Abstract
This report describes an outbreak of Clostridium difficile infection (CDI) in a nephrology ward in 2012, caused by the fluoroquinolone- and clindamycin-resistant polymerase chain reaction (PCR) ribotype 027 strains. An increase in the number of cases of diarrhoea was noted among patients hospitalised between 26 November 2012 and 17 December 2012 in a hospital in North Poland. Eight patients were on haemodialysis in the outpatient dialysis facility, while one patient was receiving peritoneal dialysis. The 027 strain could be detected in eight haemodialysis patients. One strain, isolated from the patient receiving peritoneal dialysis, belonged to PCR ribotype 001. In this study, we documented the first outbreak of CDI caused by a fluoroquinolone-resistant (FQR) C. difficile PCR ribotype 027 strain in Polish dialysis patients.
Introduction
Clostridium difficile infection (CDI) is the most common cause of infections in healthcare facilities. The clinical manifestations of CDI range from asymptomatic colonisation to toxic megacolon, which has a high risk of mortality [1]. The three main risk factors for CDI include antibiotic exposure, higher age (65 years and above) and prolonged hospitalisation [2, 3]. Additional risk factors include cancer, haematopoietic stem cell transplant (HSCT), co-morbidities associated with immunosuppression, use of gastric acid-reducing drugs, admission to the intensive care unit, use of nasogastric tubes and inflammatory bowel diseases [4–7]. Patients with kidney disease are also at an increased risk of developing CDI and represent an especially vulnerable group of patients [8–10].
C. difficile NAP1/BI/027 strains have been reported to be aetiological agents of severe infections in the USA, Canada and Europe [11]. Recently, fluoroquinolones (FQ) have been linked to CDI, particularly those caused by polymerase chain reaction (PCR) ribotype 027 [12]. In the present study, we describe the epidemiology of CDI as well as the clinical and microbiological data of dialysis patients hospitalised in the nephrology ward of a hospital in Poland, with diarrhoea, caused by the PCR ribotype 027 strain.
Materials and methods
Patients
The current study was retrospective in nature, using C. difficile isolated from stool samples that were collected and stored when an outbreak was suspected in the nephrology ward. Diagnostic testing of CDI in the surveyed ward was performed only upon the physician’s request. The study was conducted at the 620-bed Provincial Hospital (PH), located in North Poland. In 2012, the incidence of CDI at the hospital was 4.5/10,000 patient-days (results not shown). Between 1 January 2012 and 31 December 2012, CDI was suspected in 96 symptomatic patients. Of these, 53 tested positive for C. difficile toxins A/B. The CDI-positive patients were hospitalised in different wards. Between 26 November 2012 and 17 December 2012, a high incidence of diarrhoea was observed in the 32-bed nephrology unit. These cases included nine dialysis patients with diarrhoea. Our study focused only on patients hospitalised in the nephrology ward who were receiving dialysis in the outpatient haemodialysis facility.
Definitions
Diarrhoea was defined on the basis of well-known criteria [13, 14]. According to the European Centre for Disease Prevention and Control (ECDC), CDI can be classified into two types: healthcare-associated CDI, which usually occurs in a hospital after 48 h of admission, and community-associated CDI [15]. A severe case of CDI is defined by fever (>38.5 °C), decreased kidney function, high leukocyte count (>15 × 109 cells/L) or colonic wall thickening revealed by a computed tomography (CT) scan [15]. Recurrent CDI is diagnosed when CDI re-occurs within 8 weeks of the onset of a previous episode, provided the symptoms from the previous episode resolved after the completion of initial treatment. An outbreak of CDI is defined as two or more cases related in time and place over a defined period (28 days), based on the date of onset of the first case. In this study, a possible CDI outbreak was identified based on the occurrence of three cases within 30 days, a departure from the normal pattern [16].
Collection of demographic and clinical data
Medical records of the patients were studied for the presence of clinical symptoms of CDI. Data, including information on patient demographics, age, sex, admission type, diagnoses, length of stay, type of dialysis, use of antibiotics, type of discharge, recurrences and mortality, were obtained for 26 November 2012 through 17 December 2012. Stool character and frequency, and abdominal pain were also monitored. The characterisation of dialysis patients with diarrhoea due to C. difficile is shown in Table 1.
Table 1.
Characterisation of dialysis patients with diarrhoea due to Clostridium difficile strain polymerase chain reaction (PCR) ribotype 027 and a sporadic case due to PCR ribotype 001
| No./sex/age (years) | Date of hospitalisation D/Mo/Yr | PCR ribotype | Type of dialysis | AB | Date of onset of CDI | Leucocytosis ×109 L | 1.5× increase in CL | Recurrence | Fatality associated with CDI |
|---|---|---|---|---|---|---|---|---|---|
| 1/F/52 | 15/11–31/12/2012 | 027 | HD (one episode) | AMC, II° cephalosporin, II° fluoroquinolone, fluconazole | 26/11/2012 | 15.67 | Yes | No | No |
| 2/F/61 | 14/11–01/12/2012 | 027 | HD | AMC | 28/11/2012 | 22.21 | Yes | No | No |
| 3/M/61 | 15/11–31/12/2012 | 027 | HD | Aminopenicillin (amoxicillin, ampicillin) | 30/11/2012 | 15.9 | Yes | No | No |
| 4/F/77 | 02/12–17/12/2012 | 027 | HD | VA (iv), gentamicin | 03/12/2012 | 17.38 | No | Yes | Yes |
| 5/M/73 | 19/11–17/12/2012 | 027 | PD | I° and II° cephalosporin | 04/12/2012 | 10.11 | Yes | Yes | No |
| 6/M/85 | 01/12–24/12/2012 | 027 | HD | AMC, II° cephalosporin, macrolide | 04/12/2012 | 17.5 | No | No | No |
| 7/F/77 | 05/12–27/12/2012 | 027 | HD | II° cephalosporin, aminoglycoside, II° fluoroquinolone | 05/12/2012 | 17.8 | Yes | Yes | Yes |
| 8/F/89 | 07/12–24/12/2012 | 027 | HD | IP | 11/12/2012 | 15.62 | Yes | No | No |
| 9/M/72 | 01/12/2012–04/01/2013 | 001 | HD | Fluconazole | 06/12/2012 | 3.12 | No | No | No |
AB antibiotics, CDI C. difficile infection, CL creatinine level, HD haemodialysis, PD peritoneal dialysis, AMC amoxicillin and clavulanic acid, I° first generation, II° second generation, VA vancomycin, iv intravenous, IP imipenem
Microbiological methods
Detection of C. difficile toxin and culture of C. difficile strains
CDI in patients who developed diarrhoea was diagnosed by an immune-enzymatic assay for glutamate dehydrogenase (GDH; TechLab®, Inc., USA). In samples that tested positive in the GDH assay, CDI was confirmed using the C. difficile TOX A/B Test II™ kit (TechLab®, Inc., USA) and the detection of toxigenic C. difficile in culture. C. difficile was cultured from faecal samples using selective CLO Agar (bioMérieux SA; Marcy l’Etoile, France). After incubation at 35 °C for 48 h under anaerobic conditions, isolates were confirmed to be C. difficile on the basis of well-known criteria. All C. difficile strains were frozen at −70 °C in a Microbank™ bacterial storage system (Pro-Lab Diagnostics, UK). C. difficile isolates from the dialysis patients were collected for further analysis.
Real-time (RT)-PCR and PCR ribotyping
All C. difficile strains were sent to the Anaerobe Laboratory (AL) in Warsaw and tested for genetic markers of toxigenicity using the Xpert CD assay (Cepheid; Sunnyvale, CA, USA). The Xpert CD assay can be used to detect toxigenic C. difficile strains and identify the C. difficile NAP1/BI/027 strain. RT-PCR was performed to detect the tcdB, cdtA and cdtB genes (binary toxin genes), and a deletion in the tcdC gene (encodes a negative regulator of toxin A/B production). PCR ribotyping was performed as described by Stubbs et al. [17]. Binary toxin-producing control strains belonging to the Cardiff-ECDC reference strain collection were provided by Jon Brazier (Anaerobe Reference Laboratory, Cardiff, UK) and Ed Kuijper (Leiden University Medical Center, Leiden, The Netherlands), and consisted of R8637 (PCR ribotype 019), R5989 (PCR ribotype 023), R10456 (PCR ribotype 056) and strains from PCR ribotypes 045, 078, 027 and 176. The banding patterns obtained for the C. difficile strains were compared with those obtained for the reference strains.
Antimicrobial susceptibility testing
C. difficile strains were grown on Columbia Agar plates (bioMérieux SA, Marcy l’Etoile, France) and subsequent suspensions were adjusted to an optical density of 1 McF at 900 nm using a bioMérieux ATB 1550 densitometer and streaked on the surface of Brucella Agar plates. Etests (bioMérieux SA, Marcy l’Etoile, France) were placed on each plate and these contained the following antibiotic gradients: clindamycin (CL), erythromycin (EM), ciprofloxacin (CI), moxifloxacin (MX), metronidazole (MTZ) and vancomycin (VA). The plates were incubated under anaerobic conditions at 37 °C for 48 h, according to the manufacturer’s instructions. The Clinical and Laboratory Standards Institute (CLSI) recommendations were used to define antibiotic resistance [18]. Quality control strains (Bacteroides fragilis NCTC 11295, Bacteroides thetaiotaomicron ATCC 29741, Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923) were included in all experiments.
Environmental decontamination and extended cleaning programme
Chlorine bleach was used to clean the sanitary facilities in rooms occupied by patients diagnosed with CDI. Indomethacin and Octenisept were used to clean all medical equipment and furniture. Sanitary ware and hand-contact surfaces were cleaned on a daily basis until post-outbreak to maintain high standards of hygiene. An additional terminal clean with the hydrogen peroxide vapour (HPV) decontamination technology was completed to eliminate any spores left behind in the environment. Adjacent rooms received a full final cleansing upon discharge of CDI-positive patients.
Results
Epidemiological and clinical findings
CDI was observed sporadically among patients hospitalised in the nephrology unit between January 2012 and October 2012. Only seven cases of CDI were confirmed during this period. Between 26 November 2012 and 17 December 2012, an increased incidence of CDI was observed among dialysis patients hospitalised in the nephrology unit. The demographic and clinical data are presented in Table 1. The outbreak involved nine patients (five women and four men) in total, whose ages ranged from 52 to 89 years (with a median age of 70.5 years). Watery diarrhoea was observed in all patients, and was the major clinical manifestation of CDI. Three patients developed recurrent CDI. Two patients died of complications due to CDI, yielding a mortality rate of 22 % (2/9).
History of outbreak
Initially, two cases of CDI were detected in the nephrology unit. The first patient was admitted to the internal department of cardiovascular diabetology on 15 November. Subsequently, the patient was transferred to the nephrology unit. Since his risk of acquiring C. difficile was high, he was placed under isolation. CDI was diagnosed on 26 November. Patient 2 was admitted on 14 November. CDI was diagnosed after 14 days of hospitalisation. A history of the symptoms of CDI among all dialysis patients hospitalised in the nephrology unit is shown in Fig. 1.
Fig. 1.
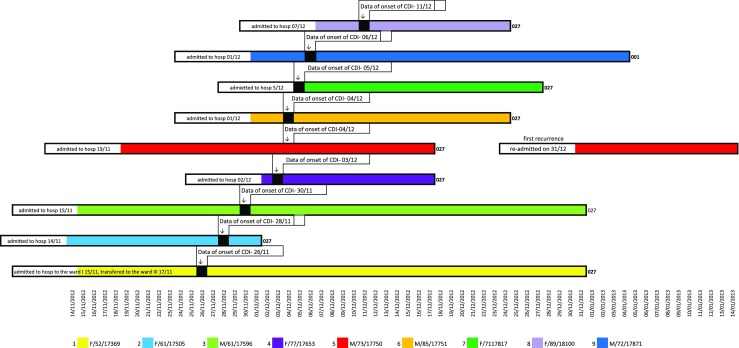
A history of symptoms of CDI among all dialysis patients hospitalised in the nephrology unit
Microbiological analysis
Of the nine C. difficile isolates, eight contained the tcdB gene, the binary toxin genes (cdtA and cdtB) and a deletion in the tcdC gene, and one strain possessed the tcdB gene. All eight isolates positive binary toxin genes were recognised by the Xpert assay as belonging to the PCR ribotype 027 strain of C. difficile. PCR ribotyping classified the nine C. difficile isolates into two ribotypes: 027 (n = 8) and 001 (n = 1). Susceptibility testing was performed for all strains. All nine strains were found to be resistant to CI [minimum inhibitory concentration (MIC) ≥ 32 mg/L], while the eight PCR ribotype 027 strains were resistant to MX (MIC ≥ 32 mg/L). Only type 001 was susceptible to all the other antibiotics. The MIC of CL was 0.38–6 mg/L, while that of EM was 0.75–256 mg/L. All PCR ribotype 027 strains were resistant to EM. Nine strains were susceptible to MTZ (MIC range 0.094–0.38 mg/L) and VA (MIC range 0.25–0.5 mg/L).
Discussion
We describe the first outbreak of CDI caused by the C. difficile PCR ribotype 027 strain in dialysis patients in Poland. In an earlier study, 175 C. difficile strains were isolated from patients hospitalised in a teaching hospital in Warsaw during 2005–2006. Of these, only one isolate belonged to PCR ribotype 027 [19]. This ribotype was not observed in Poland until 2008. In the period 2008–2010, outbreaks of antibiotic-associated diarrhoea occurred in three different hospitals in Poland [20]. We concluded that outbreaks of CDI associated with hypervirulent strains belonging to PCR ribotype 027 occurred in hospitals in Poland in 2008–2010.
All patients with diarrhoea were promptly identified and isolated. Strict isolation procedures remained in place for 30 days after the last identified case. All symptomatic patients remained in isolation with dedicated toilets and shower facilities until discharge, regardless of symptoms. In an effort to reduce the incidence of CDI, the hospital used accelerated hydrogen peroxide as the hospital disinfectant. Accelerated hydrogen peroxide has been shown to inactivate C difficile spores within a 15-min window [21, 22]. This ward was then disinfected with non-buffered hypochlorite, after which the outbreak ended. Fawley et al. compared the effects of five different cleaning agents on epidemic and non-epidemic C. difficile strains [23]. Their study showed that only chlorine-based germicides were able to inactivate C. difficile spores. Martinez et al. support the use of chlorine-based disinfectants for preventing the transmission of C. difficile [24]. The implementation of strict infection-control precautions, antimicrobial stewardship and enhanced environmental cleaning are key components in managing a C. difficile outbreak successfully. The outbreak at PH was controlled successfully by terminal cleaning of the ward and reinforcement of infection-control precautions.
Our findings suggest that dialysis patients who are treated with antimicrobial agents are at a risk of developing CDI. The outbreak at PH was controlled successfully. Our report emphasises the importance of monitoring CDIs and the early identification of symptomatic patients to prevent the spread of infection.
Acknowledgements
We would like to thank the following personnel who were involved in managing the outbreak successfully: medical and nursing staff of the nephrology unit, management of infection control and the medical microbiology department.
We would like to thank Prof. E. Kuijper and Prof. J. Brazier for the Cardiff-ECDC reference C. difficile strain collection.
This work was supported by the National Science Centre, Poland, grant no. DEC 2011/01/B/NZ7/02720.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Peniche AG, Savidge TC, Dann SM. Recent insights into Clostridium difficile pathogenesis. Curr Opin Infect Dis. 2013;26:447–453. doi: 10.1097/01.qco.0000433318.82618.c6. [DOI] [PubMed] [Google Scholar]
- 2.McFarland LV, Surawicz CM, Stamm WE. Risk factors for Clostridium difficile carriage and C. difficile-associated diarrhea in a cohort of hospitalized patients. J Infect Dis. 1990;162:678–684. doi: 10.1093/infdis/162.3.678. [DOI] [PubMed] [Google Scholar]
- 3.Bignardi GE. Risk factors for Clostridium difficile infection. J Hosp Infect. 1998;40:1–15. doi: 10.1016/S0195-6701(98)90019-6. [DOI] [PubMed] [Google Scholar]
- 4.Kamboj M, Son C, Cantu S, Chemaly RF, Dickman J, Dubberke E, Engles L, Lafferty T, Liddell G, Lesperance ME, Mangino JE, Martin S, Mayfield J, Mehta SA, O’Rourke S, Perego CS, Taplitz R, Eagan J, Sepkowitz KA. Hospital-onset Clostridium difficile infection rates in persons with cancer or hematopoietic stem cell transplant: a C3IC network report. Infect Control Hosp Epidemiol. 2012;33:1162–1165. doi: 10.1086/668023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barletta JF, El-Ibiary SY, Davis LE, Nguyen B, Raney CR. Proton pump inhibitors and the risk for hospital-acquired Clostridium difficile infection. Mayo Clin Proc. 2013;88:1085–1090. doi: 10.1016/j.mayocp.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Salva S, Duran N, Rodriguez V, Nieto L, Serra J, Rello J, CRIPS Investigators Clostridium difficile in the ICU: study of the incidence, recurrence, clinical characteristics and complications in a university hospital. Med Intensiva. 2014;38:140–145. doi: 10.1016/j.medin.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Reddy SS, Brandt LJ. Clostridium difficile infection and inflammatory bowel disease. J Clin Gastroenterol. 2013;47:666–671. doi: 10.1097/MCG.0b013e31828b288a. [DOI] [PubMed] [Google Scholar]
- 8.Keddis MT, Khanna S, Noheria A, Baddour LM, Pardi DS, Qian Q. Clostridium difficile infection in patients with chronic kidney disease. Mayo Clin Proc. 2012;87:1046–1053. doi: 10.1016/j.mayocp.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pant C, Deshpande A, Anderson MP, Sferra TJ. Clostridium difficile infection is associated with poor outcomes in end-stage renal disease. J Investig Med. 2012;60:529–532. doi: 10.2310/JIM.0b013e318242b313. [DOI] [PubMed] [Google Scholar]
- 10.Wolfgram DF, Foster D, Astor BC, Chan MR. Development of Clostridium difficile colitis in peritoneal dialysis patients treated for peritonitis. Perit Dial Int. 2012;32:666–668. doi: 10.3747/pdi.2011.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuijper EJ, Barbut F, Brazier JS, Kleinkauf N, Eckmanns T, Lambert ML, Drudy D, Fitzpatrick F, Wiuff C, Brown DJ, Coia JE, Pituch H, Reichert P, Even J, Mossong J, Widmer AF, Olsen KE, Allerberger F, Notermans DW, Delmée M, Coignard B, Wilcox M, Patel B, Frei R, Nagy E, Bouza E, Marin M, Akerlund T, Virolainen-Julkunen A, Lyytikäinen O, Kotila S, Ingebretsen A, Smyth B, Rooney P, Poxton IR, Monnet DL (2008) Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill 13(31). pii: 18942 [PubMed]
- 12.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crobach MJT, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI) Clin Microbiol Infect. 2009;15:1053–1066. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 14.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, ECDIS Study Group Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 15.Kuijper EJ, Coignard B, Tüll P, ESCMID Study Group for Clostridium difficile. EU Member States. European Centre for Disease Prevention and Control Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12(Suppl 6):2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 16.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, McFarland LV, Mellow M, Zuckerbraun BS. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478–498. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 17.Stubbs SL, Brazier JS, O’Neill GL, Duerden BI. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37:461–463. doi: 10.1128/jcm.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute (CLSI) Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11-A7. 7. Wayne: CLSI; 2007. [Google Scholar]
- 19.Pituch H, Bakker D, Kuijper E, Obuch-Woszczatyński P, Wultańska D, Nurzyńska G, Bielec A, Bar-Andziak E, Łuczak M. First isolation of Clostridium difficile PCR-ribotype 027/toxinotype III in Poland. Pol J Microbiol. 2008;57:267–268. doi: 10.1099/jmm.0.47317-0. [DOI] [PubMed] [Google Scholar]
- 20.Obuch-Woszczatyński P, Lachowicz D, Schneider A, Mól A, Pawłowska J, Ożdżeńska-Milke E, Pruszczyk P, Wultańska D, Młynarczyk G, Harmanus C, Kuijper EJ, van Belkum A, Pituch H. Occurrence of Clostridium difficile PCR-ribotype 027 and it’s closely related PCR-ribotype 176 in hospitals in Poland in 2008–2010. Anaerobe. 2014;28C:13–17. doi: 10.1016/j.anaerobe.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Perez J, Springthorpe VS, Sattar SA. Activity of selected oxidizing microbicides against the spores of Clostridium difficile: relevance to environmental control. Am J Infect Control. 2005;33:320–325. doi: 10.1016/j.ajic.2005.04.240. [DOI] [PubMed] [Google Scholar]
- 22.Dawson LF, Valiente E, Donahue EH, Birchenough G, Wren BW. Hypervirulent Clostridium difficile PCR-ribotypes exhibit resistance to widely used disinfectants. PLoS One. 2011;6(10):e25754. doi: 10.1371/journal.pone.0025754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fawley WN, Underwood S, Freeman J, Baines SD, Saxton K, Stephenson K, Owens RC, Jr, Wilcox MH. Efficacy of hospital cleaning agents and germicides against epidemic Clostridium difficile strains. Infect Control Hosp Epidemiol. 2007;28:920–925. doi: 10.1086/519201. [DOI] [PubMed] [Google Scholar]
- 24.Martinez FJ, Leffler DA, Kelly CP. Clostridium difficile outbreaks: prevention and treatment strategies. Risk Manag Healthc Policy. 2012;5:55–64. doi: 10.2147/RMHP.S13053. [DOI] [PMC free article] [PubMed] [Google Scholar]


