Abstract
cellPACK assembles computational models of the biological mesoscale, an intermediate scale (10−7–10−8m) between molecular and cellular biology. cellPACK’s modular architecture unites existing and novel packing algorithms to generate, visualize and analyze comprehensive 3D models of complex biological environments that integrate data from multiple experimental systems biology and structural biology sources. cellPACK is currently available as open source code, with tools for validation of models and with recipes and models for five biological systems: blood plasma, cytoplasm, synaptic vesicles, HIV and a mycoplasma cell. We have applied cellPACK to model distributions of HIV envelope protein to test several hypotheses for consistency with experimental observations. Biologists, educators, and outreach specialists can interact with cellPACK models, develop new recipes and perform packing experiments through scripting and graphical user interfaces at http://cellPACK.org.
INTRODUCTION
The biological mesoscale lies between the cellular and molecular scales at lengths of about 0.1 micron to 10 nanometers. Except in special cases, atomic details of the mesoscale are generally too small to resolve by microscopy and too large and heterogenous to determine with methods such as x-ray crystallography and NMR spectroscopy. Methods of cellular tomography provide the most detailed experimental view of this level with many successes in the localization of larger macromolecules (such as ribosomes) within whole cells1,2. However, an atomic resolution view of this intermediate scale, in spite of its utility in hypothesis generation, science communication, and simulation, remains sparsely modeled and visualized compared to larger and smaller scales of study3–5.
No methods currently exist to observe the mesoscale in atomic-resolution detail, however, many sources of data may be used to synthesize a view of this level. For the past twenty years, we have taken a semi-quantitative approach to integrate these diverse data into a coherent model, creating artistic depictions of cellular environments (Fig. 1a) based ultrastructural data from light and electron microscopy, atomic structures from x-ray crystallography and NMR spectroscopy, and biochemical data on concentrations and interactions6,7. More recently, we and other groups8 have developed computational approaches that automate the steps of this semi-quantitative approach to extend the results from 2D paintings into 3D models that can be explored, animated, simulated, analyzed, and easily edited and updated. This process involves two conceptual steps: gathering of data to create a recipe(s) for the model, and use of this recipe to build a virtual model.
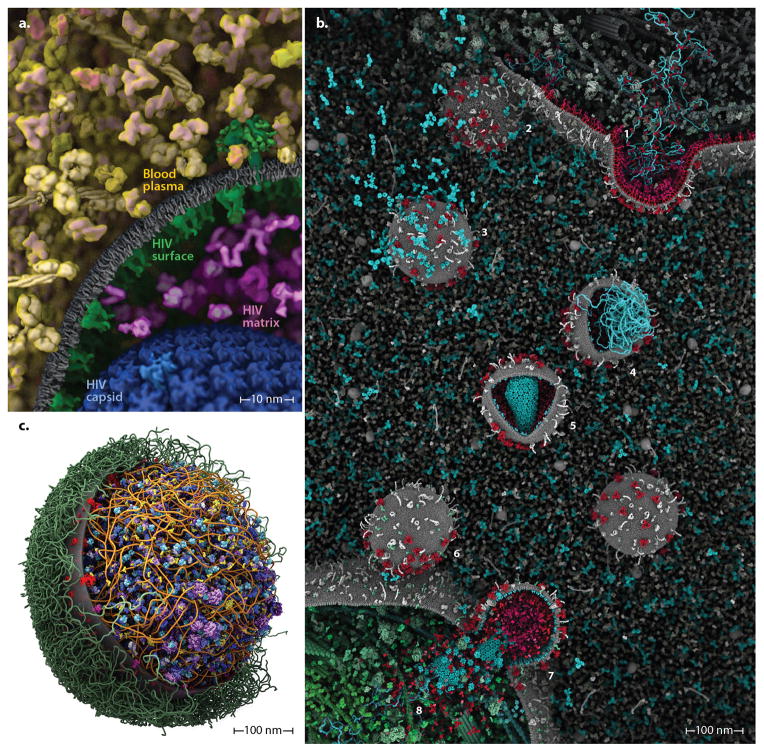
Figure 1.
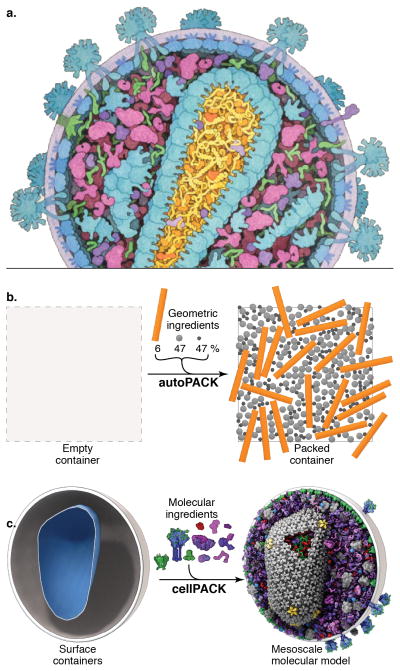
cellPACK creates 3D models of the cellular mesoscale. (a) This hand-drawn painting of HIV shows three complex packing types–volumetric, surface, and procedural (fibrous)–that must interoperate in a mesoscale modeler. (b) autoPACK is a generalized packing algorithm that positions collections of objects (ingredients) into, onto, or outside of volumes to satisfy provided constraints. It operates multiple types of packing algorithms efficiently on a single unified model using an efficient global tracking grid. (c) cellPACK is a biological extension of autoPACK optimized to pack molecular structures and other data types into biological volumes. In this image cellPACK generates an editable model of HIV by packing a from a recipe of molecular ingredients30 into the ultrastructure of an HIV envelope surface.
Since the field of structural biology is advancing so quickly, methods to automate the first step of the pipeline, the generation of a recipe based on available data, are essential. This is a challenging goal given the heterogeneous nature of the data, but we have developed automated methods for several key steps. For example, we have developed tools to integrate bioinformatics data from sources like Stanford’s WholeCellViz9 and atomic structures from the Protein Data Bank. Since much of the data that our approaches use require manual curation, we have begun projects to implement these recipes in a way that allows community experts to update and improve the recipes across all scales of detail, to extrapolate predictions where reasonable, and to vote with confidence values for all contributing parameters and resulting assemblies.
Given a molecular recipe, the construction of a quantitative 3D mesoscale model requires solving a non-trivial loose-packing problem. In biological systems, this includes packing soluble, membranous, and fibrous components with proper localizations and biologically relevant interactions. Packing problems are a popular topic of study in mathematics, engineering and biology. The non-biological methods are typically limited to simple components such as boxes and spheres10,11, or to providing one non-interacting packing-type solution at a time, such as surface packing, volumetric packing, or tree branching algorithms12,13, which can only contribute partial solutions towards recreating the organic complexity of a mesoscale model like HIV (Fig 1a,c). Other common non-biological methods pack non-discrete components that can expand to fill space or contract to avoid overlaps14 or rely on macroscale gravitational forces15,16. Physicists and engineers have applied modified molecular dynamics methods to pack spherical and cylindrical granules17,18, however, molecular dynamics functions on a scale too small and computationally expensive to provide a solution for mesoscale packing. Biological constraint modelers like IMP19, procedural and analytical modeling approaches like Molecular Silverware20,21 and relaxation approaches like Brownian Dynamics modeling22,23 are currently able to build large-scale models. However, they are designed to function locally and do not model ultrastructural features like organelle membranes or fibrous molecules like actin. To model large cellular subjects (up to and beyond tens of microns), a hybrid method is needed that combines local methods for populating defined spaces with multiscale methods that integrate ultrastructure and infrastructure while tracking and satisfying all input constraints. Such a framework must further recapitulate the complex interplay of randomness and specific interaction that lies at the heart of biology.
As the data available for biological systems at the molecular level increases in size and complexity, the creation of structurally integrative models of such systems has become a substantial bottleneck in the process of simulation and analysis. An important goal for this work is to automate the creation of these complex mesoscale models, so that large numbers of models that are statistically consistent with the known structural and compositional characteristics of a system can be easily generated.
To achieve this goal, we have developed two computational toolkits, autoPACK and cellPACK, for modeling mesoscale content (Fig. 1b–c). autoPACK is a generalized algorithm that provides a heuristic solution to the irregular loose-packing problem and simultaneously integrates regular and procedural packing solutions. autoPACK can be used, for example, to fill an architectural engineering model with concrete aggregate in preparation for earthquake simulations, or it can fill an artery with blood cells at appropriate densities to generate a histological model for a medical illustration. cellPACK is built on autoPACK, optimizing the method for biological data and generating probabilistic 3D models of large sections of cells that can contain dozens to millions of molecules. cellPACK additionally provides tools to store, visualize, analyze and interact with the results to make mesoscale models and mesoscale modeling accessible to a variety of audiences. This report describes the autoPACK and cellPACK methods, as well as several applications in education and research.
RESULTS
autoPACK
autoPACK is a generalized packing algorithm that defines a desired volume and stochastically packs it with objects, called ingredients, according to a recipe (Fig. 1b). It uses an “omniscient” grid to discretize and describe a volume, to enable multiple types of modular packing algorithms to interoperate on the same model. Ingredients of arbitrary shape are placed into the allowable 3D space with minimal overlap and with random distribution. Constraints and agent behaviors may be associated globally or with specific ingredients to add specific modes of interaction. A grid-based approach is used to generate models quickly while tracking global parameters, however, the packing algorithms explore continuous space within each grid voxel to prevent lattice regularities. In tests, autoPACK can combine several complex packing algorithms to integrate three different major localization modes–volumetric, surface, and procedural–into unified models.
The current release of autoPACK can pack ingredient objects of any size and shape into a unified model scene with complete randomness, up to and beyond biological concentrations, without requiring scene-specific adjustments (Supplementary Fig. 1 provides two examples of general object packing). autoPACK accomplishes this by sorting the ingredients in order of packing difficulty and weighting the more difficult ingredients with higher priorities, favoring them to be picked and positioned first. Procedural ingredients (e.g. fibrous or branching growth) are seeded into the volume and typically expand early in the fill, and larger surface ingredients typically deposit earlier since they are more difficult to pack compared to globular soluble ingredients. This prioritized approach is similar to packing a moving truck: it is easy to pack a truck densely by stacking small boxes around previously loaded large furniture, but difficult to position a twelve-seat dining room table into a truck jumbled with fragile knickknacks.
cellPACK
cellPACK is a biology-specific extension of autoPACK that consists of a database of mesoscale recipes (such as cytoplasm or synaptic vesicles) and examples of models for each recipe. cellPACK further extends the general packing capabilities of autoPACK with numerous modules for cell and molecule-specific packing. cellPACK also includes optional protocols and links to 3rd party algorithms that provide component solutions to the global packing problem.
autoPACK uses a cooking metaphor, building a model scene from a recipe by placing ingredients into containers defined by surface meshes. cellPACK mixes the metaphor with biologically relevant terms, where ingredients (typically molecules) are stochastically packed into containers (typically organelle ultrastructure output from surface-segmented tomograms as 3D polygonal meshes) up to densities provided by organelle-specific recipes. Each ingredient comes with associated properties, including a structural representation (such as a molecular surface or volume occupancy shell), and behaviors such as a particular collision-avoidance method or a list of binding partners that will modify the ingredient’s global and local packing. In the resultant model, each ingredient retains a connection to various other forms of data to enable deeper analysis, preparation for systems integration or large-scale simulations, or for modifications of the ingredient representations. The algorithm and data structures function recursively across scales to enable organelle recipes to be combined into larger models at the cellular and tissue scale levels. For example, container ingredients, once placed, can be subsequently packed with nested recipes of ingredients specific to that container’s surface and interior.
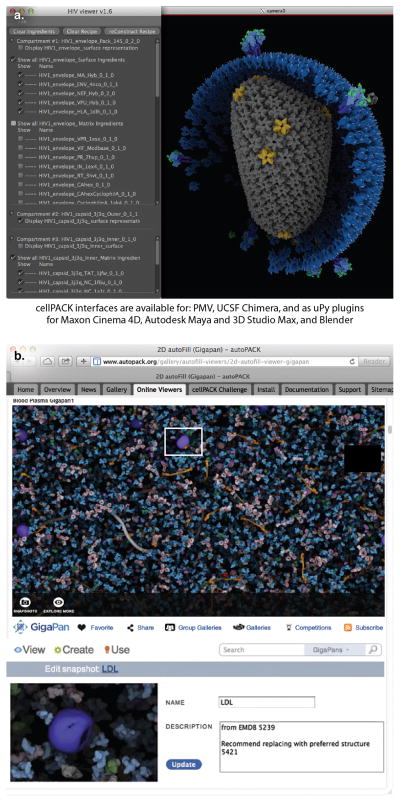
cellPACK provides scripting and graphical user interfaces (GUIs) to access viewing, analysis, and modeling modules. An online database enables invited users to critique and modify recipes and ingredients while maintaining version histories. Viewing software is built with open-source GUI plugins using our ubiquitous Python API (uPy)24 that enable any uPy-enabled host to load existing models or to run cellPACK interactively. Current hosts include molecular viewers like the Python Molecular Viewer (PMV)25 and professional animation software like (Maxon’s) Cinema4D, (Autodesk’s) Maya, and Blender. The molecular viewer UCSF Chimera26 now includes an autoPACK result file reader to load and analyze cellPACK scenes. These uPy and Chimera plugins generate representations of ingredients positioned into model scenes and allow for interaction and analysis using cellPACK analysis tools coupled with functions native to each host.
Interacting with cellPACK and cellPACK Results
cellPACK can run as a standalone Python algorithm or as a plugin to a variety of uPy-hosting software packages. With uPy, a single uPy GUI can function across multiple 3D viewers with no redundant programming. The cellPACK GUI is organized in a user-friendly format to expose the more complex options to advanced users as needed (Fig. 2a). As detailed in the methods, a novice user is initially presented with a View Tab that enables them to load cellPACK models directly from the prototype cellPACK databank. The user can switch to the Pack Tab to modify and pack models using existing recipes from the databank or local files, and they can export their modified recipes and models. Users can also switch to a Build Tab to construct recipe files and ingredient components from scratch. In all of the tabs, a user can choose from simple, intermediate, advanced, or debug versions of the GUI. The more advanced options expose more parameter widgets in the GUI.
Figure 2.
Users interact with cellPACK through a variety of different interfaces. (a) User interfaces have been developed for modeling, animation, and analysis. uPy generates consistent GUIs and models across all supported molecular viewer and professional animation software packages to provide sensible modeling, animation, and analysis interfaces rather than constructing mesoscale viewers from scratch. Users can construct models de novo as described in the online methods and in greater detail in cellPACK’s online documentation, or they can adjust the parameters of existing models to modify cellPACK results. In either case, users can modify scripts directly or interact through provided GUIs and export the modified parameters as a new recipe. (b) Several online tools are available for model viewing and interaction. cellPACK results may be displayed online as libraries of static images, movies, zoomable images, and interactive 3D models. Online multiscale viewers such as Gigapan enable users to register, critique, and annotate models and provide interactive guided walkthroughs that highlight notable details. The gallery pages for each model at cellPACK.org currently encourage visitors to suggest changes to the models by submitting emails or commenting publically on the pages.
Audiences of all levels and interests can interact with cellPACK software or with cellPACK results through the project website. The site offers visual representations of models as static images, animated movies, and interactive 2D and 3D multiscale viewers (Fig. 2b), and visitors may add comments to critique models and suggest improvements. To create the types of online representations shown on the website, users can download and install the cellPACK software to visualize and modify cellPACK ingredients and recipes, perform analysis, and generate visual output. As an open-source project, we provide both the source code and an API for easy scripting, and encourage users to modify and upload all software components to participate in development.
cellPACK Analysis Tools
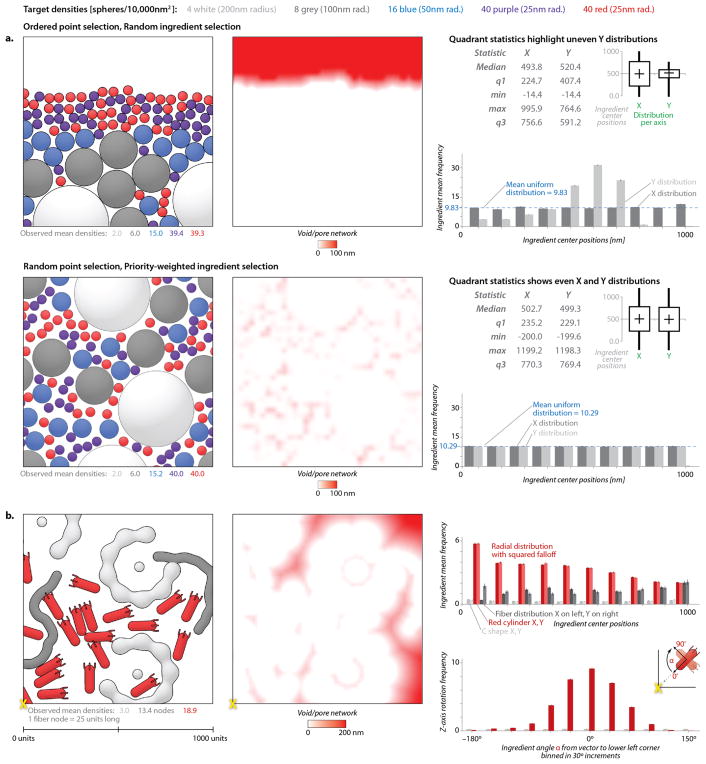
Our early prototypes of autoPACK underscored the need for effective analysis tools to quantify the success of different packing algorithms. autoPACK currently offers several basic analysis tools for assessing the positional and orientational randomness of placement in packing (Fig. 3). Position and orientation histograms are used to highlight variations about a mean uniform distribution (frequency) line for randomly distributed objects or about a target frequency curve for gradient affected objects. Box and whisker plots can reveal any global skewing across orthogonal selections from within a fill volume. Size-dependent available volume graphs quantify the distribution of negative spaces to reveal pore networks accessible to objects of different sizes.
Figure 3.
Analysis tools quantify randomness and packing bias. (a) Two methods for packing are evaluated with the autoPACK analysis tools. The upper method, using random ingredient selection, shows a clear bias, as quantified in the void-pore network, the box and whisker plot of distribution statistics, and the graph of occupancies binned along the two axes. The lower method, which uses the current default packing modes of autoPACK, shows a more uniform distribution by all of these analysis methods, and fills models up to and beyond the most common biological densities. (b) Three complex ingredients with different packing types can interact while recapitulating the intended distribution functions in this 2D packing. Two difficult ingredients (a concave ‘C’ shape and a procedurally grown flexible fiber) pack randomly amongst a red convex polarized object that is given two intentional packing biases: a distribution39–41 and orientation42,43 preference for the lower left corner of the 2D fill area. All graphs show data averaged over 1000 independent runs chosen to have an error bound <5%. Histogram bar heights indicate the average frequency per bin with error bars equal to the margin of error at 95% confidence interval (1.96*SEM) and Supplementary Fig. 4 provides larger graphs with values labeled.
We used these tools, for instance, to quantify edge effects in the packing algorithm when applied to large, homogenous fields of molecules of diverse shape, as in Fig. 3a, or to validate the construction of a scene with an artificial gradient of concentration and several ingredients with challenging shapes, as in Fig. 3b. Nearest neighbor, distance to center, and point of closest contact data can be exported to analyze mixing or interaction biases, but we have not yet implemented formal tools to output these graphs for interactive quantification of molecular interactions within cellPACK scenes. Decoupling the goals and constraints of packing from the packing algorithms will further ease recipe analysis, algorithm verification, and algorithm comparison–these are important topics for future work.
Biological Applications
We chose blood plasma (Fig 4.) as a simple initial test case. The molecular composition of blood plasma is structurally and biochemically well described and it has a relatively low protein density (~6% occupancy by volume), about 1/5th that of prokaryotic cytoplasm27. Plasma is largely homogenous, but still has numerous fibrous components, so it is an effective biological test for the combination of rigid body and procedural filling. Most of the ingredients were treated as rigid bodies based on atomic structures, and von Willebrand’s factor is modeled with a procedural snake-growth algorithm. The user can adjust the “waviness” and kinking of this structure, guided by low-resolution structures from electron microscopy.
Figure 4.
The Human Blood Plasma mesoscale recipe contains information for the most abundant macromolecules. This 1,000 × 1000 × 50 nm volume of blood plasma contains 28,755 ingredient instances that cellPACK calculated in 116 seconds on a typical laptop computer. When printed or viewed on screen with the scalebar equal to 150mm wide, the model visualization is enlarged to 1,500,000x and the inset is1,000,000x where 1mm of the image equals 1nm of the model. In the top half and inset, each ingredient has a unique color. The bottom half color-codes the ingredients by their data-source type, which highlights molecules with structural uncertainty. Interactively explore and comment on the full micron square of blood plasma online at http://gigapan.org/gigapans/85568
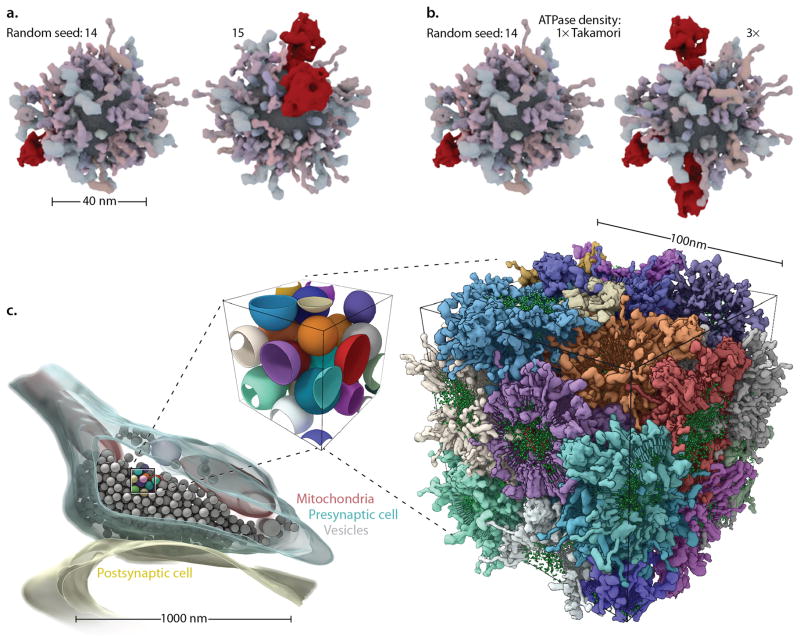
A large multidisciplinary collaboration determined the recipe for an average synaptic vesicle28. These authors kindly provided the modeled protein structures used to generate their mesoscale model. Building on this information, we used cellPACK to synthesize stochastic variations of the vesicle at a variety of sizes (Fig 5.). In addition, we used the same recipe to create a molecular model of a larger volume of closely packed vesicle surfaces segmented from an electron tomogram29. Despite the complexity of this task, even the massive surface proteins like the vesicular ATPase (vATPase) are able to interdigitate with neighboring vesicles and pack together at the high observed per-vesicle concentrations. The 100nm cube of interdigitating synaptic vesicles (Fig. 5c) was created in less than an hour on a standard laptop computer, with most of that time spent packing the lipids into the bilayers. Simpler models, for example that include only the surface proteins of one synaptic vesicle (as in Fig. 5a–b), typically require ~25 seconds to fully pack.
Figure 5.
Surface and volume packers interoperate to fill complex multi-organelle volumes. A synaptic vesicle recipe28 is used to model a single spherical vesicle with multiple stochastic variations (a) to the random seed parameter or (b) to the vesicular ATPase ingredient density parameter. (c) cellPACK scales to pack more than one interacting container. A 100nm cube of synaptic vesicle surfaces (center), isolated from the tomogram of an axon terminus29 (left) packed tightly with vesicles, reveals a complex intervesicular space that creates a difficult packing problem. The molecules belonging to each packed vesicle (right) are colored per-vesicle. Note that vesicles in c were modeled as spheres– a common technique for hand-segmenting in tomography. These estimating spheres generate a more constrained negative space in which to pack a high density of surface proteins compared to carefully traced vesicles, but cellPACK can pack the reduced tight negative space that results outside of the mathematical spheres. In this particular fill, a probabilistic soft collision was applied which very rarely allows proteins to penetrate neighboring bilayers. This option is turned off by default, but we applied it here to highlight cellPACK’s flexible ability to control interaction across scales, from proteins to organelles in this case.
The modular nature of cellPACK is designed to promote the easy combination of recipes to generate unified, hierarchical models. Fig. 6 shows two hybrid models generated from integrated recipes. The model of HIV in blood plasma combines seven different recipes into a single model, ranging from the outermost recipe for blood plasma, as described in Fig. 4, to a recipe for the distribution of envelope glycoprotein30 (Env), to recipes for host proteins and two model options for the cone-shaped capsid31,32. The Mycoplasma mycoides model packs DNA into a surface container using a fibrous-extension packing mode that is constrained by probabilistic placement of DNA-binding proteins that stochastically generate particular bend angles. The cytoplasm and surface recipes pack simultaneously, but have lower priorities. Lipopolysaccharides grow from the surface with a fibrous packing mode.
Figure 6.
cellPACK can integrate recipes into unified hybrid models that may be iteratively refined by content experts and systems-biology database improvements for each organelle system. (a) An HIV-in-BloodPlasma model shows seven different recipes (each a different color coding) unified into a single model. (b) As part of the autoPACK Visualization Challenge, J. Klusak generated this complete model of the HIV life cycle (including an animated version of most components) using the cellPACK software in conjunction with molecular modeling packages mMaya (http://www.molecularmovies.com/toolkit) and Chimera to generate additional molecular ingredients (see http://www.cgsociety.org/index.php/CGSFeatures/CGSFeatureSpecial/autopack_challenge_winners). (c) Our first model of a whole cell, Mycoplasma mycoides integrates recipes with surface, fibrous, and volumetric ingredients into a single comprehensive model. Interactive versions, updates and variations of these models can be viewed online at http://www.autopack.org/gallery/autofill-viewers.
cellPACK as a Research Tool
We have used cellPACK to simulate experimental results from fluorescence microscopy, similar to the approach described by Gardner et al33, to test six competing hypotheses for distribution of the Env glycoprotein in immature HIV-1 virions (Fig. 7). The models were compared with super resolution STED fluorescence microscopy results34, which interpreted the experimental results to support a random distribution of Env on the surface of the virion, in spite of the known asymmetric distribution of matrix protein inside the membrane and possible interaction of matrix and Env. Based on our simulated results, a model of a completely random distribution of Env gives substantially fewer single foci than is observed by fluorescence microscopy in immature virions, indicating that the asymmetric distribution of Gag in immature virions may be causing at least some measure of asymmetry in Env distribution. As expected, models with clustered Env are clearly not consistent with observations. The results, however, are not sensitive enough to distinguish between several models of interaction between Env and Gag, although three models of specific interaction (R-InMa, closeMa and closeMa-Env) show better fit to data than the model of passive incorporation (R-offMa). Within the observed ranges of Env quantification, the number of Env packed into a virion does not affect the foci count in any of the models (Supplementary Fig. 2), which agrees with the original observations. However, as described in the online methods, the distribution statistics are dependent on the technique used to distinguish the number of foci (Supplementary Fig. 3) in each STED image, and we chose a method designed to match the manual counting method used for the published experimental work.
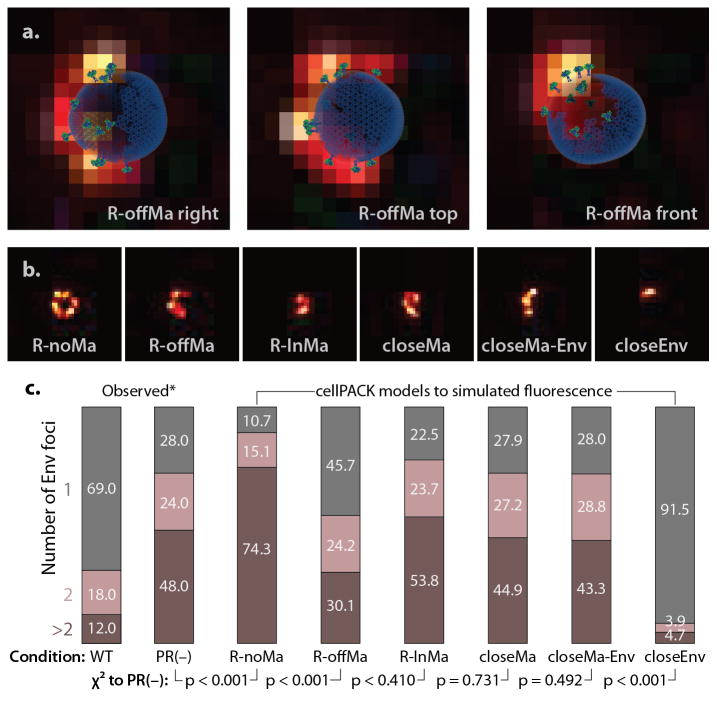
Figure 7.
cellPACK models may be used to simulate and interpret results from fluorescence microscopy. We constructed a cellPACK recipe for six hypotheses for the interaction of HIV-1 Env and Gag in immature virions. (a) Three cellPACK HIV models with Env fluorophores are superimposed over their corresponding simulated fluorescence images. (b) Samples from each of six the models are shown. (c) A direct comparison of cellPACK results to observed data identify ingredient parameter ranges that lead to specific emergent behaviors. An automated foci counting algorithm was used to analyze the 3,000 images generated for each model, with the sample number chosen to have an error bound <5%. χ2 analysis with two degrees of freedom, performed to duplicate the significance assessment reported in the original experiment, shows that several models with specific interaction between Gag and Env are consistent with the observed distribution for the immature PR(−) virion, where as a completely random distribution (R-noMa) and a model that clusters Env (closeEnv) do not.
DISCUSSION
The goal of cellPACK is to create a framework that integrates multiple types of data, across multiple scales into comprehensive spatial models for analysis, communication and simulation. On one level, cellPACK models integrate data and theory to produce editable community models, allowing an iterative crowd-sourcing process to refine the current understanding and to impel hypothesis-driven research and collaboration. The models can now be used as starting points for mesoscale algorithms like Brownian Dynamics35 and could provide foundations for whole-cell dynamics projects such as the 3D Virtual Cell http://3dvcell.org. On another level, cellPACK offers immediate applications for peer communication, education, and outreach where the concurrent development of our uPy and embedded Python Molecular Viewer software (ePMV)36 provides easy access to these tools for users from diverse backgrounds. The modular open-source architecture allows cellPACK to grow and adapt as new computational approaches and theories of mesoscale biology emerge.
We have applied cellPACK to generate a diverse collection of models based on published recipes, and we have begun to solicit community input and expert opinion on the resultant models. Thousands of researchers, scientific illustrators and digital artists have already begun using cellPACK to enhance communication, education and outreach (Battle et al.37 and http://biochem.web.utah.edu/iwasa/projects/HIV.html). As exemplified by the winners of our first annual autoPACK Visualization Challenge: Present HIV in Blood Plasma using cellPACK at http://autopack.cgsociety.org38 (Fig. 6b), accessibility to mesoscale structure has provided new mechanisms to reach broad audiences.
To view models, install the software, learn from tutorials or participate in evaluating, editing, or initiating models, please visit http://cellPACK.org. The open-source code and custom scripts used for this study are available at https://github.com/gj210/autoPACK.
METHODS
Overview
cellPACK is a toolkit that generates probabilistic 3D models based on a list of ingredients and a set of constraints, which may include 3D structures of biomolecules, three dimensional representation of membranes and compartments, and procedural description of fibrous components such as DNA or polysaccharides. The graphic user interfaces to cellPACK facilitate end-user interactions and public project software repositories facilitate open-source development.
Data Structure
cellPACK operates as a standalone Python algorithm that takes inputs from recipe files to produce result files, but most end-users currently access cellPACK through several GUIs. End-users may work with cellPACK on many levels: to view the resulting model of a previous packing; to load and modify an existing recipe in order to produce a new packing result; or to construct recipes, ingredients, and their components from scratch. In each case, the user interacts with a consistent cellPACK data structure, allowing easy transition to work at different skill levels throughout any nested recipe hierarchy and allowing cellPACK to operate across all data scales.
We have defined a generalized referential Hybrid Model Format (HMF) file type that is used throughout cellPACK’s prototype database to store recipe and ingredient data. cellPACK generates and reads the generalized hybrid model file (HMF), as a flexible referential file type that can adapt to handle multiple (and future) data types while minimizing data redundancy. cellPACK generates and reads XML, and Json versions of HMF. The cellPACK algorithm uses recipe scripts, HMF files, or recipe GUI parameters to assemble input data from a variety of formats and produces a collection of HMF output files into a directory that includes a primary recipe file with pointers to any nested recipe or ingredient files, an autoPACK Result File (.apr) for each container, intermediate construction files and analysis files. An ingredient file (actually a specific variation of the more general recipe file type) contains data or reference pointers to data in public databanks that allow the hosting software to reconstruct meshes (such as ultrastructural organelles or protein representations) for use in modeling, viewing, or analysis. Nested interactions and general HMF flexibility allows a recipe to become an ingredient in a higher-level recipe to minimize redundancy when working across multi-scale data. We described autoPACK with “recipes” and “ingredients” for semantic clarification and convenience, but from a programming perspective, autoPACK uses only the concept of “recipe” as its single universal data structure. An ingredient is a more specific recipe type, defined as a recipe that includes an associated structure (or for procedural ingredients, a potential structure) and in the simplest terms can be considered “an object to pack”. An .apr file contains a list of ingredient IDs with positions and orientations as described in the online documentation (http://www.autopack.org/documentation/-apr-autopack-result-file-anatomy) as well as a pointer back to the recipe that created the .apr. By default, the structural and other data files that the HMF files ultimately reference get downloaded into a cache directory system that reconstructs the needed portion of the online database on the fly during an interactive modeling or viewing session. The software can also access the ingredients directly from local or cloud storage via browser loading or via custom file paths to known sources.
As described in Fig. 2, the following levels of data structure interaction are visually separated into separate “tab” panels of the default cellPACK GUI.
Interaction Level 1: View the resulting model of a previous packing
At the top (and simplest) level of interaction with the data structure, a user can visualize a model that resulted from any stored previous packing via the autoPACK result file (.apr). The .apr file points to the main recipe that produced the result, followed by a list of component IDs and matrices (organized by any sub-recipes) needed to rebuild the scene (for details see http://www.autopack.org/documentation/-apr-autopack-result-file-anatomy). To visualize this model, cellPACK pre-constructs the scene components (container and ingredient representations and packing surfaces) as described in the “cellPACK Host Scene Preparation…” section of the Methods below. It then generates, organizes, positions and orients one instance (a computationally efficient copy) for every ingredient-ID per matrix line listed in the .apr file. Lastly, cellPACK produces a corresponding graphic user interface as shown in Fig. 2, all in one step. The user may additionally load analysis files associated with the .apr, such as arrays that describe the distance relationships within the volume. The “bottom level” ingredient recipes do not typically point to a default .apr file, but a properly constructed primary or midlevel recipe file includes a pointer to a default .apr file that any user can exactly recreate by packing the default recipe.
The input and output HMF files can be accessed directly for visualization and analysis, or for other uses such as Brownian Dynamic simulation. For example, we provide uPy plugins that construct cellPACK scenes into a host’s native GUI, where 3D models (coarse molecular surfaces by default) represent each ingredient. To enable standard laptop computers to handle the bulky data of a typical cellPACK model, uPy constructs cellPACK scenes into hosts with efficient sensible organizations that are native to each system, taking advantage of hardware instancing and other host optimizations that reduce data redundancy and other computational bottlenecks where available.
Displaying cellPACK results in uPy-enabled hosts provides access to the capabilities of each host for interacting with and analyzing the model. With the View Tab, entire organelles or individual ingredients can be hidden, deleted, added, colored, etc. Representations of molecular ingredients can be swapped and altered. In conjunction with native host functions, distances can be calculated or more sophisticated energetic potentials can be displayed as 3D heat maps. Points of closest contact can be illuminated, and simulations can be initiated or animations generated with a variety of techniques such as those described in our embedded Python Molecular Viewer (ePMV) paper.
Interaction Level 2: Modify an existing recipe to produce a new packing result
At this deeper interaction level, a user can work directly with the recipe file (i.e., the same “primary recipe” file and all nested recipe files that the resulting .apr file will point back to when produced), to reproduce a packing result, or modify parameters in the collected recipe(s) to produce a new packing result. When this recipe is run, it will produce only an .apr file by default, but the user may also select to write out a collection of grid arrays and other files useful in analysis. Depending on the host software used to perform the packing, cellPACK can also store a representative image or movie of the packing result (useful for automated packing while exploring parameter space). The primary recipe file can be a bottom-level recipe that sets parameters directly upon container and ingredient Python objects, or it can refer to more complex nested recipes that it unites (as in the HIV model in Fig. 6a), with the option to override particular sub-recipe ingredients or parameters (Supplementary Fig. 5). HMF flexibility allows a primary recipe to combine sub-recipe pointers with container and ingredient Python objects. The result of a new packing run can be written to an .apr and visualized from the View tab or visualized immediately from memory using the same process described in Data Structure Level 1 above.
As demonstrated in Fig. 3, randomness and biases from different packing methods can be quantified with built in analysis tools such as the planar distribution histograms (generated with mathplotlib, http://matplotlib.org) and box and whisker plots. Position and orientation histograms are used to highlight variations about a mean uniform distribution (frequency) line for randomly distributed objects or about a target frequency curve for gradient affected objects. When packing with the default benchmark goal of a uniform random distribution, spikes in the planar distribution histogram averaged over multiple runs can indicate problems in the algorithm or recipe data that warrant further manual investigation. Box and whisker plots can reveal at a glance any global skewing across orthogonal selections from within a fill volume.
Size-dependent available volume graphs mathematically track the distribution of negative spaces, to reveal what scales of protein-sized molecules could diffuse through these pore networks. Variations of these void-pore networks can be used to calculate permeability or as a starting point to simulate microscale flow (especially useful in macroscale engineering applications i.e. via the more generalized autoPACK software (http://etd.lsu.edu/docs/available/etd-08022006-140010/unrestricted/Zhang_dis.pdf).
Interaction Level 3: Construct recipes, ingredients, and components from scratch
Whether nested in other recipes or a single flat (bottom-level) recipe, a recipe structure eventually points to containers and ingredients that it assembles and can optionally modify or override with parameters. Containers and ingredients become Python objects when cellPACK operates upon them as inputs and a user can choose to generate them directly as recipe lists using HMF keywords in either XML, or Json formats (inline or with file pointers to subrecipes) or parametrically with a Python script. The uPy cellPACK GUI provides a Build tab to assist with recipe production. The current version enables a user to load a variety of data representations (e.g., the surface model of a Protein Databank File (PDB) generated with ePMV or any molecular viewer) in order to produce an ingredient-level recipe file, e.g., a container with an associated recipe of ingredients to pack, or an ingredient with associated structure and collisionTree files.
The cellPACK GUI working in uPy supported hosts also allows users to create recipes (that use only cellPACK defaults) on the fly by dragging and dropping ingredient and container geometries into provided scene groups (see a demonstration at http://www.youtube.com/watch?v=S94GUmFQ7yM). cellPACK recognizes the new ingredients placed and generates the default ingredient parameters by type. cellPACK then allows the user to modify these parameters through the standard Pack tab GUI. Recipes can be exported from the GUI or .apr files created by clicking a “pack” button.
cellPACK Host Scene Preparation for Result Visualization and Interactive Packing
To generate a cellPACK model, a user provides an environment volume, a selection volume to pack, and at least one recipe of ingredients. The environment volume may be an empty box or divided into containers that compartmentalize the space. A selection volume defines some subsection of the environment volume as the volume to be packed and is independent of container boundaries– the inset rectangle of Supplementary Fig. 6a (labeled “User selection” in Supplementary Fig. 6b) is an example of a selection volume used for packing. Environment volumes, selection volumes, containers, recipes, and ingredients and the parameters that define how these packing components interact can be declared in scripts, imported from other file types, created live in the visualization hosting software, or modified live through a cellPACK GUI. When a cellPACK recipe is run to produce a new packing result, the result is both stored in memory and written to an .apr file along with optional analysis files. To visualize the result of a previous packing from a file (via the View tab) or a current packing from memory (via a new Packing run from within the Pack tab), cellPACK first generates a representation of each unique geometry that will be represented in the final model to prepare the software. These single geometric models get efficiently instanced during visualization. Certain types of packing modules can optionally use this “visualization root” geometry directly in combination with capabilities native to the hosting visualization software to provide novel packing options, such as native collision detection or physics engines.
Definition of Scene Components
Selection volumes define the volume to pack
A selection volume can be defined mathematically in a script, manually as an interactive selection in a 3D viewport, or automatically set equal to the environment container described in the next section. A selection volume is always orthogonal to the origin of the scene, but may be set to any scale on the x, y, and z axis including scales equal to zero to produce a point, line, or 2D plane selections. Selections can have names and be looped in a Python packing script, for example, a user could build and name multiple cubic objects in their visualization host software then loop through these objects as selection boundaries to pack each in succession (or in parallel with multiprocessing) to fill a larger volume in a checkerboard fashion. With scripting, a polygonal object can be used to make complex 3D selection shapes.
Environment and ultrastructure containers
Similar to the selection volume, cellPACK requires at least one container, which is defined by default as the environment selection box. The environment container may be defined mathematically in a script, manually as an interactive selection in a 3D viewport, or automatically as a bounding box of a set of polygonal data (containers), for example, a model of organelles exported from a segmented tomogram with surface meshes centered on the bilayers.
When visualizing a result or performing an interactive packing, cellPACK first analyzes the prepared cellPACK scene in the 3D host to store any existing objects and the names of any annotated objects found. If the declared or live selection volume intersects with any found container objects, and a container’s annotation name matches a recipe name, then cellPACK associates that specific recipe to that container. For example, polygonal mesh containers can be exported from tomography software such as IMOD44 in VRML format with names annotated on each organelle mesh. These names get retained when imported into the host software and can automatically be associated with recipes if they exist when subsequently initializing a cellPACK recipe for a new packing run.
If surfaces are not annotated or associated recipes are not found in the library, a user may link to or construct them from scratch. A recipe file may also contain or reference a container object or container object file to build the containers into the scene as part of the recipe environment preparation process. Currently, containers may be imported or referenced as polyhedral meshes (vrml, collada, fbx, .c4d, .maya, .max, .blend, or .si file types), or as files of vertices and faces. The environment container may contain a recipe as well and the environment box is typically used to bound what cellPACK would otherwise consider to be an infinite “external” recipe.
Recipes
A minimal recipe HMF file contains a list of ingredients (ingredients are programmatically just sub-recipes) and a recipe name (often associated with a container name) to pack. A more typical recipe file may also include: parameter overrides for particular ingredients; pointers to other recipe files that the code recursively integrates into the model; containers or pointers to container files; a selection box or selection name (to pickup interactive selections) in which to bound ingredient packing; sub-recipe groups that combine the ingredients of multiple sub-recipes associated with each container; container or global environment modification parameters such as prohibited regions of packing or preferential packing gradient modifications; or overrides to default global packing parameters, result file paths, etc.
More sophisticated recipes can be scripted in Python to enable script level modification of cellPACK functions, for example, to loop through multiple packings, to generate multiple ingredients with just a few lines of code by iterating parameter values in loops, or to write out custom analysis files.
Ingredients
An ingredient is a specialized recipe type that has a geometric representation associated with it (this can be a single mesh or an entire packing result for example), most simply defined as an object to pack. A minimal ingredient HMF file contains an ingredient type, name, collisionTree list or collisionTree file pointer, and a representation or representation file pointer. Collision trees are used for efficient collision detection using the default jitter packing module– one of the local packing modules available in cellPACK and to update the packing grid with all other packing modules. A more typical ingredient file may also include: ingredient parameter default overrides (e.g., PriorityWeight, packing mode, rotation boundaries); interaction partners or interaction partner file pointers; data references (e.g., PDB or EMDB files used to generate the representation including pointers to custom PDB files stored in the cellPACK database, as well as non-spatial data); or alternate parameter triggers used by the code for logic operations, e.g., to alter parameters mathematically if packing in particular environments.
Currently, cellPACK recognizes a variety of types of ingredients. These include: (1) ingredients that have been manually or computationally placed within the selection volume prior to the cellPACK run; (2) soluble rigid-body ingredients that will be packed within given volumes; (3) membrane-spanning ingredients (including lipids) that orient normal and with defined directionality to a membrane surface with variable axial rotation and precession; (4) custom procedural ingredients that build an extended structure from smaller primitives, such as actin filaments and DNA; (5) procedural fiber ingredients that extend variable length wavy, kinked, and branched curves through the volume. In addition, HMF files allow an existing recipe or recipe result .apr file to be used as a nested-recipe ingredient. For example to pack a cellPACK model of HIV into Blood Plasma, the HIV HMF recipe or result can be treated as an ingredient for efficient referential reconstruction into the scene using the same methods but recursively described in Data Structure above. If treated as a result, the entire HIV model represented by a provided .apr file would pack as a single rigid body, but if treated as a recipe, the HIV envelope container would pack first into the serum before each of the HIV sub-recipe ingredients could pack into it, according to their global priority, while interacting with the blood plasma components as they simultaneously pack.
Producing a New Packing Result Step 1: Build a Grid (or load an existing grid)
With a recipe established, in order to produce a new result, cellPACK first organizes all of the containers and ingredients declared in the recipe (and recursively through any nested recipes) to create master lists. cellPACK uses these lists to subdivide the environment container into a grid that associates multiple types of data with the spatial data of the packing scene. The grid describes the space and tracks changes efficiently, for example, by enabling algebraic identification of neighboring points and previously placed ingredients for distance evaluations, collision tests, or agent interactions between objects.
To create the grid (Supplementary Fig. 7), cellPACK sorts the list of all ingredients by the packing radius parameter (by default: the radius of gyration of a molecule), and uses the smallest packing radius to establish a cubic grid with a diagonal spacing equal to that smallest packing radius (Supplementary Fig. 6c–d). By default, the grid fills the selection box, but can optionally be expanded to fill the environment box to reduce any boundary issues, to prepare for multiple consecutive packings in the same packing scene, or to incorporate grid-weighting effects from nearby containers or previous ingredients that are not already contained by the selection box.
After building the master grid, cellPACK uses the list of containers to compartmentalize the grid (Supplementary Fig. 6e). cellPACK uses a signed-distance function to assign each point in the grid to an appropriate container and to specify whether it is inside or outside of its surface. The system acts recursively, so surfaces can be nested in other surfaces, and the points inside of one surface and outside of a deeper surface remain associated with the recipe for the more superficial surface. For example, HIV in situ nests from superficial to deep as: (1) Host blood plasma (Environment recipe bound by the environment box); (2) HIV envelope (Surface recipe); (3) HIV matrix (Volume recipe); (4) HIV capsid (Surface recipe with no ingredients, only a container representation); (5) HIV genome region (Volume recipe). See more recipe interaction details in Supplementary Fig. 5.
If there are particularly small molecules (such as lipids or ions), rather than refining the entire master grid volume for one subset of very small ingredients, separate subdivisions are assigned and associated with the master grid. These finely subdivided grids are then associated with lookup tables in the master grid for fast sorting. For example, surface polygons with sides longer than the master grid spacing are subdivided to make fine surface points for lipid packing and tracking, allowing them to pack tightly in a continuous membrane. At the same time, the master grid is only as fine as it needs to be to allow all of the macromolecular ingredients to explore continuous space while avoiding collisions.
The grid is thus compartmentalized and each discrete voxel in space is associated with a container and its accompanying recipe of ingredients. In the final grid preparation step, cellPACK weights the grid with any optional container-level packing bias functions provided in the recipe (Supplementary Fig. 6f). During packing (detailed below), the signed-distance relationships along with the nearest neighbors, weighting, and other data stored with each grid point get updated to provide efficient lookup tables rather than recalculating relationships redundantly during packing.
To calculate the total number of each ingredient instance that should be placed into each compartment’s surface or interior, the area for each container surface and the volume of each compartment (including the exterior compartment) is calculated. Each ingredient’s molarity or surface density is then multiplied by the organelle volume or surface, and modified by any special localization gradient values that may affect local concentrations within a volume or surface. As noted in Fig. 5, logic or mathematical operators can modify the total number of ingredients to recapitulate observed values. To generate a whole number for the number of instances that will be packed, the numberOfInstances parameter value calculated from the concentration gets rounded up or down probabilistically. For example, if the raw numberOfInstances calculates to 14.73 molecules for the container, there is a 73% chance that the number will round up to 15, otherwise it gets rounded down to 14. This ensures that for volumes and surfaces with very low concentrations, there will still be a finite chance to include one instance of the ingredient.
Producing a New Packing Result Step 2: Ingredient Packing Priority (order is important)
Prior to packing, the master list of ingredients is sorted by a PriorityWeight ingredient parameter that reflects the difficulty of packing. Early studies revealed that larger, longer, and more branched ingredients are harder to pack homogenously than smaller globular ingredients as a volume gets crowded. We developed a simple default definition of priority weighting that gives more difficult ingredients a higher probability to pack earlier in the process:
where the surface area and volume are calculated by MSMS45, the encompassing radius is the maximal extent of the molecule from its packing center, and the constant is set to one by default. The Constant term allows an ingredient or recipe to bias the PriorityWeight of an ingredient or to tailor a Constant value across multiple packing results until desired densities are reached. A user can completely override the PriorityWeight of each ingredient with values coded into the ingredient file or broadly in the recipe file or they can override the default PriorityWeight function with a recipe script. Ingredients with user-defined negative PriorityWeights will pack first, in succession, starting from the ingredient with the most negative packing priority. Ingredients with a packing priority of zero will have the packing priority calculated by the default PriorityWeight equation shown above which will then be considered with ingredients that have user-defined positive PriorityWeights. Any manually overridden positive PriorityWeighted ingredients will be sorted with the calculated PriorityWeighted ingredients and will then pack in a random probability-biased order. For example, in the synaptic vesicle model (Fig. 5), all of the bulky vesicular ATPase proteins were given negative priority-weights, to ensure that they were given space to pack within the vesicles. Large positive priority-weights were used to “encourage” the lipids to pack as the last of the surface ingredients, while still tending to pack well before the small glutamates attempt to pack into the matrix of the vesicle. Without these empirically adjusted overrides, all ingredients would pack in a random but weighted order by default.
Producing a New Packing Result Step 3: Pack and Track
With all of the recipe and ingredient data connected to the spatial data of the containers via the grid, cellPACK uses the local packing method declared in each ingredient file, e.g., an agent packing spring method described in the Procedural Packing section below is detailed in Supplementary Fig. 8, to place recipe ingredients onto and into their appropriate containers (Supplementary Fig. 6h). Regardless of the local packing method, when a suitable place for an ingredient is found, an instance of the ingredient gets deposited and the grid is updated with knowledge of the ingredient and any pertinent data linked in from the ingredient’s file. Distances to the ingredient surfaces are updated and stored in the neighboring grid points and the ingredient identification is added to any points where the newly deposited ingredient becomes a new closest ingredient. When an ingredient gets picked, it searches through a list of grid point distances (that may be weighted globally or locally by other factors) and can only pick a point with a large enough minimum distance to accommodate the ingredient’s packing radius. A typical randomly-packing ingredient will analyze the current density of the fill, and if sparse, will pack on any random point rather than running the expensive sorting calculation. This calculation makes the filling more efficient later when the fill becomes denser and the sorting gets triggered appropriately. The filling loop continues until there are no more ingredients to add or until there are no more points available with radii large enough to accommodate more ingredients.
Procedural Packing (Branching Fibers or Crystals)
Procedural ingredients grow according to rules defined in their ingredient file (Supplementary Fig. 8). When a procedural ingredient is added, it can grow up to its maximum length immediately, or get deposited as a procedural seed for later extension (Supplementary Fig. 9). At any time in the fill, a live seed can be called upon (either initiated by an agent, random probability, or immediately upon its placement) to grow, pause, or die. As each unit is added, the total length or other property of the ingredient is evaluated and a probability is calculated to either continue or to terminate the growth. Growth can also be paused, returning into the main loop to continue packing other ingredients elsewhere, but leaving the end of this fiber or crystal as a dormant seed that could continue to grow once triggered. As the ingredient is extended, other types of seeds can be added to create branches. For example, in the construction of an actin cytoskeleton, a unit of actin with Arp2/3 could be added, creating a branch that could be extended at any time. Crystals can be grown in a similar fashion according to spacegroups or other constraints and rules provided by the crystal seed’s ingredient file. Currently, cellPACK includes procedural processes for the construction of filaments with defined helical symmetry (such as actin), as well as nucleic acid, and more general spline-based filaments (see Fig. 4 von Willebrand Factor and Fig. 6c mycoplasma lipopolysaccharides).
Preferential Packing: Gradients, interactomes, agents, density clustering
By default, cellPACK places ingredients randomly, on, in, or around polyhedral surfaces, creating a homogenous distribution. If desired, global and local controls can be applied to generate preferential and even deterministic deposition. While placing objects, a variety of probability-logic driven and agent placement systems can be applied to recapitulate current data or theories of localization or interaction in the model. This may be specified at a global or local level. For instance, a Ran-GTP ingredient can be globally biased to pick a point closer to the center of a dividing cell or closer to the cell’s chromatin46 or on a local level, an ingredient with agent behaviors can bear a list of binding partners that affect how it packs.
If not offered in the core code, special traits can be programmed into recipe scripts. For example, to match the Takamori paper’s observations when constructing the Synaptic Vesicle recipe (Fig. 5), we found it necessary to implement a method for “quantization” effects: experimental observations from Takamori et al. had revealed that, perhaps through some unknown sorting mechanism, every observed vesicle included at least one vATPase regardless of the vesicle’s diameter. Within a cellPACK Python recipe script, the default probability of placement for the vATPase ingredient was modified with a simple logic operation run after the number of vAPTases was calculated for a given vesicle (if vATPaseCount == 0: vATPaseCount=1) to ensure that at least one of these types of low-copy-number ingredients were included in each vesicle.
In the current release of cellPACK, we have implemented a few local agent methods. When an agent picks a location in which to position itself, it gets a list of neighbors in that area, scans the list for known binding partners, and probabilistically determines if it should be paired with any of them. When an agent decides to pair, one of several approaches can be used to position the new ingredient adjacent to an existing interaction partner: as demonstrated in the lipid packing of HIV in Fig. 6a, the closePacking algorithm weights the points surrounding the existing partner(s) and attempts to pack within this list of points; as demonstrated in the hexagonal packing of Gag-Ca in Fig. 7, binding transformations can be used to constrain growth from a selected interaction partner edge; or as demonstrated with the DNA binding protein HU in Fig. 6c, binding transformations can be constrained along a spline or surface, etc.; and a non-specific tight binding method generates a spring(s) to pair the two binding surfaces of the ingredients and relaxes the spring-connected system with appropriate collision detection that includes all neighboring geometry or atoms. This type of biased interaction can assist with the construction of larger cellular environments, for instance, to guide the interaction of DNA-binding proteins with DNA. Another approach modifies the weighting of the grid that surrounds particular container or ingredient surfaces to increase or decrease the probability of packing for any agent ingredient parameterized to recognize those particular grid biases. Less specifically, an ingredient can pack against itself or other ingredients using an optional “closePacking” method that biases the grid to encourage ingredients to pack tightly against previously packed ingredients. This approach serves well to pack lipid bilayers up to their correct densities from large libraries of single or patch lipid ingredients we have isolated from molecular dynamic resolved lipid PDB files. A recent experimental integration of the LipidWrapper software via a postprocessing protocol has provided a faster and more robust solution to pack lipids into bilayers. We are now working with the LipidWrapper programmers to integrate this as a new optional packing module specifically designed to pack lipids as patches onto surfaces directly in cellPACK with standard priority weighting and all other cellPACK features. The integration of 3rd party packages such as LipidWrapper for packing molecular dynamics-solved lipid patches into ultrastructure surfaces (http://nbcr.ucsd.edu/data/sw/hosted/lipidwrapper), and several previously integrated packages including panda3D with Bullet and Open Dynamics Engine physics engine support in Python (http://www.panda3d.org), RAPID for triangle intersection detection (http://gamma.cs.unc.edu/OBB), and scipy with KDTree to quickly collect neighboring molecules or polygons (http://www.scipy.org), demonstrates how the modular framework of cellPACK can expand to interoperate an extensive variety additional of packing and analysis algorithms via Python as the project continues to grow.
cellPACK models and simulated fluorescence microscopy
We tested six models for distribution of HIV-1 envelope glycoprotein (Env) for consistency with recent observations from fluorescence nanoscopy34 (FN). In the FN experiments, Env distribution was visualized with fluorescent antibodies, and virions were manually classified into three categories: particles with a single focus of fluorescence (presumed to be one cluster of Env), particles with two foci, and particles with three or more foci. For an immature virion (the PR(−) phenotype, missing the protease essential for maturation), the study observed that 28% of the virions had a single focus, 24% had two foci, and 48% had three or more foci.
cellPACK generated ten models for the distribution of the Gag protein in the immature virion to be consistent with observations reported from cryo-electron tomography47 (Gag-CET). Gag is observed to form hexamers, which assemble to form an irregular array that covers roughly 2/3 of the inner surface of the membrane. Our Gag models are generated with a cellPACK neighbor-growth packing module, which in this case extends hexagonal tiles from any existing hexagonal tile edges as described in the Preferential Packing section of the online methods. An open hexagon edge is selected randomly and a tile is hinged on that edge to an angle that is perpendicular to the packing surface. If this new hexagon does not collide with an existing tile, the position is accepted. Since it is impossible to cover perfectly a curved shape using regular hexagons, this tiling introduces similar defects as observed in the tomography models. The hexagon radii of 55 Å matches the size provided in the tomography models and the tiling is programmed to stop when the coverage reaches 60% of the packing surface’s surface area to match to observed coverage by Gag. The script is provided at http://mgldev.scripps.edu/projects/autoPACK/web/scripts/testTiling.py
cellPACK projected the Gag hexagons onto the 150 nm diameter envelope packing surface of HIV-1_0.1.6, and positioned Ma hexamers to be consistent with the Gag-CET observations.
An ingredient for Env is modeled from the trimer generated from PDB entry 4nco with the b12 Fab fragment from PDB entry 2ny7, currently stored as HIV1_Env_4nco_0_1_3 in the cellPACK database. No structural information is available for the entire C-terminal domain48–50, so it is approximated as a sphere of 450 amino acids, with a radius of 2.4 nm.
The Env model is placed randomly within the constraints of the six competing hypotheses represented in six recipes with hypothesis labels from Fig. 7: R-noMa places Env randomly on a uniform virion membrane as a control; R-offMa places Env in any empty space with no Gag hexagon (i.e., onto the 40% empty region of the envelope surface); R-inMa places in the center of any Gag hexagon (i.e, restricted to the 60% Gag covered region); Close-Ma places Env next to any of the Gag hexagons to simulate a 100% Env binding to Gag, but with some occlusion by the Env C-terminal tail; Close-Ma-Env places Env next to any of the Gag or next to any existing Env with a 50% chance to choose either; Close-Env places Env next to any existing Env.
For each hypothesis, cellPACK performed 100 models of Env distribution on each of the 10 Gag virion models. Each model included 7±5 Env trimers, with the number chosen randomly to sample the range of values provided in Fig. 2b from the FN study. The Env positions are computed using standard cellPACK recipes with the constraints provided for each hypothesis.
The resultant models are viewed from three orthogonal directions and fluorescence images are simulated using a Gaussian filter that approximates the point spread function, which assumes a 400 Å resolution of the STED setup. The 3D coordinates of the fluorophores are interpolated within a 3D grid of a with 100 Å gridpoint spacing. The interpolation to the camera resolution is done by incrementing the closest grid point to each Env positions. The grid is then convoluted with a 3D Gaussian function with HWFM = 400.0 Å to produce a simulated fluorescence image. The convoluted grid is summed individually on the three axes and interpolated to the proper resolution of 1 pixel = 200 Å. The 2D grid is then displayed using an offset “hot” color map.
Images are automatically separated into the three categories used in experimental FN work (single, two, and multiple foci) to generate the statistics for each model, for comparison with Fig. 3b from the FN paper. Before the counting, the images are filtered by removing background values below a certain threshold, and are then subject to a feature detection algorithm provided by the package scipy51 (http://www.scipy.org). This approach returns the number of connected pixels forming a blob. In order to make the distinction between the three categories, we look at the size of each of these features, assuming a small blob represents a unique foci, while a large diffuse one can represent multiple foci (Supplementary Fig. 2). Supplementary Fig. 3 describes the sensitivity of the three categories to the parameters of this counting algorithm, which was used to provide a more objective and controlled measure compared to manual human counting.
Supplementary Material
Acknowledgments
We would like to thank Takamori et al for the model of an average synaptic vesicle, C.P. Arthur for his synaptic cleft tomogram model, K. Schulten and A. Shih for their HDL models from molecular dynamics, K. Schulten and J.R. Perilla for their extra HIV capsid models, M. Yeager for his HIV capsid model, T.D. Goddard for developing Chimera’s .apr reader and for simplified HIV capsid models, J.A. Briggs and his lab for discussions on details on their HIV Ca tomography models, and J. Klusak (http://www.jiriklusak.cz) for his award-winning model of the HIV life cycle. We thank our colleagues for detailed suggestions on cellPACK experiments and extended analysis tools that can more easily bridge gaps to experimental biology. This is manuscript number 25098 from the Scripps Research Institute. This work was supported in part by a predoctoral fellowship from NSF (NSF 07576 to G.T.J.), gift donations from Autodesk, grants from the NIH (P41 GM103426 to L.A., M.F.S., and A.J.O., and P50GM103368 to D.S.G. and A.J.O.), a QB3 Fellowship grant from the California Institute for Quantitative Biosciences, qb3@UCSF to G.T.J., and a UCSF School of Pharmacy, 2013 Mary Anne Koda-Kimble Seed Award for Innovation to G.T.J.
Footnotes
AUTHOR CONTRIBUTIONS
G.T.J. conceived autoPACK and cellPACK and designed the original code, wrote the core code, designed user interfaces and edited interface code, conducted experiments and analyzed results. L.A. wrote core code, designed and wrote the interface code, wrote analysis code, designed and conducted HIV experiments and analyses. M.A. co-designed and wrote prototype autoPACK code. D.S.G. co-conceived and guided cellPACK design and designed HIV experiments. M.F.S. transposed the prototype autoPACK code to Python with G.T.J., added cellPACK packing and grid preparation features, structured and cleaned the first drafts of the current core code. A.J.O. guided code and experiment design and implementation.
The authors declare no competing financial interests.
References
- 1.Stölken M, et al. Maximum likelihood based classification of electron tomographic data. J Struct Biol. 2011;173:77–85. doi: 10.1016/j.jsb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Yu Z, Frangakis AS. Classification of electron sub-tomograms with neural networks and its application to template-matching. J Struct Biol. 2011;174:494–504. doi: 10.1016/j.jsb.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Ball P. Portrait of a molecule. Nature. 2003;421:421–2. doi: 10.1038/nature01404. [DOI] [PubMed] [Google Scholar]
- 4.Harrison SC. Whither structural biology? Nat Struct Mol Biol. 2004;11:12–5. doi: 10.1038/nsmb0104-12. [DOI] [PubMed] [Google Scholar]
- 5.Wong B, Kjaegaard RS. Pencil and paper. Nat Methods. 2012;9:1037. doi: 10.1038/nmeth.2223. [DOI] [PubMed] [Google Scholar]
- 6.Goodsell DS. Escherichia coli. Biochem Mol Biol Educ. 2009;37:325–32. doi: 10.1002/bmb.20345. [DOI] [PubMed] [Google Scholar]
- 7.Goodsell DS. Miniseries: Illustrating the machinery of life: Eukaryotic cell panorama. Biochem Mol Biol Educ. 2011;39:91–101. doi: 10.1002/bmb.20494. [DOI] [PubMed] [Google Scholar]
- 8.Vendeville A, Lariviere D, Fourmentin E. An inventory of the bacterial macromolecular components and their spatial organization. FEMS Microbiol Rev. 2011;35:395–414. doi: 10.1111/j.1574-6976.2010.00254.x. [DOI] [PubMed] [Google Scholar]
- 9.Karr JR, et al. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szpiro G. Kepler’s conjecture: how some of the greatest minds in history helped solve one of the oldest math problems in the world. John Wiley & Sons; New Jersy: 2003. [Google Scholar]
- 11.Borkovec MDPW, Peikert R. The Fractal Dimension of the Apollonian Sphere Packing. Fractals. 1994;2:521–526. [Google Scholar]
- 12.Altendorf H, Jeulin D. Random-walk-based stochastic modeling of three-dimensional fiber systems. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:041804. doi: 10.1103/PhysRevE.83.041804. [DOI] [PubMed] [Google Scholar]
- 13.Weber J, Penn J. Creation and Rendering of Realistic Trees. SIGGRAPH ‘95 Proceedings of the 22nd annual conference on Computer graphics and interactive techniques; 1995. pp. 119–128. [Google Scholar]
- 14.Löhner R, Oñate E. A general advancing front technique for filling space with arbitrary objects. International Journal for Numerical Methods in Engineering. 2004;61:1977–1991. [Google Scholar]
- 15.Lubachevsky BD, Stillinger FH. Geometric properties of random disk packings. Journal of Statistical Physics. 1990;60:561–583. [Google Scholar]
- 16.Zhang W, Thompson KE, Reed AH, Beenken L. Relationship between packing structure and porosity in fixed beds of equilateral cylindrical particles. Chemical Engineering Science. 2006;61:8060–8074. [Google Scholar]
- 17.Stannarius R. Granular materials of anisometric grains. Soft Matter. 2013;9:7401–7418. [Google Scholar]
- 18.Williams SR, Philipse AP. Random packings of spheres and spherocylinders simulated by mechanical contraction. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;67:051301. doi: 10.1103/PhysRevE.67.051301. [DOI] [PubMed] [Google Scholar]
- 19.Russel D, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco M. Molecular silverware. 1. General solutions to excluded volume constrained problems. Journal of Computational Chemistry. 1991;12:237–247. [Google Scholar]
- 21.Byholm T, Toivakka M, Westerholm J. Effective packing of 3-dimensional voxel-based arbitrary shaped particles. Powder Technology. 2009;196:139–146. [Google Scholar]
- 22.Ando TSJ. Brownian Dynamics Simulation of Macromolecule Diffusion in a Protocell. Proceedings of the International Conference of the Quantum Bio-Informatics IV. 2011;28:413–426. doi: 10.1142/9789814343763_0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludovic A. In: uPy: A Ubiquitous CG Python API with Biological-Modeling Applications. Graham J, Johan H, Arthur O, Michel S, editors. Vol. 32. 2012. pp. 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 26.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Putnam FW. The Plasma Proteins. IV. Academic Press; 1984. Progress in Plasma Proteins; pp. 1–44. [Google Scholar]
- 28.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–46. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 29.Arthur CP, Dean C, Pagratis M, Chapman ER, Stowell MH. Loss of synaptotagmin IV results in a reduction in synaptic vesicles and a distortion of the Golgi structure in cultured hippocampal neurons. Neuroscience. 2010;167:135–42. doi: 10.1016/j.neuroscience.2010.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson GT, et al. 3D molecular models of whole HIV-1 virions generated with cellPACK. Faraday Discussions. 2014 doi: 10.1039/c4fd00017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–7. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao G, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–6. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner MK, Odde DJ, Bloom K. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods. 2007;41:232–7. doi: 10.1016/j.ymeth.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Chojnacki J, et al. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science. 2012;338:524–8. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 35.Ridgway D, Broderick G, Lopez-Campistrous A, Ru’aini M, Winter P, Hamilton M, Boulanger P, Kovalenko A, Ellison MJ. Coarse-grained molecular simulation of diffusion and reaction kinetics in a crowded virtual cytoplasm. Biophys J. 2008;94:3748–3759. doi: 10.1529/biophysj.107.116053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson GT, Autin L, Goodsell DS, Sanner MF, Olson AJ. ePMV embeds molecular modeling into professional animation software environments. Structure. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battle G, Costanzo LD, Goodsell DS, Hudson B, Lawson C, Voigt M, Zardecki C. RCSB Protein Data Bank 2014 Calendar. RCSB; Piscataway: 2013. [Google Scholar]
- 38.Johnson GT. AMI Newsletter. AMI; Lexington: 2013. Announcing the Winners of the autoPACK Visualization Challenge2012: Present HIV in Blood Plasma. [Google Scholar]
- 39.Ahmed T, Shimizu TS, Stocker R. Microfluidics for bacterial chemotaxis. Integr Biol (Camb) 2010;2:604–29. doi: 10.1039/c0ib00049c. [DOI] [PubMed] [Google Scholar]
- 40.Niethammer P, Bastiaens P, Karsenti E. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science. 2004;303:1862–1866. doi: 10.1126/science.1094108. [DOI] [PubMed] [Google Scholar]
- 41.Lipkow K, Odde DJ. Model for protein concentration gradients in the cytoplasm. Cellular and Molecular Bioengineering. 2008;1:84–92. doi: 10.1007/s12195-008-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–11. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster SC, Khan S. The bacterial flagellar motor. Annu Rev Biophys Biomol Struct. 1994;23:509–39. doi: 10.1146/annurev.bb.23.060194.002453. [DOI] [PubMed] [Google Scholar]
- 44.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 45.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–20. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 46.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–77. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- 47.de Marco A, et al. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010;6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Postler TS, Desrosiers RC. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J Virol. 2013;87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos da Silva E, Mulinge M, Perez Bercoff D. The frantic play of the concealed HIV envelope cytoplasmic tail. Retrovirology. 2013;10:54. doi: 10.1186/1742-4690-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steckbeck JD, Kuhlmann AS, Montelaro RC. C-terminal tail of human immunodeficiency virus gp41: functionally rich and structurally enigmatic. J Gen Virol. 2013;94:1–19. doi: 10.1099/vir.0.046508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliphant TE. Python for scientific computing. Computing in Science & Engineering. 2007;9:10–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.