Abstract
Notch signaling is critical for skeletal muscle development and regeneration, permitting the expansion of progenitor cells by preventing premature differentiation. We have interrogated the pathways through which ligand-mediated signaling inhibits myogenesis by identifying Notch target genes and assessing their impact on differentiation in vitro. Notch activation led to the robust induction of the transcriptional repressors Hey1 and HeyL in myoblasts, but only constitutive expression of Hey1 blocked myogenesis. siRNA-mediated knockdown of Hey1 had no effect on Notch’s ability to inhibit differentiation, suggesting the existence of additional, possibly redundant pathways. We identified 82 genes whose expression was activated when C2C12 myoblasts were cultured in the presence of the Notch ligand Dll4. One of these, MyoR, is a novel Notch-responsive gene, whose protein product is known to repress myogenesis in vitro. siRNA-mediated knockdown of MyoR alone, or in combination with Hey1, was also ineffective at rescuing differentiation in the presence of Dll4. Our data support a model in which Notch signaling inhibits myogenesis through multiple pathways, two of which are defined by the Notch target genes Hey1 and MyoR.
Keywords: Notch, Hey1, MyoR, MyoD, Myf-5, Myogenin, muscle, myogenesis
INTRODUCTION
Notch signaling is necessary for the proper development of numerous cell types and tissues (Artavanis-Tsakonas et al., 1999; Lai, 2004; Lefort and Dotto, 2004; Louvi and Artavanis-Tsakonas, 2006; Maillard et al., 2005; Roca and Adams, 2007; Wilson and Radtke, 2006). It is typically viewed as a transcriptional cascade in which the cleaved intracellular domain of the Notch receptor, NICD, enters the nucleus and activates transcription of target genes through direct interactions with Mastermind (Mam) and the DNA binding protein CSL (CBF1, Suppressor of Hairless, Lag-1, RBP-J). The best-characterized targets of NICD are genes that encode DNA binding proteins of the Hes (1, 5, 7) and Hey (1, 2, L) families of bHLH transcriptional repressors (Iso et al., 2003). The induction of Hes proteins has been shown to account for some of Notch’s activities, notably in the developing nervous system (Ohtsuka et al., 1999), keratinocytes (Mammucari et al., 2005), and adipocytes (Ross et al., 2004), but it does not account for others, such as the ability of NICD to transform T lymphocytes (Kawamata et al., 2002). Hey family members, by contrast, have been linked to critical functions of Notch in the developing heart and vasculature (Fischer et al., 2004; Fischer et al., 2007). The c-Myc gene is also directly activated by Notch in mammary epithelial cells (Klinakis et al., 2006) and T cells (Weng et al., 2006), contributing to both the proliferative and oncogenic properties of NICD in those cell types. HIF-1α has been shown recently to bind directly to NICD and thereby potentiate activation of Notch target genes under hypoxia (Gustafsson et al., 2005). Indeed, Notch’s ability to induce the expression of multiple genes, with each potentially eliciting a slightly different or perhaps partial response, underscores the importance of viewing the pathway more as a transcriptional network with NICD’s activation of CSL being the primary hub.
Notch signaling was shown to inhibit muscle differentiation over a decade ago, yet the precise mechanism remains inconclusive (Kopan et al., 1994; Lindsell et al., 1995). Weintraub and colleagues showed that NICD could inhibit the activity of MyoD and Myf-5, arguing that Notch signaling targeted the muscle regulatory factors (MRFs) (Kopan et al., 1994). It was shown subsequently that overexpression of the Notch targets Hes1 or Hey1 can inhibit MyoD, possibly through formation of inactive Hes1/MyoD or Hey1/MyoD heterodimers (Sasai et al., 1992; Sun et al., 2001). However, it is not known if this occurs with physiological levels of Notch signaling and Hes1/Hey1 protein expression. In one study Notch signaling blocked differentiation of C2C12 myoblasts, but did not appreciably induce Hes1, and constitutive expression of Hes1 did not block differentiation (Shawber et al., 1996). In another study in which Notch signaling was initiated by the ligand Dll1, Hes1 expression was found to oscillate, yet signaling led to a rapid, sustained disappearance of MyoD RNA, rendering any specific effects of Notch or of Hes1 on MyoD activity irrelevant (Kuroda et al., 1999). The inhibitory effect of Notch was reversed by overexpressing MyoD, arguing further that it was the reduction of MyoD expression, not MyoD activity, that elicited Notch’s effects.
The ability of Notch to inhibit myogenesis is critical for the expansion of muscle progenitors. Mice carrying a hypomorphic allele of the Notch ligand Delta-like1 (Dll1) or a conditional deletion of CSL display severe muscle hypotrophy due to premature differentiation of the progenitor cell pool (Schuster-Gossler et al., 2007; Vasyutina et al., 2007). Notch signaling has also been linked to muscle regeneration (Conboy and Rando, 2002). Muscle injury results in the induction of Dll1, which promotes the proliferation of muscle stem cells, known as satellite cells (SCs). Subsequent induction of Numb, an inhibitor of Notch signaling, correlates with differentiation of the expanded SC pool into myotubes. Forced expression of NICD in cultured SCs led to a population of cells with down-regulated MyoD and Myf-5, again suggesting that Notch inhibits muscle cell differentiation by reducing MRF expression. Poor induction of Dll1 correlates with the reduced ability of older mice to mobilize SCs and to repair muscle (Conboy et al., 2003).
In light of the important role now appreciated for Notch in controlling the proper timing and execution of skeletal myogenesis in-vivo, we have revisited the unresolved question of how Notch activity represses the myogenic program. In this report we investigated the pathways through which Notch exerts its inhibition. Constitutive expression of Hey1, one of two Hey genes robustly induced by Notch in myoblasts, significantly repressed C2C12 differentiation. Knockdown of Hey1, however, had no impact on Notch-mediated repression, suggesting the presence of additional effectors downstream of the pathway. Using a microarray expression screen, we identified a large number of novel Notch targets in C2C12 cells. We demonstrated that constitutive expression of at least one of these, MyoR, was also sufficient to inhibit differentiation, but that knockdown of MyoR did not impair Notch activity. Simultaneous reduction of both Hey1 and MyoR expression levels also failed to rescue inhibition by Notch. These results argue that Notch acts through multiple pathways—two of which are defined by Hey1 and MyoR—to repress myogenic differentiation.
MATERIALS AND METHODS
Plasmids
Plasmids expressing the extracellular domains of Dll4 and Trail Receptor4 as Fc-fusion proteins were provided by Dr. Marion Dorsch (Millennium Pharmaceuticals, Cambridge, MA). Retroviral vectors encoding FLAG-tagged Hey1 and HeyL were generated by sub-cloning the murine cDNAs, generously provided by Dr. Eric Olson (University of Texas), into the EcoRI site of the multiple cloning site of pBABE-puro-FLAG. Retroviral vectors for Nrarp, Trib2, and MyoR were generated by PCR amplification of the respective cDNAs from Notch ligand-stimulated C2C12 myoblasts followed by insertion into pBABE-puro at the BamHI/SalI sites (Nrarp) or the BamHI/EcoRI sites (Trib2, MyoR). FLAG-tagged Nrarp and Trib2 were generated by PCR-subcloning the respective cDNAs into the SalI site (Nrarp) or EcoRI site (Trib2) of pBABE-puro-FLAG. G22Riken cDNA was obtained from Invitrogen (clone 2649431) and sub-cloned by PCR into the BamHI/EcoRI sites of pBABE-puro. All plasmids generated by PCR were verified by sequencing.
Cell culture
C2C12 myoblasts and 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) supplemented with L-glutamine and penicillin-streptomycin (growth medium, GM). Human skeletal muscle myoblasts (SkMC and HSMM) were purchased from Cambrex and maintained as directed by the manufacturer. For differentiation of myoblasts, cells were grown to near confluence on immobilized ligand (see below), and then shifted to DMEM containing 0.5% FBS (differentiation medium, DM).
Notch signaling was induced by exposing the cells to immobilized ligand. Conditioned medium was first prepared from 293T cells transfected with plasmids coding for fusion proteins between Fcγ of human IgG and either the extracellular domain of Notch ligand Delta-like-4 (Fc-Dll4) or that of Trail Receptor 4 (Fc-control). Culture plates were initially coated for 1 hour at room temperature with 10 μg/ml anti-Fc antibody (Jackson Immunoresearch, West Grove, PA). The anti-Fc PBS solution was then aspirated and replaced by filtered conditioned medium described above. Following 1 hour incubation, supernatant was aspirated and cells were plated.
Microarray expression screen
2.5×106 C2C12 cells were plated on 10cm dishes coated with either Fc-Dll4 or Fc-control ligand (2.5 ml per dish) and grown in GM for 6 hours. RNA was harvested with the RNeasy Kit (Qiagen). Three replicates were included for each condition. RNA was submitted to the University of Pennsylvania Microarray Core Facility for subsequent transcript profiling analysis on Affymetrix MOE430v2.0 GeneChip arrays. Raw data was processed at the Penn Bioinformatics Core Facility using Array Assist Lite (Stratagene), Spotfire (Tibco), and Significance Analysis of Microarrays (Stanford University).
Retroviral infections
Infections were performed as previously described (Pear et al., 1993) with minor modifications. Briefly, retroviral supernatants were harvested from 293T cells two days following FuGENE6 (Roche)-mediated transfection with 8 μg of the indicated pBABE vector and 2 μg of gag/pol and env helper plasmids. Supernatants were filtered (0.4 μm) to remove non-adherent 293T cells prior to direct use or storage at −80°C. 18–24 hours prior to infection, C2C12 cells were plated on 6-well plates at a density of ~1×105 cells/well. Each well was incubated for 4–6 hours with 1.5 mL viral supernatant supplemented with 8 μg/mL polybrene. 24–48 hours following infection and subsequent re-plating on 10cm dishes, selection was initiated with 2 μg/mL puromycin and continued for 3–5 days to obtain stable lines.
siRNA knockdown
C2C12 cells were transfected with 100–150 nM of the indicated SMARTpool siRNA oligonucleotides purchased from Dharmacon. Transfections were performed as specified by the manufacturer using the Dharmafect#3 reagent. Briefly, myoblasts were plated on 12-well dishes at a density of ~1×104 cells per well the day prior to transfection. One day post-transfection, cells were trypsinized and re-plated on ligand-coated 12-well plates. Wells were coated with 400 μl of Fc-control supernatant and either 15 μl (Fig. 3 & S1A), 100 μl (Fig. 7A & S1B), or 80 μl (Fig. 7B & S2A) of Fc-Dll4 supernatant; the total ligand volume on Fc-Dll4-coated wells was kept constant (400 μl) by mixing Fc-control supernatant as required. Cultures were switched from GM to DM one day following re-plating and harvested for RNA after an additional 24–72 hours as indicated. For the double knockdown, a mixture of 110 nM Hey1 siRNA and 40 nM MyoR siRNA was employed.
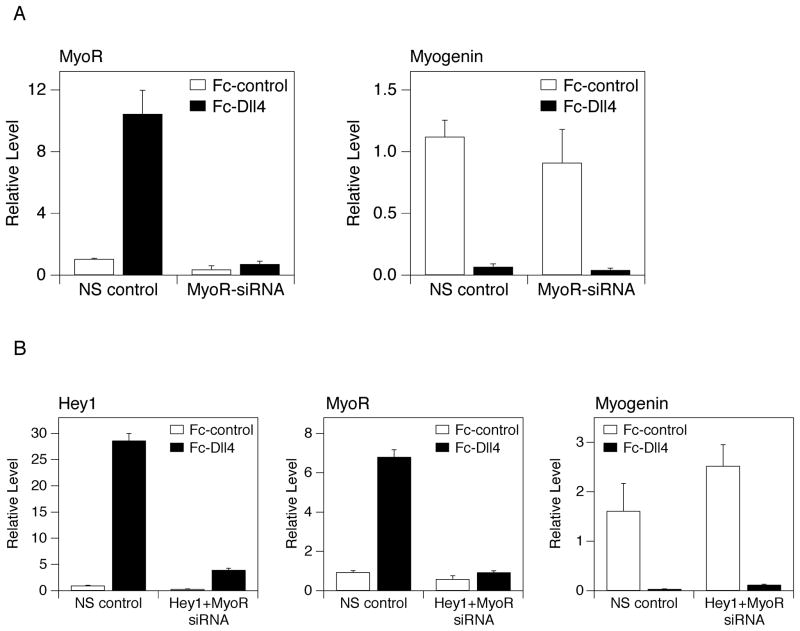
Figure 3.
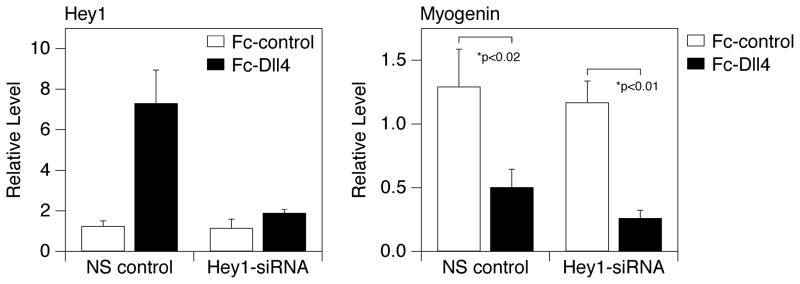
Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of Hey1. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or Hey1-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of Hey1 (left panel) and Myogenin (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. p values were computed by a standard unpaired t-test.
Figure 7.
(A) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of MyoR. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or MyoR-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of MyoR (left panel) and Myogenin (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. (B) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of Hey1 and MyoR. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or a mixture of Hey1-directed and MyoR-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of Hey1 (left panel), MyoR (middle panel), and Myogenin (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples.
Semi-quantitative and quantitative RT-PCR
Total RNA was isolated from C2C12 cultures using the RNeasy kit (Qiagen). 0.125 to 2 μg of RNA was used to generate cDNA with the High Capacity cDNA Archive Kit (Applied Biosystems). For semi-quantitative RT-PCR, 5% of the cDNA was included in each PCR reaction. Products were run out on 1.5% agarose gels and visualized by ethidium bromide staining. For quantitative PCR, TaqMan gene expression assays were employed for MyoD, Myf-5, Myogenin, Mef2C, Myh3, Hey1, Hey2, HeyL, MyoR, IL-6, Id3, ABF-1, and 18S as an endogenous control (Applied Biosystems). 1–4% of a given cDNA reaction, 10 μl of 2X Taq Universal Mastermix, and 1 μl of the indicated 20X TaqMan assay were included in a 20 μl reaction volume per well. All reactions were performed in triplicate. Results were analyzed using the SDS2.2 Software (Applied Biosystems). Primer sequences used for SQ-RT-PCR are as follows:
HPRT 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ and 5′-TGGGGACGCAGCAACTGACATTTCT-3′;
Myogenin 5′-GCGGACTGAGCTCAGCTTAAG-3′ and 5′-GCTGTCCACGATGGACGTAAG-3′;
MEF2A, 5′-TTGGAATGAACAGTCGGAAAC-3′ and 5′-CTAGTCCCTGTGGAGGCAAG-3′;
MEF2C, 5′-GAGAAGCAGAAAGGCACTGG-3′ and 5′-ATCTCACAGTCGCACAGCAC-3′;
MEF2D, 5′-AGCTCTCTGGTCACTCCTTCC-3′ and 5′-GCCCTGGCTGAGTAAACTTG-3′;
GAPDH, 5′-AACGGATTTGGTCGTATTGGG-3′ and 5′-TGGAAGGATGGTGATGGGATTTC-3′;
Hey1 5′-GAAGCGCCGACGAGACCGAATCAA-3′ and 5′-CAGGGCGTGCGCGTCAAAATAACC-3′;
HeyL 5′-GGTCCCCACTGCCTTTGAGA-3′ and 5′-TAGCTGACTGCTCAGGGAAGGCAA-3′;
Nrarp 5′-TGGTGAAGCTGTTGGTCAAG-3′ and 5′-GTAGTTGGCGGGAAGGTACA-3′;
IL-6 5′-CCGGAGAGGAGACTTCACAG-3′ and 5′-GGAAATTGGGGTAGGAAGGA-3′;
Trib2 5′-GCAACATCAACCAAATCACG-3′ and 5′-GCGTCTTCCAAACTCTCCAG-3′;
8430408G22Rik 5′-CTCCTGCCACCCTGACTG-3′ and 5′-TGGGCTGTGACCTTGTCC-3′;
MyoR (Fig. 4) 5′-GCTACGAGGACAGCTATGTGC-3′ and 5′-AGGAGGGCAAACAACACTTG-3′;
MyoR (Fig. 6) 5′-GGGAGGATGCAAGAGGAAG-3′ and 5′-CGTCCAGAGACCACGAATG-3′.
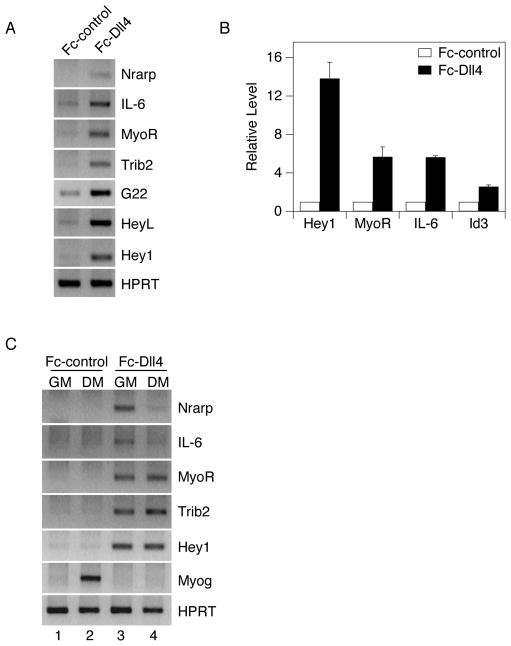
Figure 4.
Validation of microarray targets by RT-PCR. 10 cm dishes were coated with 2.5 ml of ligand-containing supernatant. C2C12 myoblasts were plated on either Fc-Dll4 or Fc-control ligand and propagated in growth medium (GM) for six hours. RNA expression of selected genes was determined by (A) RT-PCR or (B) quantitative RT-PCR using HPRT or 18S as a loading control, respectively. Quantitative RT-PCR values for individual genes are normalized to the Fc-control condition (defined as 1) and plotted as the average +/− standard deviation of three replicate samples. (C) C2C12 cells were plated on Fc-control or Fc-Dll4 ligand, propagated in GM, and switched to DM for 24 hours. RNA expression of indicated targets was analyzed by RT-PCR, using HPRT as a loading control.
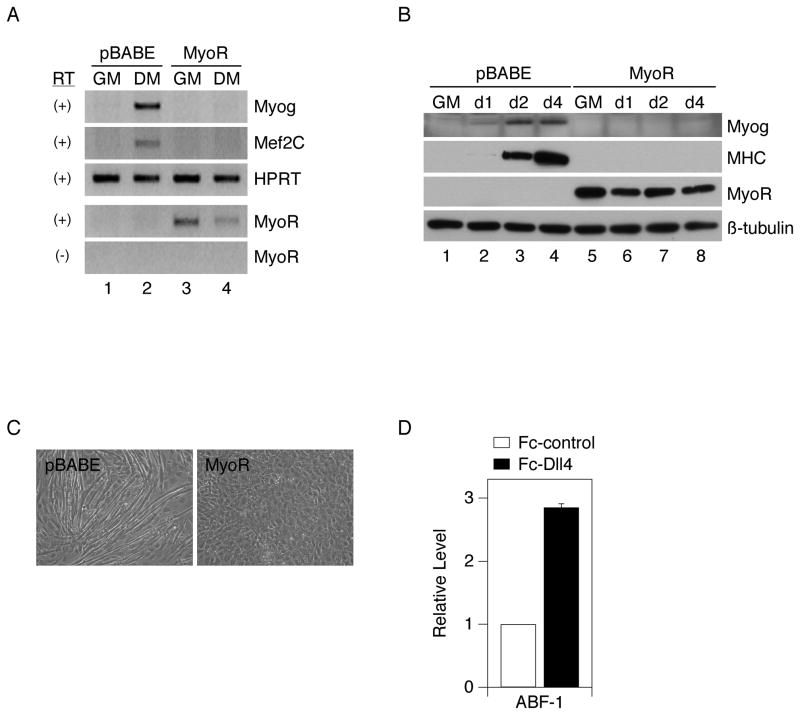
Figure 6.
Constitutive expression of MyoR inhibits myogenesis. C2C12 cells were stably transduced with parental retrovirus (pBABE-puro) or a MyoR-expressing retrovirus (pBABE-MyoR), propagated in growth medium (GM) and then shifted to differentiation medium (DM). Expression levels of the indicated differentiation markers and MyoR were determined by (A) RT-PCR, after 24 hours in DM or (B) Western immunoblot, after 1, 2, or 4 days in DM. (C) Myoblast fusion was examined in pBABE control cells or MyoR-expressing cells after three days in DM. D) ABF-1 (human MyoR) RNA was assessed by quantitative RT-PCR in human myoblasts plated on Fc-Dll4 or Fc-control and propagated in GM. The induction level represents an average +/− standard deviation obtained from two independent myoblast isolations.
Western immunoblot analysis
Protein lysates for Western blots were prepared from C2C12 cells using RIPA lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) or a modified lysis buffer (10 mM Tris pH 7.3, 150 mM NaCl, 1% NP-40) supplemented with freshly added protease inhibitor cocktail (Roche), 10mM sodium fluoride, and 400 μM sodium orthovanadate. Lysates were incubated on ice for 15 minutes and cleared by centrifugation. Protein concentrations were determined using the DC Assay (Bio-Rad). 25–50 μg of lysate was added to 2X or 6X SDS sample buffer and boiled for 5 minutes prior to analysis by SDS-PAGE. Proteins were transferred to nitrocellulose and blotted with the following antibodies at the indicated dilutions: 1:1000 β-tubulin (Sigma T-5293), 1:500 Myogenin (Santa Cruz M-225), 1:20 MHC (MF20, Developmental Studies Hybridoma Bank), 1:2000 cleaved Notch (Cell Signaling 2421), 1:500 FLAG (Abcam ab6711-200) or 1:1000 FLAG (Sigma M2), 1:500 MyoR (Santa Cruz M-20). After incubation with 1:2000 dilutions of HRP-conjugated anti-rabbit, anti-mouse, or anti-goat secondary antibodies (Amersham), bands were visualized via the LumiLight or LumiLight-plus detection system (Roche).
RESULTS
Ligand-induced Notch signaling blocks an early step in myogenesis
To obtain physiological levels of Notch signaling, we exposed cells to Notch ligands. Specifically, we grew cells in the presence of Fc-fusion proteins, containing either the extracellular domain of Delta-like4 (Fc-Dll4) or Jagged1 (Fc-Jag1), adhered to the surface of tissue culture dishes with an anti-Fc antibody (Varnum-Finney et al., 2000). Plating C2C12 myoblasts on Fc-Dll4, but not on a control Fc-fusion protein (Fc linked to a portion of the Trail receptor 4; dubbed “Fc-control” throughout), led to a robust generation of NICD (Fig. 1A). When transferred from high serum (GM, growth medium) to low serum (DM, differentiation medium), cells grown on Fc-Dll4 were unable to form myotubes (data not shown) or to induce the expression of Myogenin, an early marker of myogenesis (Fig. 1A). Similar results were obtained using Fc-Jag1 as a Notch ligand. C2C12 cells whose differentiation is blocked by Notch signaling remain myoblasts since they retain the ability to form myotubes when transferred to normal culture dishes (data not shown).
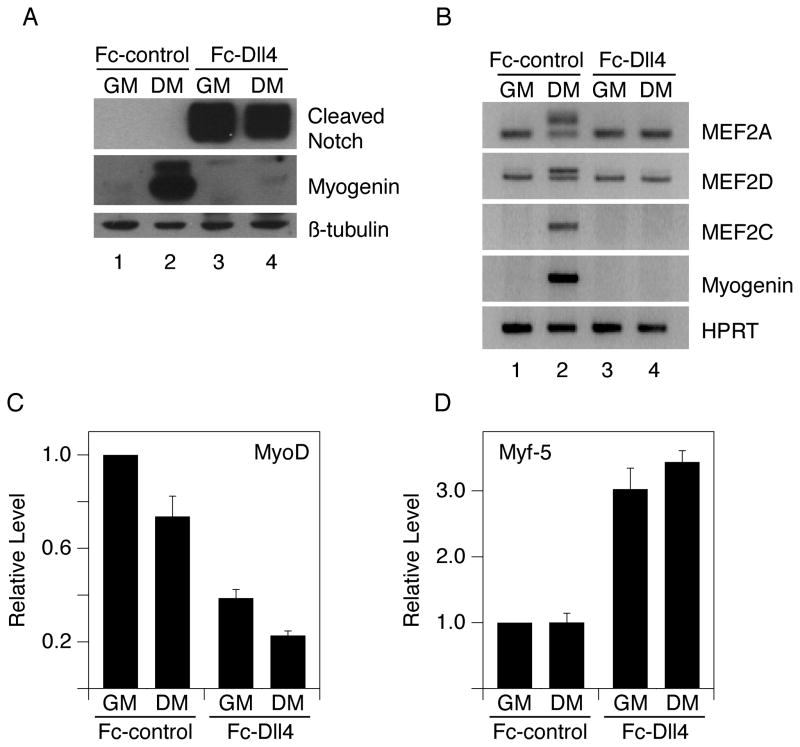
Figure 1.
Ligand-induced Notch signaling blocks myogenesis. 6-well plates were coated with 4.5 ml of ligand-containing supernatant per well. C2C12 cells were grown on Fc-Dll4-coated or Fc-control-coated plates and switched from growth medium (GM) to differentiation medium (DM) as indicated. Cells were analyzed after 24 hours for A) cleaved Notch1 and Myogenin proteins (Western); B) MEF2A, MEF2D, MEF2C and Myogenin RNAs (RT-PCR); C) MyoD and D) Myf-5 RNAs (quantitative RT-PCR). MyoD and Myf-5 levels shown in (C–D) are normalized to the Fc-control-GM condition (defined as 1) and plotted as the average +/− standard deviation of two replicate samples. The upper bands of the MEF2A and MEF2D RNA doublets are the differentiation-induced splice variants. β-tubulin protein and HPRT or 18S RNA were used as loading controls.
Notch signaling also blocked the induction of RNAs encoding Mef2C and the splicing isoforms of Mef2A and Mef2D normally induced upon muscle differentiation (Fig. 1B; (Zhu et al., 2005)). Expression of MyoD RNA was reduced in cells exposed to Notch ligand, but a significant level (30–40% of that observed in cells plated on Fc-control) still remained (Fig. 1C). Additionally, Myf-5 RNA levels were induced by approximately three-fold in cells grown on Fc-Dll4 (Fig. 1D). These data confirm the results of others that ligand-mediated Notch signaling blocks myogenesis (Lindsell et al., 1995), but challenge the notion that down-regulation of MyoD or Myf-5 expression is the primary mechanism responsible for this inhibition, as proposed previously (Conboy and Rando, 2002; Kuroda et al., 1999). They also indicate that direct inhibition of MEF2C activity by NICD (Wilson-Rawls et al., 1999) is less likely to be important, as Notch signaling acts prior to the induction of Mef2C RNA. Based on our results, we propose that Notch functions to antagonize MyoD activity and thereby repress the induction of Myogenin and Mef2C, two critical downstream mediators of MyoD in the myogenic program (Cheng et al., 1993; Wang et al., 2001).
Although Notch signaling is thought to stimulate satellite cell proliferation (Conboy and Rando, 2002), Fc-Dll4 had only a minor effect on the ability of C2C12 cells to exit the cell cycle when transferred to DM (data not shown). Furthermore, forced expression of p21 in C2C12 cells, while causing a complete block in G1, did not relieve the inhibition of myogenesis by Fc-Dll4 (data not shown). We conclude that Notch does not block differentiation simply by preventing cell cycle exit.
Constitutive expression of Hey1 recapitulates the early block to myogenesis
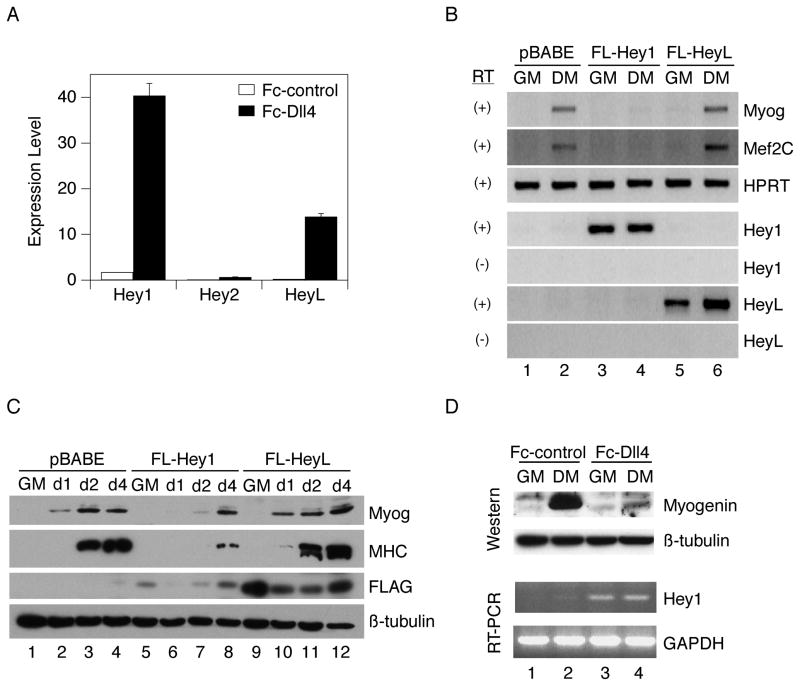
We next determined if members of the Hey or Hes family of transcriptional repressors might mediate the effects of Notch in C2C12 cells. We found that while all three Hey family members (Hey1, Hey2 and HeyL) were induced as a consequence of Notch signaling, the overall level of Hey2 was extremely low (Fig. 2A), consistent with a previous report (Iso et al., 2001). Members of the Hes family regulated by Notch in other cell types (Hes1, Hes5, Hes7) were not appreciably induced (data not shown). To determine if constitutive expression of either Hey1 or HeyL could mimic the effect of Notch on differentiation, we used retroviral vectors to express FLAG-tagged versions of these repressors in C2C12 cells. Cells expressed comparable amounts of Hey1/HeyL RNAs; however, when we assessed differentiation, only Hey1 blocked induction of Myogenin and Mef2C transcripts (Fig. 2B) and reduced myoblast fusion (data not shown). Western analysis demonstrated that FLAG-Hey1 and FLAG-HeyL proteins were indeed expressed (the indicated bands migrated at the expected mobility of 40–45 kD), albeit at a low level in the case of Hey1 (Fig. 2C). Induction of both Myogenin and Myosin heavy chain (MHC) proteins occurred normally in the presence of constitutively expressed HeyL, whereas in Hey1-expressing cells, these markers were completely repressed at early time-points and only became detectable by day 4. While we also observed that a FLAG-tagged version of Hey2 exhibited the ability, like Hey1, to inhibit Myogenin induction (data not shown), we do not consider Hey2 a major player in this system, given that ligand-mediated stimulation induces only a negligible amount of Hey2 transcription. Fc-Dll4 also induced Hey1 in primary human myoblasts, and this correlated with a block to Myogenin induction (Fig. 2D). We conclude that constitutive expression of Hey1 strongly inhibits early inductive events of myogenesis and ultimately delays the course of differentiation.
Figure 2.
Ligand-induced Notch signaling significantly induces Hey1 and HeyL expression, but only constitutively expressed Hey1 blocks myogenesis. A) 6-well plates were coated with 3 ml of ligand-containing supernatant per well. C2C12 cells were plated on Fc-Dll4 or Fc-control ligand and propagated for 48 hours in growth medium (GM). Hey1, Hey2 and HeyL RNA levels were determined by quantitative RT-PCR using 18S as a loading control. Expression levels (x) for individual genes were computed from ΔCt values (relative to 18S) according to the formula (x) * 2ΔCt = (c), where c is an arbitrary constant, and plotted as the average +/− standard deviation of three replicate samples. B) C2C12 cells were stably transduced with parental retrovirus (pBABE-puro) or retroviruses expressing FLAG-tagged Hey1 or HeyL and analyzed for expression of the indicated cDNAs by RT-PCR. RT, reverse transcriptase. C) Transduced cells were propagated in GM, shifted to DM and analyzed for expression of Myogenin, MHC or FLAG-tagged proteins after 1, 2, or 4 days by Western immunoblotting using β-tubulin as a loading control. D) Human myoblasts were grown on Fc-Dll4-coated or Fc-control-coated plates and switched from GM to DM for 24 hours as indicated. Myogenin protein was assessed by Western immunoblotting using β-tubulin as a loading control, and Hey1 RNA was determined by RT-PCR using GAPDH as a loading control.
We next asked if Hey1 induction is necessary for Notch to inhibit myogenesis. We reasoned that if we sufficiently reduced the level of Hey1 expression we might observe normal Myogenin induction despite ongoing Notch signaling. Transfection of C2C12 cells with siRNAs directed against Hey1, relative to control siRNAs, did not appreciably affect the low level of Hey1 RNA in cells plated on Fc-control (this was somewhat variable across multiple experiments; see Fig. 7), but led to a significant reduction (~75 percent) in Hey1 expression when cells were plated on Fc-Dll4 (Fig. 3, left). However, the induction of Myogenin (Fig. 3, right) and two other myogenic markers, Mef2C and Myosin heavy polypeptide 3 (Myh3) (Fig. S1A), was still inhibited. This result argues that the high levels of Hey1 expression induced by Notch are not necessary for the inhibition of myogenesis, and that other, potentially redundant pathways may contribute.
Identification of novel Notch targets in C2C12 cells
To identify additional effectors downstream of Notch, we performed a microarray-based expression screen using C2C12 cells. Myoblasts were plated on either Fc-Dll4 or Fc-control ligand and maintained in growth medium (GM) for six hours prior to isolation of RNA for expression profiling. The six-hour time-point was chosen to bias the screen towards the detection of direct (early) targets of the pathway. Activation of Notch was verified by Western blot of protein lysates harvested from a parallel set of cultures using an antibody specific for cleaved Notch (data not shown). RNA was submitted to the Penn Microarray Core Facility for processing on Affymetrix MOE430v2.0 gene chips.
With a false discovery rate set to 0% and a fold change cutoff of two, the microarray identified 82 transcripts that were induced and five transcripts that were repressed by Notch ligand stimulation. The top 30 induced genes are shown in Table 1. This observed skewing towards gene activation as opposed to repression suggests that our list may indeed contain a large number of direct targets of the pathway, since Notch is primarily thought to function as a transcriptional activator. Three known Notch targets—Hey1, HeyL, and Nrarp—were at the top of the list of induced genes. Among the most highly induced were the cytokine interleukin-6 (IL-6), the transcription factor MyoR, the kinase-like protein Tribbles2 (Trib2), and the RIKEN cDNA 8430408G22 (G22).
TABLE 1.
Genes induced by Notch in C2C12 myoblasts.
| Affymetrix ID | Fold changea | Gene Symbol | Description | ||
|---|---|---|---|---|---|
| 1 | 1417985_at | 14.02 | [C] | Nrarp | Notch-regulated ankyrin repeat protein |
| 2 | 1419302_at | 10.86 | [C] | Heyl | hairy/enhancer-of-split related with YRPW motif-like |
| 3 | 1415999_at | 7.818 | [C] | Hey1 | hairy/enhancer-of-split related with YRPW motif 1 |
| 4 | 1450297_at | 6.389 | [C] | Il6 | interleukin 6 |
| 5 | 1418417_at | 5.204 | [C] | Msc | musculin [MyoR] |
| 6 | 1426640_s_at | 4.982 | [C] | Trib2 | expressed sequence AW319517 [Tribbles2] |
| 7 | 1433837_at | 4.743 | [C] | 8430408G22Rik | RIKEN cDNA 8430408G22 [G22] |
| 8 | 1439794_at | 4.483 | [C] | Ntn4 | netrin 4 |
| 9 | 1418937_at | 4.324 | [NT] | Dio2 | deiodinase, iodothyronine, type II |
| 10 | 1421106_at | 4.066 | [NT] | Jag1 | jagged 1 |
| 11 | 1436033_at | 3.979 | [C] | BC031353 | cDNA sequence BC031353 |
| 12 | 1420757_at | 3.328 | [C] | Myf5 | myogenic factor 5 |
| 13 | 1436570_at | 3.242 | [NT] | BG143461 | EST mab56d07.x1 |
| 14 | 1434140_at | 3.151 | [C] | Mcf2l | mcf.2 transforming sequence-like |
| 15 | 1424530_at | 3.122 | [C] | Sec14l2 | SEC14-like 2 (S. cerevisiae) |
| 16 | 1448886_at | 3.104 | [NC] | Gata3 | GATA binding protein 3 |
| 17 | 1434430_s_at | 3.093 | [NT] | Adora2b | adenosine A2b receptor |
| 18 | 1440990_at | 2.994 | [C] | 4832420M10 | hypothetical protein 4832420M10 |
| 19 | 1450922_a_at | 2.954 | [NC] | Tgfb2 | transforming growth factor, beta 2 |
| 20 | 1428250_at | 2.927 | [C] | Gpr30 | G protein-coupled receptor 30 |
| 21 | 1435888_at | 2.923 | [C] | Egfr | epidermal growth factor receptor |
| 22 | 1440761_at | 2.85 | [NT] | 4833422C13Rik | RIKEN cDNA 4833422C13 |
| 23 | 1417122_at | 2.71 | [C] | Vav3 | vav 3 oncogene |
| 24 | 1416613_at | 2.699 | [NT] | Cyp1b1 | cytochrome P450, family 1, subfamily b, polypeptide 1 |
| 25 | 1438531_at | 2.656 | [NT] | A730054J21Rik | RIKEN cDNA A730054J21 |
| 26 | 1454768_at | 2.618 | [NT] | Kcnf1 | potassium voltage-gated channel, subfamily F, member 1 |
| 27 | 1429021_at | 2.601 | [NT] | Epha4 | Eph receptor A4 |
| 28 | 1425814_a_at | 2.595 | [NT] | Calcrl | calcitonin receptor-like |
| 29 | 1416630_at | 2.579 | [C] | Id3 | inhibitor of DNA binding 3 |
| 30 | 1456830_at | 2.557 | [NT] | Ppp1r2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 |
Targets subsequently confirmed by RT-PCR are marked “C”, those not confirmed are marked “NC”, and those not tested are marked “NT”.
We validated a subset of the Notch-responsive genes by RT-PCR. IL-6, MyoR, Trib2, G22, and the known targets Nrarp, Hey1, and HeyL, were all induced to varying extents in C2C12 cells exposed to Fc-Dll4 after six hours (Fig. 4A). Quantitative RT-PCR provided additional confirmation and revealed fold changes very similar to, or greater than, those reported by the array for Hey1, MyoR, IL-6, and an additional responsive gene, Id3 (Fig. 4B). In total, seventeen of the 30 most highly induced genes were confirmed by RT-PCR, two were not confirmed, and eleven were not tested (Table 1). Further studies demonstrated that the induction of three genes (MyoR, Trib2, and Hey1), was maintained upon serum withdrawal, while Nrarp and IL-6 expression was reduced (Fig. 4C). This result suggests that the expression of certain genes appears to require the combined actions of Notch and undefined factors present in serum.
Expression of a subset of Notch target genes inhibits C2C12 differentiation
We next asked if constitutive expression of any of our newly identified Notch-responsive genes could inhibit myogenesis. We chose to focus initially on a subset of those most highly induced: Nrarp, Trib2, G22, and IL-6. Nrarp, Notch-regulated ankyrin repeat protein, has been shown to function as a feedback inhibitor of the pathway in Xenopus and zebrafish, but was also reported to augment Notch-mediated transcriptional activation in cultured cells (Ishitani et al., 2005; Lamar et al., 2001). Trib2 is a kinase-like protein implicated in the pathogenesis of acute myelogenous leukemia and also reported to inhibit phosphorylation of Akt, a signaling molecule important in myogenesis (Du et al., 2003; Heron-Milhavet et al., 2007; Keeshan et al., 2006; Naiki et al., 2007; Wilson and Rotwein, 2007). Interleukin-6 is an inflammatory cytokine that is expressed in regenerating muscle and may promote satellite cell proliferation (Cantini et al., 1995; Kami and Senba, 1998).
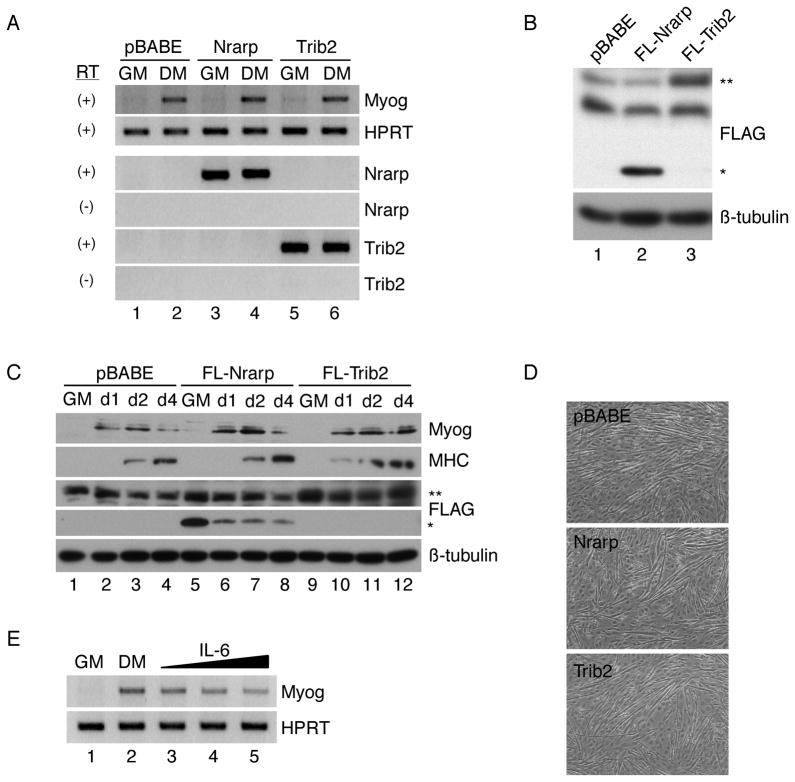
cDNAs for Nrarp and Trib2 were cloned into the pBABE-puro retroviral expression vector, and individual viruses were used to infect C2C12 myoblasts. Resulting stable cell lines clearly expressed the specified transcripts, but exhibited normal induction of the early differentiation marker Myogenin when deprived of serum for 24 hours (Fig. 5A). Similar results were obtained when cells were infected with a retrovirus expressing the G22 cDNA (data not shown). C2C12 cells were also infected with retroviruses encoding FLAG-tagged Nrarp or Trib2, and protein expression was verified by Western blot (Fig. 5B–C). Bands migrating at the appropriate mobility were detected in cells transduced with either of the two retroviruses, while the FLAG-Trib2 signal ran at the same mobility as a non-specific background band (FLAG-Nrarp, ~20 kD (*), FLAG-Trib2, ~45 kD (**)). Induction of Myogenin and MHC proteins occurred normally in FLAG-Nrarp and FLAG-Trib2 cell lines over four days in DM (Fig. 5C). Fusion of myoblasts into myotubes after three days in differentiation medium was also unimpaired relative to that observed in the pBABE control cultures (Fig. 5D). Nrarp-transduced cells were also evaluated for their ability to induce Hey1 when cultured on Fc-Dll4, testing the hypothesis that Nrarp is a feedback inhibitor of Notch signaling. While the induction of Hey1 was moderately reduced, this occurred at a level of Nrarp that exceeded that typically induced by ligand (data not shown). Thus, Nrarp does not appear to exert a significant effect on overall Notch signaling in our system.
Figure 5.
Constitutive expression of Nrarp or Trib2 does not impair myogenesis, while IL-6 inhibits at high doses. (A) C2C12 cells were stably transduced with parental retrovirus (pBABE-puro) or retroviruses expressing Nrarp or Trib2. Lines were propagated in growth medium (GM) and then shifted to differentiation medium (DM) for 24 hours and analyzed for expression of Myogenin or the indicated cDNAs by RT-PCR using HPRT as a loading control. RT, reverse transcriptase. (B) C2C12 cells were stably transduced with parental retrovirus (pBABE-puro) or retroviruses expressing FLAG-tagged Nrarp or Trib2. Lines were propagated in GM and analyzed for expression of FLAG-tagged proteins by Western immunoblotting. (*) indicates the position of FLAG-Nrarp, and (**) indicates the position of FLAG-Trib2. (C) Transduced cells were propagated in GM, shifted to DM and analyzed for expression of Myogenin, MHC or FLAG-tagged proteins after 1, 2, or 4 days by Western immunoblotting using β-tubulin as a loading control. (D) Fusion of myoblasts into myotubes was examined in the indicated cell lines after three days in DM. (E) C2C12 cells were maintained in GM and then switched to DM for 24 hours in the absence or presence of increasing concentrations of IL-6 (2.8, 10 and 100ng/mL). Myogenin RNA levels were assessed by RT-PCR using HPRT as a loading control.
We also asked if IL-6 affects myogenesis. We observed a modest dose-dependent inhibition of Myogenin induction after serum withdrawal when cells were bathed in increasing concentrations of IL-6, with maximum inhibition of approximately two-fold occurring at a dose of 100 ng/ml (Fig. 5E). Given that this effect occurred only at high concentrations of the cytokine, which are likely to be supra-physiological, we are hesitant to ascribe a major role to IL-6 in mediating the effects of Notch in muscle, but do not rule out a potential contributory influence.
In contrast to the findings for Nrarp, Trib2, and G22, forced retroviral expression of MyoR resulted in a complete block to C2C12 myogenesis, consistent with a previous report that has implicated this bHLH protein as negative regulator of MyoD and of myogenic conversion of fibroblasts (Lu et al., 1999). MyoR-expressing myoblasts failed to induce Myogenin or Mef2C transcripts at 24 hours after serum withdrawal (Fig. 6A), showed no induction of Myogenin or MHC proteins over four days in differentiation medium (Fig. 6B), and exhibited no evidence of fusion after three days in DM (Fig. 6C). Expression of MyoR in these cells was verified by RT-PCR and Western blot analysis (Fig. 6A–B). In parallel with these findings, we observed that ligand-mediated Notch signaling also induced expression of ABF-1, the human orthologue of MyoR, in primary human myoblasts (Fig. 6D). We conclude that Notch signaling strongly induces the expression of Nrarp, Trib2, G22, IL-6, and MyoR, but of these, only constitutively expressed MyoR is sufficient to recapitulate the inhibition imposed by the pathway as a whole.
The precise manner by which MyoR expression is activated by Notch signaling remains an open question. Importantly, we have excluded the possibility that MyoR induction occurs downstream of Hey1, as MyoR RNA levels were essentially unchanged under conditions of Hey1 retroviral expression (data not shown). Bioinformatic analysis failed to reveal any conserved CSL binding sites within the MyoR 2kb proximal promoter, which was found to be unresponsive to NICD in luciferase reporter assays, but a far-upstream potential enhancer region containing three conserved CSL sites was responsive to NICD. However, mutation of these sites did not compromise NICD-responsiveness (data not shown). Accordingly, MyoR could well be an indirect target of Notch, but further studies will be needed to clearly define its mode of regulation.
Knockdown of MyoR alone or in combination with Hey1 does not impair Notch activity
Given that constitutive expression of either Hey1 or MyoR mimicked the inhibitory effects of Notch in C2C12 cells, it appeared that Notch was acting through multiple pathways, and that perhaps no single gene target would be required for Notch to exert repression. To further test this notion, we performed an additional siRNA knockdown experiment to reduce the level of MyoR. Transfection of C2C12 cells with MyoR siRNAs, relative to control siRNAs, led to a significant reduction (>90 percent) in MyoR expression when cells were plated on Fc-Dll4 (Fig. 7A, left). In this experiment, a higher dose of Fc-Dll4 supernatant was employed relative to that depicted in Figure 3 to obtain robust induction of MyoR, a less sensitive Notch target than Hey1. The reduced level of MyoR, however, did not compromise the ability of Notch to repress the induction of Myogenin (Fig. 7A, right), or of two additional markers, Mef2C and Myh3 (Fig. S1B). This result is consistent with our previous data indicating that Hey1 appears sufficient to account for the effects of Notch on early myogenesis; in the absence of MyoR, Hey1 would be expected to compensate.
To investigate the existence of any additional MyoR- and Hey1-independent pathways, we employed siRNAs to simultaneously knock-down both Hey1 and MyoR expression. Transfection of C2C12 cells with this mixture of siRNAs resulted in >85 percent reduction of both Hey1 and MyoR RNA levels when cultures were plated on Fc-Dll4 (Fig. 7B, left & middle). Despite the drastically reduced levels of Hey1 and MyoR, exposure to Fc-Dll4 still effectively repressed induction of Myogenin at 24 hours in DM (Fig. 7B, right). Similar results were obtained when cultures were taken out to three days in DM and analyzed for Myh3 induction (Fig. S2A), with the caveat that knock-down efficiency had declined to ~65 percent. Myoblast fusion at 3–4 days in DM also continued to be repressed in cultures treated with Hey1/MyoR siRNAs and plated on Fc-Dll4 (Fig. S2B). While we cannot rule out the possibility that low levels of residual Hey1 or MyoR are sufficient to block myogenesis, our data suggest that Notch signaling inhibits myogenesis through multiple pathways, and that yet additional mediators beyond Hey1 and MyoR are likely to contribute to the pathway’s biological effects in our system.
DISCUSSION
Notch signaling was shown over a decade ago to inhibit myogenesis in cultured cells and more recently to prevent the premature differentiation of muscle progenitor cells and satellite cells in-vivo (Conboy and Rando, 2002; Kopan et al., 1994; Lindsell et al., 1995; Schuster-Gossler et al., 2007; Vasyutina et al., 2007). However, the molecular pathways through which Notch exerts its inhibitory effects have not been clearly defined. Here we provide evidence for the existence of multiple pathways. Notch induced the expression of a multitude of genes in cultured myoblasts, and individual constitutive expression of at least two of them, Hey1 and MyoR, was sufficient to block (or significantly delay) myogenesis. Consistent with a model in which no single factor downstream of Notch is required for myogenic inhibition, siRNA knockdown experiments directed against either Hey1 or MyoR revealed that significantly reducing the dosage of either of these factors had no appreciable effect on the ability of Notch to exert repression. Intriguingly, even simultaneous knockdown of both Hey1 and MyoR did not appear to rescue repression by Notch in any substantial fashion, suggesting the existence of additional contributory factors downstream of the pathway.
Notch affects myogenesis at an early step. This most likely reflects its ability to antagonize MyoD activity, though not simply via down-regulation of MyoD expression, as suggested previously (Kuroda et al., 1999). We observed that exposing cells to Notch ligand resulted in only a partial reduction of MyoD RNA and led to a three-fold increase in Myf-5 transcripts. Moreover, while Hey1 and MyoR both blocked myogenesis, neither significantly affected MyoD RNA levels (M.B., unpublished observation). These results argue against a model in which down-regulation of MyoD expression is the primary mechanism underlying Notch’s inhibitory activity. We are currently exploring the mechanisms by which Hey1 and MyoR interfere with myogenic transcription in cultured cells. A previous study reported the formation of inactive Hey1/MyoD heterodimers (Sun et al., 2001), but our own results contradict this finding and suggest the possibility of alternate modes of repression (M.B. & S.K., in preparation).
Members of both the Hes and Hey (HRT, HERP, CHF) families of bHLH repressors can be induced by Notch. In agreement with others (Shawber et al., 1996), we found that Hes1 was expressed in C2C12 cells, but poorly induced, and was not effective at blocking myogenesis. Other Hes family members were not appreciably induced. By contrast, all three members of the Hey family, Hey1, Hey2 and HeyL, were induced by Fc-Dll4, but the overall level of Hey2 was very low. Constitutive expression of Hey1 repressed myogenesis while, surprisingly, HeyL had no effect. This argues that, despite a high level of structural similarity, the biological activities of the Hey proteins are distinct. Hey2, when expressed as a FLAG-tagged construct, did exhibit the ability to repress myogenesis (M.B., unpublished observation); however, given the very low level of endogenous Hey2 transcript induced by Fc-Dll4, we do not consider Hey2 to be a significant player in the mediating the effects of Notch in this system. Our results do, however, implicate Hey1 as a potentially important Notch effector in myoblasts. Our data showing that a reduced level of Hey1 has no effect on the response of cells to Notch were therefore unexpected. This was true even at levels of Notch signaling that only partially induced Hey1 and partially restricted Myogenin induction.
In contrast to a simple model of Notch acting primarily to induce the transcription of Hes or Hey family members, our expression screen revealed over 82 transcripts that were upregulated after only six hours of ligand stimulation. Several strongly induced targets did not inhibit myogenesis when tested functionally by constitutive expression (Nrarp, Trib2, G22). However, whether or not these genes play any functional role in muscle remains to be determined. For example, it is possible that these genes exhibit little impact when expressed individually, but will affect myogenesis when expressed in combination. Despite this possibility, our screen suggests that Notch may generally induce a large number of target genes, but employ only specific subsets of these to execute the pathway’s effects in different cell types.
Importantly, our work has identified MyoR as a novel Notch target that appears to contribute to myogenic repression. MyoR was originally identified in a screen for cDNAs with homology to capsulin, another bHLH transcription factor (Lu et al., 1999). MyoR exhibits a skeletal muscle-specific pattern of embryonic expression and has been shown to antagonize the activity of MyoD in reporter assays and bind to E-box DNA elements in vitro. Accordingly, it was suggested that this bHLH inhibitor functions to delay the progression of myogenesis during development. MyoR was also found to be induced during muscle regeneration, arguing for an additional role in satellite cells (Zhao and Hoffman, 2006). ABF-1, the putative human orthologue of MyoR, was cloned concurrently from activated B-cells (Massari et al., 1998), suggesting muscle-independent functions.
Our data are consistent with a model in which Notch signaling acts through at least two myogenic inhibitors, Hey1 and MyoR, to repress myoblast differentiation in culture. It is tempting to speculate that these same proteins may also represent important arms of the Notch pathway during embryonic and/or post-natal myogenesis in-vivo. Impairment of Notch activity results in premature progenitor cell differentiation in the embryo and compromised satellite cell proliferation in the adult (Conboy et al., 2003; Schuster-Gossler et al., 2007; Vasyutina et al., 2007). While knockout mice deficient for either Hey1 or MyoR have been generated and do not exhibit overt embryonic muscle phenotypes (Fischer et al., 2004; Lu et al., 2002), the absence of such defects in single-knockout animals is consistent with our model in which multiple factors downstream of Notch contribute to the pathway’s phenotypic effects. It has been observed that Hey1 and MyoR collaborate with related transcription factors, Hey2 and Capsulin, respectively. Hey1 and Hey2 act redundantly in the embryonic vasculature (Fischer et al., 2004), while MyoR and Capsulin function redundantly in the formation of the facial musculature (Lu et al., 2002). Preliminary in-situ hybridization studies have revealed a striking overlap in the expression patterns of Hey1 and MyoR in E10.5 embryos (M.B & T.K., unpublished observation), a finding consistent with both distinct and coordinate regulation of these two genes.
In light of the results from our double knockdown experiment (Fig. 7B), it appears likely that additional factors beyond Hey1 and MyoR are important in contributing to Notch-mediated inhibition of myogenesis. The large number of target genes identified by our array is consistent with the notion that Notch activates an extensive gene network in order to execute its critical functions in muscle. One additional target gene, Id3, is an HLH protein shown previously to inhibit C2C12 differentiation when expressed constitutively (Atherton et al., 1996). Notch activity has also been linked to Id3 induction in Xenopus (Reynaud-Deonauth et al., 2002). While the level of Id3 activation observed in our system in response to Fc-Dll4 is relatively modest, this factor, in combination with other target genes, such as IL-6, or even the negligible level of Hey2 induced by ligand stimulation, may indeed participate in myogenic repression.
The positioning of activated Notch as the hub of a transcriptional network containing multiple effectors, many of which may contribute functionally to the pathway’s biological effects, is likely to be applicable to tissues other than muscle. Generally, such a framework may serve to render any one particular target of a signaling cascade dispensable for the overall phenotypic consequences of the pathway. The specific targets (nodes) employed by Notch may differ from tissue to tissue, but the principle of functional redundancy could represent a general feature that ensures a robust signaling response. Redundancy and associated robustness are critical attributes of complex physiological systems that enhance their capacity to evolve (Kirschner and Gerhart, 1998).
Supplementary Material
(A) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of Hey1. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or Hey1-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of Mef2C (left panel) and Myh3 (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. (B) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of MyoR. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or MyoR-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of Mef2C (left panel) and Myh3 (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. NOTE: While MHC protein levels were only appreciably induced in control cells by 48 hours in DM, Myh3 RNA was elevated ~300 fold by 24 hours (M.B., unpublished observation).
(A) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of Hey1 and MyoR. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or a mixture of Hey1-directed and MyoR-directed siRNAs were propagated on the coated plates and then shifted to DM for 72 hours. Expression of Hey1 (left panel), MyoR (middle panel), and Myh3 (right panel) RNA was determined by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. (B) Myoblast fusion in cultures treated with either non-silencing (NS) control siRNAs or Hey1/MyoR siRNAs and plated on either Fc-control or Fc-Dll4 was assessed after four days in DM.
Acknowledgments
This work was supported by funds from the Muscular Dystrophy Association (MDA 3888). M.B. was supported by NIH Training Grant 5-T32-GM-008216-20 and S.K. by NIH NRSA Training Grant F32GM068394-01A1. We would like to thank Dr. John Tobias for assistance with microarray analysis and Ruth Walmsley and Yao-juan Liu for technical support.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Atherton GT, Travers H, Deed R, Norton JD. Regulation of cell differentiation in C2C12 myoblasts by the Id3 helix-loop-helix protein. Cell Growth Differ. 1996;7(8):1059–1066. [PubMed] [Google Scholar]
- Cantini M, Massimino ML, Rapizzi E, Rossini K, Catani C, Dalla Libera L, Carraro U. Human satellite cell proliferation in vitro is regulated by autocrine secretion of IL-6 stimulated by a soluble factor(s) released by activated monocytes. Biochem Biophys Res Commun. 1995;216(1):49–53. doi: 10.1006/bbrc.1995.2590. [DOI] [PubMed] [Google Scholar]
- Cheng TC, Wallace MC, Merlie JP, Olson EN. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science. 1993;261(5118):215–218. doi: 10.1126/science.8392225. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302(5650):1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3(3):397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300(5625):1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18(8):901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Steidl C, Wagner TU, Lang E, Jakob PM, Friedl P, Knobeloch KP, Gessler M. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res. 2007;100(6):856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9(5):617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Heron-Milhavet L, Mamaeva D, Rochat A, Lamb NJ, Fernandez A. Akt2 is implicated in skeletal muscle differentiation and specifically binds Prohibitin2/REA. J Cell Physiol. 2007;214(1):158–165. doi: 10.1002/jcp.21177. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Matsumoto K, Chitnis AB, Itoh M. Nrarp functions to modulate neural-crest-cell differentiation by regulating LEF1 protein stability. Nat Cell Biol. 2005;7(11):1106–1112. doi: 10.1038/ncb1311. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21(17):6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami K, Senba E. Localization of leukemia inhibitory factor and interleukin-6 messenger ribonucleic acids in regenerating rat skeletal muscle. Muscle Nerve. 1998;21(6):819–822. doi: 10.1002/(sici)1097-4598(199806)21:6<819::aid-mus20>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Kawamata S, Du C, Li K, Lavau C. Overexpression of the Notch target genes Hes in vivo induces lymphoid and myeloid alterations. Oncogene. 2002;21(24):3855–3863. doi: 10.1038/sj.onc.1205487. [DOI] [PubMed] [Google Scholar]
- Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, Rodriguez CG, Maillard I, Tobias JW, Valk P, Carroll M, Aster JC, Delwel R, Pear WS. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10(5):401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci U S A. 1998;95(15):8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci U S A. 2006;103(24):9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120(9):2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274(11):7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131(5):965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15(15):1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort K, Dotto GP. Notch signaling in the integrated control of keratinocyte growth/differentiation and tumor suppression. Semin Cancer Biol. 2004;14(5):374–386. doi: 10.1016/j.semcancer.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80(6):909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu J, Webb R, Richardson JA, Olson EN. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci U S A. 1999;96(2):552–557. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JR, Bassel-Duby R, Hawkins A, Chang P, Valdez R, Wu H, Gan L, Shelton JM, Richardson JA, Olson EN. Control of facial muscle development by MyoR and capsulin. Science. 2002;298(5602):2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Tommasi di Vignano A, Sharov AA, Neilson J, Havrda MC, Roop DR, Botchkarev VA, Crabtree GR, Dotto GP. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev Cell. 2005;8(5):665–676. doi: 10.1016/j.devcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Massari ME, Rivera RR, Voland JR, Quong MW, Breit TM, van Dongen JJ, de Smit O, Murre C. Characterization of ABF-1, a novel basic helix-loop-helix transcription factor expressed in activated B lymphocytes. Mol Cell Biol. 1998;18(6):3130–3139. doi: 10.1128/mcb.18.6.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T, Saijou E, Miyaoka Y, Sekine K, Miyajima A. TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J Biol Chem. 2007;282(33):24075–24082. doi: 10.1074/jbc.M701409200. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18(8):2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud-Deonauth S, Zhang H, Afouda A, Taillefert S, Beatus P, Kloc M, Etkin LD, Fischer-Lougheed J, Spohr G. Notch signaling is involved in the regulation of Id3 gene transcription during Xenopus embryogenesis. Differentiation. 2002;69(4–5):198–208. doi: 10.1046/j.1432-0436.2002.690413.x. [DOI] [PubMed] [Google Scholar]
- Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21(20):2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- Ross DA, Rao PK, Kadesch T. Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol Cell Biol. 2004;24(8):3505–3513. doi: 10.1128/MCB.24.8.3505-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6(12B):2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Cordes R, Gossler A. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in Delta1 mutants. Proc Natl Acad Sci U S A. 2007;104(2):537–542. doi: 10.1073/pnas.0608281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawber C, Nofziger D, Hsieh JJ, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122(12):3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- Sun J, Kamei CN, Layne MD, Jain MK, Liao JK, Lee ME, Chin MT. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J Biol Chem. 2001;276(21):18591–18596. doi: 10.1074/jbc.M101163200. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113(Pt 23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- Vasyutina E, Lenhard DC, Wende H, Erdmann B, Epstein JA, Birchmeier C. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci U S A. 2007;104(11):4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128(22):4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rawls J, Molkentin JD, Black BL, Olson EN. Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol Cell Biol. 1999;19(4):2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Radtke F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 2006;580(12):2860–2868. doi: 10.1016/j.febslet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- Wilson EM, Rotwein P. Selective control of skeletal muscle differentiation by Akt1. J Biol Chem. 2007;282(8):5106–5110. doi: 10.1074/jbc.C600315200. [DOI] [PubMed] [Google Scholar]
- Zhao P, Hoffman EP. Musculin isoforms and repression of MyoD in muscle regeneration. Biochem Biophys Res Commun. 2006;342(3):835–842. doi: 10.1016/j.bbrc.2006.01.188. [DOI] [PubMed] [Google Scholar]
- Zhu B, Ramachandran B, Gulick T. Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J Biol Chem. 2005;280(31):28749–28760. doi: 10.1074/jbc.M502491200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of Hey1. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or Hey1-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of Mef2C (left panel) and Myh3 (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. (B) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of MyoR. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or MyoR-directed siRNAs were propagated on the coated plates and then shifted to DM for 24 hours. Expression of Mef2C (left panel) and Myh3 (right panel) RNA was assessed by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. NOTE: While MHC protein levels were only appreciably induced in control cells by 48 hours in DM, Myh3 RNA was elevated ~300 fold by 24 hours (M.B., unpublished observation).
(A) Ligand-induced Notch signaling effectively blocks myogenesis in cells expressing reduced levels of Hey1 and MyoR. Individual tissue culture wells were treated with Fc-Dll4 or Fc-control as indicated in the Methods. C2C12 cells transfected with either non-silencing (NS) control siRNA oligonucleotides or a mixture of Hey1-directed and MyoR-directed siRNAs were propagated on the coated plates and then shifted to DM for 72 hours. Expression of Hey1 (left panel), MyoR (middle panel), and Myh3 (right panel) RNA was determined by quantitative RT-PCR. Q-PCR values are presented as the average +/− standard deviation of three replicate samples. (B) Myoblast fusion in cultures treated with either non-silencing (NS) control siRNAs or Hey1/MyoR siRNAs and plated on either Fc-control or Fc-Dll4 was assessed after four days in DM.