Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal disease with a dismal prognosis. However, while most patients die within the first year of diagnosis, very rarely, a few patients can survive for more than10-years. Better understanding the molecular characteristics of the pancreatic adenocarcinomas from these very long-term survivors (VLTS) may provide clues for personalized medicine and improve current pancreatic cancer treatment. To extend our previous investigation, we examined the proteomes of individual pancreas tumor tissues from a group of VLTS patients (survival ≥10 years) and short-term survival patients (STS, survival <14 months). With a given analytical sensitivity, the protein profile of each pancreatic tumor tissue was compared to reveal the proteome alterations that may be associated with pancreatic cancer survival. Pathway analysis of the differential proteins identified suggested that MYC, IGF1R and p53 were the top three upstream regulators for the STS associated proteins, and VEGFA, APOE, and TGFβ-1were the top three upstream regulators for the VLTS associated proteins.
Immunohistochemistry analysis using an independent cohort of 145 PDAC confirmed that the higher abundance of ribosomal protein S8 (RPS8)and prolargin (PRELP)were correlated with STS and VLTS, respectively. Multivariate Cox analysis indicated that “High-RPS8 and Low-PRELP”was significantly associated with shorter survival time (HR=2.69, 95% CI 1.46-4.92, p=0.001).In addition, galectin-1, a previously identified protein with its abundance aversely associated with pancreatic cancer survival, was further evaluated for its significance in cancer-associated fibroblasts. Knockdown of galectin-1 in pancreatic cancer-associated fibroblasts dramatically reduced cell migration and invasion.The results from our study suggested that PRELP, LGALS1 and RPS8 might be significant prognostic factors, and RPS8 and LGALS1 could be potential therapeutic targets to improve pancreatic cancer survival if further validated.
Keywords: pancreatic cancer, pancreatic ductal adenocarcinoma (PDAC), prognostics, biomarker, galectin-1 (LGALS1), prolargin (PRELP), 40S ribosomal protein S8 (RPS8), proteomics, mass spectrometry, immunohistochemistry, tissue microarray
The prognosis of pancreatic ductal adenocarcinoma (PDAC) is extremely poor: the majority of patients die within the 6 months of diagnosis, and only 6% of patients survive for five years1. At the time of diagnosis, most patients have surgically unresectable disease. For the patients who undergo surgical resection, the clinical outcome is considerably better, with 5-year survival rates of up to 20% in some studies 2-11. In very rare situations, some patients can survive for a very long-term (VLTS, defined as patients with ≥10-year survival following resection) and are essentially cured of their disease. There are some published studies which have identified VLTS of histologically validated PDAC, usually at a frequency less than 5% in most series2, 3, 10, 12. Surprisingly, patients with aggressive diseases such as T3-T4 lesions and involvement of lymph node were also presented in these VLTS cohorts, suggesting that the basis of VLTS in PDAC extends beyond simply resecting node-negative localized cases, but might indicate an intrinsic molecular phenotype of such tumors13.
Several clinicopathological characteristics have been demonstrated to have significant effect on long-term survival including: small tumor size (<2 cm), negative lymph node status, negative resection margin (i.e. R0) and differentiation grade14. However, the molecular characteristics relating to long term-survival of PDAC are poorly understood. Using a PDAC survival tissue microarray, a study evaluated 13 putative biomarkers selected from the literature, and identified mucin 1 and mesothelin as biomarkers for predicting aggressiveness of pancreatic cancer biology, outperforming pathologic features and other putative biomarkers15. In our previous efforts, using pooled samples we discovered that the abundance of stromal galectin 1 (LGALS1) was adversely associated with survival from PDAC, while the abundance of prolargin (PRELP), osteoglycin (OGN) and rheumatoid factors D5 light chain (V<kappa>3) were higher in the pancreatic tumor tissues of VLTS relative to STS13. Immunohistochemistry (IHC) analysis of pancreatic tumor tissues using two independent cohorts confirmed that an increased/strong abundance of galectin-1, a T-cell immune-modulator and metastasis-related protein, in the cancer stroma was a negative indicator of pancreatic cancer survival13. For PRELP, contradictorily, a recent study suggested that the high abundance of PRELP was associated with shorter pancreatic cancer survival16. While various factors could influence the observations in different studies, the role of PRELP in pancreatic cancer survival remains uncertain and warrants further investigation.
In the current study, we sought to expand our understanding of the proteome characteristics associated with pancreatic cancer survival by analyzing the individual cancerous tissues from VLTS and STS patients using a label-free proteomics approach. Functional protein annotation and pathway analysis of the differential proteins associated with VLTS and STS aided in the characterization of the mechanisms underlying survival. While a comprehensive interpretation of the data obtained will require additional information beyond this investigation, selected exploration of a few interesting proteins are presented here. IHC validation was performed using a clinically annotated tissue microarray (TMA) to examine the significance of the protein targets in the context of PDAC survival. In addition, a functional study was performed to evaluate the effects of galectin-1 on the metastatic behavior of cancer-associated fibroblasts.
Methods and Materials
Patients and specimens
This study was approved by the Human Subjects Division of the University of Washington, the Internal Review Board (IRB) of the University of California at UCLA, and the IRB of the Johns Hopkins Medical Institutions. Very long-term survivors (VLTS) and short-term survivors (STS) were identified as survivors of PDAC for ≥10 years and <14 months, respectively, following surgical resection. Before inclusion, the histology was reviewed for all of the identified VLTS and STS cases from Johns Hopkins by two experts in pancreatic pathology (RHH and AM), in order to confirm the diagnosis of PDAC and exclude the possibility of confounding lesions like ampullary cancers and/or intraductal papillary mucinous neoplasm (IPMN). Five VLTS and five STS cases were matched by tumor stage and grade for proteomic analysis. These tumors were obtained prior to treatment and thus the proteomes represented the naïve state of the adenocarcinoma. The changing standards for adjuvant chemotherapy over the past 15 years precluded us from matching cases by type of treatment received.The validation cohort was from UCLA with 145 PDAC cases constructed on a previously described TMA17. The clinicopathological and survival information for each patient were obtained from UCLA surgical database of pancreatic patients. Survival analysis of the UCLA cohort was limited to overall survival, which was determined by review of clinical records and search of the Social Security Death Index. Survival intervals were calculated from date of surgery to date of confirmed death or last patient contact. The clinicopathological and survival information of the PDAC cases included in our study are provided in Table 1.
Table 1.
Clinicopathologic and survival information for the PDAC cases.
| Cohort | VLST | STS | UCLA TMA cohort | |
|---|---|---|---|---|
| No. of cases | 5 | 5 | 145 | |
| Age | <60 | 2 | 1 | 49 |
| ≥60 | 3 | 4 | 96 | |
| Gender | M | 3 | 2 | 75 |
| F | 2 | 3 | 70 | |
| Survival time | Mean | >10 yrs | 10 months | 34.7 months |
| Range | >10yrs | 6-14 months | 0.49-100 months | |
| Tumor size | <3cm | 2 | 1 | 87 |
| ≥3cm | 3 | 4 | 58 | |
| Tumor grade | Low grade (G1-2) | 0 | 0 | 83 |
| High grade (G3-4) | 5 | 5 | 62 | |
| AJCC stage | 1 | 1 | 40 | |
| 2 | 4 | 105 | ||
| unknown | 0 | 5 | 0 | |
| Lymph nodes | negative | 2 | 3 | 69 |
| positive | 3 | 2 | 76 | |
Protein extraction from formalin-fixed paraffin-embedded tissue
VLTS of pancreatic cancer is rare, and the availability of fresh frozen tissue specimens from these VLTS patients is therefore extremely scarce. The current study made use of the commonly preserved format of clinical specimens (i.e. formalin-fixed paraffin-embedded (FFPE)) for comparative proteomics analysis18. Extraction of proteins from FFPE tissue were carried out as described in our previous study18. Briefly, unstained 15 μm sections of FFPE tissue were deparaffinized and rehydrated. The corresponding H&E slides were examined under the microscope to delineate the areas with highest neoplastic cellularity (including both PDAC epithelium and associated stroma), while excluding obvious areas of non-neoplastic pancreatic acinar tissues and inflammatory infiltrates.An equivalent of 1 cm×1 cm×15 μm amount of deparaffinized tissue samples were scraped from each slide and transferred into 300 μl lysis solution of 70% 50 mM ammonium bicarbonate and 30% acetonitrile, using sterile blades and needle tips. The lysates were then incubated at 90°C for 30 minutes followed by 60°C for 120 minutes. The samples were sonicated for 2 minutes followed by 1 minute incubation on ice. The sonication and ice incubation process was repeated twice more. The homogenized samples were incubated at 60°C for 1 hr, followed by a second sonication step.
Sample preparation for proteomics analysis
The protein lysates were digested with sequencing-grade trypsin (Promega, Madison, WI) at a trypsin to protein ratio of 1:50 at 37°C for 18 hours. The digested samples were centrifuged at 1500×g for 10 minutes and the supernatants were collected. The protein digests were reduced with 10 mM dithiothreitol (DTT) at 60°C for 60 minutes then incubated with 25 mM iodoacetamide at room temperature in the dark for 30 minutes for cysteine alkylation. The samples were then purified with C18 columns (UltraMicroSpin Column/Vydac C18 silica, The Nest Group, Inc., Southborough, MA), dried down and stored at −20°C until mass spectrometric analysis.
LC MS/MS analysis
An LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA) coupled with a nano-flow HPLC (Eksigent Technologies, Dublin, CA) was used in this study. A 2 μg sample of each case was injected for the mass spectrometric analysis. The samples were first loaded onto a 1.5 cm trap column (IntegraFrit 100 μm, New Objective, Woburn, MA) packed with Magic C18AQ resin (5 μm, 200 Å particles; Michrom Bioresources, Auburn, CA) with Buffer A (water with 0.1% formic acid) at a flow rate of 3 uL/minute. The peptide samples were then separated by an 27 cm analytical column (PicoFrit 75 μm, New Objective) packed with Magic C18AQ resin (5 μm, 100 Å particles; Michrom Bioresources) followed by mass spectrometric analysis. A 90-minute linear LC gradient was used as follows: 5% to 7% Buffer B (acetonitrile with 0.1% formic acid) versus Buffer A over 2 minutes, then to 35% over 90 minutes, then to 50% over 1 minute, hold at 50% for 9 minutes, change to 95% over 1 minute, hold at 95% for 5 minutes, drop to 5% over 1 minute and recondition at 5%. The flow rate for the peptide separation was 300 nL/minute. For MS analysis, a spray voltage of 2.25 kV was applied to the nanospray tip. The mass spectrometry experiment was performed using data-dependent acquisition with a m/z range of 400–1800, consisting of a full MS scan in the Orbitrap followed by up to 5 MS/MS spectra acquisitions in the linear ion trap using collision induced dissociation (CID). Other mass spectrometer parameters include: isolation width 2 m/z, target value 1e4, collision energy 35%, max injection time 100 ms. Lower abundance peptide ions were interrogated using dynamic exclusion (exclusion time 45 seconds, exclusion mass width −0.55 m/z low to 1.55 m/z high). A charge state screen was used, allowing for MS/MS of any ions with identifiable charge states +2, +3, and +4 and higher. The samples were analyzed in duplicate with random order using the identical instrument conditions.
Proteomics data analysis
Raw machine output files from all MS runs were converted to mzXML files and searched with X!Tandem19 configured with the k-score scoring algorithm20, against version 3.69 of the human International Protein Index (IPI) database. The search parameters were as follows: enzyme, trypsin; maximum missed cleavages, 1; fixed modification, carboxamidomethylation on cysteine; potential modification, oxidization on methionine; parent monoisotopic mass error, 2.5Da. Peptide identifications were assigned probability by PeptideProphet 21, and all identifications with an assigned probability <0.95 were discarded (estimated false discovery rate varied per experimental run in the range 0.006-0.007). Protein inference were assigned by ProteinProphet 22 using the peptides with probability scores ≥ 0.95. Any proteins with only one spectrum identified in one group and none in the other group were excluded from ratio calculation and subsequent analysis.The comparative analysis between the VLTS and STS was achieved using spectral counts of all the peptides in each protein/protein group. Both replicate data were included in the data analysis as averaging the spectral counts from the duplicate experiments reduces the technical variations. The protein ratio between VLTS group and STS group was calculated using the total spectral counts of 5 VLTS samples divided by the total spectral count of 5 STS samples.
Functional annotation, enrichment and pathway analysis
The enrichment of the biological processes was assessed by using The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 23. The pathway analysis was performed using ingenuity pathways analysis (IPA, Ingenuity Systems). The proteins that had a minimum of 2-fold abundance change between VLTS and STS groups were uploaded to IPA to identify upstream regulators significant to the dataset.
IHC analysis
Paraffin-embedded, formalin-fixed TMA block was sectioned at 4 μm onto charged slides and deparaffinized, processed for heat-induced epitope retrieval in 0.01mol/L citric acid buffer, pH 6.0 for 15 minutes in a vegetable steamer, followed by incubation with primary antibody against prolargin (Epitomics) or RPS8 (Proteintech) with 1:100 dilution incubation at room temperature for 1 hour. The LSAB+ HRP System (Dako, Carpinteria, Calif) was used for visualization. Three separate 1.0 mm cores for each annotated tumor in the TMA were independently scored by 2 blinded observers using semiquantitative histoscores (range 0–300). Histoscores were the product of staining intensity (0–3) and the percentage of tumor cells staining at that intensity (0–100). If any core's histoscore differed by more than 30 points between observers, a revised score was assigned by consensus evaluation. Median histoscore of both observers was used for analysis.
Cell culture & stable transfection
Human cancer associated fibroblast cell line was prepared by the outgrowth method. Fresh tissue was obtained from residual pancreatic adenocarcinoma specimen from one patient undergoing primary surgical resection at University of Michigan. The human sample was obtained in accordance with the policies and practices of the Institutional Review Board of the University of Michigan. Briefly, tumor sample was minced and seeded in six-well plates containing 15% FBS/DMEM, L-glutamine(2 mmol/L), penicillin/streptomycin, and amphotericin. After 5 days, cells were able to grow out from the tissue clumps. Medium was changed every 3 days. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. When fibroblast cells grew to confluence, cells were trypsinized and passaged.The identification of fibroblast cell was determined by immunohistochemistry for αSMA, vimentin, PDGFR and FAP. The contamination of immunocytes and tumor cells were excluded by CD45 and CK19, respectively.
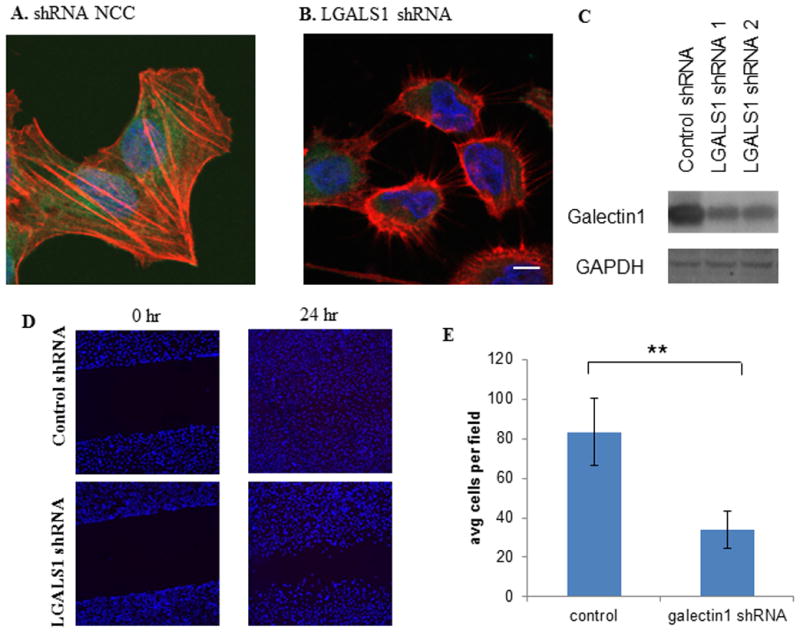
Galectin-1 knockdown cells were generated by stable transfection of galectin-1 shRNA (Qiagen #KH19360P) using Attractene Transfection Reagent (Qiagen) and selection with puromycin as per manufacturer's suggested protocol. Knockdown was confirmed by immunofluorescence and Western blot analysis using anti-galectin1 antibody (R&D AF1152) as per manufacturer's instructions. GAPDH was used as a loading control for Western blots.
Wound healing and Invasion assays
For wound healing, cells were grown to confluence on glass coverslips. Cells were manually wounded with a pipet tip. 24 hours later, cells were fixed and mounted onto slides with Prolong Gold plus DAPI (Invitrogen). Images were taken with a Leica DMLB microscope equipped with a Diagnostic Instruments Color Mosaic camera and software. Invasion assays were performed as described24.
Statistical analysis
All statistics were performed using SPSS, version 20.0, for Windows. Chi-square tests were used to compare dichotomized IHC groups against baseline clinicopathologic factors. Univariate Cox regression was performed to evaluate the prognostic significance of individual variables. Kaplan-Meier survival curves were analyzed by log rank test.Multivariate analysis was performed by stepwise Cox analysis with backward selection using the Akaike Information Criterion. The level of significance for all tests was defined as α=0.05.
Results
Proteomics analysis
Five pathologically and clinically well-characterized samples from each group of VLTS and STS were included and analyzed in duplicate. The samples were interrogated in a random order using the same reversed-phase analytical column, LC gradient and mass spectrometer parameters. Using only the peptides identified with a PeptideProphet probability score ≥0.95 (FDR < 1%), 1050 non-redundant proteins (based on gene symbol) was identified. To evaluate the reproducibility of the platform, all 10 samples analyzed were combined to assess the correlation of peptide intensity between two replicate runs. The ion intensity of the peptides in each analysis was plotted versus each other. A high correlation of the peptide intensities between the replicates was observed (Spearman R=0.95) (Supplemental Figure 1), indicating a robust technical reproducibility.
Proteins associated with pancreatic cancer survival
For each protein identified, the protein ratio between VLTS and STS were quantified by calculating the ratio of total spectral counts between all 5 VLTS cases and all 5 STS cases.The spectral count information could be found in Supplemental Table 1. For the proteins that were detected in VLTS group, but not in STS group, the ratio of VLTS vs STS was assigned 100; and for the proteins that were only detected in STS group, the ratio of VLTS vs STS was assigned 0.01. Using 2-fold ratio as a threshold, 332 differentially expressed proteins between VLTS and STS were identified, including 139 proteins with ≥2-fold increase in abundance in VLTS group (designated as “proteins associated with VLTS group”), and 193 proteins with ≥2-fold abundant increase in STS group (designated as “proteins associated with STS group”). Figure 1 shows the distribution of protein ratios (natural log-transformed) calculated between the VLTS and STS groups, excluding the proteins with a ratio with 0.01 or 100. Notably, the differentially expressed proteins identified in this study are relative between VLTS and STS, and many of them were found frequently over-expressed in pancreatic tumor tissues when compared to normal pancreas18, 25-27.
Figure 1.
Functional enrichment and upstream regulation networks
To further characterize functional clusters of the proteins associated with PDAC survival groups, we uploaded the VLTS and STS survival-associated proteins onto DAVID online database for enrichment analysis23, 28. The enriched protein groups associated with STS and VLTS are summarized in Table 2. For STS associated differential proteins, the most significantly enriched protein cluster was Cytoskeleton Proteins, with 30 cytoskeleton proteins more abundantly associated with STS group. Proteins in this cluster were involved in cellular movement, cell division, endocytosis, movement of organelles, and maintenance of cell shape. The second enriched cluster of the STS associated proteins was Ribonucleoprotein Complex/Protein Biosynthesis, with 31 differential proteins included in this functional cluster. Within this cluster, there were proteins involved in nucleoporins, translational elongation, ribosomal proteins and protein biosynthesis. For VLTS associated differential proteins, the most significantly enriched cluster was copine proteins. Copines are Ca2+-dependent phospholipid-binding proteins that are thought to be involved in membrane-trafficking in response to intracellular calcium increase29. Eight copine proteins were more abundantly expressed in VLTS group. The second enriched cluster of VLTS associated proteins was Mitochondrion. Nineteen mitochondrial proteins were more abundantly expressed in VLTS group, including mitochondrial solute carrier proteins, mitochondrial membrane and matrix proteins, and other enzymes involved in cellular respiration. Both the STS and VLTS associated proteins had an enriched cluster of Generation of Precursor Metabolites and Energy. But there were differences within this cluster regarding the survival-related proteins that were identified. The 10 VLTS associated proteins in this cluster were mostly mitochondrial proteins involved in oxidation and reduction process, while only 5 out of the 14 STS associated proteins were mitochondrial.
Table 2. Enrichment Analysis of Survival Associated Proteins.
| Enrichment category | Enrichment Score | No. of Proteins | Gene Name | p value | |
|---|---|---|---|---|---|
| Short-term survival associated | Cytoskeleton | 6.21 | 30 | ALDOA, KRT6C, KRT6A, KRT6B, PDLIM7, PDLIM3, CCT3, VCL, PNN, KRT9, PEA15, CTTN, DES, KRT5, KRT2, TUBB4, H1F0, PPP2R1A, MYO1C, ACTN4, ACTA2, FSCN1, KRT13, FLNC, PALLD, EPB41L2, NME1, CFL1, WDR1, TGFB1I1 | 6.11E-03 |
| Ribonucleoprotein complex/ protein biosynthesis/RNA processing | 5.85 | 31 | RPL18, RPL13, PABPC4, RPL27A, SYNCRIP, RPS2, PNN, HNRNPL, HNRNPA3, RPL7, HNRNPF, SND1, RPL5, RPL12, PABPC1, ACTN4, RRBP1, PTBP1, EEF2, ILF3, RBMX, HNRNPA1, NCL, RPS8, RPS7, RPL21, RBMXL2, CPSF6, RPS10, SRP72, BAT1 | 3.51E-12 | |
| Generation of precursor metabolites and energy | 3.86 | 14 | ALDOA, LDHA, ACO1, ADPGK, PGAM1, ATP6V1B2, ATP6V1B1, PPP1CB, PDHB, GAA, ENO2, PDHA1, ATP5A1, ENO1 | 1.83E-04 | |
| Long-term survival associated | Copine | 7.16 | 8 | CPNE8, CPNE9, CPNE4, CPNE5, CPNE6, CPNE7, CPNE3, CPNE2 | 4.50E-14 |
| Mitochondrion | 2.64 | 19 | CYB5R3, ATP5D, UQCRC1, SLC25A4, SLC25A5, SLC25A6, ETHE1, NDUFA13, PRDX3, IDH3A, SDHA, SLC25A31, ANXA10, PPP2CA, HEBP1, HARS2, COX6B1, ATP5H, MDH2 | 4.67E-03 | |
| Generation of precursor metabolites and energy | 2.33 | 10 | SDHA, ATP5D, UQCRC1, SLC25A4, NDUFA13, ERO1L, ATP5H, IDH3A, MDH2, MDH1 | 7.79E-04 |
Upstream regulation network
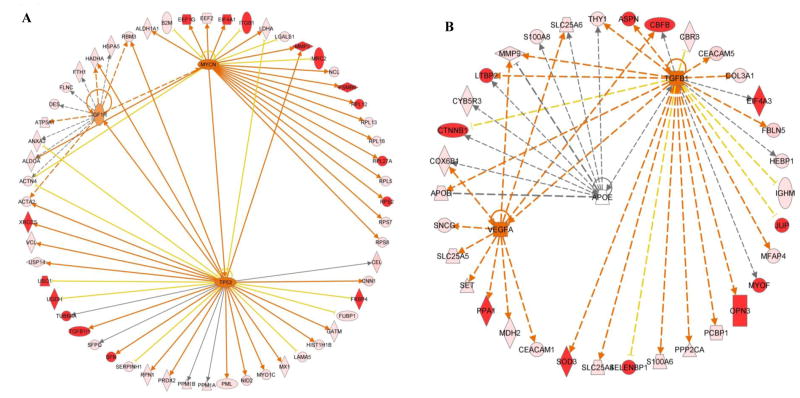
To reveal potential pathways involved in pancreatic cancer survival, the Ingenuity Pathway Analysis (IPA) was used for upstream regulator analysis. The top three upstream regulators for the STS associated proteins were MYC, insulin-like growth factor 1 receptor (IGF1R) and TP53 (Figure 2A). MYC is a transcription factor that controls expression of numerous genes that govern cell proliferation, cell growth, cell differentiation and apoptosis. In our previous tissue proteomics studies, c-MYC was identified as a significant regulator implicating in pancreatic cancer, chronic pancreatitis, as well as precancerous PanIN318, 25, 27. In the current study, 25 STS associated proteins were regulated by MYC. IGF1R plays a crucial role in tumor transformation by regulating cell growth and survival. Eleven STS associated proteins were downstream of IFG1R pathways, five of which were also regulated by MYC. Tumor suppressor gene p53 is frequently mutated or deleted in pancreatic cancer. The protein product of p53 gene plays a pivotal role monitoring cellular repair and apoptosis in response to genetic damage or metabolite disorder. Twenty-four STS associated proteins were regulated by p53 protein. In connection with the other two upstream regulators MYC and IGF1R, the top three upstream regulators and their downstream targets (48 STS associated proteins) formed a cross-talk network involving two essential traits of cancer cells: proliferation and apoptosis. The top three upstream regulators for VLTS associated proteins were vascular endothelial growth factor A (VEGFA), apolipoprotein E (APOE) and TGF-β1, regulating 10, 9, and 24 VLTS associated proteins respectively (Figure 2B). These three upstream regulators linked 35 VLTS associated proteins in a network that regulated angiogenesis (VEGFA), cholesterol regulation (APOE) and proliferation, differentiation, survival and apoptosis (TGF-β1). While the information revealed by IPA is informative and consistent with our understanding of pancreatic tumorigenesis, much work is needed to experimentally investigate these molecular events that can potentially influence pancreatic cancer survival.
Figure 2.
Correlation of prolargin with longer pancreatic cancer survival
Prolargin (PRELP) was one of the differential proteins associated with VLTS identified in our previous study13. Based on the proteomic data,overall, PRELP was 3.3 fold more abundant in the tumor tissues of the VLTS group compared to the STS group. The PRELP protein was detectable in all 5 VLTS cancer samples, with spectral counts ranging from 9-23. In contrast, PRELP was marginally detectable in 4 of the 5 STS samples with much lower spectral count number, ranging from 1-7 (Figure 3A). The spectral counts of PRELP peptides were significantly higher in the long-term survival group than short-term survival group (p=0.010).
Figure 3.

IHC analysis was performed on tissue sections from independent pancreatic cancer samples (UCLA cohort), constructed into a tissue microarray. The UCLA cohort consisted of 145 pancreatic adenocarcinoma patients with a mean survival time of 35 months, ranging from 0.45 to 100 months. Overall, the neoplastic epithelial cells displayed varying degrees of PRELP protein staining, while peritumoral stromal cells were mostly negative with rare weak positivity seen in occasional cases.The IHC scoring was therefore based on tumor cell staining. The survival time of the patients was blinded at the time of IHC scoring.Three out of the 145 specimens were excluded from the analysis because of an absence of tumor cells in the tissue cores available for analysis. The 142 cases were separated into tertiles based on the IHC scores, with top two tertiles being the high PRELP group, and the bottom tertile being the low PRELP group. No significant associations were noted between PRELP abundance and baseline clinicopathologic parameters including age, gender, tumor grade, tumor size, tumor stage, lymph node involvement, and tumor margin status. Neoplastic cells from tumor tissue of patients with longer survival displayed higher intensity PRELP protein staining than the tumor tissue of patients with shorter survival (Figure 3B and 3C). Kaplan-Meier survival analysis suggested that patients with high PRELP staining had significantly better survival time (p=0.013, Figure 4A). Overall mean survival time for patients with high versus low PRELP staining was 40.9 versus 27.0 months, respectively. Univariate Cox regression analysis confirmed that high abundance of PRELP was significantly associated with improved overall survival (hazard ratio=0.61, 95% confidence interval 0.42-0.91, p=0.015). In multivariate Cox analysis, high PRELP abundance fell just short of statistical significance (HR=0.68, 95% CI 0.46-1.02) in a model with other significant pathologic variables including high histologic grade (HR=1.51, 95% CI 1.02-2.23), positive lymph node involvement (HR=1.63, 95% CI 1.12-2.39) and low pathologic T stage (HR=0.62, 95% CI 0.41-0.92) (Table 3A).
Figure 4.

Table 3.
Multivariate COX regression analysis evaluating PRELP and RPS8 staining with PDAC survival.
| A. PRELP Bottom Tertile Staining in Multivariate Analysis | ||||
|---|---|---|---|---|
|
| ||||
| Variable | P value | HR | 95% CI Lower | 95%CI Upper |
|
| ||||
| High grade | 0.039 | 1.511 | 1.022 | 2.235 |
| pN1 | 0.012 | 1.632 | 1.116 | 2.388 |
| BottomTertile_PRELP | 0.062 | 0.684 | 0.459 | 1.019 |
| B. RPS8 Top Quartile Staining in Multivariate Analysis | ||||
|---|---|---|---|---|
|
| ||||
| Variable | P value | HR | 95% CI Lower | 95%CI Upper |
|
| ||||
| Female | 0.081 | 1.398 | 0.96 | 2.037 |
| High Grade | 0.003 | 1.788 | 1.217 | 2.627 |
| pN1 | 0.026 | 1.546 | 1.053 | 2.272 |
| TopQuartile_RPS8 | 0.012 | 1.806 | 1.139 | 2.863 |
| C. Combination of “LowRPS8 and HighPRELP” vs. all others | ||||
|---|---|---|---|---|
|
| ||||
| Variable | P value | HR | 95% CI Lower | 95%CI Upper |
|
| ||||
| Female | 0.074 | 1.407 | 0.967 | 2.049 |
| High grade | 0.035 | 1.521 | 1.03 | 2.247 |
| pN1 | 0.024 | 1.555 | 1.059 | 2.283 |
| LowRPS8_HighPRELP | 0.024 | 0.638 | 0.432 | 0.942 |
| D. Combination of “HighRPS8 and LowPRELP” vs. all others | ||||
|---|---|---|---|---|
|
| ||||
| Variable | P value | HR | 95% CI Lower | 95%CI Upper |
|
| ||||
| High grade | 0.008 | 1.686 | 1.146 | 2.481 |
| pN1 | 0.009 | 1.682 | 1.136 | 2.49 |
| HighRPS8_LowPRELP | 0.001 | 2.685 | 1.464 | 4.924 |
Correlation of 40S ribosomal protein S8 (RPS8) with shorter pancreatic cancer survival
Another protein candidate selected for IHC analysis was the 40S ribosomal protein S8 (RPS8). Overall, RPS8 protein was 7.7 fold more abundant in the tumors from the STS group compared to the VLTS group according to the proteomics data. At the individual sample level, the RPS8 protein was detected in all 5 STS tumor samples, but in only one VLTS tumor sample (Figure 3D). The spectral counts of RPS8 were significantly higher in STS than VLST (p=0.019). Moreover, protein network analysis revealed that a large group of ribosomal and ribosomal-associated proteins extensively interact with RPS8 (Supplemental Figure 2). The many proteins from this group were elevated in the tumor tissue of STS patients and highly interactive among themselves, implying that pathways impacting RPS8 and associated ribosomal proteins may play a role in pancreatic cancer survival.
IHC analysis was performed on the TMA of UCLA PDAC cohort and the scoring was based on tumor cell staining. The 141 scorable cases were separated into quartiles based on the IHC scores, with top quartile being the high RPS8 group, and the bottom three quartiles being the low RPS8 group. No significant associations were noted between RPS8 abundance and baseline clinicopathologic parameters as mentioned above. Kaplan-Meier survival analysis suggested that patients with low RPS8 staining had significantly longer survival time (p=0.011, Figure 4B). Representative IHC staining of tumor tissues from patients with longer survival and shorter survival were presented in Figure 3E and 3F, respectively. Overall mean survival time for patients with high RPS8 staining was 23.8 compared to 39.3 months for those patients with low RPS8. Univariate Cox regression analysis confirmed that high abundance of RPS8 was significantly associated with poor overall survival (hazard ratio=1.79, 95% confidence interval 1.13-2.82, p=0.012). Multivariate Cox analysis also confirmed that high RPS8 abundance was significantly associated with shorter survival time (HR=1.81, 95% CI 1.14-1.2.86) in a model with other significant pathologic variables incorporated, including high histologic grade (HR=1.79, 95% CI 1.22-2.63), and positive lymph node involvement (HR=1.55, 95% CI 1.05-2.27) (Table 3B).
Combination of PRELP and RPS8
To evaluate the combination of PRELP and PRS8 for survival prediction, we used either “High-RPS8 and Low-PRELP” or “Low-RPS8 and High-PRELP” for Kaplan-Meier survival analysis and multivariate COX analysis. As shown in Figure 4C, “High-RPS8 and Low-PRELP” was associated with worse survival (p=0.004). Overall mean survival time for cases with “High-RPS8 and Low-PRELP” status was 16.9 months, while mean survival time for cases with other staining status was 38.0 months. In a separate survival analysis, “Low-RPS8 and High-PRELP” was associated with markedly improved survival (p=0.003, Figure 4D). Overall mean survival time for cases with “Low-RPS8 and High-PRELP” status was 43.1 months, while mean survival time for cases with other staining status was 27.1 months.
Multivariate Cox regression analysis was then used to test the combination of either “High-RPS8 and Low-PRELP” or “Low-RPS and High-PRELP” along with other variables from baseline clinicopathologic parameters. As shown in Table 3C and 3D, both “High-RPS8 and Low-PRELP” and “Low-RPS and High-PRELP” were significantly associated with survival time (HR=2.69 and HR=0.64 respectively). Factors including high histologic grade and positive lymph node involvement continued to be significant pathologic variables in these multivariate analyses. The combination of “High-RPS8 and Low-PRELP” displayed improved prognostic value over any single marker alone or the combination of “Low-RPS and High-PRELP”.
Knockdown of galectin-1 in pancreatic cancer associated fibroblasts dramatically reduced cell migration and invasion
In this study we observed that galectin-1 was 7 fold higher in the STS group compared to VLTS group, confirming our previous finding that the abundance of galectin-1 was aversely associated with survival in resectable pancreatic cancer13. Since galectin-1 is primarily expressed in cancer-associated fibroblasts, we sought to determine its functional role in influencing cancer aggressiveness using cancer-associated fibroblasts (CAF). We first generated stable clones of CAF with galectin-1 knocked down via several distinct galectin-1 shRNA constructs. One of such constructs (shRNA1) resulted in substantial reduction (86% reduction) of galectin-1 abundance as confirmed by fluorescence immunostaining and western blot (Figure 5A-C). In the wound healing assay, control CAF cells usually closed the wound gap within 24 hours post-wounding (Figure 5D). In contrast, CAF with LGALS1 shRNA migrated much slower, with a still visible gap after 24 hours post-wounding. To test for invasion, the CAF control and CAF LGALS1 shRNA cells were placed into transwell coated with Matrigel. After 24 hours, the cell number of the CAF LGALS1shRNA cells that invaded though the transwell was significantly reduced (Figure 5E, p <0.001) compared to parental CAF cells. These observations suggested that knockdown of galectin-1 in pancreatic cancer associated fibroblasts can dramatically reduce cell migration and invasion capabilities.
Figure 5.
Discussion
PDACs typically have very exuberant stroma, which can make up to 90% of the tumor mass that surrounds the tumor cells, playing important mechanistic roles in the progression of PDAC. In our study, we used microscopic macro-dissection to dissect the tissue areas with highest neoplastic cellularity (including both PDAC epithelium and associated stroma), while excluding areas of non-neoplastic pancreatic acinar tissues and inflammatory infiltrates. Therefore, the tissues we analyzed were primarily comprised of neoplastic cells and stromal cells. While the mass spectrometry data may not provide us enough information to completely decode a signal from a heterogeneous population of cells, it allowed us to identify proteins with an overall abundance change in the tumor tissues. Such approach has pros and cons, reflecting our emphasis on biomarker discovery and acquisition of information regarding tumor microenvironment. The subsequent IHC analysis permitted us to further examine a particular protein candidate for its distribution among different cell types and ECM. It is also notable that very long term survivors of pancreatic cancer are rare; and inclusion of only pathologically and clinically well-defined cases compromised the number of the specimens available for our initial proteomics discovery. Nonetheless, the protein profiling data was informative; and the IHC validation of selected protein candidates using expanded number of PDAC cases (independent cohort) with various overall survival times provided a diagonal confirmation on the selected targets.
The comparison of pancreatic tumor tissues of resectable PDAC patients with very long survival times versus short survival times revealed a group of differentially expressed proteins associated with VLTS and STS patients, respectively, including PRELP and LGALS1 which were also evidenced in our previous work13. The most enriched functional cluster of STS associated proteins was Cytoskeleton, which is a dynamic cellular structure that maintains cell shape, adhesion, motility, as well as intracellular trafficking and signaling. It is now well recognized that tumorigenesis is associated with altered cytoskeletal proteins and that these proteins clearly play a role in the metastatic process 30, 31. We also noticed that several proteins associated with epithelial-mesenchymal transition (EMT), which plays a pivotal role in the tumor progression, were up-regulated in the STS compared to VLTS, including laminin, integrin beta-1, smooth muscle actin and MMP2. Two classic mesenchymal markers (vimentin and fibronectin), however, did not display significantly different abundance between VLTS and STS – which may be due to various factors and requires further confirmation.
The second enriched cluster for STS proteins was Protein synthesis /Ribonucleoprotein Complex/RNA Processing, reflecting the underlying active cellular biosynthesis that contributes to tumor growth. The third and common enriched cluster for both the STS associated proteins and the VLTS associated proteins was Generation of Precursor Metabolites and Energy. In proliferating cells, cellular metabolism provides both energy needed for maintaining homeostatic processes and the precursors for macromolecular biosynthesis needed for proliferation. Cancer cells have dysregulated cellular energy metabolism. More than fifty years ago, Warburg demonstrated that cancer cells had unusually high glycolytic activity even in aerobic condition (Warburg Effect). Today it is generally regarded that cancer cells must rewire their metabolic programming in order to meet the demands of high energy and anabolic processes necessary for rapid growth. In our study, we observed that differential metabolism-related proteins (including metabolites, energy generating, glycolysis and TCA cycle) in the pancreatic cancer tissues were related to patient survival, suggesting that metabolic programming differs between pancreatic cancer patients with long-term survival and patients with short-term survival. Our observation supports the notion that tumors display heterogeneity in regards to their metabolic reprogramming32. Such metabolic reprogramming could be driven by both genetic (mutation) and non-genetic (tumor microenvironment) factors32, and could ultimately influence cancer survival.
Using IHC analysis on tissue microarray, we demonstrated that the PRELP abundance level was statistically associated with longer survival. PRELP is a proteoglycan found in the extracellular matrix. It binds perlecan and collagens and may function as a basement membrane anchor33. PRELP was found as one of the predictor gene signatures for brain tumors34. Additionally, PRELP can act as a NF-kappaB inhibitor impairing osteoclastogenesis 35. NF-kappaB is constitutively overexpressed in PDAC and its inhibition is a mandate for a new class of therapeutic drugs. Related to this observation, we also discovered that copines were overexpressed in the VLTS proteomic analysis. While the functional role of these Ca+ dependent membrane proteins is mostly unknown, copine-1 has been identified as another NF-kappaB inhibitor36. These combined observations suggest that repression of NF-kappaB could prolong survival in pancreatic cancer.
RPS8,a component of the ribosomal 40S subunit, was found to be inversely associated with survival in our study. Increased abundance of ribosomal genes including RPS8 was previously observed in colorectal tumors and colon polyps compared to matched normal colonic mucosa37. Studies also suggested that the abundance of RPS8 could determine the susceptibility of pancreatic cancer cells to gemcitabine treatment38. In the current study, we observed that higher abundance of RPS8 was associated with worse survival of pancreatic cancer. The combination of “High-RPS8 and Low-PRELP” provided improved prognostic value over RPS8 or PRELP alone.
A third protein related to pancreatic cancer survival was galectin-1. Up-regulation of galectin-1 has been documented for several tumor types in both the cancerous epithelial cells, as well as in the stroma adjacent to cancer cells39. Galectin-1 is a β-galactoside binding protein that is involved in immunosuppression and angiogenesis. Recent study suggests that galectin-1 could bind to glycosylated VEGFR in endothelial cells to promote angiogenesis in the absence of VEGF40. In the comparison of VLTS and STS of pancreatic cancer patients, we consistently observed that the lower abundance of galectin-1 in tumor tissues of the VLTS patients.IHC analysis indicated that galectin-1 was mainly detected in the CAF, but not in cancer epithelial cells13. Knock-down of galectin-1 gene in CAF cells significantly reduced cell migration and invasion of CAF, implicating its potential role in tumor microenvironment contributing to pancreatic cancer progression and metastasis. Inhibition of galectin-1 as a potential therapeutic target has been evaluated in breast, colon, ovary, and prostate cancers 41. Our results further support that gelactin-1 could be a potent target for pancreatic cancer therapy.
Supplementary Material
Acknowledgments
We thank Dr. Ralph H. Hruban for his assistance with histopathology of the Johns Hopkins cases.
Funding source: This study was supported in part with federal funds from the National Institutes of Health under grants K07CA116296, K25CA137222, R21CA161575, R21CA149772, R01CA180949 and private funds from Gene and Mary Ann Walters Pancreatic Cancer Foundation and Donald E. Bocek Endowed Research and Development Award.
List of Abbreviation
- PDAC
pancreatic ductal adenocarcinoma
- VLTS
very long-term survivors
- STS
short-term survival patients
- IHC
Immunohistochemistry
- TMA
tissue microarray
- FFPE
formalin-fixed paraffin-embedded
Footnotes
Disclosure. The authors declare no duality of interest.
Supplementary information is available at Laboratory Investigation's website.
Reference List
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Adham M, Jaeck D, Le BJ, et al. Long-term survival (5-20 years) after pancreatectomy for pancreatic ductal adenocarcinoma: a series of 30 patients collected from 3 institutions. Pancreas. 2008;37:352–357. doi: 10.1097/MPA.0b013e31818166d2. [DOI] [PubMed] [Google Scholar]
- 3.Bradley EL., III Long-term survival after pancreatoduodenectomy for ductal adenocarcinoma: the emperor has no clothes? Pancreas. 2008;37:349–351. doi: 10.1097/MPA.0b013e31818e9100. [DOI] [PubMed] [Google Scholar]
- 4.Cleary SP, Gryfe R, Guindi M, et al. Prognostic factors in resected pancreatic adenocarcinoma: analysis of actual 5-year survivors. J Am Coll Surg. 2004;198:722–731. doi: 10.1016/j.jamcollsurg.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Conlon KC, Klimstra DS, Brennan MF. Long-term survival after curative resection for pancreatic ductal adenocarcinoma. Clinicopathologic analysis of 5-year survivors. Ann Surg. 1996;223:273–279. doi: 10.1097/00000658-199603000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han SS, Jang JY, Kim SW, et al. Analysis of long-term survivors after surgical resection for pancreatic cancer. Pancreas. 2006;32:271–275. doi: 10.1097/01.mpa.0000202953.87740.93. [DOI] [PubMed] [Google Scholar]
- 7.Mosca F, Giulianotti PC, Balestracci T, et al. Long-term survival in pancreatic cancer: pylorus-preserving versus Whipple pancreatoduodenectomy. Surgery. 1997;122:553–566. doi: 10.1016/s0039-6060(97)90128-8. [DOI] [PubMed] [Google Scholar]
- 8.Reddy S, Wolfgang CL, Cameron JL, et al. Total pancreatectomy for pancreatic adenocarcinoma: evaluation of morbidity and long-term survival. Ann Surg. 2009;250:282–287. doi: 10.1097/SLA.0b013e3181ae9f93. [DOI] [PubMed] [Google Scholar]
- 9.Riall TS, Cameron JL, Lillemoe KD, et al. Resected periampullary adenocarcinoma: 5-year survivors and their 6- to 10-year follow-up. Surgery. 2006;140:764–772. doi: 10.1016/j.surg.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Schnelldorfer T, Ware AL, Sarr MG, et al. Long-term survival after pancreatoduodenectomy for pancreatic adenocarcinoma: is cure possible? Ann Surg. 2008;247:456–462. doi: 10.1097/SLA.0b013e3181613142. [DOI] [PubMed] [Google Scholar]
- 11.Shimada K, Sakamoto Y, Nara S, et al. Analysis of 5-year survivors after a macroscopic curative pancreatectomy for invasive ductal adenocarcinoma. World J Surg. 2010;34:1908–1915. doi: 10.1007/s00268-010-0570-9. [DOI] [PubMed] [Google Scholar]
- 12.Allen PJ. Pancreatic adenocarcinoma: putting a hump in survival. J Am Coll Surg. 2007;205:S76–S80. doi: 10.1016/j.jamcollsurg.2007.06.331. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Pan S, Ottenhof NA, et al. Stromal galectin-1 expression is associated with long-term survival in resectable pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2012;13:899–907. doi: 10.4161/cbt.20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcea G, Dennison AR, Pattenden CJ, et al. Survival following curative resection for pancreatic ductal adenocarcinoma. A systematic review of the literature. JOP. 2008;9:99–132. [PubMed] [Google Scholar]
- 15.Withers SG, Aebersold R. Approaches to labeling and identification of active site residues in glycosidases. Protein Sci. 1995;4:361–372. doi: 10.1002/pro.5560040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iuga C, Seicean A, Iancu C, et al. Proteomic identification of potential prognostic biomarkers in resectable pancreatic ductal adenocarcinoma. Proteomics. 2014;14:945–955. doi: 10.1002/pmic.201300402. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen KA, Arensman M, Lay AR, et al. HOXB7 promotes invasion and predicts survival in pancreatic adenocarcinoma. Cancer. 2013;119:529–539. doi: 10.1002/cncr.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan S, Chen R, Stevens T, et al. Proteomics portrait of archival lesions of chronic pancreatitis. PLoS ONE. 2011;6:e27574. doi: 10.1371/journal.pone.0027574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 20.Keller A, Eng J, Zhang N, et al. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:2005. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller SR, Kitagawa K, Aebersold R, et al. Isolation and characterization of the 160,000-Da phosphotyrosyl protein, a putative participant in insulin signaling. J Biol Chem. 1991;266:12817–12820. [PubMed] [Google Scholar]
- 22.Nesvizhskii AI, Keller A, Kolker E, et al. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 23.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Brentnall TA, Lai LA, Coleman J, et al. Arousal of cancer-associated stroma: overexpression of palladin activates fibroblasts to promote tumor invasion. PLoS ONE. 2012;7:e30219. doi: 10.1371/journal.pone.0030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R, Yi EC, Donohoe S, et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Chen R, Brentnall TA, Pan S, et al. Quantitative Proteomics Analysis Reveals That Proteins Differentially Expressed in Chronic Pancreatitis Are Also Frequently Involved in Pancreatic Cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Pan S, Chen R, Reimel BA, et al. Quantitative proteomics investigation of pancreatic intraepithelial neoplasia. Electrophoresis. 2009;30:1132–1144. doi: 10.1002/elps.200800752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 29.Tomsig JL, Creutz CE. Copines: a ubiquitous family of Ca(2+)-dependent phospholipid-binding proteins. Cell Mol Life Sci. 2002;59:1467–1477. doi: 10.1007/s00018-002-8522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 31.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 32.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bengtsson E, Morgelin M, Sasaki T, et al. The leucine-rich repeat protein PRELP binds perlecan and collagens and may function as a basement membrane anchor. J Biol Chem. 2002;277:15061–15068. doi: 10.1074/jbc.M108285200. [DOI] [PubMed] [Google Scholar]
- 34.Castells X, Garcia-Gomez JM, Navarro A, et al. Automated brain tumor biopsy prediction using single-labeling cDNA microarrays-based gene expression profiling. Diagn Mol Pathol. 2009;18:206–218. doi: 10.1097/PDM.0b013e31818f071b. [DOI] [PubMed] [Google Scholar]
- 35.Rucci N, Rufo A, Alamanou M, et al. The glycosaminoglycan-binding domain of PRELP acts as a cell type-specific NF-kappaB inhibitor that impairs osteoclastogenesis. J Cell Biol. 2009;187:669–683. doi: 10.1083/jcb.200906014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsey CS, Yeung F, Stoddard PB, et al. Copine-I represses NF-kappaB transcription by endoproteolysis of p65. Oncogene. 2008;27:3516–3526. doi: 10.1038/sj.onc.1211030. [DOI] [PubMed] [Google Scholar]
- 37.Pogue-Geile K, Geiser JR, Shu M, et al. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toshimitsu H, Iizuka N, Yamamoto K, et al. Molecular features linked to the growth-inhibitory effects of gemcitabine on human pancreatic cancer cells. Oncol Rep. 2006;16:1285–1291. [PubMed] [Google Scholar]
- 39.Astorgues-Xerri L, Riveiro ME, Tijeras-Raballand A, et al. Unraveling galectin-1 as a novel therapeutic target for cancer. Cancer Treat Rev. 2013 doi: 10.1016/j.ctrv.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Croci DO, Cerliani JP, Dalotto-Moreno T, et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 2014;156:744–758. doi: 10.1016/j.cell.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 41.Ito K, Stannard K, Gabutero E, et al. Galectin-1 as a potent target for cancer therapy: role in the tumor microenvironment. Cancer Metastasis Rev. 2012;31:763–778. doi: 10.1007/s10555-012-9388-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.