Abstract
Purpose:
The authors have recently developed a novel 4D-MRI technique for imaging organ respiratory motion employing cine acquisition in the axial plane and using body area (BA) as a respiratory surrogate. A potential disadvantage associated with axial image acquisition is the space-dependent phase shift in the superior–inferior (SI) direction, i.e., different axial slice positions reach the respiratory peak at different respiratory phases. Since respiratory motion occurs mostly in the SI and anterior–posterior (AP) directions, sagittal image acquisition, which embeds motion information in these two directions, is expected to be more robust and less affected by phase-shift than axial image acquisition. This study aims to develop and evaluate a 4D-MRI technique using sagittal image acquisition.
Methods:
The authors evaluated axial BA and sagittal BA using both 4D-CT images (11 cancer patients) and cine MR images (6 healthy volunteers and 1 cancer patient) by comparing their corresponding space-dependent phase-shift in the SI direction and in the lateral direction , respectively. To evaluate sagittal BA 4D-MRI method, a motion phantom study and a digital phantom study were performed. Additionally, six patients who had cancer(s) in the liver were prospectively enrolled in this study. For each patient, multislice sagittal MR images were acquired for 4D-MRI reconstruction. 4D retrospective sorting was performed based on respiratory phases. Single-slice cine MRI was also acquired in the axial, coronal, and sagittal planes across the tumor center from which tumor motion trajectories in the SI, AP, and medial–lateral (ML) directions were extracted and used as references from comparison. All MR images were acquired in a 1.5 T scanner using a steady-state precession sequence (frame rate ∼ 3 frames/s).
Results:
4D-CT scans showed that was significantly greater than (p-value: 0.012); the median phase-shift was 16.9% and 7.7%, respectively. Body surface motion measurement from axial and sagittal MR cines also showed was significantly greater than . The median and was 11.0% and 9.2% (p-value = 0.008), respectively. Tumor motion trajectories from 4D-MRI matched with those from single-slice cine MRI: the mean (±SD) absolute differences in tumor motion amplitude between the two were 1.5 ± 1.6 mm, 2.1 ± 1.9 mm, and 1.1 ± 1.0 mm in the SI, ML, and AP directions from this patient study.
Conclusions:
Space-dependent phase shift is less problematic for sagittal acquisition than for axial acquisition. 4D-MRI using sagittal acquisition was successfully carried out in patients with hepatic tumors.
Keywords: 4D-MRI, tumor motion, respiratory motion, cine-MRI, liver cancer
1. INTRODUCTION
Motion management plays a critical role in radiation therapy (RT) of mobile tumors in the thorax and abdomen.1–6 To assess patients’ respiratory motion, four-dimensional computed tomography (4D-CT) is widely used in tumor motion management.7–12 However, the radiation dose delivered to the patient is one of the major concerns of 4D-CT,13,14 especially when considering the trade-off between better image quality and lower imaging dose. Many methods have been proposed to reduce imaging dose using high efficiency detectors15,16 or using iterative reconstruction algorithms.17 Moreover, soft tissue tumors in the liver can be difficult to track using 4D-CT because of its inherent low soft-tissue contrast. 3D phase contrast CT technology has been developed to differentiate interfaces in soft tissue.18 However, it has not been applied to radiation therapy for respiratory management. Conversely, MRI provides excellent soft-tissue contrast, and subjects are not exposed to ionization radiation. Therefore, 4D-MRI is highly desirable for better visualization of tumor motion, especially for cancers in the abdomen.
Several methods of 4D-MRI have been recently proposed. A detailed review of these studies can be found in the literature.19 In brief, there are two main approaches to develop 4D-MRI. (1) Prospective 4D-MRI: use fast 3D MR sequences to acquire real-time volumetric images. However, due to current technical limitations, significant compromises in image quality20 have to be made in order to achieve high temporal resolution 4D images. Typical temporal and spatial resolutions of prospective 4D-MRI are approximately 1 s and 4 mm, respectively, which are inadequate for radiotherapy. Recently, several new methods have been developed to accelerate MR image acquisition, such as compressed sensing21,22 and multiband sequences.23 However, applications of these techniques in 4D-MRI have not yet been comprehensively investigated. (2) Retrospective 4D-MRI: use fast 2D MR sequences to continuously acquire images from all respiratory phases and all slice locations, and then retrospectively sort them by respiratory phases. Compared to prospective 4D-MRI, retrospective 4D-MRI has substantially improved spatial/temporal resolution and reduced motion artifacts. However, it requires some form of surrogate to monitor the patient’s breathing during image acquisition, adding a layer of complexity and monitor equipment or technique cost to the 4D-MRI.
We have previously demonstrated a retrospective 4D-MRI technique which uses body area (BA) as an internal respiratory surrogate.19 An advantage of an internal surrogate over an external surrogate is the elimination of a breathing monitoring device and invasive procedure. Since the breathing signal is directly extracted from the images, internal surrogates have the potential to reduce cost and improve the accuracy of 4D-MRI reconstruction. In our previous study,19 image acquisition of 4D-MRI was performed in the axial plane, mimicking the multicine MR image acquisition scheme. This technique has been demonstrated in both phantoms and human subjects.19
In our recent studies, we further investigated the robustness of the axial BA surrogate in 31 lung24 and 7 liver cancer patients.25 We found that the axial BA surrogate generally matched well with the real-time position management (RPM) surrogate (Varian Medical Systems, Inc., Palo Alto, CA). It was discovered, however, that the correlation between the two was significantly better in the abdomen than in the thorax. Furthermore, it was found that the accuracy of respiratory phase calculation and 4D-MRI reconstruction can be affected by space-dependent phase shift (δSPS), i.e., different axial slice positions reach the respiratory peak at different respiratory phases. This phenomenon and its effect on 4D imaging have not been comprehensively investigated before. Tarte et al. conducted a study to measure the relative volumetric contributions of abdominal and thoracic breathing, which illustrated the difference of volume proportions per breathing cycle of thoracic and abdominal between thoracic breathing and abdominal breathing.26 In addition, Nehrke et al. also investigated the difference of respiratory motion in abdomen and thorax region, as well as the different anatomical directions with MR navigator echo methods.27 The observed space-dependent phase shift could potentially influence the reconstruction of 4D-MRI using the BA method. In this study, we investigated the feasibility of 4D-MRI using sagittal image acquisition in combination with BA surrogate. Compared to axial image acquisition, sagittal image acquisition is expected to be a more accurate and robust way of obtaining breathing signal, primarily because respiratory motion occurs mostly in the superior–inferior (SI) and anterior–posterior (AP) directions. Sagittal image acquisition is expected to be less prone to space-dependent phase shift than the axial image acquisition, as proved by 4D-CT. To demonstrate 4D-MRI with sagittal image acquisition, we have validated this technique in a physical motion phantom, a digital human phantom, healthy volunteers, and evaluated its performance in cancer patients.
2. METHODS AND MATERIALS
2.A. Comparison of axial BA surrogate and sagittal BA surrogate
Both axial and sagittal BA surrogates are potentially affected by space-dependent phase shift, but in different directions that are perpendicular to the imaging plane, i.e., axial BA is affected by phase-shift in SI direction (labeled as ) and sagittal BA is affected by phase-shift in lateral direction (labeled as ). One potential advantage of sagittal BA over axial BA is that it is less affected by space-dependent phase shift than, i.e., is less than . To demonstrate that, ideally real-time 4D scans should be performed so that and can be simultaneously determined for a direct comparison. However, it is feasible since there is currently no fast real time 4D imaging technique available. We have thus designed two different studies in a complementary manner to investigate this.
In the first study, we performed a retrospective study using selected, high quality 4D-CT scans of real cancer patients as virtual human phantoms and determine and via computer simulations. These 4D-CT images have minimal to no motion artifacts and were chosen based on multiple metrics using published criteria.24 Cine image acquisition was simulated every 2 cm in both axial and sagittal planes based on the 4D-CT images. Breathing curves were determined for these slice positions using the BA method as described in our previous publication.19 A total of 11 4D-CT datasets were studied. Figure 1 shows an example of the normalized 10-phase breathing curves of different slice locations for the axial (a) and sagittal (b) acquisitions. Different colors represent different slice positions. Respiratory phases were calculated separately for each individual breathing curve. The peaks were set to Phase 0% (or Phase 100%). The phases for other data points were calculated via linear interpolation. Phase shift was calculated as the standard deviation of the valley phases of all breathing curves. The resultant and were compared using the Wilcoxon signed rank test.
FIG. 1.
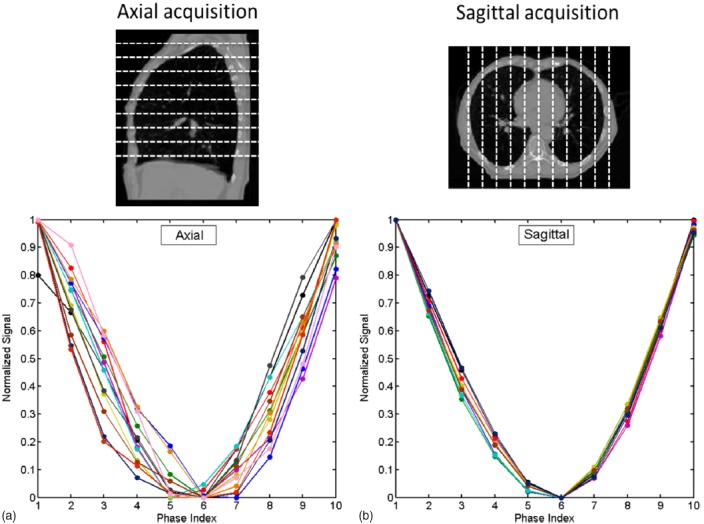
Ten-phase normalized breathing curves at different slice positions for axial acquisition (a) and sagittal acquisition (b) for a representative patient. Different curves indicate different slice positions. Large variations were observed in axial acquisition, while only minimal variation in sagittal acquisition.
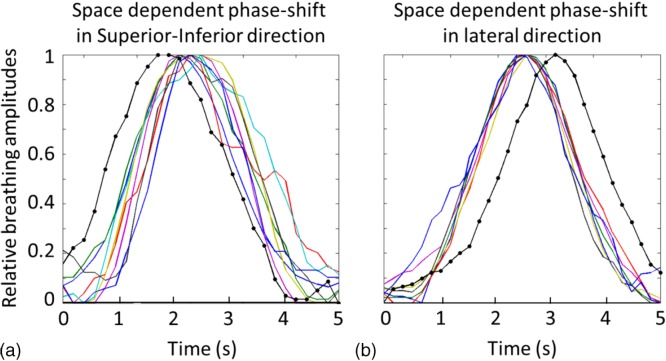
In the second study, we performed a prospective study to acquire cine MR images at multiple slice locations in both axial and sagittal planes during breathing and determine and using these images. Six healthy volunteers and one cancer patient were included in this IRB approved study. All scans were performed in a 3.0 T clinical MR scanner (TrioTim, Siemens Medical Solution, Germany). The physiologic monitoring unit (PMU) with bellows wrapped around the abdomen was used to record respiratory signal of the subjects during the scans. From the cine MR images, we determined breathing signals at different locations by tracking the motion of body surface using the BA method at the corresponding locations, as shown in Figs. 2(a) and 2(b). These breathing signals were used to calculate and for comparison. Significance was determined using the Wilcoxon–Mann–Whitney test.
FIG. 2.
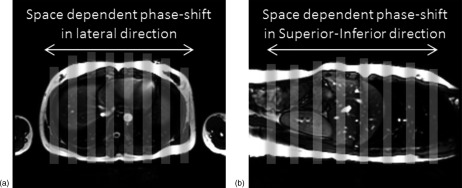
Illustration of respiratory motion tracking at different locations using the cine MR images in the axial plan (a) and sagittal plane (b). Gray stripes indicate the rectangular areas where the BA method was applied to measure the breathing signal.
2.B. Validation of sagittal BA as respiratory surrogate
To validate sagittal BA as a respiratory surrogate, we compared breathing signals determined using the sagittal BA method with those determined using a region of interest (ROI) feature-based motion tracking method.19 This study was performed on the sagittal cine MR images of ten human subjects, of which five were healthy volunteers imaged at the University of Virginia and five were cancer patients imaged at Duke University, both on 1.5 T GE scanners. All subjects signed consent forms prior to the IRB-approved studies. Subjects were imaged continuously in a single sagittal plane (for five healthy volunteers) or multiple sagittal planes (for five cancer patients) using a steady-state precession sequence (labeled as FIESTA by GE and TrueFISP by Siemens). Imaging parameters were repetition time (TR)/echo time (TE), 3.7 ms/1.21 ms; matrix, 256 × 166; field of view (FOV), 350 × 300 mm; flip angle, 52°; slice thickness, 5 mm; frame rate, ∼ 3 frames/s. Imaging time per slice was 2 min for single-slice acquisition and ∼ 10 s for multislice acquisition for all subjects.
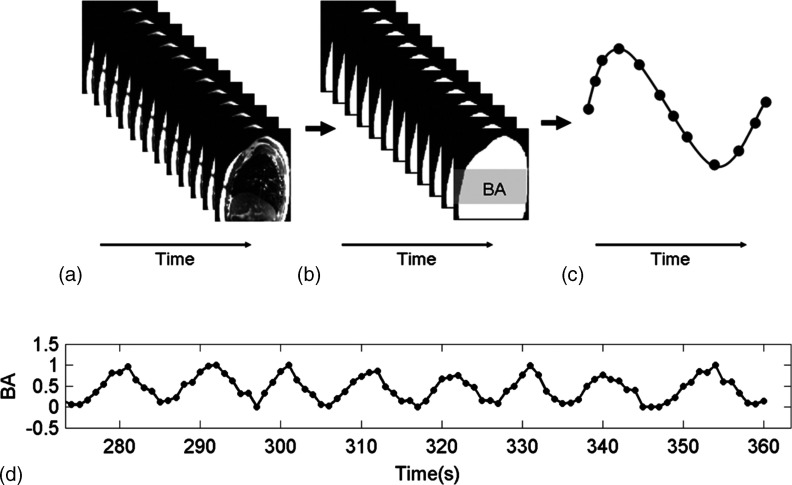
For each subject, breathing signals were determined using the sagittal BA method and the ROI motion tracking method and were compared to each other. ROI motion tracking method tracks the ROI on 2D MR images with an in-house matlab program.28 In the first frame of the MR image series, the ROI to be tracked was manually contoured, and a vicinity searching box within which the ROI was estimated to move was given. Automatic tracking of the ROI in the following MR images was achieved using the maximal cross correlation technique.29 Figure 3 illustrates the workflow of extracting breathing signals from sagittal cine MR images using the BA method, which is similar to the axial BA method as described in our previous publication.19 In short, each sagittal MR image was processed by applying an estimated image intensity threshold based on image noise to determine the sagittal body contour. Morphological operations were then performed to exclude extraneous pixels induced by noise in the image. BA was defined as the number of pixels within the body contour [white area in Fig. 3(b)]. In practice, the BA was calculated only using the central part of the image [gray area in Fig. 3(b)] where the respiratory movement is most significant. For each sagittal slice, an individual breathing curve [Fig. 3(c)] was generated by plotting the BA as the function of image acquisition time. For multiple slice acquisitions, the complete breathing curve was obtained by plotting all individual breathing curves sequentially [Fig. 3(d)].
FIG. 3.
Workflow of extracting breathing signals from sagittal MR images using the BA surrogate. (a) sagittal cine MR images at one slice position, (b) calculation of BA (white area) from sagittal cine MR images. In practice, only the middle section (gray area) was used for BA calculation, (c) the BA-derived breathing curve for a single breathing cycle, and (d) the BA-derived breathing curve for multiple breathing cycles in case of multiple slice acquisitions.
To extract the breathing signal using the ROI motion tracking method, a ROI, such as tumor, diaphragm, or pulmonary vessels, was contoured manually in the first frame of the MR image series and then the ROI was tracked automatically in the following frames by searching the vicinity of the structure and matching with a maximum cross correlation technique. For multiple slice acquisition, the ROI tracking was performed at each slice position, requiring manual contouring of the ROI in the first frame of MR images of each slice position. All breathing curves were normalized prior to the calculation of respiratory phases, using the same method as described in our previous work.19 To validate the sagittal BA as a respiratory surrogate, we compared the breathing curves determined from the two methods using a measure of peak time difference , which is defined as
| (1) |
where is the peak time difference between BA method and ROI tracking method for the ith breathing curve, is the time the ith breathing curve reaches its peak using BA method, is the time the ith breathing curve reaches its peak using ROI tracking method, periodi is the period for the ith breathing curve.
To quantify the difference between the two methods over several breathing curves, the mean and standard deviation of for several breathing curves measured over a period of time could be calculated
| (2) |
| (3) |
where N is the total number of respiratory curves, is the mean peak time difference for an individual subject, and is the standard deviation of the peak time difference for an individual subject.
2.C. Motion phantom study
The 4D-MRI technique with sagittal image acquisition was tested on an in-house constructed MRI-compatible motion phantom,24 consisting of a MRI-compatible motion stage and a motion motor (BrainLAB, Inc., Feldkirchen, Germany). The motion stage consists of a supporting platform (2 cm solid water slab), an inverse-T shaped motion stage made from styrofoam, a cylindrical gel (radius = 2.3 cm, height = 3.2 cm), and a 5 mm-thick bolus piece on a plastic flat board. The motion stage was driven by a motor on one end via a surgical low-elastic thread and attached to the other end via a rubber band. The motor was set to move in a sinusoidal wave (peak-to-peak amplitude = 2.0 cm, period = 5 s), driving the motion stage with cylindrical gel (simulating tumor) to move in the same pattern along the SI direction. Consequently, the bolus piece rotated along the fixed axis, simulating the body surface motion. The triangle area under the bolus was the area under “pseudo chest wall” mimicked the sagittal BA of motion phantom. A photo and structure illustration can be found in Ref. 19. In our study, the actual motion of the phantom is measured by tracking the motion of the cylindrically shaped gel in the multiple slice cine MR images in the coronal plane.
4D-MRI of the phantom was acquired on a clinical 1.5 T scanner (Signa, GE Healthcare, Milwaukee, WI) using a FIESTA sequence and a six channel phased array coil. Multiple slice sagittal cines were acquired. There is no gap in time between image acquisitions at consecutive slices. The breathing signal extracted from MR images is continuous. The MR sequence used does have a steady state process: the first several images show higher signals than the rest images. This phenomenon has been considered in our technique by removing the first several images from 4D-MRI reconstruction. They are still used when extracting breathing signals, so this will not influence the continuity of the breathing signal. Imaging parameters were TR/TE, 3.2 ms/1.0 ms; FOV, 300 × 300 mm; flip angle: 50°; slice thickness, 5 mm; matrix, 192 × 128. Frame rate was 3 frames/s. All images were acquired in the sagittal plane. Using cine mode, each slice was imaged for 6 s. The MR images were interpolated to 256 × 256 before further analysis. BA of the phantom was defined as the area under the bolus piece23 in the sagittal MR images.
Breathing signals were extracted from the sagittal images using the BA surrogate. Respiratory phases were calculated accordingly. Ten-phase 4D-MRI images were then reconstructed based on phase-binning. Gel motion extracted from the reconstructed 4D-MRI was compared to that from the coronal multiple slice cine MR.
2.D. Digital phantom study
The 4D-MRI technique with sagittal image acquisition was also tested on the 4D extended Cardiac Torso (XCAT) digital human phantom developed by Segars et al.30–32 The respiratory motion of the 4D-XCAT phantom was modeled using a regular breathing profile. The 4D-XCAT phantom was generated only for the abdomen region using the following parameters: in-plane resolution, 256 × 256; voxel size, 2.5 mm; maximum diaphragm motion, 2.0 cm; maximum anterior body motion, 1.0 cm; breathing period, 5 s; frames per breathing cycle, 21. The XCAT phantom was generated in the activity mode in order to produce MRI-like images. Signal intensities of organs and tissues were assigned using values derived from FIESTA/TrueFISP MR images. A spherical tumor of 3 cm in diameter was inserted into the liver.
Virtual experiments of 4D-MRI using the sagittal BA surrogate were carried out on the 4D-XCAT phantom according to the following steps: (1) mimic the image acquisition of 4D-MRI by continuously extracting images of the same sagittal slice from the 4D-XCAT phantom for more than one breathing cycle, (2) repeat step 1 for all sagittal slice positions, (3) calculate BA of each sagittal slice and determine the breathing curve for each slice position, (4) calculate respiratory phases for each sagittal slice, and (5) retrospectively sort the sagittal slices based on their respiratory phases to generate the simulated “4D-MRI”. In order to evaluate the accuracy of the simulated 4D-MRI, we also generated original 4D-XCAT images using the same respiratory motion profile as a reference for comparison. Motion trajectories of the tumor were determined from the simulated 4D-MRI, and compared to those measured from the original 4D-XCAT images.
2.E. Patient study
Six patients (four female, two male, mean age 62.0) with cancer(s) located in the liver were prospectively enrolled in an IRB-approved study. For each patient, multiple slice sagittal MR images were acquired continuously throughout the breathing cycle for 4D-MRI reconstruction. Single-slice cine MR was also acquired in the axial, coronal, and sagittal planes across the center of the tumor for 30 s. All images were acquired in a 1.5 T GE clinical scanner using the FIESTA sequence, with a frame rate of about 3 frames/s. The subjects were positioned head-first-supine with arms down, and no immobilization device was used. They were instructed to breathe normally during the scans. Each sagittal slice was imaged for approximately 8 s. Imaging parameters were summarized in Table I. Breathing signals were first generated by tracking the changes of BA in the sagittal plane, followed by manual inspection and correction for erroneous peak detections. Respiratory phases were calculated and 4D-MRI were reconstructed as described in Secs. 2.A–2.B. Tumor motion trajectories in the SI, AP, and medial–lateral (ML) directions were determined from 4D-MRI and compared to those from single-slice cine MR images, which served as references. Absolute amplitude difference between 4D-MRI and cine MRI for each respiratory phase bin has been calculated, labeled as absolute error. Furthermore, since each patient has different maximum respiratory motion amplitude for each individual cycle, the absolute error might not adequate to indicate curve differences. The relative amplitude difference, which is defined as the absolute amplitude differences divided by the total absolute amplitude of each individual respiratory cycle were also calculated, labeled as relative error (%).
TABLE I.
Summary of MR acquisition parameters.
| TR | 2.7 ms |
| TE | 1.0 ms |
| FOV | 480 × 480 mm |
| Flip angle | 50° |
| Slice thickness | 5 mm |
| Matrix | 256 × 256 |
| Bandwidth | 977 Hz/pixel |
Note: TR, repetition time; TE, echo time; FOV, field of view.
3. RESULTS
3.A. Comparison of axial BA surrogate and sagittal BA surrogate
Results of phase-shift analysis based on 4D-CT for a representative patient are shown in Fig. 1. The breathing curves for the axial acquisition have larger valley phase variation than those of the sagittal acquisitions: the valley phases were located among phase 50%, 60%, and 70%, for the axial acquisition, while they were all located in phase 60% for the sagittal acquisition. For example, for the case as shown in Fig. 1, and were 6.2% and 0%, respectively. Averaging over 11 patients, was found to be significantly (p-value: 0.012) greater than : the median was 16.9% and 7.7% for and , respectively. Table II summarizes the results for all patients.
TABLE II.
Validation of space-dependent phase shift: Summary of and measurements on 4D-CT for 11 cancer patients, along with sign test results.
| Patient # | Total number of axial slices | |||
|---|---|---|---|---|
| 1 | 28.3 | 25.6 | Yes | 76 |
| 2 | 5.8 | 3.6 | Yes | 116 |
| 3 | 37.8 | 25.0 | Yes | 112 |
| 4 | 13.4 | 7.7 | Yes | 80 |
| 5 | 29.4 | 12.3 | Yes | 128 |
| 6 | 20.5 | 19.8 | Yes | 100 |
| 7 | 4.4 | 6.1 | No | 72 |
| 8 | 16.9 | 8.9 | Yes | 108 |
| 9 | 31.8 | 7.5 | Yes | 124 |
| 10 | 9.2 | 7.5 | Yes | 68 |
| 11 | 5.9 | 2.9 | Yes | 120 |
| Median | 16.9 | 7.7 | Sign test: p = 0.012 | 100 (±22) |
Figure 4 shows normalized breathing signals at different locations from the same breathing cycle extracted from the cine MR images for a representative subject. It can be clearly seen that the breathing signals had greater variation in respiratory phase in the SI direction than in the lateral direction. In both cases, the PMU respiratory signal deviated from the BA-derived breathing signals, presumably due to the differences between external surrogate motion and internal anatomical motion.33 However, it should be noted that the calculation of phase-shift is independent of the PMU breathing signal. A total of 930 breathing cycles were analyzed for seven subjects, and the median was 11.0% and 9.2% (p-value = 0.008), for and , respectively.
FIG. 4.
Representative respiratory cycles illustrate the space dependent phase-shift in SI direction (a) and lateral direction (b). Gray curves without dots show the body surface motion extracted from MR cine and black curves with dots show the respiratory signal recorded by PMU.
3.B. Validation of sagittal BA as respiratory surrogate
Figure 5 shows an example of a good match in normalized breathing signals between the sagittal BA method and the ROI tracking method for a single-slice acquisition. Averaging over five patients, the mean (±standard deviation) of for single-slice acquisition was 0.06 (±0.02). Figure 6 shows an example of the comparison in normalized breathing signals between the sagittal BA method and the ROI tracking method for a multiple slice acquisition. Despite difference in the amplitude, the respiratory peaks generally matched well ( is 0.07 for this patient). Averaging over five patients, the mean (±SD) of for single-slice acquisition was 0.06 (±0.01). Table III summarizes the results for all comparisons.
FIG. 5.
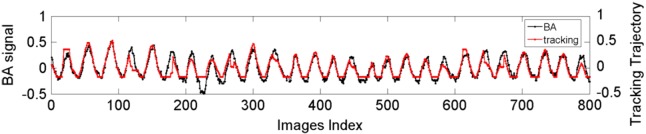
Comparison of breathing signals and respiratory phases between the BA method (black) and the ROI motion tracking method (gray) for a single-slice acquisition. Respiratory amplitudes have been normalized.
FIG. 6.
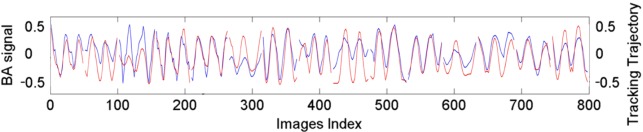
Comparison in normalized breathing signals between the sagittal BA method (black) and the ROI tracking method (gray) for a multiple slice acquisition. In this example, the imaging time per slice position is slightly more than two breathing cycles.
TABLE III.
Summary of measurements of and for each patient.
| Single-slice patients | Multiple slice patients | ||||
|---|---|---|---|---|---|
| Subject # | Subject # | ||||
| 1 | 0.059 | 0.049 | 1 | 0.069 | 0.025 |
| 2 | 0.10 | 0.050 | 2 | 0.064 | 0.022 |
| 3 | 0.022 | 0.019 | 3 | 0.056 | 0.029 |
| 4 | 0.059 | 0.038 | 4 | 0.080 | 0.035 |
| 5 | 0.063 | 0.051 | 5 | 0.040 | 0.032 |
| Mean (±SD) | 0.061 (±0.022) | — | Mean (±SD) | 0.062 (±0.011) | — |
3.C. Motion phantom study
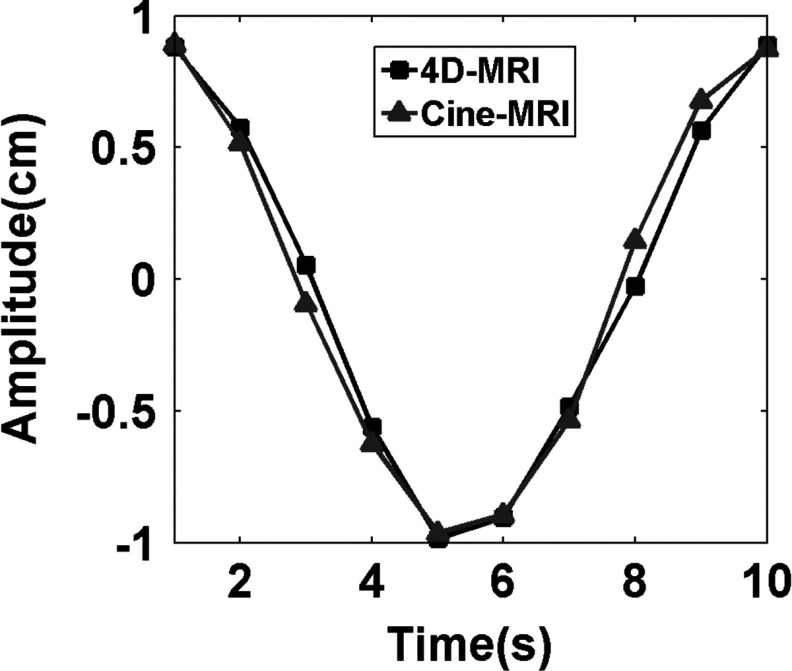
Figure 7 shows the relative location of the gel target at ten different phases in all three planes. Sagittal images are the originally acquired images, while axial and coronal images are the reconstructed images of the 4D-MRI. Clear sinusoidal motion is observed in all three planes with minimal image artifacts on the reconstructed images. Interpolation-induced blurring exists on the axial and coronal planes, without substantially degrading the overall image quality. The target motion measured from 4D-MRI is consistent with the input signal, as demonstrated in Fig. 8. The mean (±SD) absolute difference in target motion amplitude between the two is 0.70 (±0.64) mm.
FIG. 7.
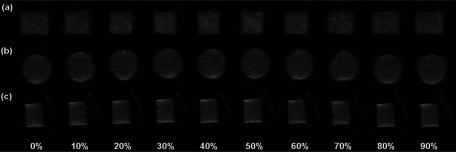
Ten-phase 4D-MRI images of a cylindrical gel phantom in (a) axial, (b) coronal, and (c) sagittal planes. Images were acquired in sagittal planes (thus it has high resolution).
FIG. 8.
Comparison of motion trajectories in SI direction of the imaging object in the phantom study between 4D-MRI and coronal cine MR.
3.D. Digital phantom study
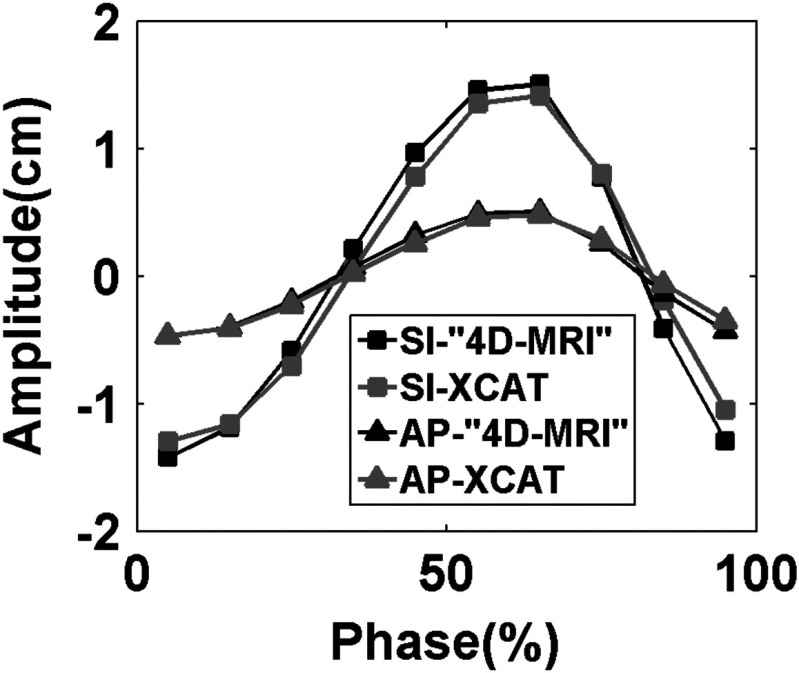
Figure 9 shows the respiratory signals and phases of the 4D-XCAT phantom that are determined using sagittal BA surrogate. Simulated 4D-MRI of the XCAT phantom matched well with the original 4D-XCAT phantom, as illustrated in Fig. 10. No apparent artifacts were observed. Figure 11 shows that the motion trajectory of the hypothesized tumor matched well with the input motion profile: the mean (±SD) absolute difference in motion amplitude is 1.3 (±0.7) mm in the SI direction and 0.4 (±0.3) mm in the AP direction.
FIG. 9.
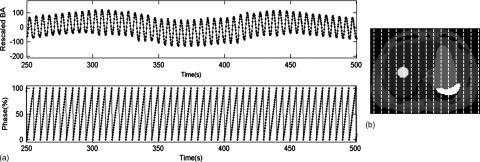
(a) Breathing signals and respiratory phases determined using the sagittal BA method in the digital phantom study. (b) Axial view of the XCAT phantom with a pseudo liver tumor. Organs’ intensities of the XCAT phantom were assigned to mimic T2-weighted MRI. Vertical dashed lines indicate the slice positions at which the sagittal cine images were acquired.
FIG. 10.
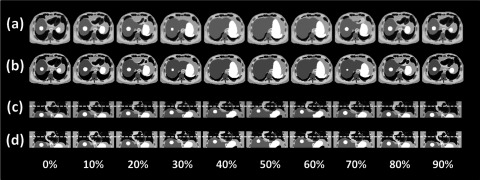
Comparison between (a) the original 10-phase 4D-XCAT phantom images and (b) the simulated 10-phase 4D-MRI images of the 4D-XCAT phantom in axial view; comparison between (c) the original 10-phase 4D-XCAT phantom images and (d) the simulated 10-phase 4D-MRI images of the 4D-XCAT phantom in coronal view; dashed lines were added for better visualization of motion. Minimal sorting artifacts were observed in certain phases of the simulated 4D-MRI images.
FIG. 11.
Compare breathing curve in SI and AP directions between reconstructed 4D-MRI and original XCAT 3D images.
3.E. Patient study
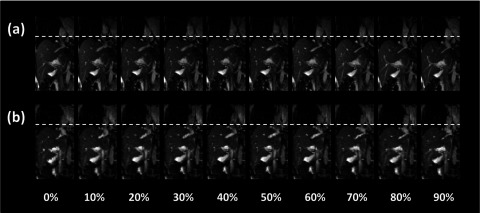
Breathing signals of the six cancer patients were successfully extracted from the sagittal MR images using the BA method. Figure 12 shows an example of the breathing curves of a representative patient (only breathing curves of the right side of the body are shown). 4D-MRI images successfully revealed respiratory motion of all six patients. Figure 13 shows the 10-phase 4D-MRI images in the axial and coronal views of a representative patient. Reconstructed coronal 4D-MRI images are largely consistent with coronal cine MR images, as illustrated in Fig. 14 for the same subject. Furthermore, tumor motion trajectories determined from 4D-MRI matched well with those from cine MR, as summarized in Table IV. Averaging over six patients, the mean (±SD) absolute differences in tumor motion amplitude between 4D-MRI and cine MR were 1.5 (±1.6), 2.1 (±1.9), and 1.1 (±1.0) mm in the SI, ML, and AP directions, respectively. Figure 15 shows an example of the comparisons of the tumor motion trajectories.
FIG. 12.
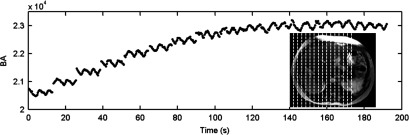
Breathing signals (for only the right side of the body) extracted from sagittal MR images using the BA surrogate. Dash lines indicate the imaged sagittal planes.
FIG. 13.
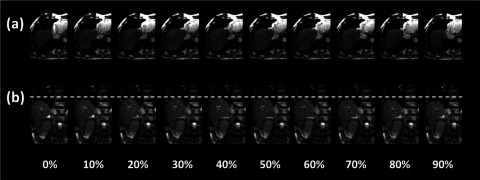
Ten-phase axial and coronal 4D-MRI of a representative patient.
FIG. 14.
Ten-phase coronal images of the liver in cine MR images (a) and 4D-MRI images (b) of a representative patient.
TABLE IV.
Tumor motion trajectories comparison in patient study, 4D reconstructed images with reference from single-slice cine MR. Measurements of tumor CNR comparing CT and MRI fast sequence FIESTA.
| Mean absolute error (mm) | Amplitude from cine (mm) | Relative error (%) | CNR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient # | Age | Gender | Cancer site | SI | AP | ML | SI | AP | ML | SI | AP | ML | CT | FIESTA |
| 1 | 64 | M | Liver Mets | 4.7 | 2.4 | 5.5 | 16 | 15 | 9.9 | 30 | 16 | 56 | 3.5 | 7.8 |
| 2 | 76 | F | HCC | 0.25 | 0.12 | 2.0 | 3.6 | 1.7 | 1.8 | 6.8 | 7.2 | 108 | 0.12 | 4.1 |
| 3 | 68 | M | Liver Mets | 1.0 | 2.4 | 0.96 | 12 | 12 | 6.4 | 8.9 | 19 | 15 | 2.9 | 2.0 |
| 4 | 70 | F | HCC | 1.5 | 0.63 | 0.56 | 8.2 | 4.8 | 2.1 | 18 | 13 | 27 | 2.9 | 56 |
| 5 | 58 | M | HCC | 0.42 | 0.38 | 0.37 | 6.1 | 1.9 | 1.3 | 6.9 | 20 | 28 | 0.15 | 3.1 |
| 6 | 34 | F | HCC | 1.2 | 0.87 | 3.0 | 8.1 | 1.7 | 4.3 | 14 | 52 | 69 | 2.8 | 17 |
| Mean (±SD) | 65.0 (±9.7) | — — | — — | 1.5 (±1.6) | 1.1 (±1.0) | 2.1 (±2.0) | — — | — — | — — | 14 (±9.1) | 21 (±16) | 50 (±35) | 2.1 (±1.5) | 15 (±21) |
Note: F, female; M, male; SD, standard deviation; SI, superior–inferior; AP, anterior–posterior; ML, medial–lateral; CNR, contrast-to-noise ratio; CT, computed tomography; FIESTA, fast imaging employing steady-state acquisition; HCC, Hepatocellular carcinoma; Liver Mets, liver metastasis.
FIG. 15.
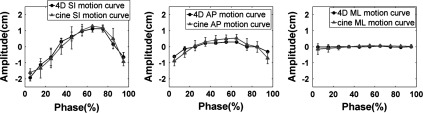
Comparison of tumor motion trajectories between 4D-MRI and cine MR.
4. DISCUSSION
In this study, we investigated a retrospective 4D-MRI technique that uses sagittal image acquisition with body area as an internal respiratory surrogate. Our results demonstrated that sagittal BA can provide comparable accuracy to ROI motion tracking in extracting breathing signals.19 We have found that for the BA respiratory surrogate, sagittal image acquisition is less prone to, and almost unaffected by, space-dependent phase shift than axial image acquisition. We have also demonstrated that 4D-MRI can be successfully generated using sagittal image acquisition together with BA as respiratory surrogate in phantom and patient studies. Compared to our previous work on 4D-MRI using axial acquisition,19 this work is different in the following aspects: (1) it demonstrated the feasibility of 4D-MRI using BA with sagittal acquisition, which was fully validated by motion phantom, digital phantom, and patient study. Although similar in principle, the physiological meaning of sagittal acquisition is fundamentally different from that of axial acquisition. The validation and feasibility of sagittal acquisition for 4D-MRI required entirely different sets of experiments. (2) Space-dependent phase shift is an important physiological phenomenon that affects 4D imaging. It has not yet been thoroughly studied before. This study for the first time approved the difference in space-dependent phase shift between sagittal and axial acquisitions, which was significant and can provide useful information for future 4D studies. (3) This study included 4D-MRI results of six cancer patients, which by itself is very new as there are only very limited publications on 4D-MRI with cancer patients data. In our previous work,19 only two healthy volunteers were included.
There are several potential advantages of 4D-MRI with sagittal image acquisition over 4D-MRI with axial image acquisition. First, the raw sagittal images have better spatial resolution and fewer artifacts than the reconstructed axial and coronal images and contain the most important respiratory motion information in the SI and AP directions. This makes the 4D-MRI images, and subsequent tumor motion measurement and tumor volume delineation, less affected by reconstruction artifacts and errors. Second, sagittal image acquisition can generate more accurate breathing signals and thus more accurate 4D-MRI than axial image acquisition. This has been partly validated in this study which shows that sagittal image acquisition is less affected by space-dependent phase shift than axial image acquisition. Third, sagittal image acquisition allows for potential further improvement in the accuracy and robustness of the breathing signal determination by combining with other image-based respiratory surrogates, such as the Fourier transform (FT) surrogate. In a previous study, we have34 shown that the FT surrogate is an accurate and robust surrogate for extracting breathing signals from sagittal cine MR images. In our future study, we will attempt to combine the BA and FT surrogates using a measure of spatial coherence35 to improve the accuracy of the surrogate. Finally, sagittal image acquisition may reduce imaging time as compared to axial image acquisition when a large volume for 4D-MRI is imaged.
It should be noted that the fast MR sequences used for retrospective 4D-MRI should have high temporal resolution in order to produce enough phase bins.19,20,36 The frame rate of the fast MR sequence should be greater than the ratio of the patient’s average breathing period over the total number of phase bins of 4D-MRI. Low frame rate of the MR sequence will result in low temporal resolution of 4D-MRI. It is also important to balance the frame rate and the image quality in 4D-MRI. Generally, the higher the frame rate, the shorter repetition time, and subsequently the noisier the MR images,19,20 which will propagate into the final 4D-MRI images. Future work should aim to optimize imaging parameters such as total number of phase bins, total number of slices, and the frame rate of the MR sequences based on patient’s breathing pattern in order to achieve best possible image quality for 4D-MRI.
There are several limitations in the current study. First, comparison between the sagittal BA surrogate and the axial BA surrogate were performed only between each other. Ideally, the evaluation should be performed directly between the sagittal approach and axial approach. As an example, a direct comparison between the two for a hepatic cancer patient was shown in Fig. 16. Less stripe sorting artifacts (red arrows) were observed in sagittal 4D-MRI than in axial 4D-MRI. However, in practice, such comparison faces significant practical challenges and may not provide more information than what has presented in the current study. The major challenge is that the direct comparison would require two separate but identical breathings for axial and sagittal 4D-MRI acquisitions, respectively. This is very challenging in practice to control the breathing of the subject to be exactly the same in two separate time periods. As we know from previous studies, patients breathe highly irregularly, even with the help of audio/video control system. This challenge, in principle, might be potentially resolved in the following two ways. First, by including a large number of patients, among those only the ones with very similar respiratory motion during axial and sagittal image acquisitions will be selected for comparison. Second, by using active breathing control with audio/video coaching to control patient breathing to minimize the difference in breathing between the two scans. Those two approaches would require thoughtfully designed prospective patient studies, which is out of the scope of our current study. In addition, we believe the main reason why sagittal acquisition is superior to axial acquisition is that axial 4D-MRI will be adversely influenced by space-dependent phase shift, while sequential 4D-MRI would not. This has been demonstrated with statistical significance in our current study. The space-dependent phase shift was defined as the standard deviations of the valley phases of all breathing curves at different slice locations. Its value will not be affected by including a ground truth signal in the equation.
FIG. 16.
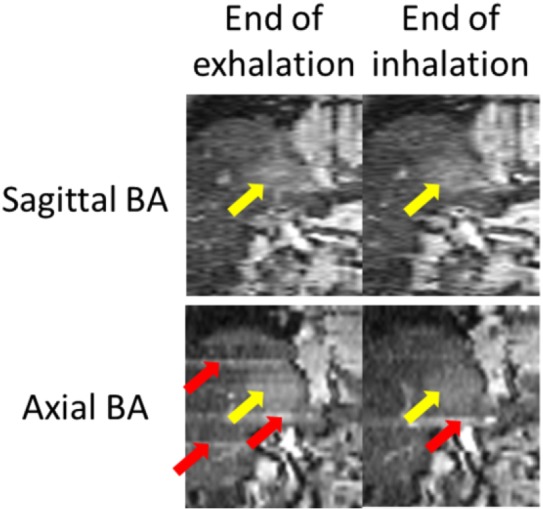
Comparison of 4D-MRI images generated using sagittal BA (top) and axial BA (bottom) for a liver cancer patient. Tumors were indicated by white arrows. Sorting image artifacts were observed in the axial BA 4D-MRI (gray arrows).
Second, a sagittal and an axial 4D-MRI on the same patient would have allowed for a more direct comparison of the breathing signal and image quality. However, due to time and logistical constraints only one set of 4D-MRI, either with sagittal image acquisition or axial image acquisition, was acquired for our studied patients. Third, only a small number of patients (n = 6) were studied. The robustness of the sagittal image acquisition for 4D-MRI needs to be further investigated for a future study with a larger patient sample size.
Finally, a slice thickness of 5 mm in sagittal acquisition was used for patient study. When reformatting into axial slices, this 5 mm in-plane resolution is not acceptable in clinical routine. However, the patient study we conducted is a feasibility study to test the sagittal BA technique. The slice thickness could be further decreased to 3 mm if desired. For most of abdominal cancer delineation, 3 mm in plane resolution should be adequate in most of the cases. Also, many of the current treatment planning software also allow for nonaxial plane contouring.
5. CONCLUSION
It is feasible to extract breathing signals from sagittal cine MRI images using BA as the respiratory surrogate. 4D-MRI using sagittal image acquisition in combination with the BA surrogate demonstrated good accuracy in respiratory motion measurement in our preliminary study. Further investigation is warranted to assess the robustness of the 4D-MRI technique with the sagittal BA surrogate.
ACKNOWLEDGMENTS
This work was partly supported by funding from NIH (1R21CA165384) and a research grant from the Golfers Against Cancer (GAC) Foundation.
REFERENCES
- 1.Keall P. J., Mageras G. S., Balter J. M., Emery R. S., Forster K. M., Jiang S. B., Kapatoes J. M., Low D. A., Murphy M. J., Murray B. R., Ramsey C. R., Van Herk M. B., Vedam S. S., Wong J. W., and Yorke E., “The management of respiratory motion in radiation oncology report of AAPM Task Group 76,” Med. Phys. 33, 3874–3900 (2006). 10.1118/1.2349696 [DOI] [PubMed] [Google Scholar]
- 2.Rietzel E., Chen G. T. Y., Chol N. C., and Willet C. G., “Four-dimensional image-based treatment planning: Target volume segmentation and dose calculation in the presence of respiratory motion,” Int. J. Radiat. Oncol., Biol., Phys. 61(5), 1535–1550 (2005). 10.1016/j.ijrobp.2004.11.037 [DOI] [PubMed] [Google Scholar]
- 3.Ozhasoglu C. and Murphy M. J., “Issues in respiratory motion compensation during external-beam radiotherapy,” Int. J. Radiat. Oncol., Biol., Phys. 52(5), 1389–1399 (2002). 10.1016/S0360-3016(01)02789-4 [DOI] [PubMed] [Google Scholar]
- 4.Xing L., Thorndyke B., Schreibmann E., Yang Y., Li T., Kim G., Luxton G., and Koong A., “Overview of image-guided radiation therapy,” Med. Dos. 31(2), 91–112 (2006). 10.1016/j.meddos.2005.12.004 [DOI] [PubMed] [Google Scholar]
- 5.Ford E. C., Mageras G. S., Yorke E., and Ling C. C., “Respiration-correlated spiral CT: A method of measuring respiratory-induced anatomic motion for radiation treatment planning,” Med. Phys. 30, 88–97 (2003). 10.1118/1.1531177 [DOI] [PubMed] [Google Scholar]
- 6.Dawson L. A. and Jaffray D. A., “Advances in image-guided radiation therapy,” J. Clin. Oncol. 25(8), 938–946 (2007). 10.1200/JCO.2006.09.9515 [DOI] [PubMed] [Google Scholar]
- 7.Pan T., Lee T., Rietzel E., and Chen G. T. Y., “4D-CT imaging of a volume influenced by respiratory motion on multi-slice CT,” Med. Phys. 31, 333–340 (2004). 10.1118/1.1639993 [DOI] [PubMed] [Google Scholar]
- 8.Keall P. J., Starkschall G., Shukla H., Forster K. M., Ortiz V., Stevens C. W., Vedam S. S., George R., Guerrero T., and Mohan R., “Acquiring 4D thoracic CT scans using a multislice helical method,” Phys. Med. Biol. 49(10), 2053–2067 (2004). 10.1088/0031-9155/49/10/015 [DOI] [PubMed] [Google Scholar]
- 9.Lu W., Parikh P. J., Hubenschmidt J. P., Bradley J. D., and Low D. A., “A comparison between amplitude sorting and phase-angle sorting using external respiratory measurement for 4D CT,” Med. Phys. 33, 2964–2974 (2006). 10.1118/1.2219772 [DOI] [PubMed] [Google Scholar]
- 10.Wink N. M., Panknin C., and Solberg T. D., “Phase versus amplitude sorting of 4D-CT data,” J. Appl. Clin. Med. Phys. 7(1), 77–85 (2006). 10.1120/jacmp.2027.25373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss E., Wijesooriya K., Dill S. V., and Keall P. J., “Tumor and normal tissue motion in the thorax during respiration: Analysis of volumetric and positional variations using 4D CT,” Int. J. Radiat. Oncol., Biol., Phys. 67(1), 296–307 (2007). 10.1016/j.ijrobp.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 12.Beddar A. S., Kainz K., Briere T. M., Tsunashima Y., Pan T., Prado K., Mohan R., Gillin M., and Krishnan S., “Correlation between internal fiducial tumor motion and external marker motion for liver tumors imaged with 4D-CT,” Int. J. Radiat. Oncol., Biol., Phys. 67(2), 630–638 (2007). 10.1016/j.ijrobp.2006.10.007 [DOI] [PubMed] [Google Scholar]
- 13.Murphy M. J., Balter J., Balter S., J. A. BenComo, Jr., Das I. J., Jiang S. B., Ma C., Olivera G. H., Rodebaugh R. F., Ruchala K. J., Shirato H., and Yin F., “The management of imaging dose during image-guided radiotherapy: Report of the AAPM Task Group 75,” Med. Phys. 34, 4041–4063 (2007). 10.1118/1.2775667 [DOI] [PubMed] [Google Scholar]
- 14.Rietzel E., Pan T., and Chen G. T. Y., “Four-dimensional computed tomography: Image formation and clinical protocol,” Med. Phys. 32, 874–889 (2005). 10.1118/1.1869852 [DOI] [PubMed] [Google Scholar]
- 15.Kalender W. and Yiannis K., “Flat-detector computed tomography (FD-CT),” Eur. Radiol. 17(11), 2767–2779 (2007). 10.1007/s00330-007-0651-9 [DOI] [PubMed] [Google Scholar]
- 16.Seppi E., Munro P., Johnsen S., Shapiro E., Tognina C., Jones D., Pavkovich J., Webb C., Mollov I., Partain L., and Colbeth R., “Megavoltage cone-beam computed tomography using a high-efficiency image receptor,” Int. J. Radiat. Oncol., Biol., Phys. 55(3), 793–803 (2003). 10.1016/S0360-3016(02)04155-X [DOI] [PubMed] [Google Scholar]
- 17.Jonathon L., LaBounty T., Heilbron B., Min J., Mancini G., Lin F., Taylor C., Dunning A., and Earls J., “Estimated radiation dose reduction using adaptive statistical iterative reconstruction in coronary CT angio-graphy: The ERASIR study,” Am. J. Roentgenol. 195(3), 655–660 (2010). 10.2214/AJR.10.4288 [DOI] [PubMed] [Google Scholar]
- 18.Endrizzi M., Diemoz P., Munro P., Hagen C., Szafraniec M., Millard T., Zapata C., Speller R., and Olivo A., “Edge illumination and coded-aperture x-ray phase-contrast imaging: Increased sensitivity at synchrotrons and lab-based translations into medicine, biology and materials science,” Proc. SPIE 8, C05008 (2013). 10.1117/12.2007893 [DOI] [Google Scholar]
- 19.Cai J., Chang Z., Wang Z., Segars W. P., and Yin F., “Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: A feasibility study,” Med. Phys. 38(12), 6384–6394 (2011). 10.1118/1.3658737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Von Siebenthal M., Székely G., Gamper U., Boesiger P., Lomax A., and Cattin Ph., “4D MR imaging of respiratory organ motion and its variability,” Phys. Med. Biol. 52(6), 1547–1564 (2007). 10.1088/0031-9155/52/6/001 [DOI] [PubMed] [Google Scholar]
- 21.Urs G., Boesiger P., and Kozerke S., “Compressed sensing in dynamic MRI,” Magn Reson Med. 59(2), 365–373 (2008). 10.1002/mrm.21477 [DOI] [PubMed] [Google Scholar]
- 22.Ricardo O., Kim D., Axel L., and Sodickson D., “Combination of compressed sensing and parallel imaging for highly accelerated first pass cardiac perfusion MRI,” Magn. Reson. Med. 64(3), 767–776 (2010). 10.1002/mrm.22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David F. and Ugurbil K., “Multiplicative increase in MRI data acquisition with multi-band RF excitation pulses in a simultaneous image refocusing pulse sequence,” U.S. patent application 13/118, 179 (Mar 8, 2012).
- 24.Chang Z., Cai J., Wang Z., and Yin F., “Evaluating dynamic magnetic resonance imaging using a new design of phantom: Validation with fast megavoltage fluoroscopic imaging,” in Proceedings of AAPM 52nd Annual Meeting, Philadelphia, PA, July 18, 2010. [Google Scholar]
- 25.Yang J., Cai J., Wang H., Chang Z., Czito B., Bashir M., and Yin F., “Four-dimensional magnetic resonance imaging using axial body area as respiratory surrogate: Initial patient results,” Int. J. Radiat. Oncol., Biol., Phys. 88(4), 907–912 (2014). 10.1016/j.ijrobp.2013.11.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarte S., McClelland J., Hughes S., Blackall J., Landau D., and Hawkes D., “A non-contact method for the acquisition of breathing signals that enable distinction between abdominal and thoracic breathing,” Radiother. Oncol. 81, S209 (2006). 10.1016/S0167-8140(06)81019-4 [DOI] [Google Scholar]
- 27.Nehrke K., Bor¨nert P., Manke D., and Boc¨k J., “Free-breathing cardiac MR imaging: Study of implications of respiratory motion-initial results,” Radiology 220(3), 810–815 (2001). 10.1148/radiol.2203010132 [DOI] [PubMed] [Google Scholar]
- 28.Cai J., Sheng K., Sheehan J., Benedict S., Larner J., and Read P., “Evaluation of thoracic spinal cord motion using dynamic MRI,” Radiother Oncol. 84(3), 279–282 (2007). 10.1016/j.radonc.2007.06.008 [DOI] [PubMed] [Google Scholar]
- 29.Forsyth D. A. and Ponce J., in Computer Vision: A Modern Approach, 1st ed. (Prentice-Hall, Upper Saddle River, NJ, 2005). [Google Scholar]
- 30.Segars W. P., Sturgeon G., Mendonca S., Grimes J., and Tsui B. M. W., “4D XCAT phantom for multimodality imaging research,” Med. Phys. 37, 4902–4915 (2010). 10.1118/1.3480985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segars W. P. and Tsui B. M. W., “MCAT to XCAT: The evolution of 4-D computerised phantoms for imaging research,” Proc. IEEE. 97, 1954–1968 (2009). 10.1109/JPROC.2009.2022417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segars W. P., Mahesh M., Beck T. J., Frey E. C., and Tsui B. M. W., “Realistic CT simulation using the 4D XCAT phantom,” Med. Phys. 35, 3800–3808 (2008). 10.1118/1.2955743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch N., Liu H. H., Starkschall G., Jacobson M., Forster K., Liao Z., Komaki R., and Stevens C. W., “Evaluation of internal lung motion for respiratory-gated radiotherapy using MRI: Part I. Correlating internal lung motion with skin fiducial motion,” Int. J. Radiat. Oncol., Biol., Phys. 60(5), 1459–1472 (2004). 10.1016/j.ijrobp.2004.05.055 [DOI] [PubMed] [Google Scholar]
- 34.Cai J., Chang Z., Wang Z., and Yin F., “Extracting breathing signal from image Fourier transform for developing 4D-MRI,” in Proceedings of 2010 ASTRO Annual Meeting, October 31–November 4, 2010. [Google Scholar]
- 35.Li R., Lewis J. H., Cerviño L. I., and Jiang S. B., “4D CT sorting based on patient internal anatomy,” Phys. Med. Biol. 54, 4821–4833 (2009). 10.1088/0031-9155/54/15/012 [DOI] [PubMed] [Google Scholar]
- 36.Herborn C. U., Vogt F., Lauenstein T. C., Goyen M., Debatin J. F., and Ruehm S. G., “MRI of the liver: Can true FISP replace HASTE?,” J. Magn. Reson. Imaging 17(2), 190–196 (2003). 10.1002/jmri.10248 [DOI] [PubMed] [Google Scholar]