Abstract
Recent evidence suggests antimicrobial peptides protect the urinary tract from infection. Ribonuclease 7 (RNase 7), a member of the RNase A superfamily, is a potent epithelial-derived protein that maintains human urinary tract sterility. RNase 7 expression is restricted to primates, limiting evaluation of its antimicrobial activity in vivo. Here we identified Ribonuclease 6 (RNase 6) as the RNase A Superfamily member present in humans and mice that is most conserved at the amino acid level relative to RNase 7. Like RNase 7, recombinant human and murine RNase 6 has potent antimicrobial activity against uropathogens. Quantitative real-time PCR and immunoblot analysis indicate that RNase 6 mRNA and protein are up-regulated in the human and murine urinary tract during infection. Immunostaining located RNase 6 to resident and infiltrating monocytes, macrophages, and neutrophils. Uropathogenic E. coli induces RNase 6 peptide expression in human CD14+ monocytes and murine bone marrow derived macrophages. Thus, RNase 6 is an inducible, myeloid-derived protein with markedly different expression from the epithelial-derived RNase 7 but with equally potent antimicrobial activity. Our studies suggest RNase 6 serves as an evolutionarily conserved antimicrobial peptide that participates in the maintenance of urinary tract sterility.
Keywords: antimicrobial peptide, ribonuclease, urinary tract infection, pyelonephritis, cystitis
INTRODUCTION
The urinary tract is one of the most common targets of bacterial infection afflicting humans. An estimated 11% of women in the United States report at least one physician-diagnosed urinary tract infection (UTI) per year, and the lifetime probability that a woman will develop a UTI is 60%.1 The clinical management of UTI is complicated by the increasing incidence of infections caused by strains of Escherichia coli (E. coli) that are resistant to commonly used antimicrobial agents. Recently, antimicrobial peptides (AMP), a fundamental component of the innate immune response, have been shown to possess antimicrobial activity against uropathogenic bacteria.2,3 Thus, AMPs may represent new classes of antibiotics against drug resistant uropathogens. AMPs have many desirable features of a novel antibiotic class. (1) AMPs display antimicrobial activity at low micromolar concentrations. (2) AMPs overcome the shortfalls of conventional antibiotics given their ability to permeabilize microbial membranes. (3) Because AMPs target cell wall components and microbes cannot readily alter their cell wall composition, development of resistance to AMPs is limited. (4) AMPs show synergy with conventional antibiotics.4 Thus, the study of AMPs as therapeutic targets is indicated.
To date, very few AMPs have been described in the human kidney and urinary tract. Recently, our research group has shown that Ribonuclease 7 (RNase 7) is an epithelial-derived AMP that helps maintain human urinary tract sterility.5–7 RNase 7 belongs to the RNase A superfamily, a vertebrate specific group of genes encoding proteins with a characteristic signal peptide, ribonuclease catalytic motif (CKXXNTF), and six to eight conserved cysteine residues for disulfide bridging.8,9 RNase 7 exhibits broad-spectrum antimicrobial activity against Gram-positive and Gram-negative uropathogenic bacteria at micromolar concentrations.6,10 The uroepithelium of the lower urinary tract and the intercalated cells of the renal collecting tubules constitutively express RNase 7 and secrete it into the urinary stream.5,6 When urinary RNase 7 is neutralized in human urine specimens in vitro, urinary bacterial growth increases.5
Three lineages of the RNase A superfamily encode proteins associated with host defense: 1) angiogenins; 2) eosinophil RNases; and 3) RNase 7 and RNase 8, which exist uniquely in primates.9,11,12 The eosinophil RNases exhibit rapid rates of non-silent amino acid substitutions, with a high degree of interspecies divergence.13–15 This rapid molecular evolution has occurred through gene duplication and deactivation in a pattern similar to the human major histocompatibility complex and T cell receptor loci. These observations attest to a potential central role for rapid evolutionary tailoring of RNase A superfamily members toward host specific microbes.
While RNase 7 is most closely related in amino acid sequence to RNase 8, the mouse genome does not encode an orthologue of either protein, thereby limiting our understanding of their roles in urinary tract host defense in vivo. Certain members of the Ribonuclease A Superfamily, with potential antimicrobial function, are conserved between humans and lower order vertebrates. Rosenberg and Dyer identified orthologous sequences of RNase 6 in both primate and non-primate mammalian species.16 Human RNase 6 displays 56% amino acid identity to RNase 7 and its mRNA expression has been identified in the human kidney by Northern blot.17 At the cellular level, transcripts encoding RNASE6 have been detected in human peripheral blood monocytes and neutrophils, suggesting a role for this ribonuclease in host defense.16 In the mouse, Rnase6 transcripts have been detected by RT-PCR in kidney, bone marrow, and spleen.18 Murine Rnase6 is syntenic with human RNASE6 and their encoded proteins display 85% amino acid identity.16,18 Thus, this study was designed with three objectives: (1) to determine if mouse and human RNase 6 exhibit antimicrobial activity against uropathogenic bacteria; (2) to compare the urinary tract expression and antimicrobial activity of human RNase 6 and RNase 7; and (3) to evaluate urinary tract expression of murine RNase 6 at baseline and following experimental UTI. The results from this study develop a foundation to evaluate RNase 6’s role in maintaining urinary tract sterility in vivo.
RESULTS
Primary and predicted tertiary structures of RNase 6 and RNase 7
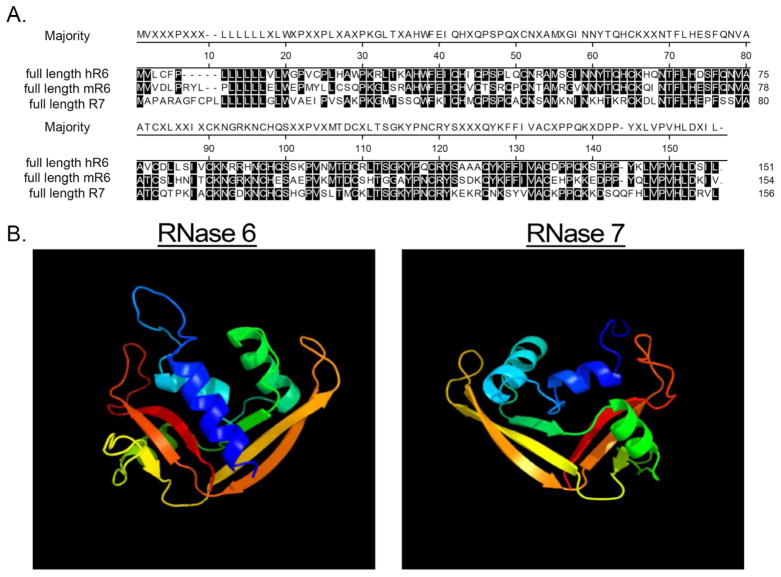
To identify murine proteins with greatest identity to human RNase 7, we queried the non-redundant Mus musculus protein database with the mature human RNase 7 peptide sequence (amino acids 29–156 of NP_115961 encoding the C-terminal 128 amino acids of RNase 7).17 This identified RNase 6 as the most similar mouse protein (52% identity and 71% similarity). Alignment of the predicted human and mouse RNase 6 mature peptides demonstrated 85% amino acid identity (Figure 1A). A protein-modeling algorithm identified similarities between the predicted tertiary structure of human RNase 6 and the NMR solution structure of RNase 7 (Figure 1B).19
Figure 1. Amino acid homology and predicted structural similarities between mouse RNase6, human RNase 6, and RNase 7.
(A) Alignment of full length proteins by Clustal W method.36 Residues matching the consensus (“majority”) are shaded. (B) Predicted solution structure of human RNase 6 and RNase 7.19,37
RNase 6 exhibits broad-spectrum antimicrobial activity against uropathogenic bacteria
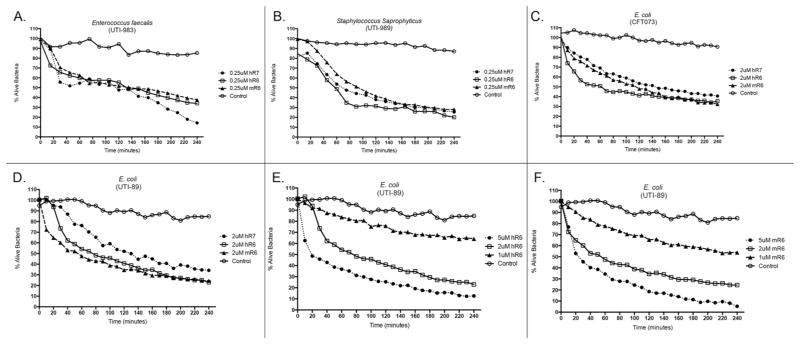
We performed Live/Dead bacterial viability assays to define the antimicrobial activity of recombinant human and mouse RNase 6 against Gram-positive and Gram-negative uropathogenic bacteria. Our results demonstrate that both human and mouse RNase 6 exhibit rapid, dose-dependent bactericidal activity toward uropathogenic Escherichia coli (UPEC), Enterococcus faecalis, and Staphylococcus saprophyticus at micromolar concentrations – comparable to RNase 7 (Figure 2).6 To evaluate if recombinant human RNase 6 and RNase 7 have synergistic antimicrobial activity against UPEC, we used the broth microdilution checkerboard antibacterial assay.20 Synergistic antimicrobial activity was not detected between these two peptides (data not shown). Recombinant human and mouse RNase 6 (0.1–10 μM) did not demonstrate cytotoxic activity toward immortalized human uroepithelial cells (UROtsa cells) or cause hemolysis of human red blood cells (data not shown).
Figure 2. RNase 6 and RNase 7 rapidly kill uropathogenic bacteria at micromolar concentrations.
Uropathogenic E. faecalis (A), S. saprophyticus (B), and E. coli strains CFT073 and UTI89 (C,D) were stained using a 1:1 mixture of SYTO®9, which labels live bacteria, and propidium iodide, which labels killed bacteria. Bacteria were incubated with fixed concentrations of recombinant murine RNase 6, human RNase 6, or RNase 7. (E/F) E. coli strain UTI89 was stained as outlined above and incubated with increasing concentrations of recombinant human or mouse RNase 6. Untreated bacteria served as the control. Bacterial viability over time was analyzed integrating fluorescent changes in SYTO®9 dye and propidium iodide dye. Values are the average of three replicates.
RNase 6 expression in the human urinary tract differs from RNase 7
To investigate the tissue distribution of RNASE6 and RNASE7 mRNA expression, we analyzed cDNA obtained from various human body tissues by quantitative real-time PCR (Figure 3 and Supplemental Figure 1). RNASE7 mRNA expression is significantly greater than RNASE6 mRNA expression in bladder (n=4, p=0.0389, unpaired students t-test) and kidney (n=10, p=0.0068, unpaired students t-test). In kidney samples with chronic pyelonephritis (n=6), RNASE6 and RNASE7 expression increases (Figure 3A). When comparing expression in other tissues, RNASE6 mRNA expression was significantly higher than RNASE7 mRNA in human spleen and thymus (Figure 3A, p=0.0243, unpaired students t-test).
Figure 3. RNase 6 and RNase 7 mRNA and peptide expression in the human urinary tract.
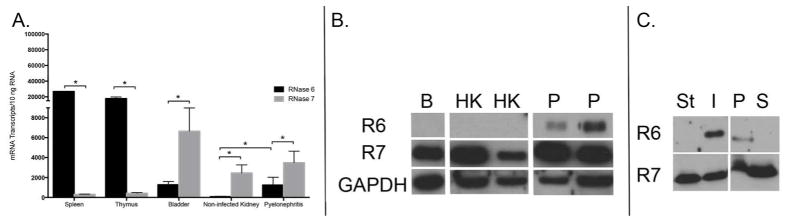
(A) Quantification of human RNASE6 and RNASE7 mRNA in non-infected tissues and pyelonephritis kidney samples by qRT-PCR. (B) Western immunoblot analysis of human RNase 6 (R6) and RNase 7 (R7) protein in bladder (B), non-infected kidney (HK) and pyelonephritis (P) lysates. GAPDH is included as a loading control. (C) Immunoblot analysis of human RNase 6 (R6) and RNase 7 (R7) in sterile (St) or UPEC infected (I) human urine (left). Infected human urine was subjected to centrifugation and immunoblot analysis was repeated using the cellular pellet (P) and supernatant (S) fractions (right).
Western immunoblot analysis was performed to evaluate RNase 6 and RNase 7 protein production in the human urinary tract. RNase 7 peptide was detected in human bladder lysates (n=4), non-infected kidney lysates (n=6), and chronic pyelonephritis kidney lysates (n=6). In contrast, RNase 6 peptide was detected only in pyelonephritis kidney lysates and was absent in non-infected kidney and bladder lysates (Figure 3B). Western immunoblot analysis was also performed on non-infected and UPEC infected human urine samples. Western immunoblot analysis routinely detected RNase 7 in sterile and infected urine samples (n=4). In contrast, RNase 6 was only detected in infected urine samples. When infected urine was cleared by centrifugation, RNase 6 reactivity was found exclusively in the pellet of urinary sediment. In contrast, RNase 7 protein was detected in both the sediment and the supernatant fraction (Figure 3C and Supplemental Figure 2).
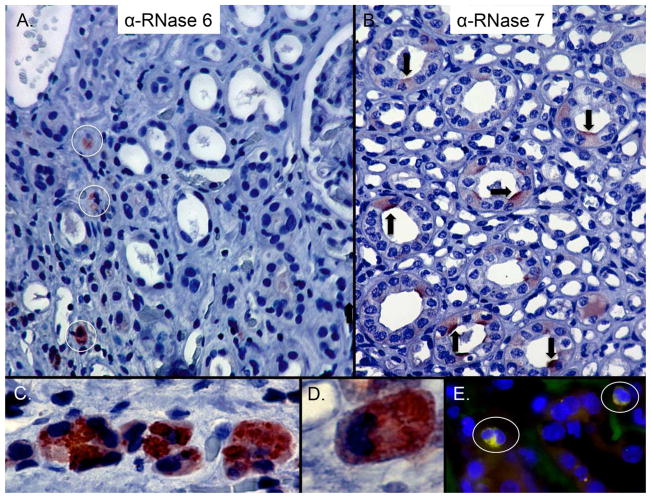
Immunohistochemistry was performed to localize RNase 6 and RNase 7 protein in the human urinary tract. As previously reported, RNase 7 production localized to the collecting tubules of non-infected kidney specimens (n=3, Figure 4).5,6,21 In contrast, epithelial RNase 6 production was not routinely detected in the kidney specimens (n=3). Instead, RNase 6 production localized to interstitial leukocytes with granulocyte and macrophage morphology in chronic pyelonephritis kidney tissues (n=3). RNase 6 reactivity was concentrated in cytoplasmic granules (Figure 4). RNase 6 expression by renal macrophages was confirmed by co-labeling with anti-CD68, a marker of the monocyte/macrophage lineage (Figure 4).
Figure 4. Expression of RNase 6 and RNase 7 in the human kidney.
(A) Immunohistochemistry demonstrates RNase 6 production (brown/circles) in isolated, interstitial leukocytes in the renal parenchyma (B) and RNase 7 production (brown/arrows) in isolated renal tubules. Epithelial expression of RNase 6 was not routinely detected. Original Magnification 20×. (C/D) High-power magnification identifies RNase 6 production by granulocytes with greatest expression in the cytoplasmic granules. Original magnification 40× and 100×, respectively. (E) Human pyelonephritis samples were labeled with RNase 6 (green), CD68 (red), and nuclei (blue). RNase 6 expression was detected in CD68-positive cells. Original magnification 40×.
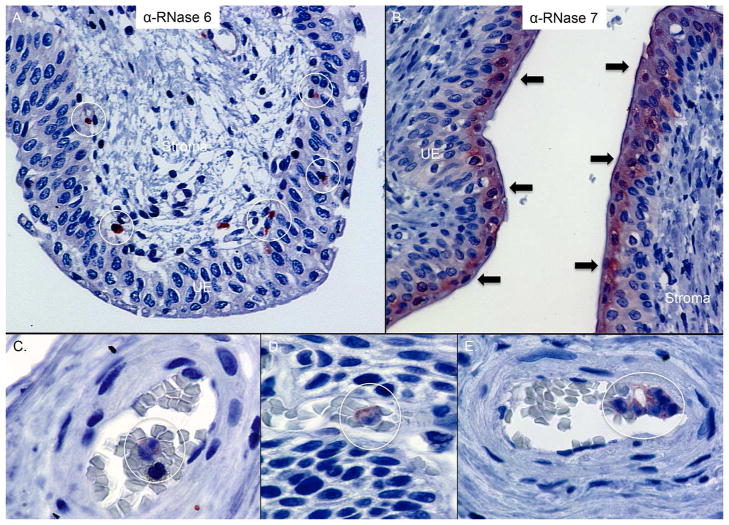
In the lower urinary tract, RNase 7 production localized to the apical uroepithelium in non-infected human bladder and ureter tissues (n=3, Figure 5).21 RNase 6 production was absent in uroepithelium but was detected in rare submucosal cells with leukocyte morphology (n=3, Figure 5).
Figure 5. Expression of RNase 6 and RNase 7 in the human lower urinary tract.
Immunohistochemistry demonstrates RNase 6 production (brown/circles) in isolated urothelial and submucosal leukocytes in the human ureter (A) whereas RNase 7 (brown/arrows) is produced by the apical uroepithelium (UE) of the lower urinary tract (B). Original magnification 40×. (C/D/E) Immunohistochemistry demonstrates RNase 6 production (red/circles) in intravascular leukocytes of human bladder. Original Magnification 100×.
Murine RNase 6 expression is similar to human RNase 6 expression in the urinary tract
We next evaluated whether mouse RNase 6 mRNA and protein expression parallels human RNase 6 in sterility and following experimental UTI. As described below, we transurethrally inoculated C57BL/6 mice with 108 colony-forming units (CFU) of UPEC strain UTI89 or CFT073 to evaluate RNase 6 expression during cystitis and pyelonephritis, respectively. In non-infected kidney and bladder homogenates, mouse RNase6 mRNA was detected by quantitative real-time PCR. Similar to humans, mouse RNase 6 mRNA expression was significantly higher in the spleen than kidney or bladder (Figure 6A and Supplemental Figure 1, p=0.006, unpaired students t-test). With cystitis, bladder RNase6 mRNA levels increased 3-fold 48 hours post infection (hpi) (2-way ANOVA with Tukey’s multiple comparisons test, p < 0.002). In contrast, with pyelonephritis, kidney RNase 6 mRNA remained constant (Figure 6B). Comparable to humans, immunoblot analysis routinely detected mouse RNase 6 peptide in pyelonephritis kidney lysates and not in non-infected kidney lysates. RNase 6 peptide was not routinely detected in non-infected or cystitis bladder lysates. As in humans, RNase 6 peptide was absent in sterile urine but detectable following UPEC infection (Figure 6C and Supplemental Figure 2).
Figure 6. RNase 6 mRNA and RNase 6 peptide expression in the murine urinary tract.
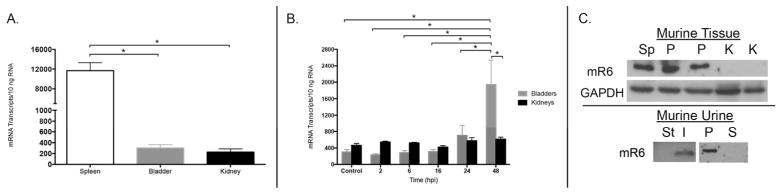
(A) Quantification of mouse Rnase6 mRNA in sterile tissues by qRT-PCR. Mean ± S.E.M. are shown from 4 bladders and kidneys. (B) Rnase6 mRNA expression over time in bladder and kidneys of UPEC infected mice. Time in hours post infection (hpi) is indicated on the X-axis. Mean ± S.E.M. are shown from 4 bladders and kidneys at each time point. (C, top) Western immunoblot of RNase 6 protein in mouse spleen (Sp), non-infected kidney (K), and pyelonephritis (P) kidney lysates. GADPH is included as a loading control. (C, bottom) Western immunoblot analysis of mouse RNase 6 peptide in sterile (St) and UPEC infected (I) urine samples (left). Infected mouse urine was subject to centrifugation and immunoblot analysis was repeated using the cellular pellet (P) and supernatant (S) fractions (right).
Although we could not routinely detect RNase 6 peptide via Western immunoblot in cystitis bladder lysates, immunohistochemistry revealed RNase 6(+) leukocytes in the bladder submucosa, uroepithelium, and urinary lumen (Figure 7A). Immunohistochemistry did not routinely detect RNase 6 protein in uninfected murine kidney and bladder (data not shown). With pyelonephritis, aggregates of RNase 6(+) leukocytes were visualized in the renal pelvis, papillary collecting tubules, perihilar fat, and renal uroepithelium (Figure 8A/D).
Figure 7. RNase 6 peptide localization in the lower murine urinary tract.
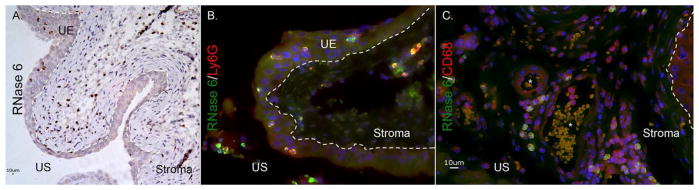
(A) Immunohistochemistry demonstrates RNase 6 production (brown) in isolated uroepithelial and stromal leukocytes in UPEC-infected mouse bladder. Original magnification 20×. (B/C) Infected mouse bladders labeled for RNase 6 (green), the neutrophil marker Ly6g (red) or the monocyte marker CD68 (red), and nuclei (blue). RNase 6 expression was detected in Ly6g and CD68-positive cells. Original magnification 40×. The dashed line demarcates the uroepithelium (UE) and the stromal layers of the bladder. US: urinary space.
Figure 8. RNase 6 peptide localization in the murine kidney.
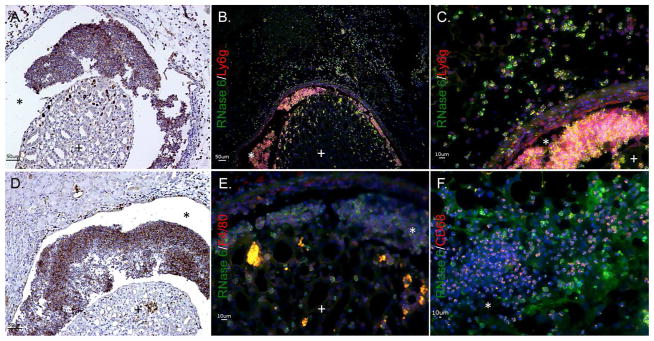
(A/D) Immunohistochemistry identifies RNase 6 production (brown) in isolated leukocytes in UPEC infected murine kidneys. Original Magnification 20×. (B/C) UPEC infected mouse kidneys were labeled with RNase 6 (green), the neutrophil marker Ly6g (red), and nuclei (blue). RNase 6 was detected in Ly6G positive neutrophils. (E/F) UPEC infected mouse kidneys were labeled with RNase 6 (green), the monocyte/macrophage markers F4/80 or CD68 (red), and nuclei (blue). RNase 6 was detected in CD68 positive cells. Original Magnification 40×. (+) identifies the renal parenchyma and (*) identifies the urinary space.
To localize the production of RNase 6 in infiltrating leukocytes, double labeling immunofluorescence was performed using leukocyte specific antigens CD68, CD11c (expressed by myeloid dendritic cells, macrophages, and natural killer cells); Ly6g (expressed by granulocytes); and F4/80 (expressed by monocytes and macrophages). Immunofluorescent detection of RNase 6 in leukocyte populations was optimized in spleen (data not shown) – where RNase6 mRNA and RNase 6 protein are abundantly expressed. In the infected bladder, RNase 6 expression co-localized with Ly6g and CD68, but was absent in cells expressing CD11c and F4/80 (Figure 7 and Supplemental Figure 3). With pyelonephritis, RNase 6 localized to infiltrating monocyte/macrophage and neutrophils, based on its co-expression with Ly6g and CD68. RNase 6 was not detected in cells expressing F4/80 (Figure 8).
Uropathogens induce RNase 6 peptide in human and murine macrophages
To investigate the cellular production of RNase 6, we exposed primary human and mouse monocyte/macrophage populations to UPEC. Using the peripheral blood from 2 healthy human donors, we enriched CD14(+) monocytes to >90% purity, exposed them to UPEC strain CFT073, and evaluated RNase 6 protein expression by Western immunoblot analysis. The RNase 6 antibody showed no cross-reactivity with UPEC (not shown). RNase 6 protein was detected as early as 30 minutes following UPEC incubation, and its presence and secretion increased over time (Figure 9A). Comparable results were obtained using non-enriched peripheral blood mononuclear cells (data not shown). Similarly, murine bone marrow derived macrophages (BMDM) did not express RNase 6 prior to CFT073 incubation. RNase 6 peptide was detected in cellular lysates and culture supernatants as early as 30 minutes following co-culture with CFT073 and increased over time (Figure 9B). RNase 6 protein induction was not observed following incubation of BMDM with lipopolysaccharide (LPS) plus interferon gamma (IFN-γ)(data not shown). Similar results were obtained in the human monocyte cell line U-937 and the murine macrophage-like cell line RAW 264.7 (not shown).
Figure 9. RNase 6 peptide production by monocyte/macrophages following bacterial exposure.
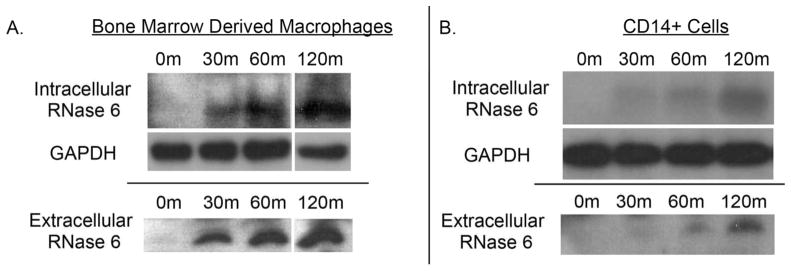
(A) Murine bone marrow derived macrophages and (B) human CD14(+) monocytes, were incubated with UPEC strain CFT073 for the indicated times and RNase 6 peptide was detected in cellular pellets and supernatants by Western immunoblot analysis.
DISCUSSION
Previously, we have demonstrated that RNase 7 is an epithelial-derived AMP that contributes to sterility in the human urinary tract.5,6 In vivo evidence to support the significance of RNase 7 is lacking, due to the absence of an orthologous gene in the laboratory mouse. To begin to evaluate the in vivo functions of antimicrobial ribonucleases, we chose to study RNase 6 given the high degree of amino acid conservation between its human and mouse orthologues, together with its mRNA expression in human and mouse kidney.16,18 Here, we provide the characterization of RNase 6 in the murine and human urinary tract and compare its function and expression to RNase 7. Our results identify RNase 6 as a novel leukocytic AMP that has potent bactericidal activity against Gram-positive and Gram-negative uropathogens at micromolar concentrations – comparable to RNase 7. To our knowledge, this is the first study to identify a potential biological role for RNase 6, namely, to kill invading uropathogens.
RNase 6 is the fifth member of the human RNase A Superfamily with experimentally proven antibacterial activity. In addition to RNase 6 and 7, human angiogenin (RNase 5), RNase 8, and the eosinophil cationic protein (ECP/RNase 3) demonstrate antimicrobial activity.9,22–24 In mice, Hooper and colleagues found that angiogenin 1 and 4 (RNase 1 and 4) have limited antibacterial activity against specific intestinal microbes.24 Like the other antimicrobial ribonucleases, RNase 6 is a highly cationic protein (PI= 9.49).16 This positive net charge may enhance binding to the negatively charged microbial membrane, promote membrane permeabilization, and facilitate cell death.
Even though RNase 6 and RNase 7 did not synergistically suppress UPEC growth in the checkerboard assay, we cannot exclude additive effects of these AMP in UTI. Additional future studies are required to investigate the antibacterial interactions and potential synergy between RNase 6, RNase 7, other bactericidal RNase A superfamily members, and conventional antibiotics for UTI treatment. RNase 3, for example, exhibits potent microbicidal activity toward E. coli at low micromolar concentrations and kills it more rapidly than RNase 7 when evaluated by Live/Dead assays.25,26 Additionally, N-terminal peptides derived from RNase 3 are 3-fold more potent than the corresponding region of RNase 7.27 Given its antimicrobial activity and limited toxicity toward human uroepithelial cells and red blood cells, the bactericidal mechanism(s) of RNase 6, RNase 7, and other bactericidal RNase A superfamily members warrants in-depth studies and supports the idea one or more of these peptides might be used as a novel pharmaceutical to treat infections or as an endogenous antibiotic that can be induced through immunomodulation.
Our results demonstrate that RNase 6 and RNase 7 expression differ in the urinary tract. In humans, bladder and kidney RNase 7 expression was significantly greater than RNase 6 expression in non-infected samples. However, with pyelonephritis, RNase 6 peptide production significantly increased. These results suggest that RNase 7 provides a front-line antimicrobial barrier that permits uropathogenic organisms from invading the urothelium. If this barrier is breached during microbial assault, other defenses like RNase 6 are activated to combat infection.
Similarly, in the murine urinary tract, RNase 6 peptide was not routinely detected in non-infected kidney and bladder tissues. With experimental UTI, bladder RNase6 mRNA significantly increased. Although we were unable to detect RNase 6 protein via Western immunoblot analysis in infected murine bladder lysates, immunostaining supported these results by demonstrating increases in RNase 6 peptide production that paralleled leukocyte infiltration. Our inability to detect RNase 6 peptide via Western immunoblot may be explained by differences in the rates of protein translation, degradation, antibody sensitivity, or the cellular origin of peptide production. In the kidney, we did not detect a significant increase in RNase6 expression with pyelonephritis yet we detected a robust increase in RNase 6 peptide with infection. These increases in RNase 6 protein expression, despite constant mRNA levels, suggest post-transcriptional control of RNase 6 expression, or, alternatively, recruitment of RNase 6 protein expressing cells to the infected kidney.
Consistent with our previously published studies, immunostaining localized RNase 7 production to the uroepithelium and intercalated cells of the renal tubules.5,21 In contrast, RNase 6 expression localized to resident urinary tract leukocytes or infiltrating granulocytes and macrophages in mouse cystitis bladders, mouse pyelonephritis kidneys, and human pyelonephritis kidneys. These findings support the original observations of Rosenberg and Dyer, who detected human RNASE6 mRNA in extracts from peripheral blood granulocytes and monocytes.16 Since myeloid cells express RNase 6 protein in humans and mice, we evaluated the effects of UPEC on RNase 6 expression by isolated monocyte/macrophage populations in culture. Whereas Western immunoblot analysis did not detect RNase 6 peptide in non-stimulated CD14(+) monocytes and murine BMDM RNase 6 protein was robustly induced by UPEC stimulation. Thus, we have identified myeloid cells as evolutionally conserved sources of RNase 6 peptide production in the context of UPEC infection. Future experiments will be directed toward characterizing the regulation of RNase 6 expression following UPEC exposure.
Within the myeloid cell populations, we observed RNase 6 localization to cytoplasmic granular structures. The distribution of RNase 6 within these cells is suggestive of subcellular localization to an intracellular organelle, such as the phagosome or lysosome. Sleat et al recently identified RNase 6 in a proteomics screen for lysosomal proteins and demonstrated partial co-localization of an RNase 6-mCherry fusion protein with the lysosomal resident protein, Npc2, in transfected COS cells (kidney fibroblasts).28 This is consistent with recent reports that RNase 6 is associated with mannose 6-phosphoryled proteins in human serum and urine29. Sleat et al also found that three mouse eosinophil associated ribonuclease (Ear) proteins (Ear1, Ear2, and Ear6) are enriched in lysosomal fractions, raising the possibility that lysosomal targeting of ribonucleases is a generalizable strategy employed by the host cell to sequester RNase proteins until their secretion is triggered by bacteria or, alternatively, to directly attack bacteria within the phagosome-lysosome system.28
The concept of active, subcellularly targeted RNase 6 is intriguing given the observation that UPEC can survive for up to 24 hours inside macrophages, where UPEC localize to Lamp1(+) structures interpreted as representing phagosomes or lysosomes.30 Together with our finding that RNase 6 expression in infected urine is restricted to leukocytes, these observations give rise to the hypothesis that myeloid cells attack UPEC harbored within phagosomes or lysosomes by specifically targeting antimicrobial peptides such as RNase 6 to these structures. Alternatively, RNase 6 may serve to degrade microbial RNA, as has been postulated in the case of Ear11, a macrophage resident RNase A superfamily member in the lung.31 Future experiments will evaluate the consequence of altered RNase 6 expression and/or subcellular localization on antibacterial activity of myeloid cells. This will establish the degree to which the antibacterial activity of macrophages and neutrophils is reliant on adequate RNase 6 levels and/or appropriate targeting of RNase 6 to subcellular structures.
In conclusion, in this study we identify RNase 6 as a novel broad-spectrum antimicrobial protein in the mammalian urinary tract with bactericidal activity comparable to RNase 7. In the urinary tract, RNase 6 and RNase 7 proteins differ in their cellular source and expression. Specifically, RNase 7 is constitutively expressed by parenchymal cells and secreted into urine, whereas RNase 6 is induced by UPEC and expressed by recruited leukocytes during pyelonephritis. These results are in line with the hypothesis that RNase 7 provides a front-line shield or barrier to protect the urinary tact and if this barrier is breached, AMPs like RNase 6 are important to help eradicate infection. Consequently, elucidation of the factors that regulate RNase 6 production and its antimicrobial mechanisms may lend novel insight into the pathogenesis and prevention of UTIs.
METHODS
Study approval
Informed written consent was obtained from all patients participating in this study. For subjects less than 18 years of age, written parental/guardian consent was obtained. The Nationwide Children’s Hospital Institutional Review Board approved this study along with the consent process and documents (IRB07-00383). The mouse experimental UTI model was approved (AR06-00119) by The Research Institute at Nationwide Children’s Hospital Institutional Laboratory Animal Care and Use Committee (Welfare Assurance Number A3544-01). Uropathogenic E. coli (UTI-89) was recovered from the urine of a patient with cystitis.32 E. coli CFT073 was isolated from the blood and urine of a woman with pyelonephritis.33
Human samples
As previously described, non-infected kidney samples and bladder tissues were obtained from adults undergoing nephrectomy for renal tumors. Tissue samples were free of microscopic signs of disease or inflammation. Infected kidney tissue was provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute.6 Non-infected and infected urine samples were obtained from children presenting to Nationwide Children’s Hospital.6
Murine UTI model
8-week-old C57BL/6 female mice (n=4 per group) were purchased (Harlan Laboratories, Indianapolis, IN) and permitted to recover for 7–10 days after delivery. The mice were maintained on a 12-hour light-dark cycle and fed standard chow (Harlan). On the day prior to inoculation, UTI89 (cystitis) or CFT073 (pyelonephritis) was inoculated into Luria Broth and cultured statically at 37°C for 16 hours. On the day of inoculation, bacteria were harvested by centrifugation and resuspended in phosphate buffered saline (PBS). Next, mice were anesthetized with isoflurane and the urethra was catheterized as described.34,35 108 colony-forming units (CFU) of UPEC strain UTI89 or CFT073 were transurethrally introduced in 50 ul PBS. The inoculum was confirmed by serial dilution and plating on LB agar for 16 hours at 37°C. Once UPEC infections had progressed for 2, 6, 16, 24, or 48 hours, animals were re-anesthetized to recover urine and sacrificed by cervical dislocation. Bladders and kidneys were snap frozen in liquid nitrogen and stored at −80°C until mRNA isolation. Alternatively, tissues were fixed in 4% paraformaldehyde for 48 hours followed by transfer to 70% ethanol and embedded in paraffin.
Protein alignment and structural predictions
The mature protein sequences used for analysis are as follows: mouse RNase 6 accession number NP_084374 (residues 28–153); human RNase 6 accession number NP_005606 (residues 24–150); human RNase 7 accession number NP_115961 (residues 29–156). Alignments were performed using MegAlign (DNAStar, Madison, WI) and Clustal W alignment method.36 The same protein sequences were uploaded to the Phyre serve for structural predictions.19 RNase 6 structures were compared to the NMR solution structure of RNase 7.37
Expression of recombinant human and mouse RNase 6
Using PCR, the human and mouse RNASE6 mature protein coding sequence was amplified from kidney cDNA and cloned into pDNR21 (Invitrogen) then mobilized to pDEST17 after sequencing to confirm absence of PCR errors.16 The mouse Rnase6 primers used were: forward 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC T TC cag cct aag ggt ctc tcc a-3′ where attB1 sequence is underlined and Rnase6 sequence is in lowercase; and reverse 5′-GGGG AC CAC TTT GTA CAA GAA AGC TGG GTC CTA cac aat ctt atc taa gtg ta-3′ where attB2 sequence is underlined. The human RNASE6 primers used were: forward 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC T TC tgg cct aag cgt ctc acc a-3′ and reverse 5′GGGG AC CAC TTT GTA CAA GAA AGC TGG GTC CTA gag aat act atc taa gtg ta-3′. The resulting constructs encode N-terminal histidine tag followed in-frame by 128 amino acids encoding the mature human or mouse RNase6 peptide. Expression was induced in E. coli BL21 AI (Invitrogen) with 0.2% L-arabinose treatment and recombinant RNase6 protein was purified from inclusion bodies, refolded and dialyzed as previously described.6,10 Protein concentrations were determined by Bradford protein assay (Bio-Rad, Hercules, CA). The presence of the full-length RNase 6 was visualized by SDS-polyacrylamide gel electrophoresis, followed by silver staining or western immunoblot analysis.
Live/Dead bacterial viability assay
Bacterial viability assays were performed using a Live/Dead BacLight bacterial viability kit (BacLight; Molecular Probes) as previously described.6,21 105 CFU/mL of stained bacteria were incubated with RNase 7, human RNase 6, or mouse RNase 6. Changes in fluorescent intensity were measured using the Spectramax M2 multimode microplate reader (Molecular Devices, Sunnyvale, CA).6,21 These results were compared with a standard curve that was generated using increasing concentrations of live:dead bacteria.
Checkerboard Assay
To evaluate if recombinant human RNase 6 and RNase 7 have synergistic antimicrobial activity against UPEC strain UTI89, we used the broth microdilution checkerboard method described in The Clinical Microbiology Procedures Handbook (http://www.ncbi.nlm.nih.gov/nlmcatalog/101528193). Briefly, the peptides were combined with 1×105 CFUs/mL UTI89 at varying fractional concentrations of their mean inhibitory concentration (MIC) in 1% peptone water. Results were expressed as the fractional inhibitory concentration index (FICI) according to the following formula: FICI = (A)/MICA + (B)/MICB, where MICA and MICB are the MICs of peptides tested alone and (A) and (B) are the MICs of the two peptides tested in combination. Synergy was defined as an FICI ≤0.5 and antagonism was defined as FICI >4.20 The MIC of RNase 6 and RNase 7 wad determined using the broth microdilution MIC test described in The Clinical Microbiology Procedures Handbook.
Analysis of cytotoxic activity
To determine if recombinant human or murine RNase 6 has cytotoxic activity, immortalized human uroepithelial cells (UROtsa cells, kidney provided by D. Sens) were seeded into a 96-well tissue culture pate (105 cells/well) and incubated with increasing concentrations of recombinant RNase 6 (0.1–10mmol/l) for 3h at 37°C. After incubation, cell viability was determined using a Live/Dead mammalian cell viability/cytotoxicity kit.6 The ability of RNase 6 to induce hemolysis was also evaluated as previously described.6
Quantitative real-time PCR
Human mRNA was isolated using the TRIzol® method. RNase 7 quantitative real-time PCR was performed as previously described.5,6,21 Human RNase 6 mRNA was amplified by RT-PCR from kidney cDNA using the following primers: h6F: 5′-AGCCCCAACACTGAGACCAGAAAA-3′ and h6R: 5′-GGTGGCAGTTGTGCCGACGA-3′. The 305 bp PCR product was gel purified and cloned into pCR4-TOPO (Invitrogen) and this was used to generate a standard curve as described.5 In brief, cDNA corresponding to 10ng RNA served as the template in a 25ml reaction of 0.1mmol/l of each primer and 1xLight-Cycler-Fast Start DNA Master SYBR green mix. The PCR conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles with each cycle consisting of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30s. Representative products were subjected to agarose gel electrophoresis with ethidium bromide staining to confirm the specificity of the PCR reactions. A melting temperature profile curve was performed for each reaction.
Mouse mRNA was isolated using the TRIzol® Plus RNA Purification System (Invitrogen) as described and complementary (c)DNA was used as a template in a quantitative (q)RT-PCR reaction.34 A 205 bp mouse Rnase6 RT-PCR product was amplified from kidney cDNA using primers 5′-TGG CCC TGT TCA CCA TAG GAG CC-3′ and Reverse 5′-GCG CAT GGC TGT GTT GCA TGG-3′, cloned into pCR4-TOPO, and was used as a standard along with an Rnase6 primer/probe set (Mm01319576_m1, ABI).
SDS-PAGE Immunoblot
Murine and human tissues were processed as previously described.6 Equal concentrations of tissue lysate tissue or urinary protein were loaded onto 15% sodium dodecyl sulfate (SDS) gels and subjected to electrophoresis. After electrophoretic separation, proteins were transferred onto a polyvinylidene difluoride membrane. The membranes were blocked and incubated with rabbit polyclonal anti-RNase 6 antibody (Abgent). After washing, a horseradish peroxidase–conjugated anti-rabbit antibody was applied for detection (Cell Signaling Technology, Danvers, MA). Proteins were visualized using an ECL detection system.
Immunohistochemistry
Four micron-thick paraffin sections were prepared. Following deparaffinization, rehydration, and antigen retrieval, a biotin block and a serum-free protein block were performed (Superblock or Mouse-to-Mouse Block; ScyTek Laboratories, Logan, UT). The slides were incubated overnight at 4°C with polyclonal rabbit RNase 6 antibody (Abcam) or polyclonal RNase 7 antibody (Sigma), washed, and incubated with antipolyvalent biotinylated antibody and UltraTek Streptavidin/HRP (ScyTek Laboratories). Sections were developed using 3-Amino-9-Ethylcarbazole and counterstained with hematoxylin. Negative-control sections were incubated with non-immune serum in place of RNase 6 or RNase 7 antibody.
Immunofluorescence
Double-labeled immunofluorescence was performed to localize RNase 6 expression in the human and murine urinary tract. Human kidney slides were incubated overnight at 4°C with a polyclonal rabbit RNase 6 antibody (1:150, Abcam), and an anti-CD68 antibody (1:200, Abgent, San Diego, CA, USA). Mouse kidney and bladder slides were incubated overnight at 4°C with a polyclonal rabbit RNase 6 antibody (1:150, Abcam), polyclonal rat Ly6g antibody (1:200, Biolegend, San Diego, CA), polyclonal rat anti-CD68 antibody (1:100, AbD Serotec, Raleigh, NC, USA), polyclonal rat anti-F4/80 antibody (1:100, AbD Serotec), or a biotinylated hamster anti-CD11c antibody (1:500, BD Pharmingen, San Jose, CA, USA). Reactivity was visualized using biotinylated or fluorophore conjugated secondary antibodies. Negative-control sections were incubated with non-immune serum in place of RNase 6 and RNase 7 antibodies. Images were obtained using the BZ-9000 series fluorescence microscope (KEYENCE, Osaka, Japan).
Human myeloid cells
Human peripheral blood mononuclear cells was collected from two healthy donors. Monocytes were enriched by positive selection using CD14 microbeads (Miltenyi Biotech, Auburn, CA). Purity of CD14(+)-selected monocytes was confirmed by staining 100,000 positively selected cells with 5 μl CD14-PE (BD Biosciences, San Jose, CA, USA) and flow cytometry (BD FacsDiva, San Jose, CA, USA), comparing to staining of unselected PBMC. Purity was > 90%.
Bone marrow derived macrophages
Bone marrow-derived macrophages (BMDMs) were generated from femurs and tibias of C57BL/6 mice. Bone marrow cells were cultured in RPMI1640 supplemented with 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, with the addition of 20% L929-cell–conditioned medium. After 5–7 days of culture, cells were seeded into 6-well tissue culture treated plates at a density of 2×106 cells/well in antibiotic-free medium overnight prior to infection. Macrophages were infected with live or heat–inactivated E. coli CFT073 at a multiplicity of infection (MOI) of 10 bacteria/macrophage. After washing, infected cells were incubated in fresh medium containing gentamicin (100 μg/ml) to limit the growth of extracellular bacteria. When indicated, macrophages were stimulated with 100 ng/ml LPS E. coli O111:B4 (Sigma-Aldrich, Saint Louis, MO) and IFN-γ (20 ng/ml, BioVision). After incubation for the indicated time points, culture supernatants and cells pellets were collected and assayed as indicated.
Supplementary Material
Acknowledgments
We would like to acknowledge the Human Cooperative Human Tissue Network for providing the human tissue samples. We would also like to acknowledge Dr. Sheryl Justice for providing the bacterial isolates, and Dr. Dan Cohen and the NCH Emergency Department staff for the infected urine samples. SPS is supported by the National Institute of Health Grant (NIAID) grant R01AI092117. JDS is supported by the National Institute of Health Grant (NIDDK) K08 DK094970-02.
Footnotes
DISCLOSURE
None of the authors declares a financial interest in the content of this manuscript.
References
- 1.Foxman B, Barlow R, D’Arcy H, Gillespie B, Sobel JD. Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol. 2000;10:509–15. doi: 10.1016/s1047-2797(00)00072-7. [DOI] [PubMed] [Google Scholar]
- 2.Spencer JD, Schwaderer AL, Becknell B, Watson J, Hains DS. The innate immune response during urinary tract infection and pyelonephritis. Pediatr Nephrol. 2013 doi: 10.1007/s00467-013-2513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zasloff M. Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. J Am Soc Nephrol. 2007;18:2810–6. doi: 10.1681/ASN.2007050611. [DOI] [PubMed] [Google Scholar]
- 4.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 5.Spencer JD, Schwaderer AL, Dirosario JD, et al. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011;80:174–80. doi: 10.1038/ki.2011.109. [DOI] [PubMed] [Google Scholar]
- 6.Spencer JD, Schwaderer AL, Wang H, et al. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013;83:615–25. doi: 10.1038/ki.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zasloff M. The antibacterial shield of the human urinary tract. Kidney Int. 2013;83:548–50. doi: 10.1038/ki.2012.467. [DOI] [PubMed] [Google Scholar]
- 8.Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. 2005;85:208–20. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg HF. RNase A ribonucleases and host defense: an evolving story. J Leukoc Biol. 2008;83:1079–87. doi: 10.1189/jlb.1107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Schwaderer AL, Kline J, Spencer JD, Kline D, Hains DS. Contribution of structural domains to the activity of ribonuclease 7 against uropathogenic bacteria. Antimicrob Agents Chemother. 2013;57:766–74. doi: 10.1128/AAC.01378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta SK, Haigh BJ, Griffin FJ, Wheeler TT. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. 2013;19:86–97. doi: 10.1177/1753425912446955. [DOI] [PubMed] [Google Scholar]
- 12.Simanski M, Koten B, Schroder JM, Glaser R, Harder J. Antimicrobial RNases in cutaneous defense. J Innate Immun. 2012;4:241–7. doi: 10.1159/000335029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg HF, Dyer KD, Tiffany HL, Gonzalez M. Rapid evolution of a unique family of primate ribonuclease genes. Nat Genet. 1995;10:219–23. doi: 10.1038/ng0695-219. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc Natl Acad Sci U S A. 2000;97:4701–6. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhania NA, Dyer KD, Zhang J, et al. Rapid evolution of the ribonuclease A superfamily: adaptive expansion of independent gene clusters in rats and mice. J Mol Evol. 1999;49:721–8. doi: 10.1007/pl00006594. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg HF, Dyer KD. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res. 1996;24:3507–13. doi: 10.1093/nar/24.18.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Wootton JC, Gertz EM, et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–9. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyer KD, Rosenberg HF, Zhang J. Isolation, characterization, and evolutionary divergence of mouse RNase 6: evidence for unusual evolution in rodents. J Mol Evol. 2004;59:657–65. doi: 10.1007/s00239-004-2657-0. [DOI] [PubMed] [Google Scholar]
- 19.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 20.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 21.Spencer JD, Schwaderer AL, Eichler T, et al. An endogenous ribonuclease inhibitor regulates the antimicrobial activity of ribonuclease 7 in the human urinary tract. Kidney Int. 2013 doi: 10.1038/ki.2013.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudolph B, Podschun R, Sahly H, Schubert S, Schroder JM, Harder J. Identification of RNase 8 as a novel human antimicrobial protein. Antimicrob Agents Chemother. 2006;50:3194–6. doi: 10.1128/AAC.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J Immunol. 1989;142:4428–34. [PubMed] [Google Scholar]
- 24.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–73. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg HF. Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J Biol Chem. 1995;270:7876–81. doi: 10.1074/jbc.270.14.7876. [DOI] [PubMed] [Google Scholar]
- 26.Torrent M, Badia M, Moussaoui M, Sanchez D, Nogues MV, Boix E. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010;277:1713–25. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 27.Torrent M, Pulido D, Valle J, Nogues MV, Andreu D, Boix E. Ribonucleases as a host-defence family: evidence of evolutionarily conserved antimicrobial activity at the N-terminus. Biochem J. 2013;456:99–108. doi: 10.1042/BJ20130123. [DOI] [PubMed] [Google Scholar]
- 28.Sleat DE, Sun P, Wiseman JA, et al. Extending the mannose 6-phosphate glycoproteome by high resolution/accuracy mass spectrometry analysis of control and acid phosphatase 5-deficient mice. Mol Cell Proteomics. 2013;12:1806–17. doi: 10.1074/mcp.M112.026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubke T, Lobel P, Sleat DE. Proteomics of the lysosome. Biochim Biophys Acta. 2009;1793:625–35. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bokil NJ, Totsika M, Carey AJ, et al. Intramacrophage survival of uropathogenic Escherichia coli: differences between diverse clinical isolates and between mouse and human macrophages. Immunobiology. 2011;216:1164–71. doi: 10.1016/j.imbio.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Cormier SA, Yuan S, Crosby JR, et al. T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am J Respir Cell Mol Biol. 2002;27:678–87. doi: 10.1165/rcmb.4882. [DOI] [PubMed] [Google Scholar]
- 32.Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc Natl Acad Sci U S A. 2000;97:8829–35. doi: 10.1073/pnas.97.16.8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mobley HL, Jarvis KG, Elwood JP, et al. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1–4) beta Gal binding in virulence of a wild-type strain. Mol Microbiol. 1993;10:143–55. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 34.Becknell B, Spencer JD, Carpenter AR, et al. Expression and antimicrobial function of Beta-defensin 1 in the lower urinary tract. PLoS One. 2013;8:e77714. doi: 10.1371/journal.pone.0077714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung CS, Dodson KW, Hultgren SJ. A murine model of urinary tract infection. Nat Protoc. 2009;4:1230–43. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang YC, Lin YM, Chang TW, et al. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J Biol Chem. 2007;282:4626–33. doi: 10.1074/jbc.M607321200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.