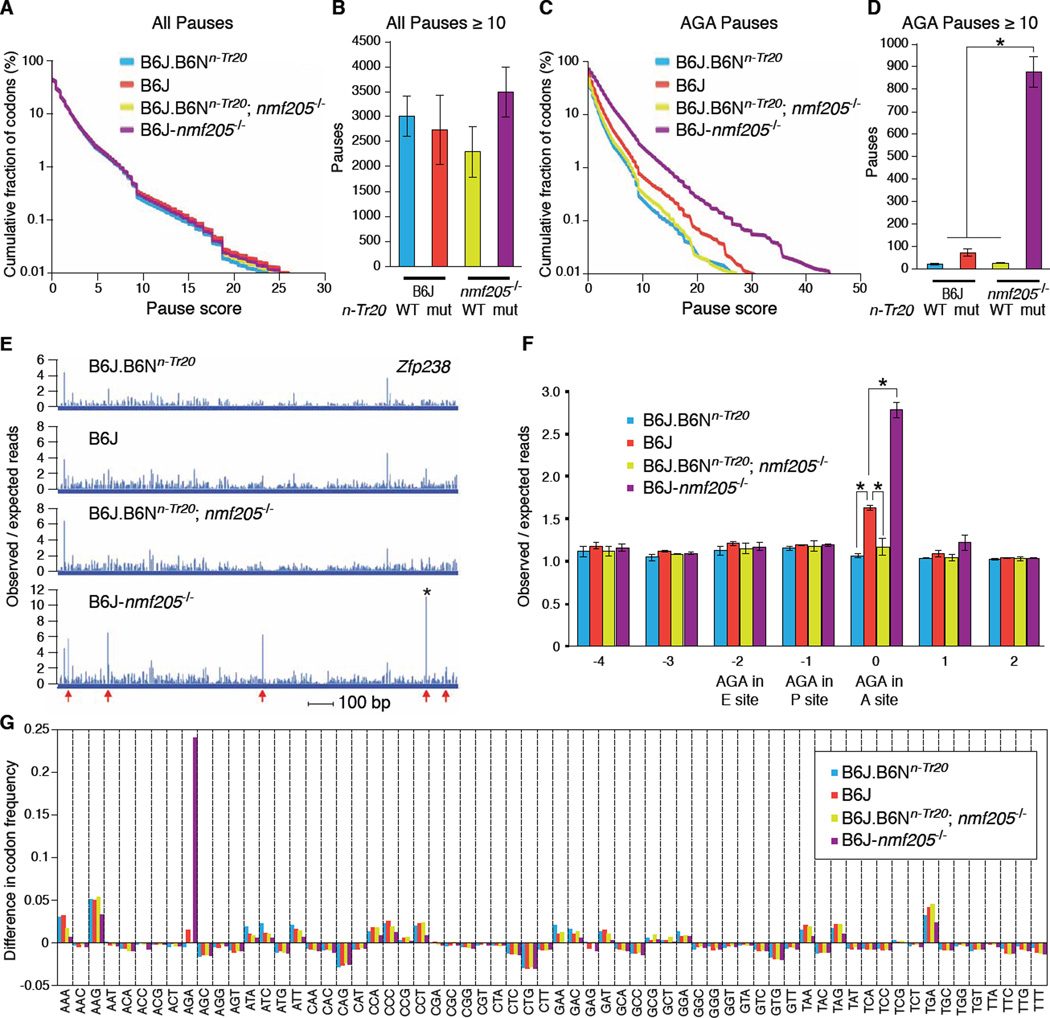
We hypothesized that the n-Tr20 mutation causes ribosome stalling at AGA codons that is exacerbated in the absence of Gtpbp2. To test this we generated ribosome profiling libraries from cerebella of 3-week-old B6J (n-Tr20 mutant), B6J.B6Nn-Tr20 (n-Tr20 wild-type), B6J-nmf205−/− (Gtpbp2−/−; n-Tr20 mutant) and B6J.B6Nn-Tr20; nmf205−/− (Gtpbp2−/−; n-Tr20 wild-type) mice (14–16). We calculated the pause strength for each codon in the ribosome A site for every gene (figs. S10 to S14). Consistent with prior studies, we observed thousands of strong pauses (P ≥10 standard deviations above background) including a well-studied pause in Xbp1, in each genotype (Fig. 4, A and B, fig. S15A, tables S2, A to H, and table S3) (16, 17). However no significant differences in pause number occurred between genotypes.
Fig. 4. The n-Tr20 mutation induces ribosome stalling which is resolved by GTPBP2.
(A) Cumulative distribution of pauses at all codons averaged between replicates. (B) The mean number of pauses ≥10 standard deviations above the background translation levels of their genes (P≥10). (C) Cumulative distribution of pause scores at AGA codons averaged between replicates. (D) The mean number of pauses (P≥10) at AGA codons. (E) Read profile for Zfp238 CDS. * indicates an AGA pause with P=45. Arrows indicate AGA codons. (See fig. S16). (F) Average pausing magnitude at AGA codons, calculated by dividing genome-wide observed reads at AGA codons by expected reads. Expectations are based on read densities in genes containing an AGA. Error bars indicate standard deviations across biological replicates. *p < 0.05 (two-tailed Student's t test). (G) Difference in the codon frequency observed in the A site at pauses (P≥10) versus the genome-wide codon frequency.
We then determined the number of pauses at AGA codons (Fig. 4C, and fig. S15B). In the B6J.B6Nn-Tr20 and B6J.B6Nn-Tr20; nmf205−/− cerebellum few strong AGA pauses were observed (Fig. 4D, and tables S2, I to P). Demonstrating the effect of impaired n-Tr20 processing, a 3-fold increase in strong AGA pauses was observed in the B6J cerebellum. However, the number of AGA pauses increased dramatically in the B6J-nmf205−/− cerebellum (Fig. 4D and E, and fig. S16). While only a limited number of AGA codons exhibited strong pausing (P ≥10), strong pause sites and scores overall showed significant concordance in biological replicates (18) (fig. S1, 4, and table S4). Gene ontology analysis of transcripts with strong pauses in the B6J-nmf205−/− cerebellum showed enrichment for translation-related genes, among others (table S2Q).
Reads at AGA codons in B6J and B6J-nmf205−/− cerebella were 1.6 and 2.8 times background expectations, respectively, whereas unusual AGA pausing was not observed in cerebellar libraries from B6J.B6Nn-Tr20 and B6J.B6Nn-Tr20; nmf205−/− mice (Fig. 4F). To determine if the increase in ribosome pausing in the B6J and B6J-nmf205−/− mice is specific to AGA codons, we compared codon frequency at (P ≥ 10) pause sites to the overall codon usage in transcripts. Although minor deviations were observed for several other codons, the strain-specific AGA effect was much larger than any other effect, demonstrating that the increase in ribosome pausing during translation in the B6J and B6J-nmf205−/− cerebellum occurs specifically at AGA codons (Fig. 4G).
Our data demonstrate that loss of function of a nuclear encoded tRNA induces ribosome stalling that is normally resolved by GTPBP2 (fig. S17). Interestingly, Hbs1l, another ubiquitously expressed Pelota-binding partner, does not rescue neurodegeneration in the absence of GTPBP2, which is consistent with non-overlapping functions of these proteins (fig. S4B) (19). In addition, the tissue-specific expression of n-Tr20 suggests that the regulation of individual isodecoder tRNAs may enable translational regulation in mammals. Further, our finding of a pathogenic mutation in one isodecoder of a five-member gene family underlines the possible deleterious consequences of epistatic mutations in individual members of cytoplasmic tRNA genes that could impact the readout of other mutations, including synonymous SNPs. Finally, these data also emphasize the potential for regulation and disease of mutations in individual members of multicopy gene families.
Supplementary Material
Acknowledgements
Data is deposited in GenBank (GSE56127). We thank The Jackson Laboratory sequencing, histology, microinjection, and multimedia services for their assistance. We also thank Krystal Brown for mouse husbandry assistance and Dr. Wayne Frankel for comments on the manuscript. This work was supported in part by an institutional CORE grant CA34196 (JAX) and a National Science Foundation Award 0850155 to JHC as part of the American Recovery and Reinvestment Act. S.L.A. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Genomic tRNA database. ( http://gtrnadb.ucsc.edu).
- 2.Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–6146. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell. 2007;99:475–487. doi: 10.1042/BC20070037. [DOI] [PubMed] [Google Scholar]
- 4.Moore SD, Sauer RT. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 2005;58:456–466. doi: 10.1111/j.1365-2958.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- 5.Rooijers K, Loayza-Puch F, Nijtmans LG, Agami R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013;4:2886. doi: 10.1038/ncomms3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldowitz D, et al. Large-scale mutagenesis of the mouse to understand the genetic bases of nervous system structure and function. Mol. Brain Res. 2004;132:105–115. doi: 10.1016/j.molbrainres.2004.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levinger L, Serjanov D. Pathogenesis-related mutations in the T-loops of human mitochondrial tRNAs affect 3' end processing and tRNA structure. RNA Biol. 2012;9:283–291. doi: 10.4161/rna.19025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levinger L, et al. Sequence and structure requirements for Drosophila tRNA 5'- and 3'- end processing. J. Biol. Chem. 1995;270:18903–18909. doi: 10.1074/jbc.270.32.18903. [DOI] [PubMed] [Google Scholar]
- 12.Kudo H, Senju S, Mitsuya H, Nishimura Y. Mouse and human GTPBP2, newly identified members of the GP-1 family of GTPase. Biochem. Biophys. Res. Commun. 2000;272:456–465. doi: 10.1006/bbrc.2000.2763. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe M, et al. Cloning, expression analysis, and chromosomal mapping of GTPBP2, a novel member of the G protein family. Gene. 2000;256:51–58. doi: 10.1016/s0378-1119(00)00346-2. [DOI] [PubMed] [Google Scholar]
- 14.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supplementary material on Science Online
- 19.Guydosh NR, Green R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.