Cohesin is implicated in establishing and maintaining pluripotency. Lavagnolli et al. demonstrate that cohesin depletion enhanced the ability of ES cells to initiate somatic cell reprogramming in heterokaryons. Increased c-Myc (Myc) expression in cohesin-depleted ES cells promoted DNA replication-dependent reprogramming of somatic fusion partners. In contrast, cohesin-depleted somatic cells were poorly reprogrammed in heterokaryons. Pluripotency gene induction was rescued by Myc and nuclear transfer, whereas reprogramming does not require DNA replication.
Keywords: cohesin, pluripotency, reprogramming, embryonic stem cells, heterokaryon, nuclear transfer
Abstract
Cohesin is implicated in establishing and maintaining pluripotency. Whether this is because of essential cohesin functions in the cell cycle or in gene regulation is unknown. Here we tested cohesin’s contribution to reprogramming in systems that reactivate the expression of pluripotency genes in the absence of proliferation (embryonic stem [ES] cell heterokaryons) or DNA replication (nuclear transfer). Contrary to expectations, cohesin depletion enhanced the ability of ES cells to initiate somatic cell reprogramming in heterokaryons. This was explained by increased c-Myc (Myc) expression in cohesin-depleted ES cells, which promoted DNA replication-dependent reprogramming of somatic fusion partners. In contrast, cohesin-depleted somatic cells were poorly reprogrammed in heterokaryons, due in part to defective DNA replication. Pluripotency gene induction was rescued by Myc, which restored DNA replication, and by nuclear transfer, where reprogramming does not require DNA replication. These results redefine cohesin’s role in pluripotency and reveal a novel function for Myc in promoting the replication-dependent reprogramming of somatic nuclei.
Cohesin is a highly conserved multiprotein complex that facilitates homology-based repair of DNA lesions and the faithful transmission of chromosomes through meiosis and mitosis by providing cohesion between sister chromatids (Nasmyth and Haering 2009). In addition, cohesin has a role in DNA replication (Terret et al. 2009; Guillou et al. 2010; Tittel-Elmer et al. 2012; Tedeschi et al. 2013) and contributes to the regulation of gene expression (Dorsett and Merkenschlager 2013) at least in part by forming long-range chromatin interactions (Hadjur et al. 2009; Mishiro et al. 2009; Nativio et al. 2009; Hou et al. 2010; Kagey et al. 2010; Seitan et al. 2011; Apostolou et al. 2013; Merkenschlager and Odom 2013; Wei et al. 2013; Zhang et al. 2013) that contribute to the establishment and maintenance of cell type-specific gene expression patterns (Bickmore and van Steensel 2013; Merkenschlager and Odom 2013).
Cohesin was identified by several RNAi screens aimed at uncovering factors that promote the self-renewal of embryonic stem (ES) cells (Ding et al. 2009; Hu et al. 2009; Kagey et al. 2010). This may not be surprising, as self-renewal requires cell proliferation, and cohesin is essential for DNA repair and chromosome segregation in cycling cells. Indeed, after prolonged cohesin depletion, ES cells show grossly aberrant gene expression that reflects DNA damage and repair functions (see below). As the activation of stress responses interferes with the expression of pluripotency genes (Lin et al. 2005; Maimets et al. 2008), the impact of cohesin on ES cell self-renewal may well be indirect. Similar considerations apply to reports that cohesin facilitates the reprogramming of somatic cells toward induced pluripotent stem (iPS) cells Apostolou et al. 2013; Wei et al. 2013; Zhang et al. 2013) because iPS cell reprogramming requires multiple rounds of cell division. Furthermore, reprogramming is sensitive to the activation of stress responses (Banito et al. 2009; Li et al. 2009; Utikal et al. 2009).
It therefore remains to be established whether cohesin contributes to reprogramming directly by regulating the expression of pluripotency genes or indirectly via its essential cell cycle-related functions. To address this question, we set out to dissociate the role of cohesin in gene regulation from its functions in the cell cycle. We made use of reprogramming systems that can reactivate the expression of pluripotency genes in the absence of proliferation or DNA replication; namely, ES cell heterokaryons and nuclear transfer into Xenopus oocytes.
The fusion of somatic cells with ES cells initiates the expression of pluripotency genes and the extinction of lineage-specific genes in somatic nuclei (Pereira et al. 2008). Heterokaryon-mediated reprogramming is facilitated by DNA replication, presumably by allowing access for reprogramming factors to cis-regulatory regions Tsubouchi et al. 2013). Cell division does not occur for several days, when nuclear fusion marks the transition from heterokaryon to hybrid cell (Fig. 1A; Pereira et al. 2008). We combined heterokaryon-mediated reprogramming with the targeted deletion of cohesin from either the ES cell or the somatic cell fusion partner to examine cohesin’s role in pluripotency and reprogramming in the absence of cell proliferation. The reported collapse of pluripotency in the absence of cohesin (Ding et al. 2009; Hu et al. 2009; Kagey et al. 2010) predicts that cohesin-depleted ES cells are ineffective at driving the reprogramming of somatic cells. Conversely, given cohesin’s role as a genome organizer (Phillips-Cremins et al. 2013; Seitan et al. 2013; Sofueva et al. 2013; Zuin et al. 2014), somatic cells without cohesin might be more susceptible to reprogramming than their wild-type counterparts.
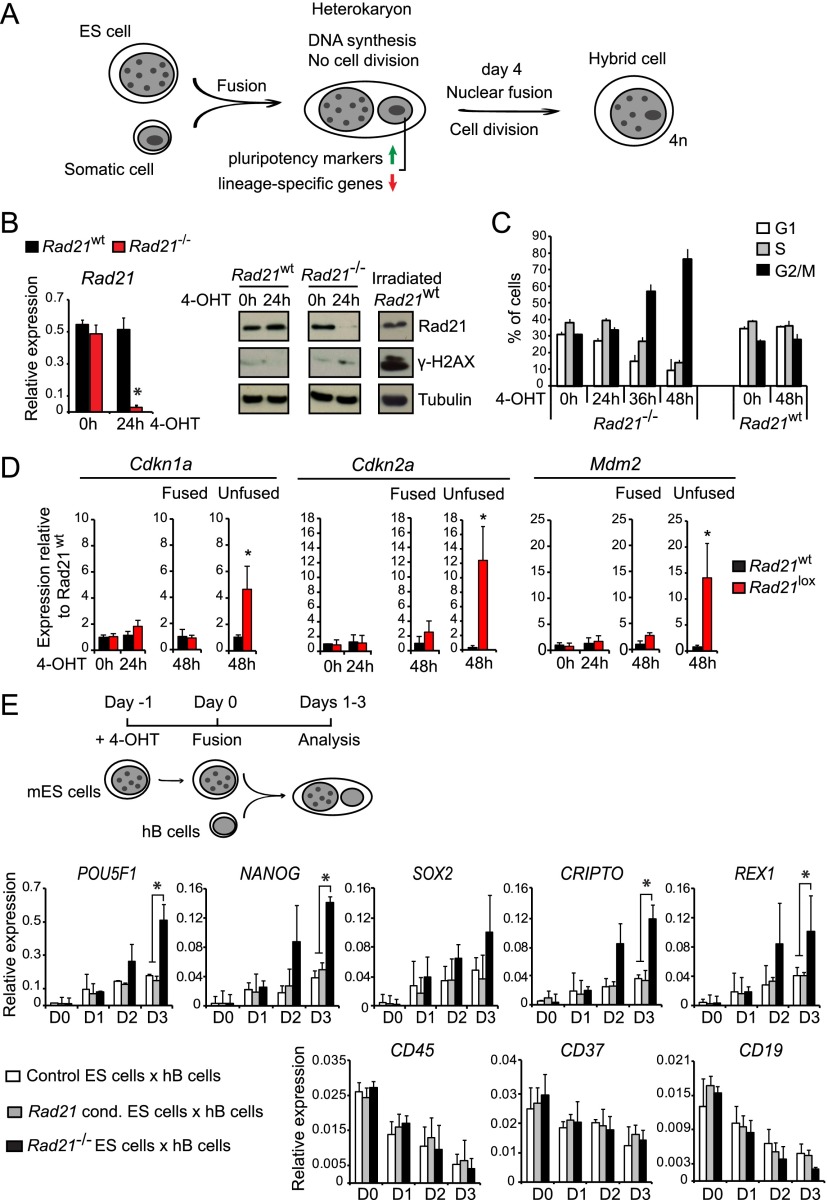
Figure 1.
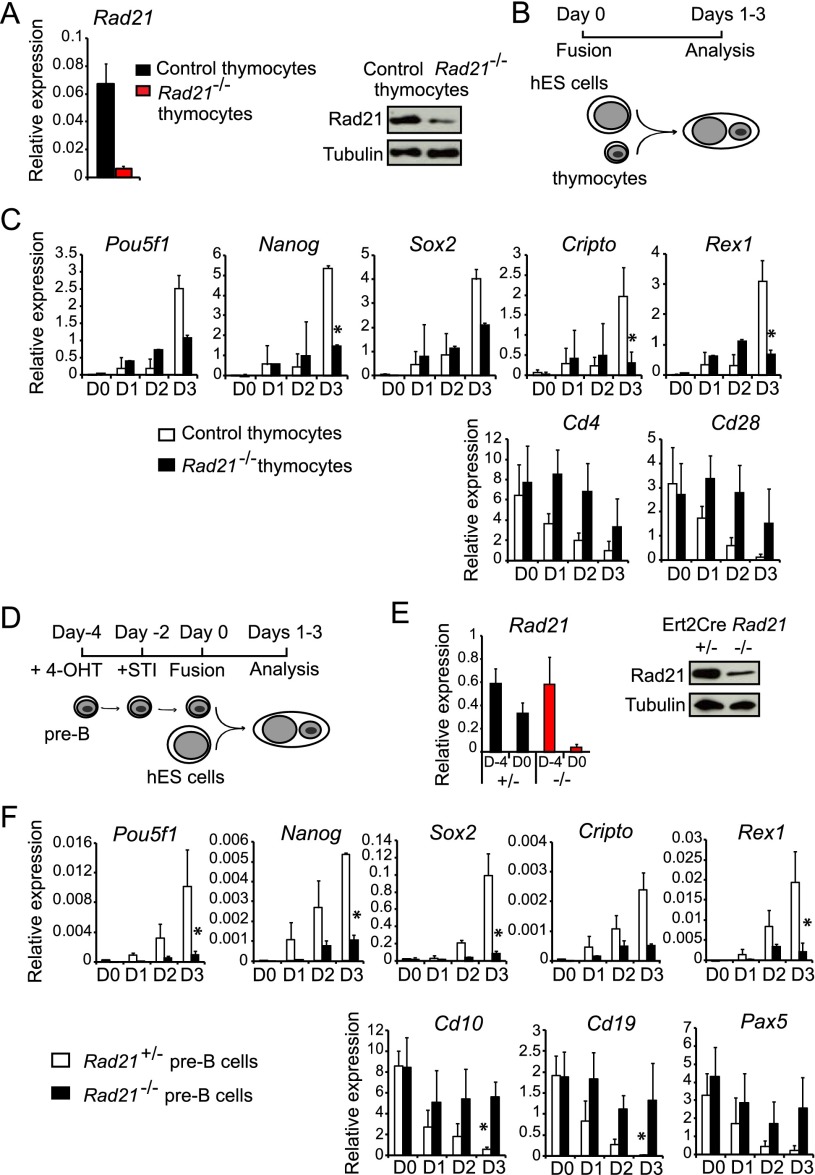
Cohesin-deficient ES cells have increased reprogramming ability. (A) The fusion between ES cells and somatic cells results in heterokaryons, where the two nuclei remain discrete within a shared cytoplasm. The presence of separate ES cell and somatic nuclei shows that cell division has not yet occurred. After 3–4 d, nuclear fusion occurs and gives rise to proliferating hybrid cells. (B) Genetic depletion of Rad21 mRNA (left) and protein (right) from mES cells. (Left) Quantitative RT–PCR analysis of Rad21 mRNA expression in control (black) and Rad21−/− (red) ES cells after 4-OHT-mediated activation of ERt2Cre for 0 or 24 h (normalized to Ubc and Ywhaz; mean ± SD; n = 3). (Right) Immunoblot analysis showed substantially reduced Rad21 protein expression but no significant induction of the DNA damage response marker γ-H2AX in control or Rad21−/− ES cells 24 h after ERt2Cre induction. Irradiation of Rad21WT ES cells served as a positive control for γ-H2AX induction. Tubulin was used as the loading control. (C) The cell cycle profile of Rad21−/− ES cells as assessed by propidium iodide staining for DNA content and flow cytometry was unchanged 24 h after ERt2Cre induction, but later time points saw an accumulation of cells in the G2/M phase of the cell cycle. (D) Quantitative RT–PCR was used to monitor the expression of the p53 target genes Cdkn1a (p21), Cdkn2a (p16), and Mdm2 at the indicated times after the 4-OHT-induced deletion of Rad21 in ES cells. Twenty-four hours after ERt2Cre activation, ES control (black) and Rad21−/− (red) ES cells were either fused with human B (hB) somatic cells to halt their proliferation (fused) or allowed to continue proliferating (unfused) (normalized to Ubc and Ywhaz and shown relative to control ES cells; mean ± SD; n = 3). (*) P < 0.05 (t-test). (E) Experimental outline of heterokaryons between mES cells and hB cells. ES cells used were Rad21 wild type (Rad21+/+) treated with 4-OHT, Rad21lox/lox (undeleted Rad21 conditional) treated with ethanol, or Rad21−/− (ERt2Cre Rad21lox/lox) treated with 4-OHT. Quantitative RT–PCR with primers that selectively amplify human transcripts was used to monitor the activation of the key pluripotency-associated factors POU5F1 (OCT4), NANOG, SOX2, CRIPTO, and REX1 (top row) and the B-cell-specific genes CD45, CD37, and CD19 (bottom row) on days 1–3 (normalized to GAPDH; mean ± SD; n = 3). (*) P < 0.05 (t-test).
Contrary to expectations, we found that acute cohesin depletion enhanced, rather than abolished, the ability of ES cells to induce the expression of pluripotency genes by somatic cells in heterokaryons. Detailed examination revealed that acute cohesin depletion did not result in the predicted global collapse of ES cell pluripotency (Ding et al. 2009; Hu et al. 2009; Kagey et al. 2010; Nitzsche et al. 2011) but rather affected the expression and chromatin conformation of a select number of target genes in a highly locus-specific fashion.
Mechanistically, enhanced reprogramming by cohesin-deficient ES cells was mediated by c-Myc (Myc), one of the original Yamanaka factors (Takahashi and Yamanaka 2006). Myc facilitates iPS cell-mediated reprogramming via transcriptional effects (Soufi et al. 2012) and by increasing the proliferation rate of iPS cells (Hanna et al. 2009). Myc can also enhance DNA replication directly (Dominguez-Sola et al. 2007). In heterokaryons, ES cell-expressed Myc enhanced reprogramming in the absence of cell proliferation at least in part by promoting DNA replication.
Cohesin depletion did not render somatic cells more susceptible to reprogramming in ES cell heterokaryons. Instead, cohesin-deficient somatic cells showed impaired reactivation of endogenous pluripotency genes. Defective reprogramming was linked to inefficient DNA replication: First, both DNA replication and reprogramming were partially rescued by increasing the expression of Myc in the ES cell fusion partner. Second, cohesin-deficient somatic cells reactivated endogenous pluripotency genes at least as efficiently as wild-type cells after injection into Xenopus oocytes, where reprogramming occurs without DNA replication (Gurdon 1976; Gurdon et al. 1976; Jullien et al. 2012). These results provide a clear separation of cohesin’s canonical role in chromosome segregation from an emerging role in DNA replication and a contribution to the regulation of gene expression. They redefine cohesin’s role in pluripotency and reveal a novel function for Myc in promoting the replication-dependent reprogramming of somatic nuclei.
Results
ES cells lacking cohesin are efficient initiators of reprogramming
The fusion of ES cells with somatic cells generates heterokaryons, where ES and somatic nuclei remain discrete within a shared cytoplasm for a period of 3–4 d. Early events in heterokaryon-mediated reprogramming include the activation of pluripotency gene expression in somatic nuclei and the extinction of the somatic gene expression program and are facilitated by the ES cell-induced replication of the somatic genome (Pereira et al. 2008; Tsubouchi et al. 2013). Eventually, nuclear fusion occurs and gives rise to proliferating hybrid cells. We focused our analysis on the heterokaryon stage to obviate the requirement for cohesin in cell division-related functions (Fig. 1A). By separating reprogramming from cell division, heterokaryons provide an opportunity to investigate the role of cohesin in the resetting of gene expression programs without interference from essential cohesin functions in chromosome segregation. We made heterokaryons in which either the somatic partner or the ES cell partner was genetically deficient in the cohesin subunit Rad21.
We established ERt2Cre Rad21lox/lox ES cells that are inducibly depleted of cohesin by the 4-hydroxytamoxifen (4-OHT)-mediated activation of ERt2Cre. ERt2Cre activation measurably reduced Rad21 protein expression by 18 h (data not shown), and, after 24 h, there was near-complete depletion of Rad21 mRNA (Fig. 1B, left) and Rad21 protein (Fig. 1B, right). At this time, we found no substantial induction of the DNA damage marker γ-H2AX in cohesin-depleted ES cells (Fig. 1B, right, irradiation served as a positive control for γ-H2AX induction). The cell cycle distribution of cohesin-depleted ES cells was unchanged 24 h after ERt2Cre activation (Fig. 1C), and the expression of the p53 target genes Cdkn1a (p21), Cdkn2a (p16), and Mdm2 remained low (Fig. 1D). Cohesin-depleted ES cells that were allowed to proliferate for an additional 12–24 h (for a total of 36 or 48 h after ERt2Cre activation) showed G2/M arrest (Fig. 1C) and substantially elevated expression of the p53 target genes Cdkn1a (p21), Cdkn2a (p16), and Mdm2 (Fig. 1D), as expected based on the essential role of cohesin in DNA damage repair and chromosome segregation. The expression of p53-responsive genes remained significantly lower when the proliferation of cohesin-depleted ES cells was halted by the formation of heterokaryons 24 h after ERt2Cre activation (Fig. 1D). This provided a window for testing the reprogramming ability of cohesin-depleted ES cells in heterokaryons.
We fused control or conditionally cohesin-depleted mouse ES (mES) cells 24 h after ERt2Cre induction with puromycin-resistant human EBV-transformed human B (hB)-cell lines (Fig. 1E; Pereira et al. 2008). We used RT–PCR with primers that selectively amplify human transcripts to monitor the expression of lineage-specific and pluripotency-associated genes in somatic nuclei. Both cohesin-depleted and control mES cells silenced the B-cell genes CD45, CD37, and CD1 (Fig. 1E). Unexpectedly, cohesin-depleted mES cells consistently induced the human pluripotency-associated genes POU5F1 (OCT4), NANOG, SOX2, REX1, and CRIPTO more strongly and with faster kinetics than control mES cells (Fig. 1E). We conclude that cohesin is not required for the ability of ES cells to induce pluripotency gene expression by somatic nuclei in heterokaryons and that cohesin-deficient ES cells initiate the reprogramming of somatic cell nuclei more potently than wild-type ES cells.
A locus-specific role for cohesin in regulating pluripotency genes in ES cells
Previous reports had indicated that RNAi-mediated knockdown of cohesin subunits for 4–5 d interferes with ES cell self-renewal (Ding et al. 2009; Hu et al. 2009; Kagey et al. 2010; Nitzsche et al. 2011). These studies gave rise to a prevailing model in which cohesin is required for the continued expression of pluripotency genes by connecting their enhancers and promoters in three-dimensional nuclear space (Kagey et al. 2010). However, 4–5 d of cohesin depletion corresponds to numerous cell cycles in rapidly proliferating ES cells. This raises the question of whether the loss of ES cell self-renewal observed in previous studies could have been secondary and due to essential cohesin functions in the cell cycle. This possibility is supported by our reanalysis of gene expression data (Kagey et al. 2010), which showed a prominent enrichment of gene ontology (GO) terms related to DNA damage, cell cycle checkpoints, etc. among genes that were deregulated 5 d after knockdown of the cohesin subunit SMC1A (Fig. 2A).
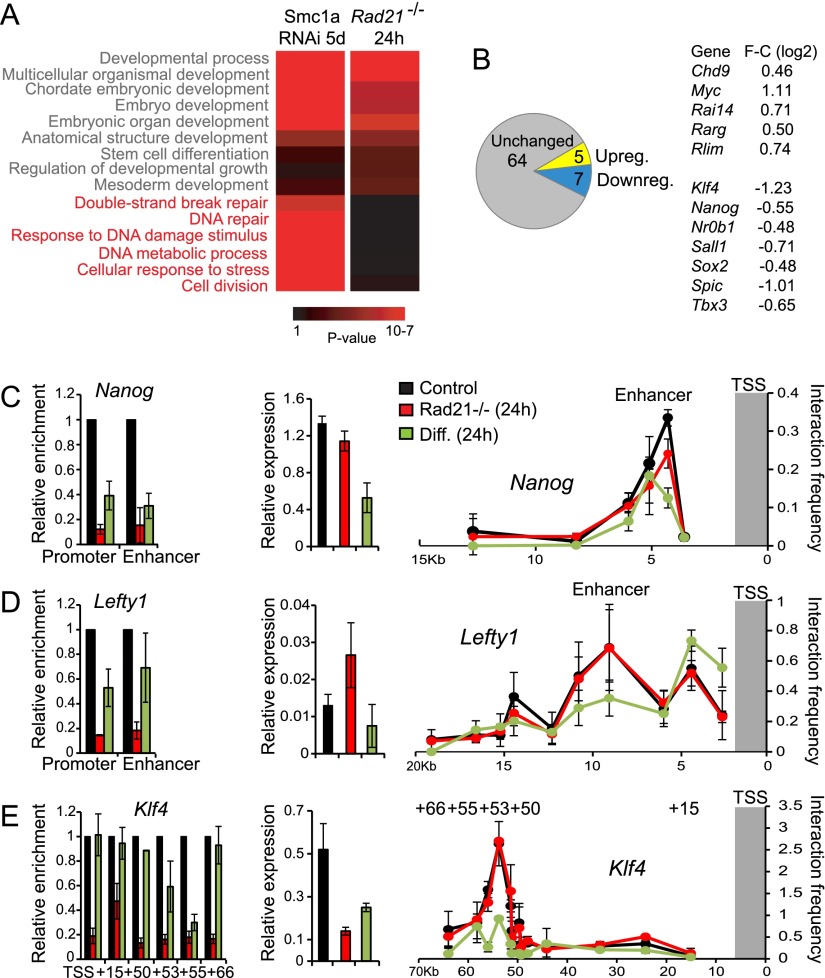
Figure 2.
Acute depletion of Rad21 triggers selective changes in gene expression and chromatin conformation but not a global collapse of pluripotency gene expression. (A) Genes related to DNA damage and chromosome missegregation are preferentially induced by prolonged knockdown of cohesin subunits but not by acute cohesin deletion. P-values (log10) are indicated by a color scale from black to red. (B) Most pluripotency genes remain expressed normally in ERt2Cre Rad21lox/lox ES cells 24 h after ERt2Cre induction. Pluripotency genes that do change expression are listed along with the log2 fold change (F-C). (C) Rad21 ChIP (left; normalized to input; mean ± SD; n = 3), mRNA expression (middle; normalized to Ubc and Ywhaz; mean ± SD; n = 3), and chromatin conformation (right; relative interaction frequency; mean ± SD; n = 3) of Nanog in control (black) and acutely cohesin-depleted (red) ES cells. Differentiating ES cells (green) provide a reference for the state of the Nanog locus during differentiation. (D) Analysis of the Lefty1 locus as detailed in C for Nanog. Note the persistence of Lefty1 promoter–enhancer interactions in cohesin-depleted ES cells. (E) Analysis of the Klf4 locus as detailed in C for Nanog. Note the persistence of Klf4 promoter–enhancer interactions in cohesin-depleted ES cells.
We analyzed gene expression by conditionally cohesin-deficient ES cells 24 h after ERt2Cre induction, at a time when Rad21 expression was substantially reduced but before the onset of G2/M arrest or the activation of p53 target genes (see Fig. 1). Transcriptional profiling showed that 598 genes were deregulated (P-value < 0.05 with Benjamini-Hochberg correction; 247 down and 341 up). Deregulated genes were enriched for developmental functions (GO terms in gray typeface in Fig. 2A, right). In contrast to prolonged knockdown of Smc1a (Fig. 2A, left), there was no enrichment of cell cycle and DNA damage-related genes (GO terms in orange typeface in Fig. 2A, right). Deregulation just hours after the onset of Rad21 depletion suggests that many of these genes were directly regulated by cohesin. This was supported by integration with published chromatin immunoprecipitation (ChIP) deep sequencing (ChIP-seq) data (Kagey et al. 2010), which showed that the binding of cohesin, the cohesin loading factor NIPBL, Mediator12, and RNA polymerase 2 was highly correlated with the deregulated expression of genes by acute cohesin depletion (4.44 × 10−50, Wilcoxon gene set test).
We assessed the impact of acute cohesin depletion on the expression of genes assigned to an extended pluripotency network (Kim et al. 2008) and a curated list of genes whose promoters are targeted by at least five of the pluripotency factors Nanog, Dax1, Sox2, Nac1, Oct4, and Klf4 (see Table 1 in Kim et al. 2008). Although statistically significant (P = 7.37 × 10−7), the frequency (12 out of of 76 vs. 2.4 out of 76 expected), direction (five up-regulated: Chd9, Myc, Rai14, Rarg, and Rlim; seven down-regulated: Klf4, Nanog, Nr0b1, Sall1, Sox2, Spic, and Tbx3), and extent (maximal log2 fold change +1.14 to −1.26) of deregulation were modest (Fig. 2B). Analysis of all genes that are targeted by Nanog, Dax1, Sox2, Nac1, Oct4, and Klf4 in ES cells (Kim et al. 2008) revealed a similar picture in that, of 379 potential pluripotency genes, 19 (5%) were up-regulated and 27 (7%) were down-regulated in Rad21-depleted ES cells (Supplemental Table 1). We conclude that cohesin depletion does not cause a global collapse of pluripotency gene expression in acutely cohesin-depleted ES cells.
To explore the relationship between cohesin, enhancer–promoter interactions, and active transcription of pluripotency genes, we selected Nanog, Lefty1, and Klf4 as three pluripotency-related factors whose expression was deregulated in acutely cohesin-depleted ES cells. In addition to mRNA expression, we determined cohesin binding by ChIP for Rad21 and long-range interactions by chromosome conformation capture (3C). For each locus examined, the impact of cohesin depletion was compared with the induction of differentiation by withdrawal of 2i inhibitors from the culture medium.
Nanog behaved as predicted (Fig. 2C; Kagey et al. 2010): Rad21 binding to the Nanog promoter and enhancer was reduced in parallel with Nanog expression, and enhancer–promoter interactions were weakened as judged by 3C. Induced ES cell differentiation also reduced cohesin binding to Nanog regulatory elements, Nanog expression, and Nanog enhancer–promoter interactions (Fig. 2C).
In contrast to Nanog, Lefty1 expression was increased in Rad21-deficient ES cells. Lefty1 enhancer–promoter interactions were maintained at wild-type levels despite efficient depletion of cohesin from the Lefty1 promoter and enhancer. This constellation argues against a model in which cohesin is required for Lefty1 promoter–enhancer interactions (Kagey et al. 2010), since efficient long-range interactions were maintained in the absence of cohesin (Fig. 2D). Importantly, interactions between the Lefty1 promoter and enhancer were reduced under differentiation conditions, demonstrating the validity of the 3C assay.
Lefty1 was not the only example that dissociated cohesin binding from promoter–enhancer interactions. At the Klf4 locus, cohesin binding was decreased in Rad21-deficient ES cells, and, as described for Lefty1, enhancer–promoter interactions were maintained (Fig. 2E). In contrast to Lefty1, the expression of Klf4 was reduced in cohesin-depleted ES cells. ES cell differentiation abolished Klf4 promoter–enhancer interactions (Fig. 2E). Thus, while it is clear that cohesin can mediate long-range chromosomal interactions (Hadjur et al. 2009; Mishiro et al. 2009; Nativio et al. 2009; Hou et al. 2010; Kagey et al. 2010; Seitan et al. 2011, 2013; Apostolou et al. 2013; Merkenschlager and Odom 2013; Wei et al. 2013; Zhang et al. 2013), this function may not be required for all enhancer–promoter interactions, as illustrated here for Lefty1 and Klf4.
Taken together, the gene expression, ChIP, and 3C data presented here call into question a simple relationship between cohesin binding, long-range interactions, and gene expression. Instead of the expected global collapse of pluripotency gene expression, we found that the impact of cohesin is highly locus-specific in the sense that not all pluripotency genes were affected by cohesin depletion. Genes that were deregulated just hours after cohesin depletion and before the onset of DNA damage responses were highly enriched for direct binding by cohesin. We used heterokaryon formation to temporarily lock in this state and assess the ability of cohesin-depleted ES cells to reactivate the expression of pluripotency genes in somatic cells.
Myc accounts for the enhanced reprogramming ability of cohesin-deficient ES cells
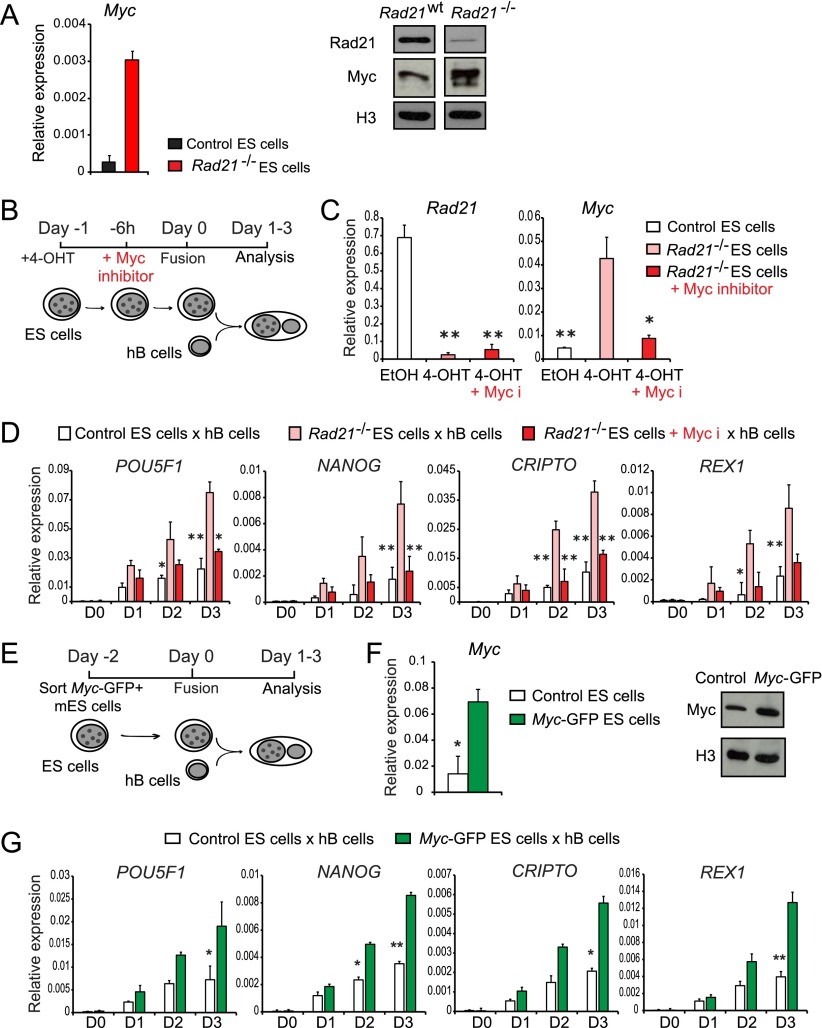
To explore how the reprogramming ability of ES cells is not only preserved but even enhanced by acute cohesin depletion, we focused on Myc, as it can facilitate reprogramming (Takahashi and Yamanaka 2006; Hanna et al. 2009; Soufi et al. 2012), and global gene expression profiling suggested that Myc transcripts are up-regulated in acutely cohesin-depleted ES cells (log2 fold change of 1.14; adjusted P < 0.05). Increased Myc mRNA expression was confirmed by quantitative RT–PCR (Fig. 3A, left), and Western blotting showed elevated Myc protein expression in cohesin-depleted ES cells (Fig. 3A, right). To test whether Myc activity was required for increased reprogramming, we treated acutely cohesin-depleted ES cells with the Myc inhibitor 10058-F4 for 6 h prior to fusion with hB cells (Fig. 3B). Inhibition of Myc abrogated the enhanced reprogramming ability of cohesin-depleted ES cells (Fig. 3C,D). Conversely, forced Myc expression in control ES cells was sufficient to enhance their reprogramming ability in heterokaryons (Fig. 3E–G). We previously demonstrated that DNA replication in the somatic fusion partner is required for efficient reactivation of pluripotency genes in ES cell heterokaryons (Tsubouchi et al. 2013). To address whether the expression of Myc in ES cells facilitates the activation of pluripotency genes by a mechanism that involves DNA replication, we fused hB cells and mES cells and then monitored human pluripotent gene induction following treatment of heterokaryons with aphidicolin, an inhibitor of DNA polymerase activity. Addition of aphidicolin abrogated the reprogramming ability of ES cells transduced with Myc (Supplemental Fig. S1). We conclude that Myc activity is necessary and sufficient for enhanced heterokaryon-mediated reprogramming.
Figure 3.
Myc drives reprogramming of somatic nuclei in ES cell heterokaryons. (A) Increased Myc mRNA (left) and protein expression (right) by cohesin-deficient ES cells shown by quantitative RT–PCR and Western blotting, respectively. Histone H3 (H3) was used as a loading control. Data are representative of three independent experiments. (B) Schematic of the fusion of puromycin-resistant hB cells with control mES cells, cohesin-deficient mES cells, or cohesin-deficient mES cells pretreated with Myc inhibitor for 6 h prior to the fusion. (C) Pretreatment of cohesin-deficient ES cells with Myc inhibitor compromised the expression of Myc itself by disrupting autoregulation by Myc but did not affect the expression of Rad21. (D) Pharmacological inhibition of Myc restrains the reprogramming potential of cohesin-deficient ES cells. Quantitative RT–PCR was used to monitor the ability of cohesin-deficient ES cells pretreated with Myc inhibitor to induce the expression of POU5F1 (OCT4), NANOG, CRIPTO, and REX1 in heterokaryons with hB cells (normalized to GAPDH; mean ± SD; n = 5). (*) P < 0.05; (**) P < 0.001 (t-test). (E) Schematic of the fusion of hB cells with control ES cells or ES transduced with Myc expression vector. (F) Retroviral expression of Myc mRNA (left) and protein (right) in mES cells shown by RT–PCR and Western blotting, respectively. Histone H3 (H3) was used as a loading control. Data are representative of five independent experiments. (G) Myc expression is sufficient to confer enhanced reprogramming activity to ES cells. Quantitative RT–PCR was used to monitor the ability of control mES cells (IRES-GFP) and mES cells transduced with Myc expression vector (Myc-IRES-GFP) to induce the expression of POU5F1 (OCT4), NANOG, CRIPTO, and REX1 in heterokaryons with hB cells (normalized to GAPDH; mean ± SD; n = 5). (*) P < 0.05; (**) P < 0.001 (t-test).
Myc facilitates reprogramming by driving somatic DNA replication in ES cell heterokaryons
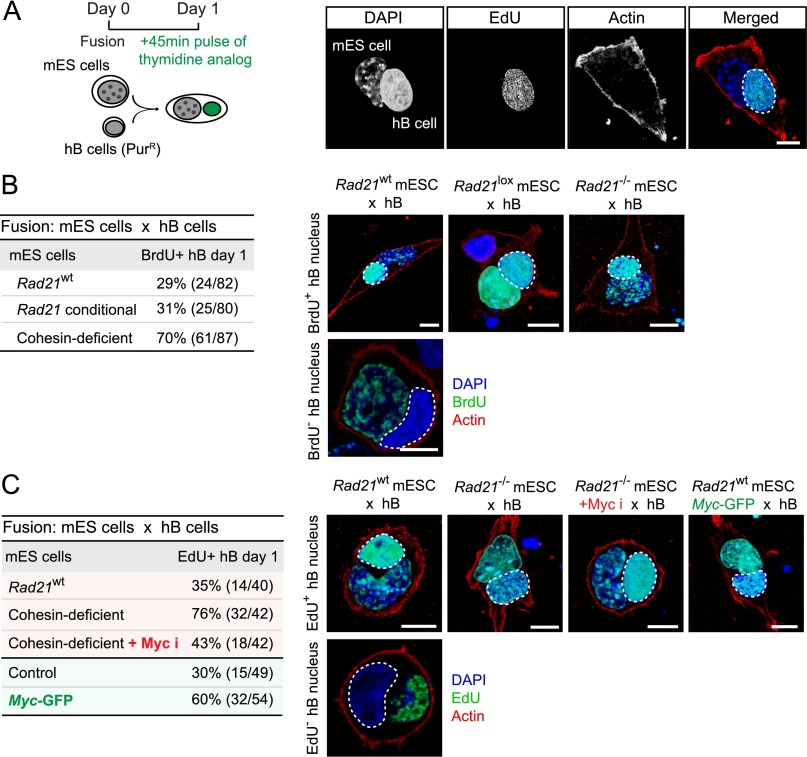
These experiments showed that Myc enhanced the efficiency of heterokaryon-mediated reprogramming but did not reveal the underlying mechanism. In addition to its transcriptional effects (Soufi et al. 2012; Valovka et al. 2013) and its ability to drive cellular proliferation (Hanna et al. 2009; Kim et al. 2010; Lin et al. 2012), Myc can also enhance DNA replication in the absence of proliferation (Dominguez-Sola et al. 2007) by influencing the assembly and activity of replication origins via physical interaction with proteins of the prereplication complex (CDC6 and MCMs), recruitment of CDC45 to chromatin, and regulation of CDK activity at replication origins (Dominguez-Sola and Gautier 2014). We fused control and acutely cohesin-depleted ES cells with hB cells and pulsed the resulting heterokaryons with thymidine analogs to enumerate hB-cell nuclei that underwent DNA synthesis (Fig. 4A). Consistent with a role for DNA replication in heterokaryon-mediated reprogramming (Tsubouchi et al. 2013), we found a higher percentage of BrdU+ hB-cell nuclei in heterokaryons with Rad21-deficient ES cells (70% BrdU+) than control ES cells (Rad21+/+ mES cells treated with 4-OHT and 29% BrdU+ and Rad21lox/lox mES cells treated with 31% BrdU+) (Fig. 4B). To ask whether elevated Myc expression facilitated reprogramming through enhanced DNA replication, we pretreated acutely cohesin-depleted mES cells with the Myc inhibitor 10058-F4 prior to fusion with hB cells. To assess DNA replication, we pulsed the resulting heterokaryons with EdU. Myc inhibition reduced the induction of DNA replication in somatic nuclei by cohesin-depleted ES cells from 76% to 43%, close to the 35% of hB-cell nuclei that incorporated EdU in heterokaryons with control mES cells (Fig. 4C). Conversely, forced expression of Myc was sufficient to enhance somatic DNA replication by control ES cells to that of acutely cohesin-depleted ES cells from 30% to 60% of hB-cell nuclei (Fig. 4C).
Figure 4.
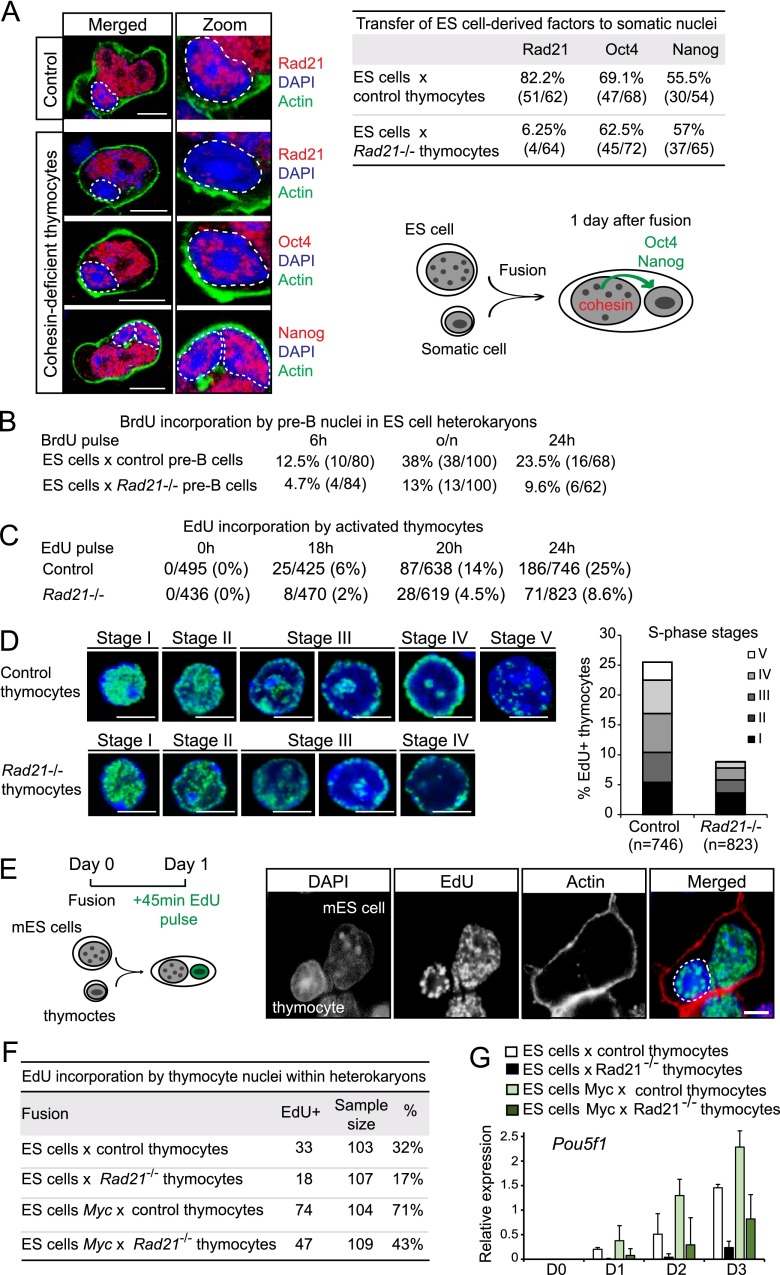
Myc drives heterokaryon-mediated reprogramming by promoting DNA replication in somatic nuclei. (A) Efficient induction of DNA replication in somatic nuclei fused with cohesin-deficient ES cells. Confocal analysis of DNA replication in heterokaryons between hB cells (encircled) and mES cells (45-min EdU pulse on day 1 after fusion). (Green) EdU; (red) actin; (blue) DAPI. Bar, 10 μm. (B) Summary of percentages and numbers (BrdU+/total; 45-min BrdU pulse on day 1 after fusion) of BrdU+ hB cells fused with Rad21 wild-type (Rad21wt), undeleted Rad21lox/lox (Rad21 conditional), or Rad21−/− (cohesin-deficient) ES cells from three independent experiments. Representative images of BrdU-positive (top) and BrdU-negative (bottom) hB-cell nuclei in the indicated heterokaryons (45-min BrdU pulse on day 1 after fusion) are shown. (Green) BrdU; (red) actin; (blue) DAPI. Bar, 10 μm. (C) Pharmacological inhibition of Myc restrains the ability of cohesin-deficient ES cells to trigger DNA replication in somatic nuclei. DNA replication in heterokaryons between hB cells and Rad21 wild-type (Rad21WT) ES cells, Rad21-deleted (cohesin-deficient) ES cells, or Rad21-deleted ES cells pretreated with Myc inhibitor for 6 h prior to fusion. Myc expression enhances the ability of ES cells to trigger DNA replication in somatic nuclei. DNA replication in heterokaryons between hB cells and wild-type ES cells transduced with control IRES-GFP vector (Control) or Myc-IRES-GFP (Myc-GFP). Representative images of EdU-positive (top) and EdU-negative (bottom) hB-cell nuclei in the indicated heterokaryons (45-min EdU pulse on day 1 after fusion) are shown. (Green) EdU; (red) actin; (blue) DAPI. Bar, 10 μm.
These results link potent reprogramming by acutely cohesin-depleted ES cells to the efficient initiation of somatic DNA replication and show that the mechanism of Myc-enhanced heterokaryon-mediated reprogramming involves DNA replication in somatic nuclei.
Heterokaryon-mediated reprogramming requires cohesin expression by somatic cells
We next assessed the role of cohesin expression by somatic cells in heterokaryon-mediated reprogramming. Depletion of cohesin from somatic thymocytes was achieved by combining conditional Rad21 alleles with developmentally regulated CD4Cre transgenes to coordinate Rad21 deletion with the developmentally programmed exit from the cell cycle at the CD4+ CD8+ double-positive stage of mouse thymocyte differentiation (Fig. 5A; Seitan et al. 2011, 2013). This approach generates cohesin-deficient somatic cells that are noncycling and therefore do not rely on cell cycle-related cohesin functions. Rad21-deficient thymocytes do not show p53 activation or DNA damage responses and have a normal life span (Seitan et al. 2011, 2013).
Figure 5.
Heterokaryon-mediated reprogramming requires cohesin expression by somatic cells. (A) Genetic depletion of Rad21 mRNA (left) and protein (right) from nonproliferating thymocytes was achieved by combining developmentally regulated CD4Cre with a conditional Rad21 allele. (B) Experimental outline and analysis of reprogramming in heterokaryons between hES cells and control (CD4Cre Rad21+/lox or CD4Cre Rad21+/+) or cohesin-deficient (CD4Cre Rad21lox/lox) mouse thymocytes. (C) Quantitative RT–PCR with primers that selectively amplify mouse transcripts was used to monitor the expression of the key pluripotency-associated factors Pou5f1 (Oct4), Nanog, Sox2, Cripto, and Rex1 (top row) and the thymocyte-specific genes Cd4 and Cd28 (bottom row) on days 1–3 (normalized to Gapdh; mean ± SD; n = 3). (*) P < 0.05 (t-test). (D) Experimental outline of heterokaryons between hES cells and control (Ert2Cre Rad21+/lox) or cohesin-deficient (Ert2Cre Rad21lox/lox) pre-B cells. (E) Genetic depletion of Rad21 mRNA (left) and protein (right) from mouse pre-B cells. (F) Quantitative RT–PCR with primers that selectively amplify mouse transcripts was used to monitor the activation of key pluripotency markers as in C (top row) and the pre-B cell-specific genes Cd10, Cd19, and Pax5 (bottom row) (normalized to Gapdh; mean ± SD; n = 3). (*) P < 0.05.
We fused human ES (hES) cells with either control (CD4Cre Rad21+/lox or CD4Cre Rad21+/+) or cohesin-deficient (CD4Cre Rad21lox/lox) mouse thymocytes (Fig. 5B; Seitan et al. 2011). To monitor the activation of the pluripotency-associated factors Pou5f1 (Oct4), Nanog, Sox2, Cripto, and Rex1 in somatic nuclei, we used quantitative RT–PCR with primers that selectively amplify mouse transcripts (Fig. 5C). Compared with control thymocytes, cohesin-deficient thymocytes were significantly less able to induce the expression of pluripotency-associated factors. In addition, cohesin-deficient thymocytes showed defective extinction of the thymocyte-specific genes Cd4 and Cd28 (Fig. 5C).
To distinguish whether reprogramming of cohesin-deficient thymocytes in ES cell heterokaryons was inefficient or simply delayed, we allowed heterokaryons to form hybrid colonies and scored their frequency. For this experiment we fused ZHBTc4 mES cells and control (Rad21lox/lox) or cohesin-deficient (CD4Cre Rad21lox/lox) mouse thymocytes harboring an Oct4-GFP transgene (Supplemental Fig. S2A; Yoshimizu et al. 1999). As ZHBTc4 ES cells lack endogenous Pou5f1 and instead express an Oct4βgeo transgene (Niwa et al. 2000), we could selectively determine the expression of thymocyte-derived endogenous Pou5f1 and Oct4-GFP mRNA in heterokaryons with ZHBTc4 ES cells. As noted above (Fig. 5C), the endogenous Pou5f1 (Oct4) locus was poorly induced in cohesin-deficient thymocytes (Supplemental Fig. S2B), but control and cohesin-deficient thymocytes activated Oct4-GFP transgene expression with similar efficiency (Supplemental Fig. S2B). This allowed the isolation of GFP+ cells by flow cytometry. Subsequent enumeration of hybrid colonies showed that fusions with cohesin-deficient thymocytes resulted in a lower number of colonies, consistent with less efficient reprogramming. Hybrid colonies generated from fusions with cohesin-deficient thymocytes also showed qualitative differences, as illustrated by reduced expression of alkaline phosphatase (Supplemental Fig. S2C).
To validate these results in a different somatic cell type, we used Abelson-transformed mouse pre-B cells. In these cells, we blocked proliferation with the Abelson kinase inhibitor STI571 (Bredemeyer et al. 2006) and coordinated the deletion of conditional Rad21 alleles with STI517-induced cell cycle arrest by the administration of 4-OHT to activate ERt2Cre (Fig. 5D,E; Supplemental Fig. S3).
In heterokaryons with hES cells, control (Ert2Cre Rad211+/lox) pre-B cells efficiently activated the expression of the pluripotency-associated factors Pou5f1 (Oct4), Nanog, Sox2, Cripto, and Rex1 and extinguished the expression of the pre-B-cell genes Cd10, Cd19, and Pax5. In contrast, the expression of pluripotency genes and the down-regulation of pre-B-cell genes were compromised in cohesin-deficient (Ert2Cre Rad21lox/lox) pre-B cells (Fig. 5F). We confirmed these results in pre-B cells deficient in another cohesin subunit, Smc3 (Supplemental Fig. S4).
Inefficient DNA replication in cohesin-deficient somatic nuclei
To explore the inefficient reprogramming of cohesin-deficient somatic cells in ES cell heterokaryons, we first analyzed the activation of the p53 pathway, which interferes with iPS cell-mediated reprogramming (Banito et al. 2009; Li et al. 2009; Utikal et al. 2009). We found no induction of the p53 target genes Cdkn1a, Mdm2, Cdkn2b, and Cdkn2a in heterokaryons between ES cells and control or cohesin-deficient thymocytes (Supplemental Fig. S5A), indicating that the observed differences in reprogramming efficiency were not due to cellular stress.
We next examined the transfer of mES cell-derived protein to mouse somatic nuclei within heterokaryons. Compared with proliferating hB cells, which showed rapid remodeling and enlargement of their nuclei in ES cell heterokaryons (Fig. 4A; Pereira et al. 2008), the nuclei of G1-arrested somatic cells remained smaller than ES cell nuclei (identified by the incorporation of EdU prior to fusion) and retained their distinctive distribution of pericentromeric heterochromatin foci 1 d after fusion (Supplemental Fig. S5B). At this time, immunofluorescence staining readily detected Oct4 and Nanog protein in thymocyte nuclei fused with ES cells (Fig. 6A). The cohesin subunit Rad21 was abundant in ES cell nuclei and the nuclei of wild-type thymocytes (Fig. 6A; Supplemental Fig. S5B). However, in contrast to Oct4 and Nanog, ES cell-derived Rad21 was not efficiently transferred to genetically Rad21-deficient thymocyte nuclei, presumably because cohesin is largely chromatin-associated in ES cell nuclei (Fig. 6A).
Figure 6.
Compromised DNA replication in cohesin-deficient somatic nuclei and rescue by Myc. (A) Transfer of ES cell-derived factors to somatic nuclei in heterokaryons. (Left) Confocal analysis of control (top row) and cohesin-deficient (rows 2–4) thymocytes 1 d after fusion with ES cells shows efficient transfer of ES cell-derived pluripotency factors, but not of cohesin, to somatic cell nuclei in experimental heterokaryons. Thymocyte nuclei were distinguished from ES cell nuclei by size and DAPI pattern (as described in Supplemental Fig. 3B) and are outlined. (Blue) DAPI; (green) actin; (red) Rad21, Oct4, or Nanog. Bar, 10 μm. (Right) Oct4 and Nanog, but not Rad21, are efficiently transferred from ES cell to somatic nuclei 1 d after fusion. (B) Compromised DNA replication in cohesin-deficient pre-B cells fused with mES cells. mES cells were pre-labeled with 10 μM EdU overnight prior to fusion with mouse pre-B cells to discriminate ES cell and somatic nuclei. Heterokaryons were pulsed with BrdU for 45 min at 6 or 24 h after fusion or overnight (o/n), and BrdU labeling was scored in (EdU-negative) pre-B-cell nuclei. Quantitative evaluation shows compromised DNA replication by cohesin-deficient pre-B-cell nuclei in ES cell heterokaryons at all time points analyzed. Data are from five independent experiments. (C) DNA replication is compromised in cohesin-deficient thymocytes stimulated to undergo DNA replication in vitro. Results of confocal analysis of activation-induced DNA synthesis (EdU incorporation from two independent experiments) are shown. (D, left) Confocal analysis of sequential EdU incorporation patterns during the progression from early (stage I) to late (stage V) S phase in thymocytes stimulated to undergo DNA replication in vitro. (Green) EdU; (blue) DAPI:. Bar, 5 μm. Control (top) and cohesin-deficient (bottom) thymocytes show comparable patterns of EdU incorporation, indicating that the global organization of DNA replication is preserved in cohesin-deficient thymocytes (no stage V nuclei were observed for cohesin-deficient thymocytes). (Right) Cumulative frequencies of S-phase stages in control and cohesin-deficient thymocytes. (E) Myc rescues impaired DNA replication in cohesin-deficient somatic cells. Efficient induction of DNA replication in cohesin-deficient somatic nuclei fused with Myc-expressing ES cells. Confocal analysis of DNA replication in heterokaryons between control (Rad21lox/lox) or cohesin-deficient (Rad21−/−) mouse thymocytes (encircled) and ZHBTc4 ES cells transduced with either Myc (Myc-IRES-GFP) or control vector (IRES-GFP; 45-min EdU pulse on day 1 after fusion). (Green) EdU; (red) actin; (blue) DAPI. Bar, 10 μm. (F) Numbers and percentages of EdU-positive control (Rad21lox/lox) or cohesin-deficient (Rad21−/−) thymocytes fused with ZHBTc4 ES cells transduced with either Myc (Myc-IRES-GFP) or control vector (IRES-GFP). Data are from three independent experiments. (G) Myc rescues reprogramming of cohesin-deficient somatic cells in ES cell heterokaryons. Expression of thymocyte-derived Pou5f1 in heterokaryons between control (Rad21lox/lox) or cohesin-deficient (Rad21−/−) thymocytes and ZHBTc4 ES cells transduced with either Myc (Myc-IRES-GFP) or control vector (IRES-GFP; normalized to Gapdh; mean ± SD; n = 3).
We conclude that the inefficient heterokaryon-mediated reprogramming of cohesin-deficient somatic cells is not due to the activation of stress responses or the inefficient transfer of ES cell-derived pluripotency factors.
As heterokaryon-mediated reprogramming is thought to involve the ES cell-mediated initiation of somatic DNA replication (Tsubouchi et al. 2013) and cohesin has been linked to efficient DNA replication (Terret et al. 2009; Guillou et al. 2010; Tittel-Elmer et al. 2012; Tedeschi et al. 2013), we considered whether cohesin-dependent alterations in DNA replication might underlie impaired reprogramming of cohesin-deficient somatic nuclei. We examined the initiation of DNA replication in wild-type and cohesin-deficient pre-B-cell nuclei in heterokaryons with ES cells (Fig. 6B). To comprehensively assess DNA replication, heterokaryons were pulsed with BrdU for 45 min at 6 h or 24 h after fusion, and cumulative BrdU incorporation was assessed by addition of BrdU overnight. While the overall frequency of DNA replication observed after fusion of G1-arrested mouse somatic cells was lower than that observed for proliferating hB cells (see Fig. 4), cohesin-deficient pre-B-cell nuclei displayed a marked reduction in the frequency of DNA replication compared with control pre-B-cell nuclei at all time points analyzed (Fig. 6B). Cohesin-deficient thymocytes showed a similar reduction in DNA synthesis in ES cell heterokaryons (data not shown). To address whether inefficient DNA replication is restricted to heterokaryons or a general feature of cohesin-deficient thymocytes, we stimulated CD4 single-positive thymocytes to enter the cell cycle (using antibodies to the T-cell receptor and the costimulatory receptor CD28) (Seitan et al. 2011) and scored activation-induced entry into and progression through S phase prior to the first mitosis. Cohesin-deficient thymocytes showed a lower frequency of EdU labeling than control thymocytes at all time points tested (18, 20, and 24 h after activation) (Fig. 6C). To analyze whether S-phase progression was delayed, we compared the patterns of EdU incorporation between control and cohesin-deficient thymocytes at 24 h after activation. The distribution of EdU patterns among cohesin-deficient thymocyte nuclei was skewed toward early stages of S phase (I: 43%; II: 26%; III: 20%; IV: 11%; and V: 1.2%) compared with wild-type thymocytes (I: 21%; II: 19%; III: 26%; IV: 23%; and V: 11%) (Fig. 6D), indicative of inefficient progression as well as initiation of S phase in cohesin-deficient thymocytes.
Hence, somatic cells lacking cohesin show inefficient initiation and progression of DNA replication. These data link poor reprogramming of cohesin-deficient somatic cells with impaired initiation and progression of DNA replication.
Taken together, these results suggest a model in which the heterokaryon-mediated reprogramming of cohesin-deficient somatic cells is impaired due to defective initiation and progression of DNA replication. This makes two testable predictions. First, if Myc can alleviate the replication defect of cohesin-deficient somatic cells, this should improve their reprogrammability in ES cell heterokaryons. Second, if reprogramming of cohesin-deficient somatic cells is defective as a result of impaired DNA replication, then cohesin-deficient somatic cells should be less disadvantaged in a replication-independent reprogramming system.
To test the first prediction, we asked whether Myc would alleviate the replication defect of cohesin-deficient somatic cells and improve their reprogrammability in heterokaryons with mES cells. We fused control (Rad21lox/lox) or cohesin-deficient (Rad21−/−) mouse thymocytes with ES cells transduced with either Myc (Myc-IRES-GFP) or control vector (IRES-GFP). To enable detection of thymocyte-derived Pou5f1 (Oct4), we used ZHBTc4 ES cells (Niwa et al. 2000). Myc expression in the ES cell fusion partner improved the efficiency with which control and cohesin-deficient thymocytes initiated DNA replication (Fig. 6E,F) and Pou5f1 (Oct4) expression (Fig. 6G), indicating that Myc-induced DNA replication can at least partly rescue the ability of cohesin-deficient somatic cells to be reprogrammed in ES cell heterokaryons.
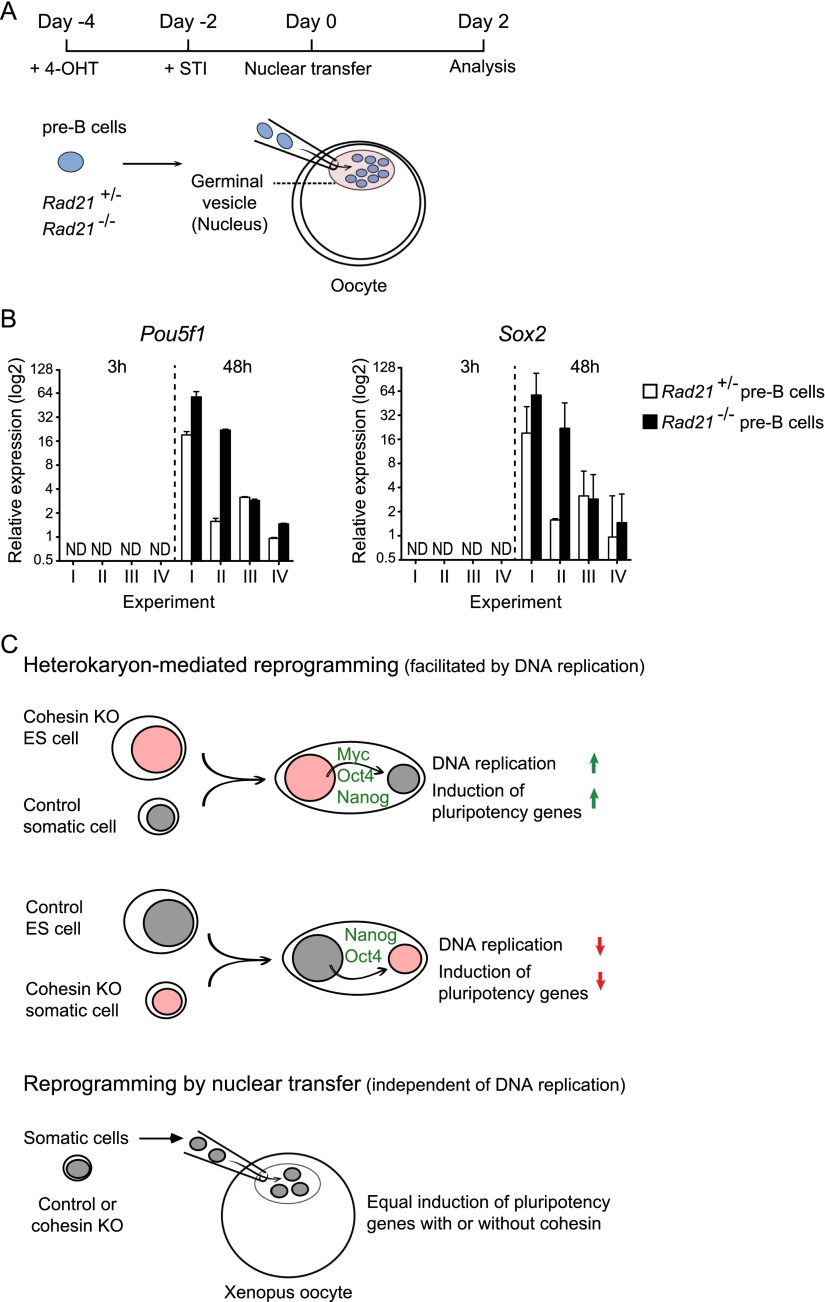
Finally, we asked whether cohesin was required for the reprogramming of somatic nuclei by Xenopus oocytes, which is known to occur in the absence of DNA replication (Gurdon 1976; Gurdon et al. 1976; Jullien et al. 2012). Somatic control or cohesin-deficient pre-B cells were injected into the germinal vesicle of Xenopus oocytes, and the expression of Pou5f1 (Oct4) and Sox2 was assessed by quantitative RT–PCR at the start of the experiment (3 h after injection) and after 2 d (Fig. 7A). In this replication-independent setting, cohesin-deficient pre-B cells initiated the expression of endogenous Pou5f1 (Oct4) and Sox2 at least as efficiently as control pre-B cells (Fig. 7B).
Figure 7.
Reprogramming of cohesin-deficient somatic cells in Xenopus oocytes in the absence of DNA replication. (A) Nuclear transfer into Xenopus oocytes. Cohesin depletion and cell cycle arrest of pre-B cells were induced as described in Figure 5D. Permeabilized control or cohesin-deficient pre-B cells were transplanted into the germinal vesicles of stage V–VI Xenopus oocytes and incubated at 16°C, and samples were collected after 3 h and 48 h for quantitative RT–PCR analysis of pluripotency gene expression. (B) Cohesin is not required for DNA replication-independent reprogramming of somatic nuclei in Xenopus oocytes. Each experiment comprised 24–40 oocytes injected with control pre-B cells and 24–40 oocytes injected with cohesin-deficient pre-B cells. Groups of six oocytes were pooled and each group was analyzed for the expression of Sox2 and Pou5f1 (Oct4) by quantitative RT–PCR analysis (two groups of six oocytes at 3 h and four to eight groups of six oocytes at 48 h, normalized to Gapdh). The mean expression for each group ±SD for each experiment is presented. At the 3-h time point, no Sox2 and Pou5f1 transcripts were detected within 40 amplification cycles of the RT–PCR reaction. (ND) Not detectable in 40 PCR cycles. After 48 h, transcriptional activation of Sox2 and Pou5f1 (Oct4) was observed in both control and cohesin-deficient pre-B cells. (C) The impact of cohesin on reprogramming in ES cell heterokaryons and nuclear transfer experiments. (Top) Acute cohesin deletion increases the reprogramming ability of ES cells by a Myc-dependent mechanism. (Middle) Cohesin depletion of somatic cells impairs DNA replication and the induction of pluripotency gene expression in ES cell heterokaryons. (Bottom) Pluripotency gene expression in cohesin-deficient somatic nuclei is induced efficiently by nuclear transfer experiments where reprogramming occurs in the absence of DNA replication.
Discussion
We addressed the role of cohesin in reprogramming and pluripotency gene expression by combining genetic cohesin depletion with reprogramming systems that dissociate cohesin functions in proliferation, DNA replication, and gene regulation. Unexpectedly, we found that the requirement for cohesin in heterokaryon-mediated reprogramming differs between somatic and ES cells (Fig. 7C). Loss of cohesin from somatic cells impaired their reprogrammability in heterokaryons, but loss of cohesin from ES cells enhanced their reprogramming ability (Fig. 7C). Reprogramming experiments with control and cohesin-deficient somatic cells in Xenopus oocytes indicate that cohesin is not required for the reactivation of the pluripotency factors Pou5f1 (Oct4) and Sox2 in the absence of DNA replication. This is in sharp contrast to the failure of cohesin-deficient somatic cells to reactivate Pou5f1 (Oct4), Sox2, and other pluripotency factors in heterokaryons, where DNA replication is required for reprogramming (Fig. 7C). The role of cohesin in somatic cell reprogramming is therefore at least in part to facilitate DNA replication in heterokaryon-mediated reprogramming (this study) and cell division in iPS cell-mediated reprogramming (Apostolou et al. 2013; Wei et al. 2013; Zhang et al. 2013).
Cohesin and the chromatin landscape
To explore the relationship between cohesin, enhancer–promoter interactions, and active transcription of pluripotency genes in ES cells, we selected Nanog, Lefty1, and Klf4 as examples of pluripotency genes whose expression was deregulated in acutely cohesin-depleted ES cells. We used ChIP and 3C to determine cohesin binding and long-range interactions. At the Nanog locus, reduced promoter–enhancer interactions mirrored reduced Rad21 occupancy as predicted (Kagey et al. 2010). In contrast, Lefty1 and Klf4 showed a clear dissociation between cohesin occupancy and long-range interactions, which were maintained in spite of reduced cohesin occupancy. Thus, while cohesin can undoubtedly mediate long-range chromosomal interactions (Hadjur et al. 2009; Mishiro et al. 2009; Nativio et al. 2009; Hou et al. 2010; Kagey et al. 2010; Seitan et al. 2011, 2013; Apostolou et al. 2013; Merkenschlager and Odom 2013; Wei et al. 2013; Zhang et al. 2013), this function is clearly not required for all enhancer–promoter interactions and may be dispensable at loci where regulatory elements are arranged in close proximity, with just a few kilobases of linear DNA sequence between them.
Analysis of ES cell heterokaryons with cohesin-deficient somatic cells suggest that the function of cohesin in the activation of pluripotency genes may be more complex than anticipated: Although cohesin-deficient somatic cells were less able to activate endogenous pluripotency genes, including Pou5f1, the Gof18 Oct4-GFP transgene was more efficiently activated by cohesin-deficient somatic cells than endogenous Pou5f1. The Oct4-GFP transgene has the same spacing between the Pou5f1 promoter and the distal Pou5f1 enhancer element (∼2 kb) as the endogenous Pou5f1 locus but evidently differs in chromosomal position and chromatin environment. While Oct4-GFP transgenes may be reactivated more easily than endogenous pluripotency genes (Tada et al. 2001), to our knowledge, this is the first example that the requirement for cohesin in the activation of a target gene depends on the gene’s chromatin environment.
In re-examining the relationship between cohesin and pluripotency gene expression, our data call into question simple models of cohesin binding, long-range interactions, and gene expression. Instead, the impact of cohesin is highly locus-specific.
Cohesin, Myc, DNA replication, and reprogramming
The expression of Myc and cohesin is positively correlated in biological systems ranging from flies to humans (Rhodes et al. 2010). Exceptions include resting thymocytes (where Myc expression is low and not affected by cohesin depletion) (Seitan et al. 2013), fibroblasts deficient in the cohesin unloading factor Wapl (which show increased cohesin association with chromatin yet fail to up-regulate Myc in response to activation with growth factors) (Tedeschi et al. 2013), and ES cells acutely depleted of cohesin under 2i conditions (this study). Hence, as noted previously (Schaaf et al. 2009), the direction of cohesin target gene deregulation in response to cohesin depletion is not always predictable. Our experiments reveal a proliferation-independent role for Myc in heterokaryon-mediated reprogramming. Myc is one of the four original Yamanaka factors (Takahashi and Yamanaka 2006) and facilitates reprogramming by increasing the proliferation of iPS cells (Hanna et al. 2009) and cooperating with other reprogramming factors in the transcriptional regulation of target genes (Soufi et al. 2012). While proliferation is not a contributing factor in heterokaryons, our data show that in addition to transcriptional effects (Kim et al. 2010; Lin et al. 2012; Soufi et al. 2012), Myc’s ability to promote DNA replication (Dominguez-Sola et al. 2007) contributes to heterokaryon-mediated reprogramming. Myc’s ability to drive DNA replication may deserve additional attention in the context of reprogramming.
In thymocytes and pre-B cells, the two somatic cell types that we investigated, cohesin facilitates the initiation and progression of DNA replication. Cohesin and replication are intimately linked, since cohesion between sister chromatids is established during replication in S phase (Gerlich et al. 2006; Nasmyth and Haering 2009; Terret et al. 2009; Gause et al. 2010). Cohesin associates with origins of DNA replication in Xenopus (Gillespie and Hirano 2004; Takahashi et al. 2004), Drosophila (MacAlpine et al. 2010), and mammalian genomes (Guillou et al. 2010), where cohesin affects the spacing of replication origins in cohesin-depleted cells (Guillou et al. 2010), the progression of replication forks (Terret et al. 2009), and the restart of stalled forks (Tittel-Elmer et al. 2012). As a result, altering the dosage or function of cohesin can affect DNA replication (Terret et al. 2009; Guillou et al. 2010; Tedeschi et al. 2013). Similar to the cohesin-deficient thymocytes and pre-B cells described here, fibroblasts that have irreversible cohesin binding as a result of Wapl mutations show defects in progression to S phase (Tedeschi et al. 2013). Myc expression was reduced in these cells, and exogenous Myc promoted S-phase entry and progression (Tedeschi et al. 2013).
Separating cohesin functions in chromosome segregation, DNA replication, and gene regulation
Our experiments delineate distinct cohesin functions in chromosome segregation, DNA replication, and the regulation of gene expression, all of which contribute to pluripotency and reprogramming in different experimental systems as outlined below.
Canonical cohesin functions
Previous studies on the role of cohesin in pluripotency (Kagey et al. 2010) and reprogramming (Apostolou et al. 2013; Wei et al. 2013; Zhang et al. 2013) used prolonged cohesin depletion over the course of several cell cycles. Cell division in the absence of cohesin results in DNA damage responses that are detrimental to pluripotency gene expression (Lin et al. 2005; Maimets et al. 2008) and reprogramming (Banito et al. 2009; Li et al. 2009; Utikal et al. 2009). Our findings do not support models in which cohesin is required for pluripotency gene expression (Kagey et al. 2010) and iPS cell-mediated reprogramming because it mediates long-range chromosomal interactions (Apostolou et al. 2013; Wei et al. 2013; Zhang et al. 2013). Rather, our data suggest that the key role of cohesin in these experiments is to preserve genome integrity. Specific conclusions about a role for cohesin in pluripotency gene expression and reprogramming should not be drawn from experiments that involve cell division in the absence of cohesin.
Emerging cohesin functions in DNA replication
Our experiments with heterokaryon-mediated reprogramming support studies that have shown a role for cohesin in DNA replication (Terret et al. 2009; Guillou et al. 2010), and this emerging role is important for the activation of pluripotency gene expression by somatic cells in ES cell heterokaryons, where the depletion of cohesin results in slow S-phase progression and impaired pluripotency gene expression. This role is mechanistically distinct from chromosome segregation in mitosis (Nasmyth and Haering 2009; Terret et al. 2009; Guillou et al. 2010). Operationally, in contrast to cell division in the absence of cohesin, DNA replication in the absence of cohesin does not trigger DNA damage responses (Guillou et al. 2010) that would dominantly block reprogramming. Importantly, we showed that heterokaryon-mediated reprogramming of cohesin-deficient somatic cells is efficiently rescued by Myc.
Specific functions of cohesin in the regulation of gene expression
Acute cohesin depletion in ES cells showed that cohesin has a role in the regulation of a specific subset of pluripotency-related genes. The expression of Pou5f1 (Oct4) and Sox2 expression was unaffected by acute depletion of cohesin in ES cells. Consistent with these data in ES cells, nuclear transfer experiments show that the expression of Pou5f1 (Oct4) and Sox2 can be readily activated in cohesin-deficient cells by nuclear transfer into Xenopus oocytes. One of the pluripotency genes actually regulated by cohesin is Myc. Up-regulated by cohesin depletion, Myc was critical for the increased reprogramming ability of cohesin-depleted ES cells in heterokaryons. As heterokaryon-mediated reprogramming is facilitated by DNA replication in the absence of proliferation, these data not only demonstrate a critical role for cohesin in regulating Myc expression in ES cells but also delineate a novel, proliferation-independent function of Myc in reprogramming.
In conclusion, these data link the loss of cohesin in somatic cells to inefficient DNA replication and reprogramming in experimental heterokaryons. They reveal novel, proliferation-independent contributions of Myc to replication-dependent reprogramming and redefine cohesin’s role in pluripotency.
Materials and methods
Mice, flow cytometry, cell sorting, and culture
Mouse work was performed under a UK Home Office Project Licence according to the Animals (Scientific Procedures) Act, UK. Mice carrying floxed Rad21 or Smc3 alleles in combination with the transgene CD4Cre (Seitan et al. 2011), ERt2Cre in the Rosa26 locus (Seibler et al. 2003), or human Eμ-bcl-2-36 Bcl2 (Strasser et al. 1991) on a mixed C57BL/129 background were maintained under specific pathogen-free conditions. Cells were stained, analyzed, and sorted on Becton Dickinson Calibur, DIVA, or Aria flow cytometer.
Control (Rad21lox/lox) and cohesin-deficient (CD4Cre Rad21lox/lox) CD4 single-positive thymocytes were isolated by flow cytometry and activated with antibodies to the T-cell receptor and the costimulatory receptor CD28 as described (Seitan et al. 2011).
Abelson-transformed pre-B-cell lines were established from mouse fetal liver cells that carried Ert2Cre (Seibler et al. 2003) and Bcl2 (Strasser et al. 1991) transgenes and Rad21lox/wt or Rad21lox/lox alleles (Seitan et al. 2011) as described (Bredemeyer et al. 2006) and maintained in IMDM, 2 mM L-glutamine, 50 µM β-mercaptoethanol, and antibiotics (10 μg/mL penicilin and streptomycin) (Invitrogen). Cohesin depletion was induced by addition of 400 nM 4′OHT and G1 arrest by addition of 2 μM STI 571 (Sigma).
Mouse ERt2-Cre Rad21wt/wt and Rad21lox/lox ES cells were derived in dual inhibition 2i medium (Ying et al. 2008). Eight-cell embryos were cultured for 2 d in KSOM medium (Millipore) with 1 µM PDO325901 (MEK inhibitor; Stemgent) and 3 µM CHIR99021 (GSK3 inhibitor; Stemgent) (KSOM + 2i) and then transferred to N2B27 medium (neurobasal medium supplemented with 50% [v/v] DMEM/F12, 0.5% [v/v] N2 supplement, 1% [v/v] B27 supplement, 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 100 µM β-mercaptoethanol, 1000 U/mL LIF, 1 µM PD032590, 3 µM CHIR99021). Outgrowths were trypsinized and transferred to laminin-coated 96-well plates for 3–7 d in N2B27 + 2i + leukaemia inhibitory factor (LIF) until ES-like colonies emerged. Cohesin depletion was induced by addition of 100 nM 4′OHT to cell cultures.
E14Tg2a Hprt−/− and ZHBTc4 mES cell lines were maintained in knockout Dulbecco’s modified Eagle medium (KO DMEM) (Invitrogen) supplemented with 10% (v/v) fetal calf serum (FCS) (Invitrogen), 2 mM L-glutamine, antibiotics (100 U/mL penicillin/100 µg/mL streptomycin), 50 µM β-mercaptoethanol, 1 mM nonessential amino acids (NEAA) (Invitrogen), and 1000 U/mL LIF. These cells were cultured in flasks coated with 0.1% gelatine (Sigma).
Human H7 ES cells were maintained in mTESR-1 serum-free medium and passaged 1:4 every 5–6 d by incubation with 0.02% EDTA (Invitrogen), and colonies were broken into small cell clusters (50–200 cells). Cells were replated on tissue culture dishes coated overnight with 0.5 mg/mL growth factor-reduced Matrigel (BD Biosciences).
Human EBV-transformed B cells were maintained in RPMI-1640 (Invitrogen) supplemented with 10% FCS, 2 mM L-glutamine, and antibiotics.
Heterokaryons between mES cells and puromycin-resistant hB cells were generated as described (Pereira et al. 2008). Unfused mES cells were eliminated by 1 μg/mL puromycin (Sigma) or HAT (20 μM hypoxanthine, 0.08 μM aminopterin, 3.2 μM thymidine; Sigma).
Heterokaryons between hES cells and mouse thymocytes or mouse pre-B cells were generated using PEG-mediated fusion as described (Pereira et al. 2008). Unfused hES cells were eliminated by the addition of 10−5 M ouabain (Sigma) and unfused nonadherent somatic cells were washed away during sample collection. Hybrids between ZHBTc4 mES cells and Oct4-GFP transgenic thymocytes (control or Rad21−/−) were sorted by GFP expression 9 d after fusion, and colonies were assayed for alkaline phosphatase expression on day 15 after fusion (86R-1KT, Sigma).
Nuclear transfer to Xenopus oocytes was carried out as described (Halley-Scott et al. 2010). Oocytes were defolliculated with liberase (05401127001, Roche) and kept in modified Barth’s solution (1× MBS; 88 mM NaCl, 1 mM KCl, 1 mM MgSO4, 10 mM Hepes, 2.5 mM NaHCO3, 0.7 mM CaCl2 at pH 7.4.) at 16°C before use. Pre-B cells were permeabilized with 40–100 μg/mL digitonin in SuNaSp (0.25 M sucrose, 75 mM NaCl, 0.5 mM spermidine, 0.15 mM spermine) for 3 min on ice. The reaction was stopped by the addition of BSA, and cells were washed and resuspended in SuNaSp and 1% BSA at a concentration of 30 million cells per milliliter. Four-hundred to 500 cells were injected into a Xenopus oocyte nucleus. The injected oocytes were cultured at 16°C in 1× MBS supplemented with 0.1% BSA and antibiotics.
RNA and protein analysis
RNA was extracted with QIAshredder and RNeasy minikits (Qiagen), and residual DNA was eliminated using DNA-free kit (Ambion) and reverse-transcribed using the SuperScript first-strand synthesis system (Invitrogen). Real-time PCR of cDNA was carried out on a Opticon DNA engine using Opticon Monitor 3 software (MJ Research, Inc.) under the following cycling conditions: 15 min at 95°C and 40 cycles of 15 sec at 94°C, 30 sec at 60°C, and 30 sec at 72°C, at which point the fluorescence was read at 72°C, 75°C, 78°C, and 83°C. The melting curve was determined from 70°C to 90°C at 0.2°C intervals. RT–PCR primer sequences are shown in Supplemental Table S1.
Transcripts derived from mouse nuclei injected into Xenopus oocytes were detected as described (Halley-Scott et al. 2010). Three hours and 48 h after nuclear transfer, six injected oocytes (equivalent to 2400–3000 injected nuclei) were pooled as one sample, and two (3 h) and four to eight (48 h) samples were analyzed per condition. RNA was extracted using Qiagen RNeasy columns. After RNA extraction, including on-column DNase I digestion, reverse transcription was performed using SuperScript III with oligo dT primers. Real-time PCR was performed as SYBR Green assays on an ABI 7300 Cycler. Quantification was done with a mix of ES cell and mouse embryonic fibroblast cDNA within the log-linear phase of the amplification curve. Primers are listed in Supplemental Table S1.
Samples for microarray analysis were prepared for hybridization to Affymetrix mouse GeneChIP 1.0 ST arrays as advised by the manufacturers (Affymetrix). Raw microarray data sets were processed using the VSN Bioconductor R package, and differential expression was determined using the Limma Bioconductor R package.
Western blot analysis of whole-cell extracts used the following antibodies: rabbit polyclonal to Rad21 (1:1000; ab992, Abcam), rabbit polyclonal to Smc3 (1:1000; ab9263, Abcam), rabbit polyclonal to Myc N-262 (1:1000; sc-764, Santa Cruz Biotechnology), mouse monoclonal anti-α tubulin (1:1000; T9026, Sigma), and rabbit polyclonal antibody to histone H3 (1:10,000; ab1791, Abcam). The secondary antibodies used were anti-mouse HRP (1:2000, GE Healthcare) and anti-rabbit HRP (1:5,000; GE Healthcare). The Amersham ECL plus kit (GE Healthcare) was used for detection.
ChIP and 3C
ChIP was done as described (Kagey et al. 2010) with the following modifications: Fifty million formaldehyde cross-linked cells were sonicated in a Diagenode Bioruptor for 20 min with 30-sec on/off cycles. Chromatin was quantified and diluted to a final concentration of 150 µg in 500 µL of sonication buffer for each ChIP reaction. For Rad21 ChIP, 500 µL of the diluted chromatin was incubated overnight at 4°C, with 100 µL of Dynal Protein G beads preincubated with 20 µg of antibody (ab992, Abcam). ChIP-PCR primers are listed in Supplemental Table S2.
3C was performed as described (Seitan et al. 2011) except that ES cell nuclei were permeabilized with 0.3% SDS, and chromatin was digested with 1500 U of DpnII (R0543M, New England Biolabs ) overnight at 37°C for Nanog and Lefty1 and 750 U of PvuII (R0151M, New England Biolabs) for 2 h followed by an additional 750 U overnight at 37°C for Klf4. BAC templates for Nanog and Lefty1 were digested with the DpnII isoschizomer Sau3AI. Digestion efficiencies in chromatin were between 90% and 100% for Nanog and Lefty1 and >80% for Klf4, except for two restriction sites outside the enhancer region, where digestion efficiencies were between 55% and 70%. The efficiency of 3C primers was verified on BAC templates as described (Seitan et al. 2011). 3C primers are listed in Supplemental Table S3.
Immunofluorescence staining and confocal microscopy
Cells or heterokaryons were plated on poly-L-lysine-coated coverslips (Sigma), fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.5% Triton X-100 (Sigma) for 5 min, treated with blocking solution (3% goat serum, 0.1% Triton X-100), and incubated with primary antibodies to Rad21 (1:500; ab154769, Abcam), Oct-3/4 (1:200; mouse monoclonal BD611203, BD Biosciences), Nanog (1:200; rabbit polyclonal REC-RCAB0001P, Cosmo Bio), or actin (1:500; mouse monoclonal MAB1501, Millipore) for 1 h at room temperature or overnight at 4°C and with secondary antibodies (Molecular Probes) for 1 h at room temperature. Coverslips were mounted in VectaShield with 0.1 µg/mL DAPI (Vector Laboratories).
For BrdU detection, heterokaryons were pulse-labeled with 100 µM BrdU (Sigma) for 45 min or 10 µM BrdU overnight, fixed with 4% PFA for 20 min at room temperature, and washed three times in 3% BSA in PBS. Samples were incubated for 30 min in blocking buffer (3% BSA, 0.05% Triton in PBS) in a humid chamber and then incubated for 45 min with anti-BrdU-FITC antibody (Beckton Dickinson 347583) diluted 1:4 in blocking buffer. Coverslips were mounted in VectaShield with DAPI.
For EdU/BrdU double staining, ES cells were incubated with 10 µM EdU for an overnight pulse. The next day, cell fusion was performed, and heterokaryons were plated on gelatinized coverslips. One day after fusion, heterokaryons were pulse-labeled with 100 µM BrdU. Samples were fixed in 4% PFA for 10 min and permeabilized with 0.5% Triton X-100 (Sigma). Washing steps were performed in 3% BSA in PBS. EdU was detected with the Click-iT EdU Alexa Fluor 647 HCS assay (Molecular Probes, Invitrogen) as advised by the manufacturer. After one wash in 3% BSA/PBS, samples were incubated with 5 µg/mL DNase (Sigma) for 10 min at room temperature followed by 10 min of incubation at 37°C. Samples were washed three times in 3% BSA/PBS and incubated with anti-BrdU-FITC antibody (1:4 in 3% BSA, 0.05% Tween, 10% normal goat serum in PBS) for 1 h at room temperature. Cells were washed three times in 3% BSA/PBS and mounted in VectaShield with DAPI.
Samples were visualized using a TCS SP5 Leica laser-scanning confocal microscope. Images were processed using Leica Confocal software and Adobe Photoshop CS5. Microscope settings and laser power were kept constant between the controls and samples.
Retroviral transduction of mES cells
MSCV retrovirus vector containing Myc cDNA (Myc-IRES-GFP) or IRES-GFP control vector was produced by calcium phosphate cotransfection with the replication-incompetent helper vector pCL-Eco (Addgene) into 293T cells. DNA-containing precipitates were formed by slowly adding 500 µL of 2× HEBS buffer (280 nM NaCl, 10 mM KCl, 1.5 mM Na2HPO4.2H2O, 12 mM glucose, 50 mM HEPES-free acid at pH 7.05) to 500 µL of 4 µg of c-Myc-MIG DNA and 4 µg of helper vector DNA in 0.4 M CaCl2. One milliliter of DNA precipitate was slowly added to a 10-cm dish of 40%–60% confluent 293T cells, and the medium was replaced after 24 h. Virus-containing supernatant was harvested 48, 60, and 72 h post-transfection. mES cells were plated at 106 cells per well of a six-well dish, retroviral supernatant was added, and the cells were centrifuged at 2500 rpm for 45 min at room temperature. The retroviral supernatants were removed, and the cells were resuspended in ES cell medium and plated onto gelatinized dishes. Three days after infection and 2 d before fusion experiments, GFP-positive mES cells were sorted by flow cytometry.
Data access
Microarray gene expression data for control and 24-h Rad21-deleted mES cells have been deposited at Gene Expression Omnibus under accession number GSE62325.
Supplementary Material
Acknowledgments
We thank Dr. J. Elliott for cell sorting, Dr. L. Game of the MRC Clinical Sciences Centre genomics facility, and the members of the Lymphocyte Development Group for discussion and advice. This work was supported by the Medical Research Council, UK (M.M. and A.G.F.); the Wellcome Trust (WT099276/Z/12/Z to M.M. and WT101050/Z/13/Z to J.B.G.); a predoctoral fellowship from Boehringer Ingelheim (T.L.); a Commonwealth Scholarship (P.G.); a long-term EMBO fellowship (E.H.); and the European Research Council (H.M.-B. and A.G.F.). E.H. and J.B.G. acknowledge the core support provided by Cancer Research UK C6946/A14492 and WT092096.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.251835.114.
References
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi T, Borowsk ML, et al. 2013. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 12: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. 2013. Genome architecture: domain organization of interphase chromosomes. Cell 152: 1270–1284. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442: 466–470. [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. 2009. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4: 403–415. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Gautier J. 2014. MYC and the control of DNA replication. Cold Spring Harb Perspect Med 4: a014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Gallowa DA, Gu W, Gautie J, Dalla-Favera R. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451. [DOI] [PubMed] [Google Scholar]
- Dorsett D, Merkenschlager M. 2013. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol 25: 3217–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D. 2010. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. 2006. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. 2004. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 14: 1598–1603. [DOI] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J. 2010. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. 1976. Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J Embryol Exp Morphol 36: 523–540. [PubMed] [Google Scholar]
- Gurdon JB, Partington GA, De Robertis EM. 1976. Injected nuclei in frog oocytes: RNA synthesis and protein exchange. J Embryol Exp Morphol 36: 541–553. [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley-Scott RP, Pasque V, Astrand C, Miyamoto K, Simeoni I, Jullien J, Gurdon JB. 2010. Mammalian nuclear transplantation to germinal vesical stage Xenopus oocytes—a method for quantitative transcriptional reprogramming. Methods 51: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. 2010. Cell types pecificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci 10: 3651–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. 2012. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chrom 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. 2005. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 7: 165–171. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimets T, Neganova I, Armstrong L, Lako M. 2008. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene 27: 5277–5287. [DOI] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. 2013. CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152: 1285–1297. [DOI] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. 2009. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 28: 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. 2009. Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558. [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al. 2011. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS ONE 6: e19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. 2008. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet 4: e1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA. 2010. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol 344: 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. 2009. Regulation of the Drosophila enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS ONE 4: e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Zevnik B, Kuter-Luks B, Andreas S, Kern H, Hennek T, Rode A, Heimann C, Faust N, Kauselmann G, et al. 2003. Rapid generation of inducible mouse mutants. Nucleic Acids Res 31: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. 2013. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 23: 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. 2013. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 32: 3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Cory S. 1991. Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 67: 889–899. [DOI] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol 11: 1553–1558. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. 2004. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 6: 991–996. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. 2013. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, François P, Hohl M, Petrini JH, Pasero P, Cobb JA. 2012. Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol Cell 48: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG. 2013. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell 152: 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valovka T, Schönfeld M, Raffeiner P, Breuker K, Dunzendorfer-Matt T, Hartl M, Bister K. 2013. Transcriptional control of DNA replication licensing by Myc. Sci Rep 3: 3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. 2013. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 13: 36–47. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T, Sugiyama N, De Felice M, Yeom YI, Ohbo K, Masuko K, Obinata M, Abe K, Schöler HR, Matsui Y. 1999. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ 41: 675–684. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, Xu X, Shen A, Li T, Niu B, et al. 2013. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell 13: 30–35. [DOI] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van Ijcken WF, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci 111: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.