Abstract
Background
Information about differences in immunogenicity of various HLA antigens may help guide donor selection and identify mismatches to avoid for patients likely to need retransplantation. To date, antibody responses to a wide array of individual mismatched antigens have not been evaluated.
Methods
Frequencies of antibodies to mismatched HLA-A, HLA-B, HLA-DR, and HLA-DQ antigens were determined for 703 renal transplant patients who had no detectable donor-specific antibody before transplantation. The impact of cross-reactive group matching and production of antibodies cross-reactive with mismatched antigens were also assessed. Antibodies were identified using multiplexed bead assays.
Results
The overall mean frequencies were similar for HLA-A (53.2%), HLA-DR (52.6%), and HLA-DQ (59.0%) antibodies, but significantly lower for HLA-B antibodies (42.4%). However, the response to individual antigens ranged from 15.0% to 76.2%. Antibody frequencies were reduced significantly for 54 of 62 specificities when the patient possessed an antigen cross-reactive with the donor mismatch, but the magnitude of the effect was variable and ranged from 8% to 83%. Moreover, there was directionality in the protective effect of cross-reactive group matching. Overall mean donor-specific antibody frequencies were comparable for men and women except for a significantly higher frequency of antibodies to HLA-DR among men (56.6% vs. 47.8%, P=0.004). Overall mean frequencies in blacks were higher than, or comparable to those of, whites, but differences were not significant.
Conclusion
There is considerable variability in the immunogenicity of different HLA antigens that is impacted by the presence or absence of cross-reactive antigens in the patient’s phenotype. This information can be used to augment the immunologic evaluation of donor-recipient pairs.
Supplemental digital content is available in the text.
Most renal transplants and nearly all nonrenal transplants involve mismatched HLA antigens. Ongoing improvements in immunosuppression and treatment of antibody-mediated rejection have resulted in improved short-term survival of mismatched renal transplants, but improvement in long-term graft survival is questionable. One factor affecting long-term graft survival is the development of antibodies to mismatched HLA antigens. There are substantial data that antibodies to mismatched HLA antigens develop after transplantation (reviewed in reference1). More sensitive and specific antibody testing and improvements in biopsy interpretation have increased the appreciation of the role of donor-specific antibodies in chronic rejection and their impact on long-term graft survival.2-4
It may be argued that any graft survival benefit of HLA matching is outweighed by increased waiting time and diminished access of some ethnic groups to transplantation. However, sensitization to HLA antigens is a major impediment to retransplantation.5,6 We have shown that even a single mismatched antigen can result in sensitization.7 Others have shown a correlation between the number of amino acid differences between donor and recipient HLA molecules and antibody production.8,9 However, although the number of amino acid differences between donor and recipient may show some correlation with overall antibody response, it does not address the issue of the immunogenicity and humoral response to specific mismatched antigens. Antibodies react with conformational epitopes, which are not necessarily defined by a linear sequence and can be affected by noncontiguous residues.10,11 Immunogenicity of an epitope is affected by the physiochemical properties of the amino acids—their electrostatic charge and hydrophobicity.12
At present, there are very few epitopes that have been defined by stringent criteria, including adsorption studies and single residue substitutions that occur not only at the site of the putative epitope but also at other sites that may affect the conformation of the epitope. Further, different sets of epitopes have been defined by different groups.13-16 Clear identification of epitope mismatches requires HLA typing of donor and recipients at the allele level, which is not the current practice for solid organ transplantation. Moreover, identification of epitopes does not reveal their immunogenicity, and multiple epitopes may contribute to the immunogenicity of an antigen. Therefore, assessing the frequency of antibody response to different HLA antigens mismatched in transplantation could provide an assessment of their immunogenicity and of the effect of the recipient’s phenotype on that response. We present here data on the relative immunogenicity of different HLA antigens, derived from donor-specific antibody frequency data from 703 patients who did not have antibody to donor antigens before transplantation.
RESULTS
We examined the incidence of antibodies specific for a mismatched antigen in patients who had received a kidney transplant and assessed the potential impact of various factors on sensitization. Only patients known to be sensitized after transplantation were considered on the premise that if a patient is sensitized, antibodies to mismatched antigens should occur with equal frequency if all antigens are equally immunogenic. For each specificity, we excluded cases in which the donor’s phenotype included both the mismatched antigen and an antigen cross-reactive with the mismatched antigen. We also evaluated the impact of cross-reactivity, both the effect of an antigen in the patient’s phenotype that is cross-reactive with the mismatched antigen (XRAg) and the extent to which a mismatched antigen results in antibodies to antigens cross-reactive with the mismatched antigen. Tables 1–4 show the frequencies of antibodies present after mismatches of different antigens. The overall mean frequencies were similar for HLA-A (53.2±14.4), HLA-DR (52.6±10.4), and HLA-DQ (59.0±13.0) antigens but the mean frequency of antibodies to HLA-B antigens (42.4±12.6) was significantly lower than for those to HLA-A (P=0.005), HLA-DR (P=0.009), or HLA-DQ (P=0.004) antigens. The mean frequency of antibodies to DR51, DR52, or DR53 was significantly higher than that of antibodies to antigens of the DRB1 locus (66.2 vs. 49.5, P=0.007). The mean frequencies of antibodies to mismatched antigens were lower when the patient’s phenotype included an antigen cross-reactive with the mismatched antigen, compared to when no XRAg was present in the patient, for all the loci considered. When individual specificities were examined, the antibody frequencies were lower when there was an XRAg in the patient for 54 of 62 specificities. Of the remaining eight cases, there were five with less than 25 patients in the group suggesting perhaps insufficient numbers to obtain meaningful results. For the 54 specificities with reduced frequencies, the reduction in antibody frequency was significant for HLA-A (P=0.000004), HLA-B (P=0.00004), HLA-DR (P=0.0003), and HLA-DQ (P=0.02) antibodies.
TABLE 1.
Frequency of antibody to mismatches: HLA-A
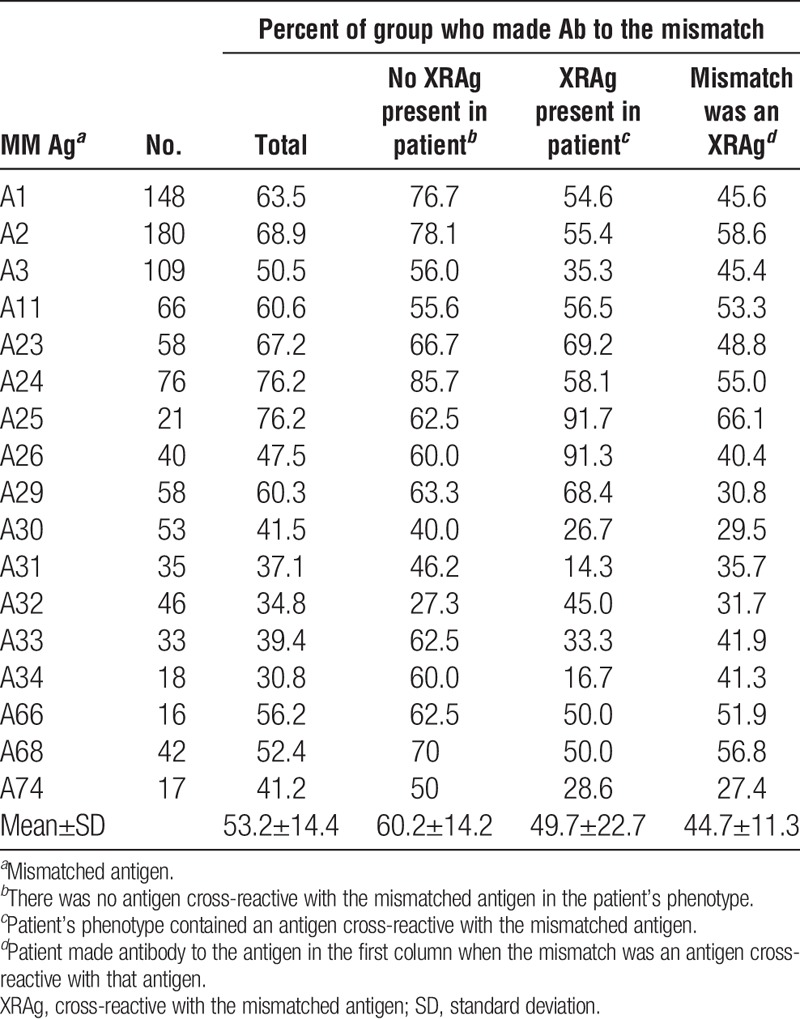
TABLE 4.
Frequency of antibody to mismatches: HLA-DQ
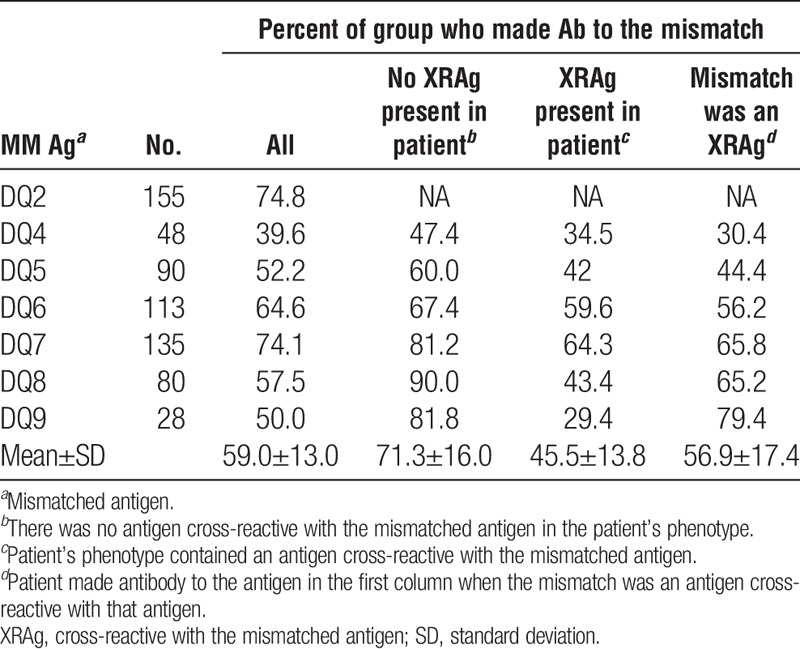
TABLE 2.
Frequency of antibody to mismatches: HLA-B
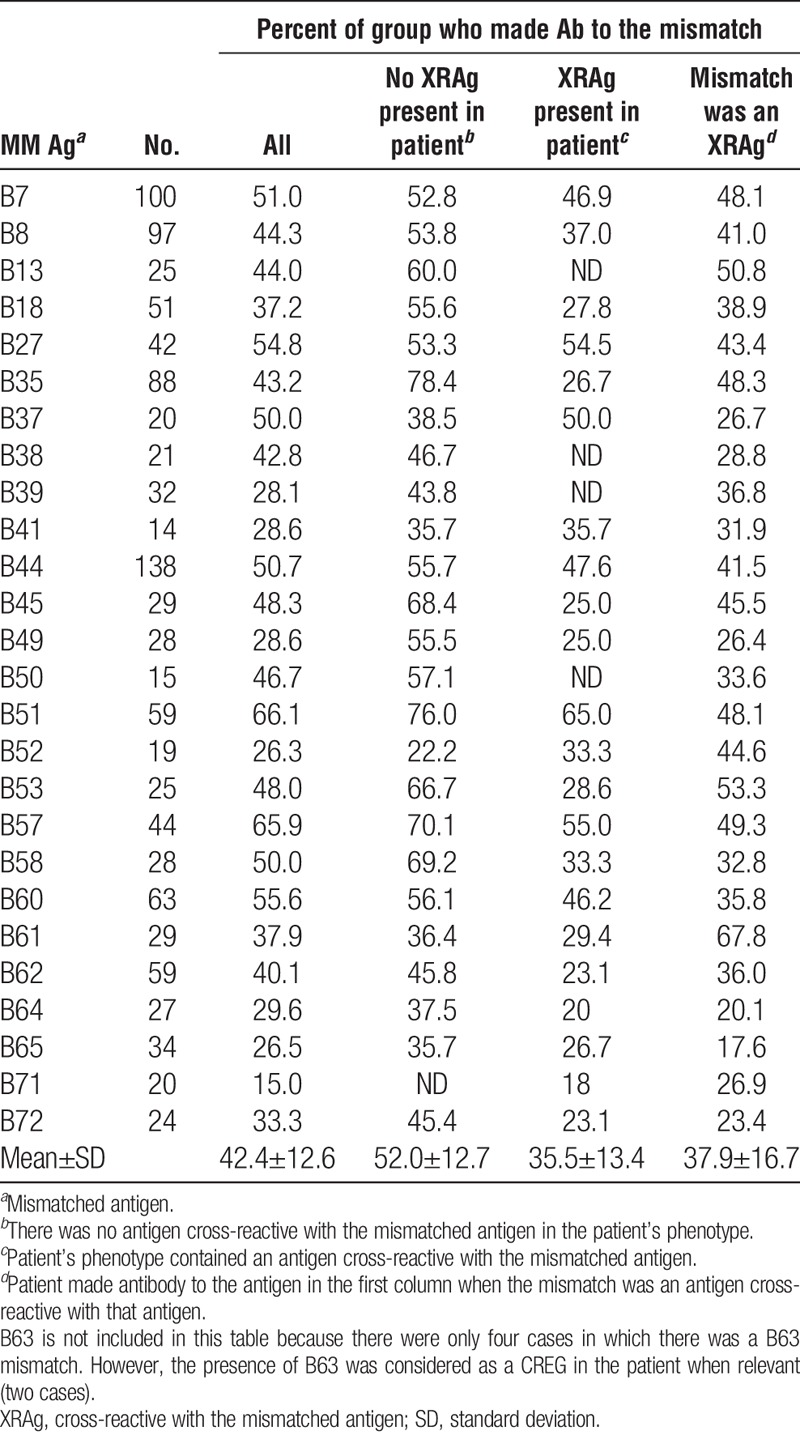
TABLE 3.
Frequency of antibody to mismatches: HLA-DR
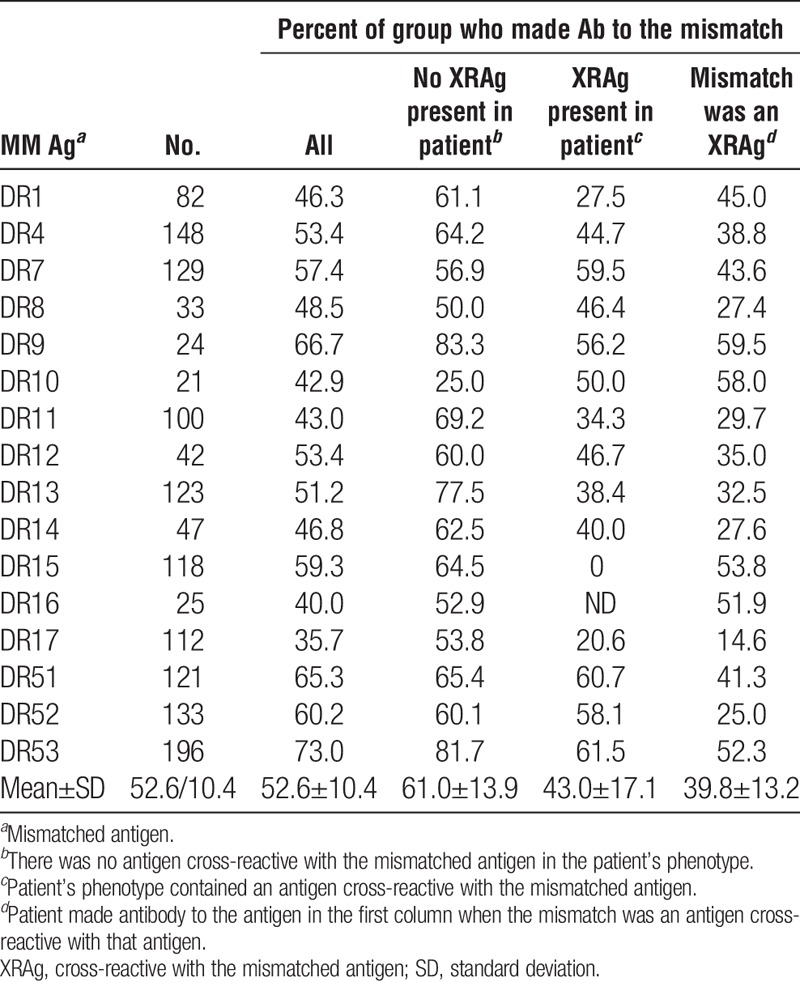
Tables 1–4 also show the frequency with which an antibody to one antigen was associated with a mismatch of an XRAg. For example, antibody to HLA-A3 occurring after a mismatch of HLA-A1 or HLA-A11. In nearly all cases when there was antibody to a mismatched antigen, antibody to cross-reactive antigens was also present. In all but four cases that showed a low numbers of patients, the frequency of antibody to an antigen was significantly lower when it was because of the mismatch of an XRAg compared to a mismatch of the specific antigen. The mean frequencies for HLA-A, HLA-B, HLA-DR, and HLA-DQ antibodies associated with XRAg mismatches were 44.7% (P=0.0002), 35.5% (P=0.03), 43.0% (P=0.000003), and 45.5% (P=0.005), respectively, and were significantly lower than the mean frequencies for antibodies to a mismatch when there was no XRAg in the patient’s phenotype.
The magnitude of the effect of an XRAg in the patient’s phenotype varied among different cross-reactive pairs (Table 5). For example, compared to the frequency of antibody to HLA-A3 among patients who were mismatched for HLA-A3 and had no XRAg in their phenotype, there was a 43.6% reduction in antibody frequency among patients with an HLA-A1 in their phenotype but only an 18.9% reduction among patients with HLA-A11 in their phenotype. Further, there was a directionality of the effect for certain cross-reactive pairs (shown in bold). For example, there was a 28.8% reduction in the frequency of antibody to an A1 mismatch when the patient had an A23 antigen but there was no reduction in antibody to an A23 mismatch when the patient had an A1 antigen. There are three specificities in the table for which there were less than 10 instances, A23-A24, A24-A23, and DR12-DR13. These were included because, despite the low numbers, they show a dramatic effect. Similarly, there was variability among antigens in the frequency with which they were associated with production of cross-reactive antibodies (Table 6). In some cases, the frequency of an antibody specific for an antigen cross-reactive with the mismatched antigen was much lower than that of the antibody to the mismatch itself, such as the 41.9% frequency of antibody to A23 and the 78.1% frequency of antibody to A2 after an A2 mismatch. In other cases, the frequencies were comparable, such as the frequencies of antibody to A68 of 70.0% and 71.6% after mismatches of A68 and A2, respectively. One would expect that an antigen that reduced the response to a cross-reactive antigen appreciably when it was present in the patient’s phenotype would be comparably effective in inducing antibodies to cross-reactive antigens when present in the donor. This was true in some but not all cases, as shown in Figure 1. For example, there was a 43.6% reduction in antibody to A3 when the patient had A1 in their phenotype and when the donor mismatch was A1, 44.7% of the patients made antibody to A3. In contrast, there was only a 28.8% reduction in the antibody to A1 when the patient was an A23, but when the mismatch was A23, 61.5% of patients made antibody to A1.
TABLE 5.
Variable reduction in Ab to MM with CREG in patient’s phenotype and directionality of effect
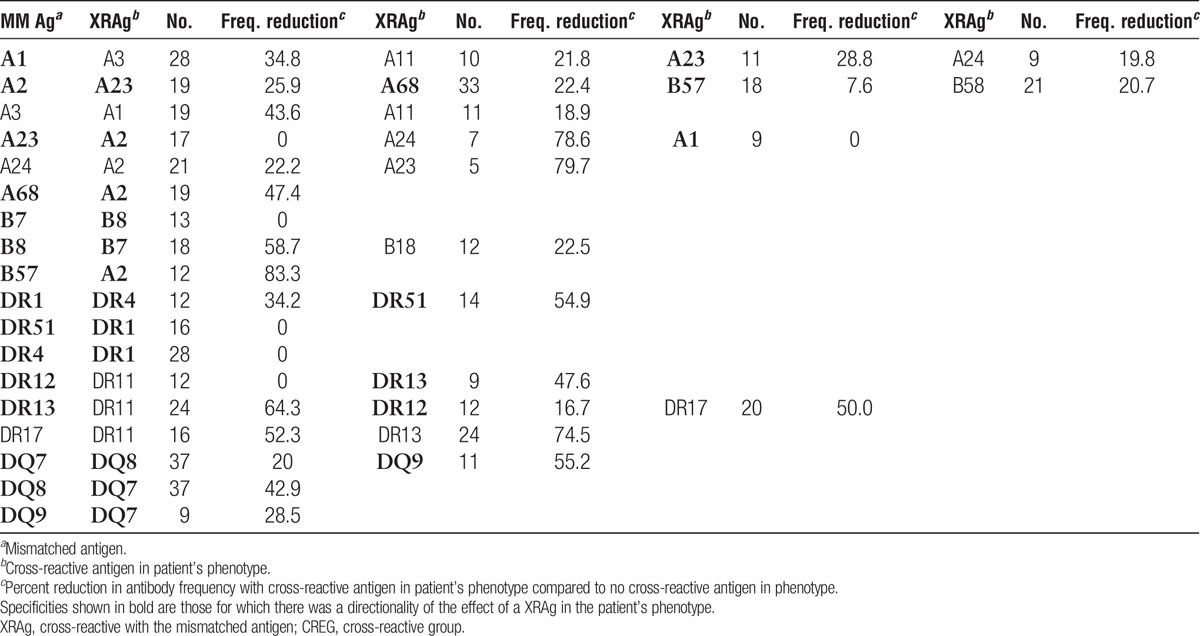
TABLE 6.
Frequencies of antibodies to antigens cross-reactive with the mismatched antigen
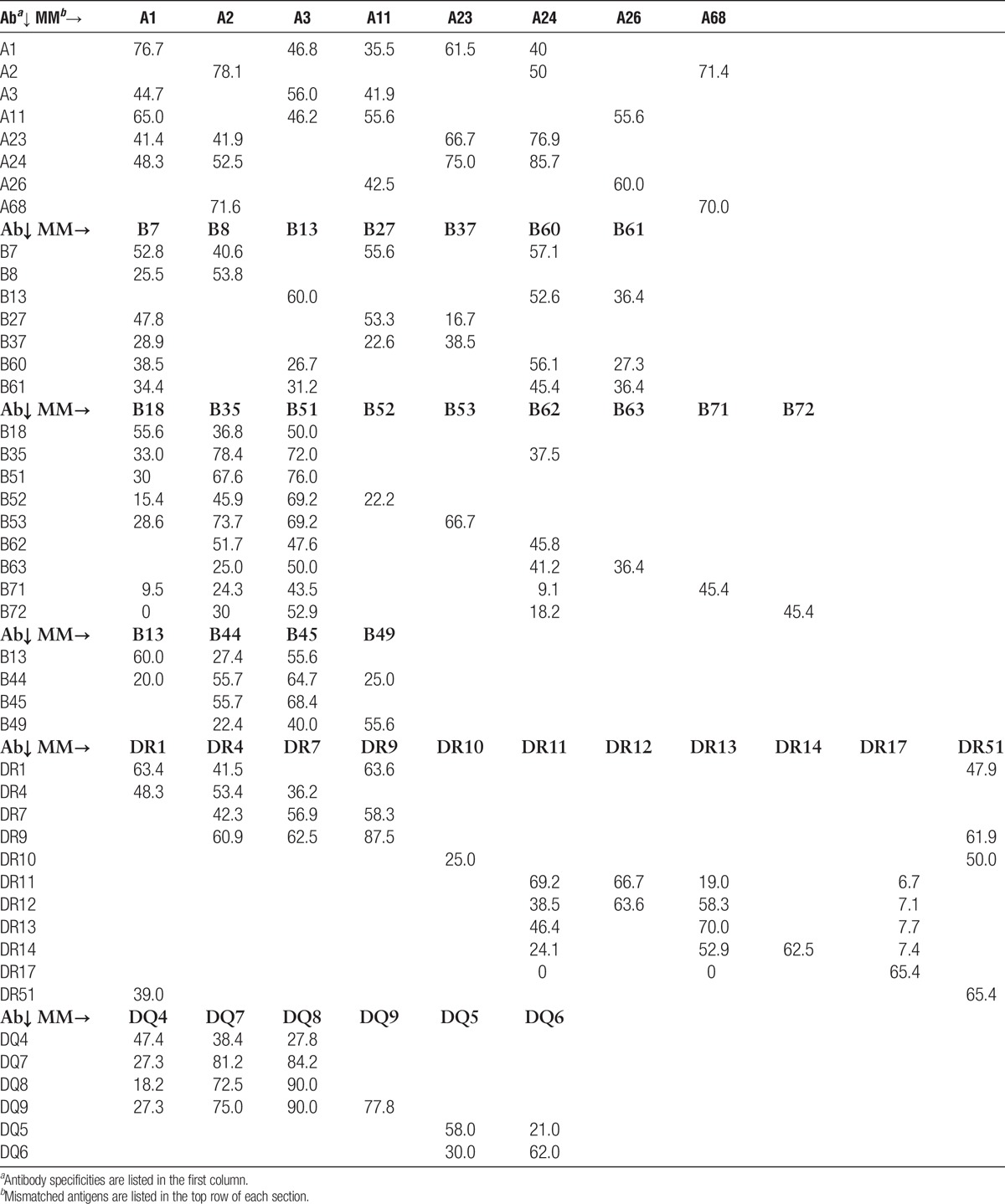
FIGURE 1.
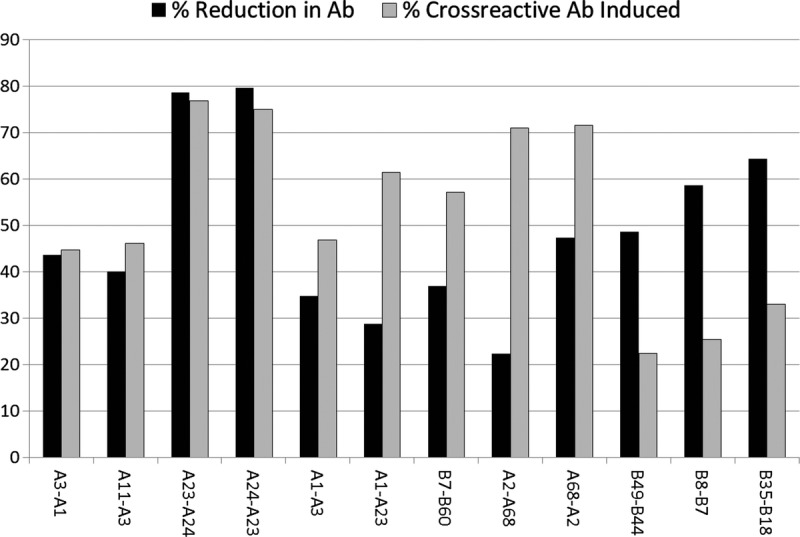
Examples of the impact of cross-reactivity both in reducing the antibody response when a mismatch is cross-reactive with an antigen in the patient and in inducing antibodies to an antigen cross-reactive with the mismatch. The data for each pair of specificities shown are represented as follows. The percent reduction in the frequency of antibody made to a mismatch of the first antigen in the pair, when the second antigen in the pair is in the patient’s phenotype (black bars). The frequency of antibody made to the first antigen in the pair when the donor mismatch is the second antigen in the pair (gray bar).
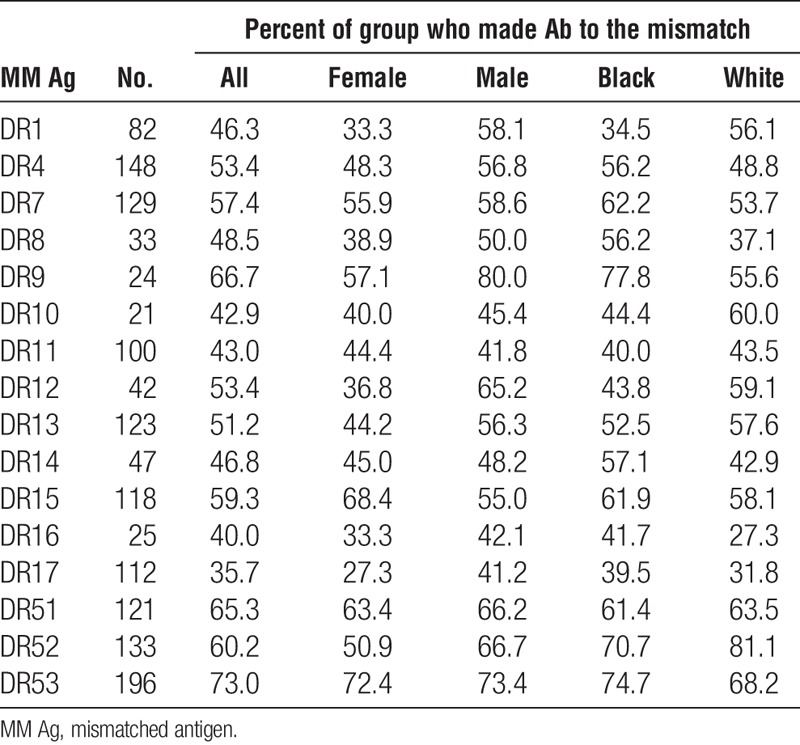
Considering only those specificities for which there were 20 or more cases, the frequencies of antibodies to mismatched antigens was comparable for men and women for antibodies specific for HLA-A (53.5 vs. 54.2), HLA-B (40.4 vs. 46.1), and HLA-DQ (59.6 vs. 58.6) antigens but there was a significantly higher frequency of HLA-DR–specific antibodies (56.6 vs. 47.8, P=0.004) among men compared to women (Tables 7–10). On average, frequencies of antibodies among blacks were higher than or comparable to those among whites for all loci (HLA-A, 59.2 vs. 53.3; HLA-B, 42.8 vs. 44.7; HLA-DR, 54.7 vs. 52.8; HLA-DQ, 60.7 vs. 57.7), and the differences were not significant. Because the presence of an antigen in the patient’s phenotype that was cross-reactive with the mismatched antigen reduced the frequency of response to the mismatched antigen, we looked to see if this could account for differences between men and women and between blacks and whites in the frequencies of antibody responses. We found that differences between demographic groups in the frequencies of antibodies to different antigens did not correlate with the frequencies of cross-reactive antigens in the patient’s phenotypes. Comparing those who made antibody to a mismatch and those who did not showed no significant difference in the total number of mismatched antigens within a class of HLA antigens except for A29 with mean numbers of mismatched antigens of 4.2 and 3.3 (P=0.03) for antibody positive and negative, respectively, and B7 with average mismatches of 3.8 and 4.6 (P=0.03) for antibody positive and negative, respectively. We found no substantial differences in the frequencies of different class II antigens in the patients’ phenotypes between those who made antibody and those who did not for any HLA-A or HLA-B specificity (data not shown). We also examined if the extent of DR match affected the frequency of response to a mismatched antigen. We eliminated from this analysis 58 patients who had received more than one transplant because this could be a confounding factor. Among the remaining 644 patients, there were 47, 347, and 250 two-DR, one-DR, and zero-DR matches. To have sufficient numbers for analysis, we assessed only those cases for which there were at least 75 instances of a mismatch with no XRag in the patient or donor. There was no consistent pattern of response. That is, in some cases, there was a higher frequency of response in 2DR matches but in others, the highest frequency occurred in one or zero DR match. We analyzed the summary data for A and B locus mismatches by chi squared analysis, and there was no significant difference in the distribution of response frequency among the different DR match groups. When we included cases with an XRAg in the patient, there was still no significant difference in the distribution of response frequency.
TABLE 7.
Association of race and gender with antibody frequencies: HLA-A antigens
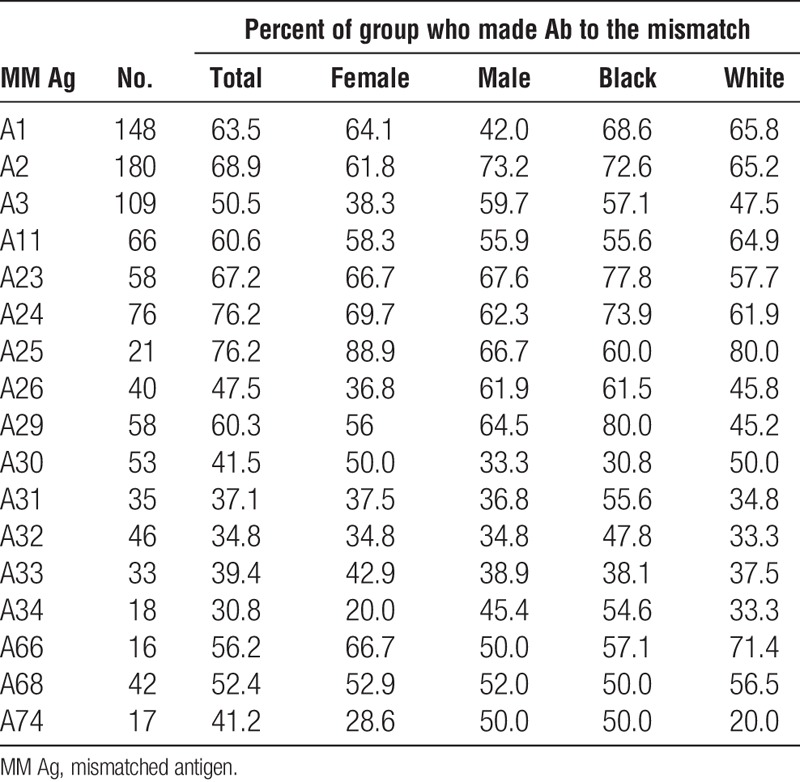
TABLE 10.
Association of race and sex with antibody frequencies: HLA-DQ antigens
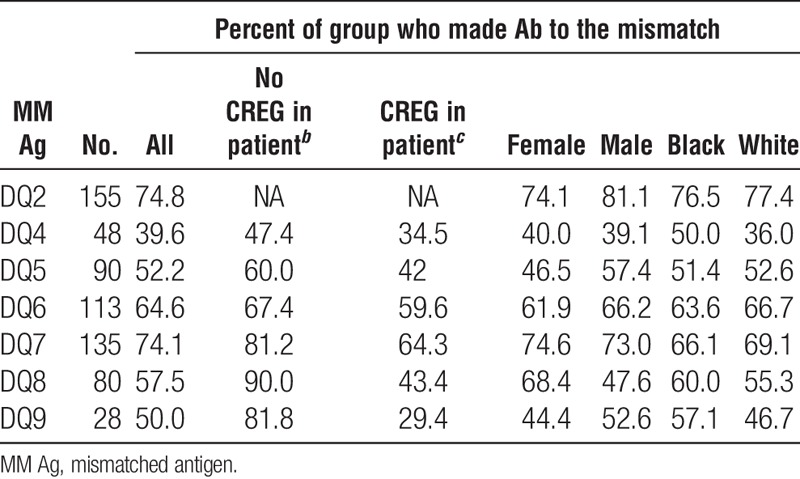
TABLE 8.
Association of race and sex with antibody frequencies: HLA-B antigens
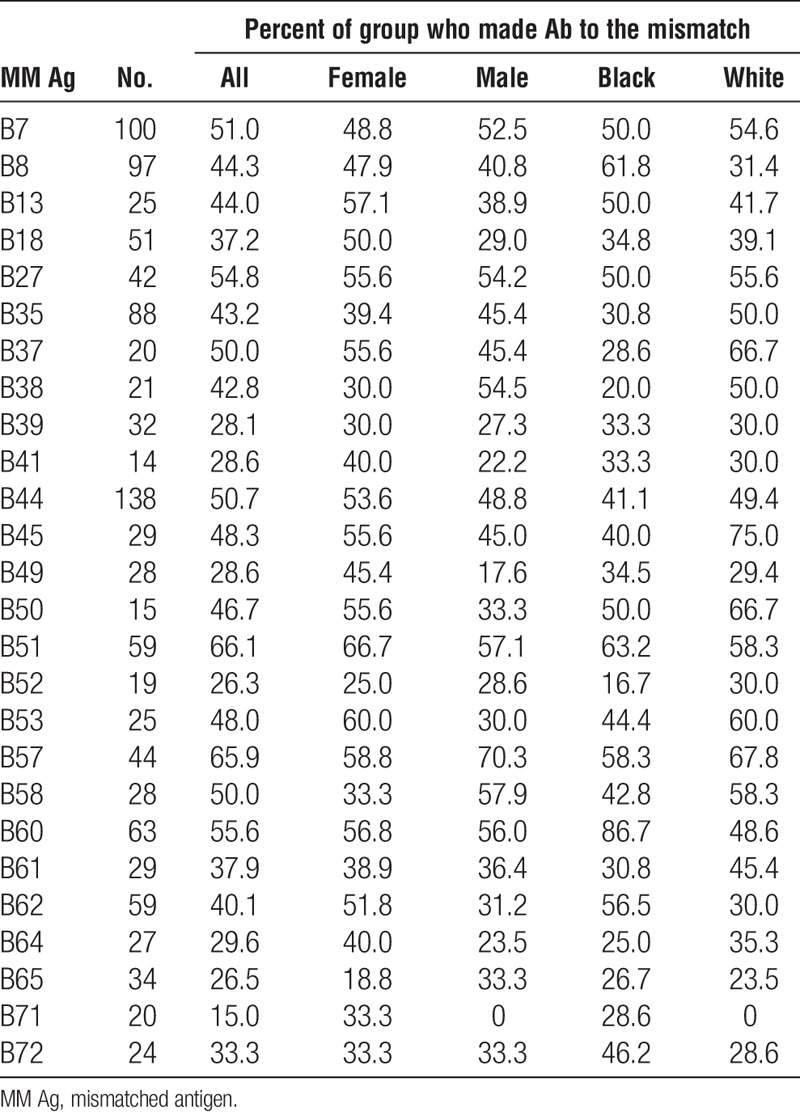
TABLE 9.
Association of race and sex with antibody frequencies: HLA-DR antigens
Patients homozygous at a locus made antibodies to a significantly greater number of antigens at the locus of homozygosity than did patients heterozygous at that locus (HLA-A: 5.6 vs. 3.8, P=0.001; HLA-B: 7.0 vs. 4.8, P=0.02; HLA-DR: 3.4 vs. 2.4, P=0.004; HLA-DQ: 2.1 vs. 1.4, P=0.0003). We examined the distribution of antibodies by strength for specificities for which the number of mismatched patients was 40 or more. Of antibodies to HLA-A, HLA-B, HLA-DR, and HLA-DQ, 53%, 47%, 45%, and 47%, respectively, were strong and 81%,75%,78%, and 79%, respectively, were of moderate or high strength. There were no significant differences in strength between antibodies categorized by specificity.
DISCUSSION
Knowing if immunogenicity varies among HLA antigens and how a patient’s phenotype may impact the antibody response to various HLA antigens is important for patients who are likely to need another transplant in the future and for selecting the immunologically optimal donor among two or more available donors. The data presented here suggest major differences in immunogenicity among different HLA antigens. The shortcomings of this study include the absence of information about sensitizing events other than transplantation, such as pregnancy and transfusion, which may contribute to the antibody response to a transplant mismatch; differences in sensitivity and specificity of various antigen-bearing beads used to assess the presence of antibody; and lack of information about antibodies to DQA. The data suggest a limited impact of sensitizing events other than transplantation. First, if other events contributed to sensitization, one would expect the frequencies of antibodies to be proportional to the frequencies of antigens in the population because a patient would be exposed to a common antigen by transfusion or pregnancy more frequently than to a rare one. We found no correlation between the frequencies of HLA antigens and the frequencies of antibodies of various specificities, which we believe is a further indication of differences in immunogenicity as, in the absence of such difference, antibody frequencies would be proportional to the frequency of mismatch. Second, the frequencies of antibodies among women were not consistently greater than those among men, which would be expected if pregnancy had contributed significantly to sensitization. Additionally, as with many studies, we have not been able to test patients routinely after transplantation, and the sera available for this study may not include antibodies that occurred only historically.1 However, the clinical relevance of historic antibodies is questionable.17-19 Interestingly, the collective effect of mismatches of the HLA-A, HLA-DR, and HLA-DQ loci were similar but that of the HLA-B locus was significantly lower than the others. This is difficult to explain. Although the number of HLA-B locus serologically defined antigens is greater than for the other loci, this would not impact the individual patient as; for each patient, there can be only one or two HLA-B locus mismatches. Further, because of the increased number of HLA-B antigens, the likelihood of a mismatch is greater. Although the difference observed may be a statistical anomaly, it is interesting to conjecture that the extent of cross-reactivity of HLA-B antigens is under-recognized. If true, the presence of an XRAg in the patient would have been underestimated and could account for a lower frequency of response to HLA-B mismatches.
Previously, Crowe20 reported multicenter data showing that cross-reactive group (CREG) matching, that is, having an antigen in the patient’s phenotype that is cross-reactive with the mismatched antigen, led to a reduced frequency of sensitization to HLA. The data presented here substantiate that finding and importantly, using the most sensitive techniques available, show an overall decrease in the frequency of the antibody response more than five times that reported by Crowe and that was significant for antibodies at all loci. We were also able to show that the effect varied among different cross-reactive antigens, with some having no effect and others reducing the frequency of response to a mismatch by as much as 80%. In an analysis of HLA-A2 and HLA-A28, Dankers et al.21 showed directionality in the protective effect of a cross-reactive antigen in the patient’s phenotype. They showed that HLA-A2 females did not make antibody to HLA-A28 offspring but that 32% of HLA-A28 mothers made antibody to the HLA-A2 of their offspring. We have shown directionality to the protective effect for 10 different pairs of cross-reactive antigens including HLA-A2 to HLA-A68, although we did not see the total nonresponsiveness seen in the Dankers et al.’s study, most likely because a more sensitive antibody test was used in our study. This directionality is a further indication that the number of amino acid differences is not, necessarily, an indication of immunogenicity because the number of differences would be the same in both directions.
The corollary to this protective effect of CREG matching is that mismatches of one antigen can lead to production of antibodies to the other cross-reactive antigens. As with the protective effect of CREG matching, we saw variability among antigens in the induction of cross-reactive antibodies. One might expect that the more a cross-reactive antigen in the patient’s phenotype reduced the response to a mismatch, the more likely as a mismatch it would be to induce antibodies to cross-reactive antigens. Interestingly, we found this to be true in some, but not all, cases.
Data on the correlation between total amino acid differences and production of donor-specific antibody suggest that the greater the number of mismatched antigens, the greater the likelihood of an immune response. However, in this study, the number of mismatched antigens at a locus was significantly different between patients who made antibody to an antigen at that locus and those who did not in only 2 of 61 comparisons, which could occur by chance. Dankers et al.8 reported that antibody production after transplantation correlated with the number of mismatched epitopes but in that study, they did not look at the number of mismatched antigens. Although epitope analysis can provide insight into the degrees of relationship between antigens, we do not believe it provides as accurate an assessment of relative antigen immunogenicity as can be obtained by analysis of the antibody patterns evoked by different antigen mismatches. First, as shown elegantly by Kosmoliaptsis et al.,22 not all epitopes are created equal and differ according to their physiochemical properties. Second, several different epitope sets have been proposed that overlap only in part.13-16 Interestingly, Hwang et al.23 reported that the 5-year graft survival correlated with matching of epitopes by one schema but not by another. Also, there are very few epitopes that have been defined by stringent criteria including adsorption studies and single residue substitutions that occur not only at the site of the epitope but also at other sites that may affect the conformation of the epitope, changing its specificity. Further, to be useful clinically, assessing epitope mismatches may require a level of HLA typing that exceeds time and cost constraints. For these reasons, this study intentionally focused on the humoral response evoked by different antigen mismatches, considering relative immunogenicity in the entire antigen molecules, rather than by specific epitopes.
Finally, our data substantiate what has been accepted anecdotally, but not always shown with methodical, controlled studies. The humoral response to HLA mismatches is, overall, greater in blacks than in whites and homozygosity at a locus is a risk factor for a broad response to other antigens at that locus. Regarding the effect of race on the humoral response, it is important to note that when one considers the response to individual antigens, there is variability such that for some antigens, the response is greater in whites than in blacks.
In summary, we believe the data presented here provide an opportunity to select donors according to the mismatched antigens, to alter immunosuppression protocols according to the immunogenicity of the mismatch, and to identify those patients for whom posttransplantation monitoring is critical.
MATERIALS AND METHODS
Subjects
Sera from 703 renal patients who had been transplanted and subsequently found to be sensitized were tested by multiplexed bead assays for HLA antibody. The criterion for inclusion of subjects was the development of HLA-specific antibodies posttransplant in patients with no donor specific antibodies before transplantation. All patients were followed up at the Johns Hopkins Comprehensive Transplant Center. Adequate patient and donor HLA phenotype data were available for HLA-A and HLA-B loci from 703 pairs, for HLA-DRB1 and HLA-DRB3 to HLA-5 loci from 699 pairs, and for HLA-DQ antigens from 525 pairs. One hundred seventy-eight pairs were omitted from the HLA-DQ analyses because of insufficient resolution of the DQ typing. Among the recipients, there were 311 women and 392 men, 246 blacks, 372 whites, 46 Hispanics, 32 Orientals, and 7 other. The distribution of racial groups, defined as black, white, and other, was comparable in men and women.
Cross-Reactivity
Table S1 (see SDC, http://links.lww.com/TP/B36) shows the cross-reactivity scheme followed here which was modified from published schemes according to our own observations.
Antibody Analysis
The most broadly reacting serum from each patient was evaluated for specificity and strength. Sera were tested on multiplexed bead assays that were primarily single antigen panels (LABScreen Single Antigen Class I—Combi and LABScreen Single Antigen Class II Antibody Detection Test—Group 1; One Lambda, Thermo Fisher Scientific, Canoga Park, CA) supplemented with phenotype panels (LIFECODES class I ID, LIFECODES class II IDv2; Immucor, Stamford, CT). Three levels of antibody strength, strong, moderate, and weak, were assigned for MFI values of 15,000 or higher, 9,000, and 2,000 MFI, respectively for HLA-A, HLA-B, and HLA-DR, and 20,000 or higher, 16,000, and 4,000 for HLA-DQ.
The frequencies of antibodies to each mismatched antigen were determined. Cross-reactive antigens present in the patient’s phenotype and the antibody response to antigens cross-reactive with the mismatched antigen were assessed. The term CREG mismatch is used to refer to a mismatched antigen that is cross-reactive with an antigen in the patient’s phenotype.
Statistical Analysis
Comparisons of antibody frequencies were performed with a Student’s t test appropriate for the data set.
Supplementary Material
Footnotes
The authors declare no conflicts of interest.
D.P.L. and A.A.Z. participated in research design, writing of the article, performance of the research, and analysis of data. M.S.L. participated in research design and writing of the article.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1. Akalin E, Pasqual M. Sensitization after kidney transplantation. Clin J Am Soc Nephrol 2006; 1: 443. [DOI] [PubMed] [Google Scholar]
- 2. Valenzuela NM, McNamara JT, Reed EF. Antibody-mediated graft injury: complement-dependent and complement-independent mechanisms. Curr Opin Organ Transplant 2014; 19: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamanaga S, Watarai Y, Yamamoto T, et al. Frequent development of subclinical chronic antibody-mediated rejection within 1 year after renal transplantation with pre-transplant positive donor-specific antibodies and negative CDC crossmatches. Hum Immunol 2013; 74: 1111. [DOI] [PubMed] [Google Scholar]
- 4. Lachmann N, Terasaki PI, Budde K, et al. Anti-human leukocyte antigen and donor-specific antibodies detected by luminex posttransplant serve as biomarkers for chronic rejection of renal allografts. Transplantation 2009; 87: 1505. [DOI] [PubMed] [Google Scholar]
- 5. Iyer HS, Jackson AM, Zachary AA, et al. Transplanting the highly sensitized patient: trials and tribulations. Curr Opin Nephrol Hypertens 2013; 22: 681. [DOI] [PubMed] [Google Scholar]
- 6. Gralla J, Tong S, Wiseman AC. The impact of human leukocyte antigen mismatching on sensitization rates and subsequent retransplantation after first graft failure in pediatric renal transplant recipients. Transplantation 2013; 95: 1218. [DOI] [PubMed] [Google Scholar]
- 7. Zachary AA, Hart JM, Lucas DP, et al. The cost of mismatching. In: Cecka JM, Terasaki PI, eds. Clinical Transplants 2007. Los Angeles: Terasaki Foundation; 2008: 261. [Google Scholar]
- 8. Dankers MKA, Witvliet MD, Roelen DL, et al. The number of amino acid triplet differences between patient and donor is predictive for the antibody reactivity against mismatched human leukocyte antigens. Transplantation 2004; 77: 1236. [DOI] [PubMed] [Google Scholar]
- 9. Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol 2006; 67: 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akkoc N, Scornik JC. HLA epitope matching. Contribution of matched residues to epitopes recognized by alloantibodies. Transplantation 1991; 52: 903. [PubMed] [Google Scholar]
- 11. Mariuzza RA, Phillips SE, Poljak RJ. The structural basis of antigen-antibody recognition. Annu Rev Biophys Biophys Chem 1987; 16: 139. [DOI] [PubMed] [Google Scholar]
- 12. Kosmoliaptsis V, Dafforn TR, Chaudhry AN, et al. High-resolution, three-dimensional modeling of human leukocyte antigen class I structure and surface electrostatic potential reveals the molecular basis for alloantibody binding epitopes. Hum Immunol 2011; 72: 1049. [DOI] [PubMed] [Google Scholar]
- 13. Duquesnoy RJ, Claas FHJ. 14th International HLA and Immunogenetics Workshop: report on the structural basis of HLA compatibility. Tissue Antigens 2007; 69s1: 180. [DOI] [PubMed] [Google Scholar]
- 14. El-Awar N, Terasaki PI, Cai J, et al. Epitopes of the HLA-A, B, C, DR, DQ and MICA antigens. In: Cecka JM, Terasaki PI, eds. Clinical Transplants 2007. Los Angeles: Terasaki Foundation; 2008: 175. [PubMed] [Google Scholar]
- 15. Duquesnoy RJ, Marrari M. Correlations between Terasaki’s HLA class I epitopes and HLAMatchmaker-defined eplets on HLA-A, -B and -C antigens. Tissue Antigens 2009; 74: 117. [DOI] [PubMed] [Google Scholar]
- 16. Rodey GE, Revels K, Fuller TC. Epitope specificity of HLA class I alloantibodies: I. Frequency analysis of antibodies to private versus public specificities in potential transplant recipients. Transplantation 1994; 39: 272. [DOI] [PubMed] [Google Scholar]
- 17. Singh D, Kiberd BA, West KA, et al. Importance of peak PRA in predicting the kidney transplant survival in highly sensitized patients. Transplant Proc 2003; 35: 2395. [DOI] [PubMed] [Google Scholar]
- 18. Baron C, Pastural M, Lang P, et al. Long-term kidney graft survival across a positive historic but negative current sensitized cross-match. Transplantation 2002; 73: 232. [DOI] [PubMed] [Google Scholar]
- 19. Avlonitis VS, Chidambaram V, Manas DM, et al. The relevance of donor T cell-directed immunoglobulin G in historic sera in the age of flow cytometry. Transplantation 2000; 70: 1260. [DOI] [PubMed] [Google Scholar]
- 20. Crowe DO. The effect of cross-reactive epitope group matching on allocation and sensitization. Clin Transplant 2003; 17s9: 13. [DOI] [PubMed] [Google Scholar]
- 21. Dankers MKA, Roelen DL, van der Meer-Prins EMW, et al. Differential immunogenicity of HLA mismatches: HLA-A2 versus HLA-A28. Transplantation 2003; 75: 418. [DOI] [PubMed] [Google Scholar]
- 22. Kosmoliaptsis V, Sharples LD, Chaudhry AN, et al. Predicting HLA class II alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation 2011; 91: 183. [DOI] [PubMed] [Google Scholar]
- 23. Hwang S-H, Oh H-B, Shin E-S, et al. Influence of mismatching of HLA cross-reactive groups on cadaveric kidney transplantation. Transplant Proc 2005; 37: 4194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


