Deep sequencing of embryonic stem cell RNA revealed many specific internal introns that are significantly more abundant than the other introns within polyadenylated transcripts. Boutz et al. identified thousands of these “detained” introns (DIs) in human and mouse cell lines as well as the adult mouse liver. Drug inhibition of Clk, a stress-responsive kinase, triggered rapid splicing changes for a specific subset of DIs, altering transcript pools of >300 genes. Srsf4 regulates the splicing of some DIs, particularly in genes encoding RNA processing and splicing factors.
Keywords: Clk kinase, detained introns, post-transcriptional splicing
Abstract
Deep sequencing of embryonic stem cell RNA revealed many specific internal introns that are significantly more abundant than the other introns within polyadenylated transcripts; we classified these as “detained” introns (DIs). We identified thousands of DIs, many of which are evolutionarily conserved, in human and mouse cell lines as well as the adult mouse liver. DIs can have half-lives of over an hour yet remain in the nucleus and are not subject to nonsense-mediated decay (NMD). Drug inhibition of Clk, a stress-responsive kinase, triggered rapid splicing changes for a specific subset of DIs; half showed increased splicing, and half showed increased intron detention, altering transcript pools of >300 genes. Srsf4, which undergoes a dramatic phosphorylation shift in response to Clk kinase inhibition, regulates the splicing of some DIs, particularly in genes encoding RNA processing and splicing factors. The splicing of some DIs—including those in Mdm4, a negative regulator of p53—was also altered following DNA damage. After 4 h of Clk inhibition, the expression of >400 genes changed significantly, and almost one-third of these are p53 transcriptional targets. These data suggest a widespread mechanism by which the rate of splicing of DIs contributes to the level of gene expression.
A large body of evidence supports the close coupling of splicing and transcription, and many if not most mammalian introns are spliced prior to the transcriptional termination and polyadenylation of the pre-mRNA; i.e., cotranscriptionally (for review, see Perales and Bentley 2009; Pandya-Jones 2011). However, numerous studies examining the subcellular localization and splicing kinetics of individual introns have pointed to the existence of a population that is processed more slowly—in some cases, post-transcriptionally (Bauren and Wieslander 1994; Smith et al. 1999; Melcak et al. 2001; Hao and Baltimore 2013). Most introns detected by in situ hybridization are gene-proximal, probably in nascent transcripts, but others are dissociated from the site of transcription (Vargas et al. 2011; Girard et al. 2012). More recently, high-throughput sequencing studies in multiple organisms have detected the presence of introns in poly(A)+ or fully transcribed, chromatin-associated RNA populations, indicating that some fraction of splicing occurs post-transcriptionally (Ameur et al. 2011; Khodor et al. 2011, 2012; Rabani et al. 2011; Bhatt et al. 2012; Tilgner et al. 2012; Windhager et al. 2012; Pandya-Jones et al. 2013; Shalgi et al. 2014; Braunschweig et al. 2014). Prior studies have observed that the kinetics of removal of alternatively spliced introns are slower than those of the typical constitutive intron (Pandya-Jones and Black 2009; Khodor et al. 2011; Vargas et al. 2011; Pandya-Jones et al. 2013). Here, it has been suggested that the kinetics may be intrinsic to the splicing outcome; i.e., that constitutive splicing is accomplished cotranscriptionally, while regulated variants are spliced more slowly.
Splicing can respond to both intracellular and extracellular signals. For example, the splicing of particular introns is inhibited by cellular stresses such as heat shock as well as during mitosis (Yost and Lindquist 1986; Bond 1988; Shin and Manley 2002; Shin et al. 2004). A handful of publications describe post-transcriptional, regulated splicing triggered by environmental signals. Apoe4 and Cyr61 transcripts each contain an intron that is altered in splicing kinetics in response to excitotoxicity and hypoxia, respectively (Xu et al. 2008; Hirschfeld et al. 2009). Polyadenylated pre-mRNAs encoding the SR protein kinase Clk1 contain two unspliced introns flanking a cassette exon. These intron-containing transcripts are retained in the nucleus until the cells experience heat or osmotic shock or treatment with a chemical inhibitor of the Clk kinases, at which point splicing occurs, allowing production of more protein (Ninomiya et al. 2011). Intriguingly, Clk kinases are also implicated in the control of splicing of retained introns in platelets. Although they are annucleate, platelets contain particular pre-mRNAs that are fully spliced with the exception of a single intron. The remaining intron is spliced out of the message in response to platelet activation to produce translatable mRNA (Denis et al. 2005; Schwertz et al. 2006). These examples hint at a process by which gene expression can be modulated through the regulated, post-transcriptional splicing of one or a few introns.
The cyclical addition and removal of phosphates from SR proteins drives the splicing reaction (Mermoud et al. 1992; Tazi et al. 1993; Roscigno and Garcia-Blanco 1995; Shi et al. 2006). SR proteins are a family of RNA-binding factors involved in both constitutive and alternative splicing (for review, see Zhou and Fu 2013) that are phosphorylated in the cytoplasm by SRPK kinase family members and in the nucleus by the Clk family kinases (Gui et al.1994; Colwill et al. 1996; Aubol et al. 2013). Dephosphorylation of SR proteins and other spliceosomal components by the PP1/2A phosphatases is required for the two steps in splicing (Mermoud et al. 1994; Roscigno and Garcia-Blanco 1995; Cao et al. 1997; Kanopka et al. 1998; Xiao and Manley 1998; Shi et al. 2006). Early recognition of exonic enhancers by phosphorylated SR proteins is important in spliceosome assembly and nuclear retention of intron-containing transcripts, but dephosphorylation of certain SR proteins during the splicing process allows nuclear export of spliced mRNAs (Chang and Sharp 1989; Legrain and Rosbash 1989; Huang et al. 2004; Taniguchi et al. 2007; Dias et al. 2010; Takemura et al. 2011). Thus, it is likely that functional prespliceosomes can be assembled on pre-mRNAs but remain poised and inactive through control of the phosphorylation state of associated factors such as SR proteins and small nuclear ribonucleoprotein (snRNP) components (Girard et al. 2012).
Autoregulation of mRNA processing or translation by the encoded protein product is a common mechanism for homeostatic control. In some cases, alternative splicing directly impacts protein expression through coupling to nonsense-mediated decay (NMD). A mature mRNA containing an in-frame premature termination codon (PTC) more than ∼50 nucleotides (nt) upstream of the last exon–exon junction is typically degraded by NMD (for review, see Popp and Maquat 2013). The most common configuration of these genes involves cassette exon splicing, where either skipping or inclusion of the cassette exon leads to the introduction of a PTC, while the other splice isoform is a coding sequence; we refer to these as “NMD switch” exons. Splicing factors, including SR proteins and hnRNPs, have been shown to autoregulate their levels by binding to their pre-mRNA and controlling the splicing of NMD switch exons, and the resulting networks of splicing regulators in turn exert combinatorial splicing control on many more downstream target exons. (Wollerton et al. 2004; Boutz et al. 2007; Lareau et al. 2007; Ni et al. 2007, Jangi et al. 2014).
Mdm4 negatively regulates p53, acting with the related protein Mdm2 as an E3 ubiquitin ligase that induces p53 degradation (for review, see Wade et al. 2013). It has previously been observed that a small-molecule Clk inhibitor triggered an increase in Mdm4 NMD isoform splicing, a corresponding decrease in Mdm4 protein, and up-regulation of p53 protein levels (Allende-Vega et al. 2013; Bezzi et al. 2013). This effect was attributed to induction of stress due to a general inhibition of splicing; however, to date, there has been no global analysis of splicing in cells treated with Clk inhibitors.
Through deep sequencing of poly(A)-selected RNA from mouse embryonic stem cells (mESCs), we discovered a striking number of nuclear transcripts containing introns that are spliced more slowly than other introns within the same gene. Many such introns are also present and often conserved in human cell lines as well as in differentiated, adult mouse livers, hinting at a previously unappreciated and widespread mechanism of “detained” introns (DIs). As an example of this phenomenon, we demonstrated that in addition to its known autoregulatory role, Clk kinase activity affects the detention of a specific subset of DIs, and a small-molecule Clk inhibitor can promote the post-transcriptional splicing or increased detention of DIs in hundreds of genes, likely through modulation of SR protein phosphorylation. Gene ontology (GO) analysis indicates that many DNA damage response genes contain DIs, and we found that specific DIs undergo changes in splicing following doxorubicin-induced DNA damage. We propose that transcripts containing DIs form a nuclear-detained pool of mostly processed pre-mRNA, subsets of which undergo regulated changes in splicing in response to changes in the cellular environment.
Results
A subset of introns is abundant in mouse and human poly(A)+ RNA
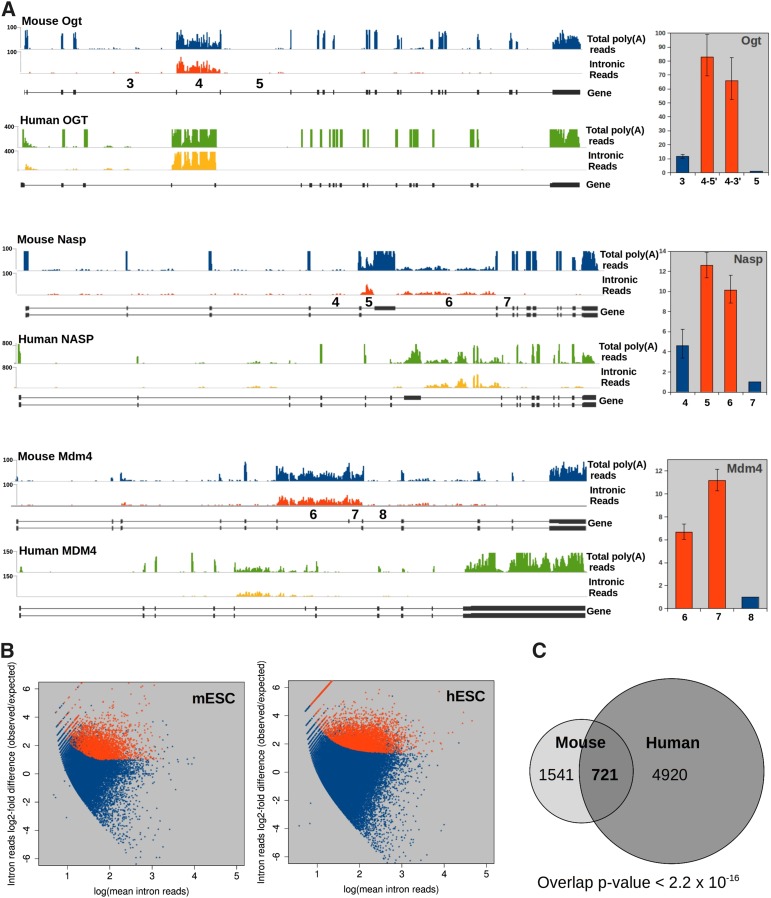
Surprisingly, deep sequencing of poly(A)-selected RNA from mESCs revealed many transcripts in which specific internal introns were significantly more abundant than other introns within the same transcripts. When compared with mapped reads from human ESC (hESC)-derived poly(A)+ RNA sequencing (RNA-seq) libraries (The ENCODE Project Consortium 2012), in most cases, the orthologous human introns also exhibit unusually high read coverage, suggesting that this property is biologically conserved (Fig. 1A). These intron reads are not from RNAs that are inherently unspliceable or misannotated, since, in all cases, we can detect reads mapping to the junctions between the flanking spliced exons. To verify the elevated abundance of high-read-coverage introns in the RNA-seq data sets using an independent method, we measured the intron levels from a number of loci by real-time quantitative RT–PCR (qRT–PCR) using primer sets spanning the exon–intron junctions of high-coverage introns as well as the nearest flanking introns. In all cases tested, the high-coverage introns identified by RNA-seq (Fig. 1A, red bars) were markedly more abundant than their flanking neighbors (Fig. 1A, blue bars and right panels; data not shown). While present at higher levels than other introns in the same transcript, even the most abundant of the high-coverage introns assayed (Ogt intron 4) was estimated to be present in only 30%–50% of poly(A) transcripts based on a comparison with exon–exon signal; measurement of high-coverage introns in fibronectin (Fn1) transcripts showed them to be present in 4%–7% of poly(A) transcripts at steady state (Supplemental Fig. S1A).
Figure 1.
Mouse and human poly(A)+ RNA contain an enriched subset of introns. (A) Total mapped read densities from poly(A)+ RNA-seq libraries from mESCs (blue tracks) or hESCs (green tracks) and the same data sets with exonic reads subtracted (intronic reads; red in mESCs and yellow in hESCs). Positions of exons are shown below (gene). Intron numbers for mESC introns assayed by qRT–PCR are shown above the gene schematic. (Right panels) qRT–PCR of high-coverage introns shown in red, normalized to the downstream flanking intron (mean n = 3–6 ± SEM). (B) Quantification of introns in mESC and hESC poly(A) RNA-seq. Introns with a greater than twofold ratio of observed/expected read count, FDR < 0.01, are shown in red; all other expressed introns are in blue. (C) Overlap of introns showing statistically significant high coverage in mice and humans for which an orthologous relationship could be determined. P-value was determined by two-sided hypergeometric. See also Supplemental Figure S1 and Supplemental Data S1 and S2.
To determine the prevalence of high-coverage introns genome-wide, we developed a novel computational pipeline using the R package DESeq (Anders and Huber 2010) to quantitatively identify introns whose read coverage exceeds the sampling noise present in RNA-seq data sets. We identified 3150 such introns (4.2% of internal introns in expressed genes) in mESCs at a false discovery rate (FDR) of 1% as well as thousands more (5%–15% of expressed introns) in four human cell lines (hESCs, HeLa, HepG2, and human umbilical vein endothelial cells [HUVECs]) (Fig. 1B; Supplemental Fig. S1B; Supplemental Table S1; Supplemental Data S1, S2; The ENCODE Project Consortium 2012). The differences in DI frequencies were not due to the number of reads mapped (data not shown). Of the 2262 high-coverage mouse introns that could be mapped directly to orthologous human introns, 721 (32%) were also significantly enriched in one or more human cell lines (Fig. 1C). This degree of overlap is highly statistically significant (P < 2.2 × 10−16), indicative of a phylogenetically conserved regulatory process. This conservation strongly suggests that the property of post-transcriptional splicing for specific introns has a biological function that underlies their selection over time. Because these introns remain present in polyadenylated transcripts in which all other introns have been spliced out and are retained in the nucleus (see below), we refer to them as DIs.
Properties of DIs
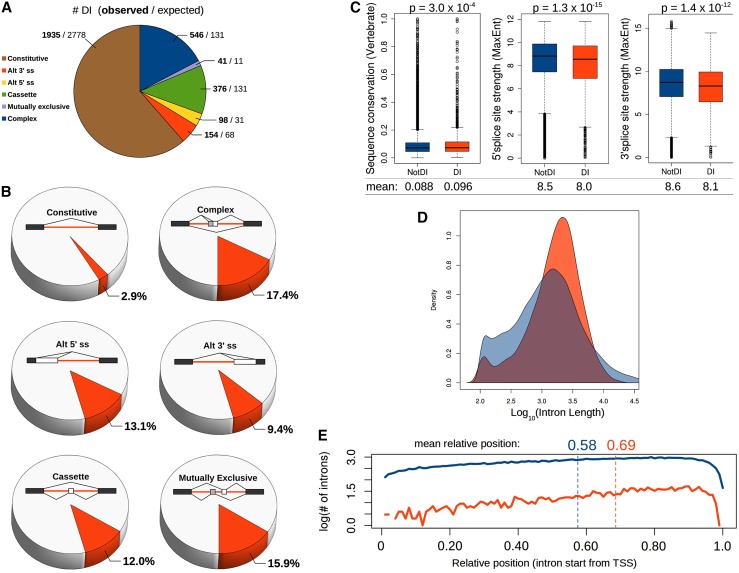
It has been observed that introns flanking alternatively spliced exons are removed more slowly than the constitutive introns in the same transcript (Pandya-Jones and Black 2009; Khodor et al. 2011; Vargas et al. 2011; Pandya-Jones et al. 2013). To determine whether the presence of a DI is indicative of alternative splicing, we developed a computational method to classify introns by splicing type based on the splice junctions identified in the RNA-seq data using the sequence aligner TopHat (Trapnell et al. 2009). Each intron was assigned to one of six classes: constitutive (no evidence for alternative splicing), alternative 5′ or 3′ splice sites (Alt 5′ ss/Alt 3′ ss), cassette, mutually exclusive, or complex (Fig. 2A). The complex group includes introns for which the difference between associated splice isoforms cannot be assigned to only one of the above classes (e.g., a cassette exon that also has alternative 3′ splice sites). Rather than being exclusive to alternatively spliced regions, 61% of DIs are constitutive. Even though they represent the majority of DIs, constitutive introns are ∼1.5-fold less likely to be detained than expected by chance (1935 were observed, while 2778 would be expected, as indicated in Fig. 2A). Conversely, DIs are about threefold more likely to flank alternative exons than would be expected by chance. However, if the slower splicing of alternative exons accounted for the presence of DIs, we would expect a high percentage of such exons to be flanked by DIs. As shown in Figure 2B, although DIs range in prevalence, they constitute only a small minority (9%–17%) of each type of alternative splicing event. That DIs are frequently found between constitutive exons and are present in only a small number of alternative events suggests that DIs represent a distinct set of specifically regulated splicing events.
Figure 2.
Detained introns are found in a minority of all types of splicing events and exhibit distinct structural properties. (A) Number of DIs within each class of constitutive/alternative splicing in mESCs. The first number in bold is the observed number of DIs per class, and the second number indicates how many of each class would be expected if DIs were randomly distributed among all introns. (B) For each class of constitutive and alternative splicing in mESCs, the percentage of events in each class in which the regulated exon is directly flanked by a DI is represented in red. A schematic for each class is shown with constitutive exons in black, alternative exons in white or gray, DIs in red, and splicing patterns indicated by lines connecting splice sites. (C) Comparison among all constitutively spliced introns expressed in mESCs between non-DI (blue) and DI (red) subsets for vertebrate conservation and 5′ and 3′ splice site strength determined by MaxEnt (Yeo and Burge 2004) (indicated on the Y-axes). P-values indicate a significant difference between the two population means (Welch’s t-test), shown below the box plots. Whiskers indicate 1.5× interquartile range. (D) Kernel density plot of the length distribution of constitutive non-DIs (blue) compared with DIs (red). DIs have a slightly longer median length (1789 compared with 1251 for non-DIs) but a shorter average length (2277 compared with 2930 for non-DIs). (E) Relative positions of the 5′ ends of constitutive DIs (red) and constitutive non-DIs (blue) within the gene. The mean relative position for the group is indicated above. See also Supplemental Figure S2 and Supplemental Table S1.
To determine whether the DI-containing genes expressed in mESCs share related biological functions, we tested whether they were enriched for particular GO categories (Supplemental Fig. S2A). Interestingly, the most highly represented GO categories are dominated by genes encoding nuclear proteins involved in RNA metabolism (splicing, RNA processing and nucleocytoplasmic transport, transcription, and chromatin modification and organization), many of which contain RNA-binding domains. Additional highly represented biological processes include DNA damage/repair, apoptosis, cellular response to stress, cell cycle, metabolism, and ubiquitin-mediated protein catabolism. Similar GO categories are significantly enriched for the set of ∼1600 DI-containing genes that is common to all four human cell lines (Supplemental Fig. S2A).
We next asked whether structural properties distinguish DIs as a class. We reasoned that, unlike introns flanking alternative exons, which contain information required to dictate choices between alternative splice sites that might confound our analysis, constitutive DIs would provide a better comparison between the DI and non-DI subsets. DIs in the constitutive class exhibit higher mean sequence conservation scores when compared with non-DIs across vertebrate genomes (Fig. 2C); a significantly higher percentage (19.4%) of constitutive introns with an identifiable human ortholog are DIs, compared with 6.3% of constitutive introns with no identifiable homologous intron in humans (P < 2.2 × 10−16). Furthermore, even within the nonconserved constitutive intron set, DIs have a significantly higher average phastCons score compared with non-DIs (P = 3.0 × 10−4). Again, these phylogenetic properties indicate that DIs in the constitutive intron class, unrelated to known alternative splicing, have a conserved and important function. The mean 5′ and 3′ splice site scores are significantly lower among the DI group (Fig. 2C). Although DIs are detected in genes expressed at different levels, the mean FPKM (fragments per kilobase of exon per million fragments mapped) of DI-containing genes is somewhat higher than controls, possibly suggesting a slight attainment bias (Supplemental Fig. S2B). DIs show a slight but statistically significant positional bias toward the 3′ end of genes, and although the median length of DIs is longer than that of non-DIs, on average, they are shorter, likely due to the lack of extremely long DIs, as the longest DI is ∼17 kb versus >300 kb for the longest non-DI (Figs. 2D,E; Supplemental Fig. S2C). These differences in the properties of DIs compared with non-DIs, particularly splice site strength and higher nucleotide-level conservation, suggest that they are a structurally distinct class, a subset of which is functionally conserved across evolutionary time.
DIs have longer half-lives than neighboring introns and are not NMD substrates
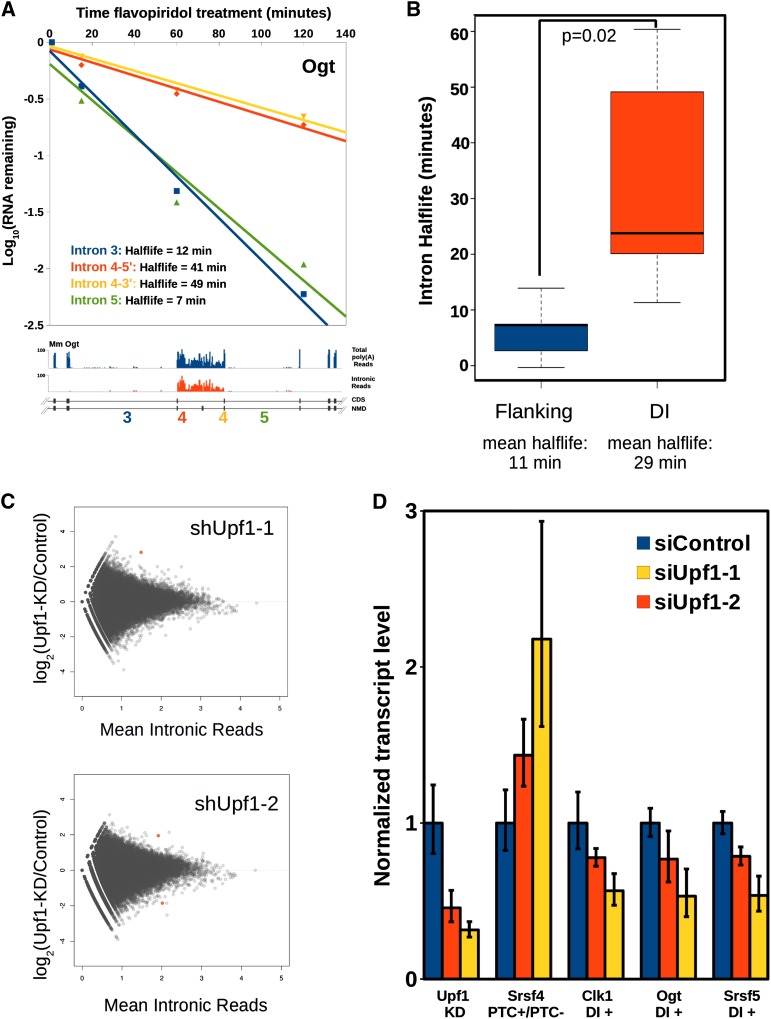
The presence of DIs in a minority of poly(A) transcripts indicates that they either are produced in very small numbers but are highly stable or are initially present in a larger percentage of transcripts and are turned over by splicing or degradation, generating the steady-state level. To distinguish between these two possibilities, we measured the rate of disappearance of several DIs and their flanking neighbors after inhibiting transcription with flavopiridol. For all introns assayed, the DIs exhibited an overall logarithmic decay pattern similar to their flanking neighbors but with slower kinetics (Fig. 3A; data not shown). When compared as two populations, the mean half-life of DIs was 29 min, compared with a mean of 11 min for flanking introns (Fig. 3B). This indicates that either the DIs are spliced out, but with slower kinetics than the other introns in the transcript, or the transcripts containing them are degraded.
Figure 3.
Detained introns have longer half-lives than other introns and are not NMD substrates. (A) Half-lives of DIs and flanking introns from Ogt determined by treating mESCs with 1 μM flavopiridol and harvesting total RNA at the indicated time points followed by qRT–PCR with the indicated primer sets; data were averaged from three independent experiments. Primer sets at both the 5′ (red) and 3′ (yellow) exon–intron junctions of intron 4 were used, and the measured half-lives are indicated for each intron assayed. (B) Half-lives for six detained and nine flanking introns were estimated from flavopiridol time courses as in A, and the population mean half-life was calculated. Whiskers indicate 1.5× interquartile range. (C) Log2 fold change in intron reads between control and Upf1 knockdowns in mESCs calculated from poly(A)+ RNA-seq data (Hurt et al. 2013) for all expressed introns (blue). Introns showing significant change with an adjusted P-value < 0.05 are indicated in red. (D) RNA from mESCs treated with one of two independent siRNAs targeting Upf1 or a control siRNA was assayed by qRT–PCR for the indicated transcripts and normalized to the control siRNA treatment (mean n = 3 ± SEM).
We next addressed the possibility that DI-containing transcripts are degraded through NMD, as has recently been reported for sets of introns during granulocyte differentiation (Wong et al. 2013). Indeed, 99.5% of all introns in the Ensembl annotation of mESC-expressed genes contain in-frame stop codons or introduce frameshifts, making them predicted NMD targets (data not shown). We examined poly(A)-selected RNA-seq data from mESCs in which the essential NMD factor Upf1 has been knocked down, resulting in the stabilization of hundreds of mRNAs that are normally NMD targets (Hurt et al. 2013). In independent Upf1 knockdowns using two different shRNAs, only three normal introns were affected, and there were no significant changes in any DI levels (adjusted P < 0.05) (Fig. 3C) compared with the control hairpin-treated cells. The fact that DIs are not up-regulated in the Upf1 knockdowns indicates that transcripts containing them do not normally undergo degradation through the NMD pathway. We also tested by qRT–PCR whether DIs that directly flank known or predicted NMD switch exons are subject to NMD degradation. Both Srsf5 and Clk1 NMD isoforms show measurable stabilization in the Upf1 knockdown RNA-seq data (Hurt et al. 2013), and the DI in Ogt contains a predicted poison exon. Knockdown of Upf1 using two different siRNAs stabilized the known NMD isoform of Srsf4 (Hurt et al. 2013) relative to the coding isoform (Fig. 3D). In contrast, none of the DI-containing transcripts from Sfrs5, Ogt, or Clk1 showed an increase in stability, indicating that they are not normally subject to NMD. These experiments demonstrate that even in genes in which DIs directly flank NMD switch exons, the DI-containing transcripts are not subject to NMD despite the presence of PTCs.
DIs are present in nuclear poly(A)+ transcripts distinct from NMD isoforms
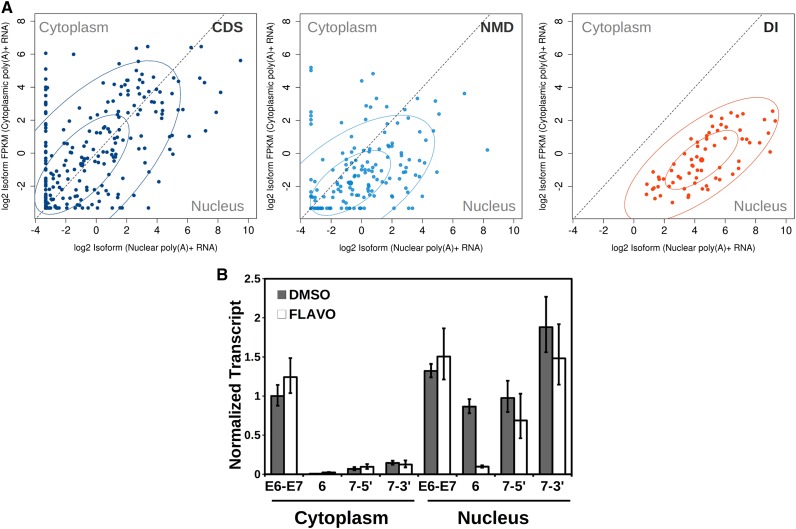
As described above, it is common, particularly among genes encoding RNA processing factors, to find DIs directly flanking NMD switch exons. We reasoned that examining the relationship between DIs, NMD regulation, and subcellular localization of transcripts might help to explain why DIs are not subject to NMD. By focusing on a set of 64 human RNA processing genes (Supplemental Table S2) that contain NMD switch exons directly flanked by at least one DI, we were able to comprehensively annotate >100 splice isoforms in 33 genes based on RNA-seq data, with each variant classified as coding substrate (CDS), DI-containing substrate (DI), or predicted NMD substrate (NMD) (Supplemental Data S3). We then used Cufflinks (Trapnell et al. 2010) to quantify the levels of all isoforms for these 33 genes in poly(A)+ RNA from nuclear and cytoplasmic fractions using the ENCODE data sets (The ENCODE Project Consortium 2012) from three human cell lines. The distribution of the three classes of transcript between the nuclear and cytoplasmic compartments showed a striking predominance of DI-containing transcripts in the nucleus (Fig. 4A; Supplemental Fig. S3A). Coding transcripts, as expected, were primarily in the cytoplasm. NMD substrates were slightly enriched in the nuclear compartment compared with cytoplasm, consistent with their rapid degradation upon export and translation. The distinct localization and higher abundance of DI-containing transcripts in the nucleus indicates that they belong to a separate pool from the NMD substrates. Their nuclear localization provides an explanation for why they are not subject to NMD, as NMD requires translation in the cytoplasm.
Figure 4.
Detained intron-containing transcripts are localized in the nucleus and constitute a separate population from NMD substrates. (A) Nuclear and cytoplasmic poly(A)+ RNA-seq reads from HeLa cells, HepG2 cells, and HUVECs (The ENCODE Project Consortium 2012) were assigned to transcripts classified as CDS (dark blue), NMD substrates (NMD; light blue), or DI-containing substrate (DI; red) for 33 genes, and FPKMs were determined for each variant in each compartment. Fifty percent and 95% concentration ellipses are shown for each class. (B) mESCs were treated with either DMSO (black bars) or flavopiridol (white bars) for 30 min and then fractionated into nucleus and cytoplasm. Relative RNA levels for each intron (6, 7-5′, and 7-3′) or exon–exon (E6–E7) junction were measured by qRT–PCR and normalized to give cell equivalents (mean of two independent fractionations ± SEM). See also Supplemental Figure S3, Supplemental Table S2, and Supplemental Data S3.
To verify that DIs are contained in nuclear localized transcripts, we followed the fate of transcribed introns by blocking transcription with flavopiridol for 30 min to allow completion of transcription, processing, and export of nascent transcripts. RNA was then isolated from the nuclear and cytoplasmic compartments, and the abundance of DIs and their flanking neighbors was determined by qRT–PCR. Exon–exon primer sets showed a varying distribution of spliced message between the nucleus and cytoplasm depending on the gene, and, as expected, the majority of intronic signals were restricted to the nucleus, as exemplified by intron 7 of glutaminase (Gls) (Fig. 4B; Supplemental Fig. S3B). Consistent with previously determined half-lives, the introns neighboring high-coverage introns largely disappeared in the flavopiridol-treated cells. In contrast, after 30 min of transcriptional inhibition, the DI was only slightly less abundant and was almost entirely present in the nuclear fraction. Together, the biochemical fractionation and RNA-seq data suggest that transcripts containing DIs are detained in the nucleus until splicing of all introns is complete or the transcript is degraded, explaining why even though most DIs contain PTCs, they are not subject to NMD. We therefore made the distinction between DIs and “retained” introns, the latter being the result of alternative splicing in which a sequence can be either spliced out as an intron or retained as an exon in the mature mRNA that is transported to the cytoplasm and either translated into protein or, if it contains a PTC, degraded by NMD.
Clk kinase activity regulates a specific set of DIs
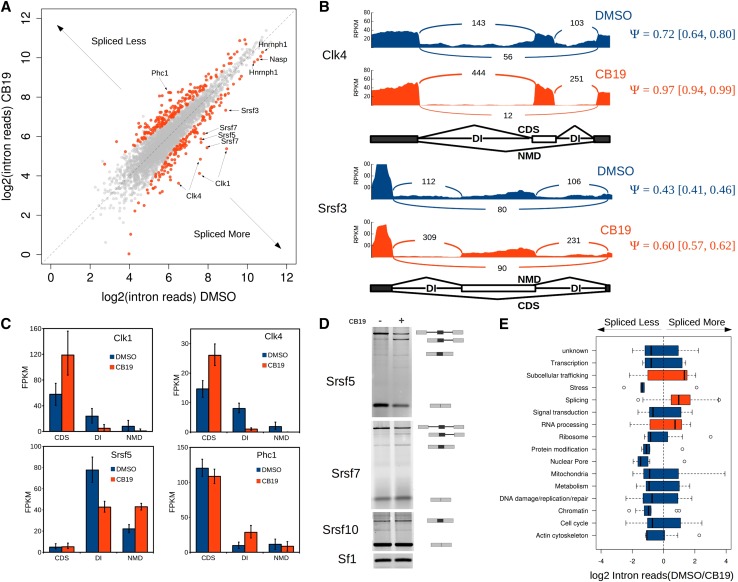
DIs are contained in predominantly nuclear transcripts and are turned over by a mechanism other than NMD, suggesting that the rate at which they are spliced may be the key determinant of their abundance. Transcripts encoding the SR protein kinase Clk1 were previously shown to accumulate in the nucleus, fully spliced except for two introns; these introns were rapidly spliced to produce coding mRNA in response to heat or osmotic shock as well as upon inhibition of Clk kinase activity by a small-molecule inhibitor (Ninomiya et al. 2011). Clk kinase activity thus appears to promote the detention of these introns by inhibiting their splicing. Consistent with this, we identified these introns as well as the homologous introns in the three other Clk family members as DIs in our computational pipeline. To determine whether the effect of Clk kinase activity is specific to these DIs and not due to a general decrease in splicing efficiency, we treated mESCs with the potent Clk1/Clk4-specific inhibitor CB19 (Fedorov et al. 2011) and assayed the Clk1 DIs and normal flanking intron levels by qRT–PCR. Clk1 DIs, but not normally spliced flanking introns on either side, were removed more rapidly in response to the drug, indicating that CB19 has the same effect on Clk1 splicing as was seen in the previous study with a related compound, TG003 (Ninomiya et al. 2011), and that this effect is specific to the DIs (Supplemental Fig. S4A). These results suggest that control of splicing of DIs could be a common point of regulation of gene activity.
The effect of Clk kinase inhibitors on splicing regulation has only been tested on a handful of individual splicing substrates to date. We therefore sought to determine the genome-wide effect of Clk kinase inhibition by treating mESCs with CB19 or DMSO as a control and performing RNA-seq on poly(A)-selected RNA. We generated libraries from RNA collected at two time points after treatment: at 2 h to identify splicing events that are most likely to be directly regulated by Clk inhibition and at 4 h to allow sufficient time to detect early primary and secondary effects on the transcriptome. After 2 h of treatment, we observed 325 DIs in 295 different genes that underwent a statistically significant change in levels (Fig. 5A). Of these introns, 137 decreased in abundance, including the two previously assayed DIs in Clk1 as well as the homologous introns in Clk4, with increases in the inclusion of the associated cassette exons, suggesting that the decrease in intron levels is due to increased splicing (Fig. 5B,C). Interestingly, slightly more than half (188) of the DIs that responded to Clk inhibition increased in intron detention levels, as exemplified by intron 20 of Phc1 (Fig. 5C; Supplemental Fig. S4B), but the majority of DIs (2822) showed no detectable change. This suggests that Clk controls, probably directly given the 2-h time point, only a subset of DIs.
Figure 5.
Inhibition of Clk kinase alters splicing of a subset of DIs. (A) Hundreds of DIs are differentially spliced in response to Clk kinase inhibitor. The scatter plot shows the log2 of the intron read density for each intron after 2 h in DMSO (X-axis) versus CB19 (Y-axis). Introns changing significantly (P < 0.05) in either direction are plotted in red, and nonresponsive introns are in gray. Introns chosen for further analysis are labeled. (B) Sashimi plots showing read density and number of splice junctions, with the percentage of spliced-in (Ψ) values determined by MISO (Katz et al. 2010) indicated in red or blue numbers at the right of the plots (±95% confidence intervals in square brackets) for cassette exons in Clk4 and Srsf3 upon CB19 treatment in DMSO control (blue) compared with CB19 treatment (red). Schematics indicating the isoform produced by splice variants are shown below. (C) FPKMs determined by Cufflinks (Trapnell et al. 2010) for summed isoforms classified as CDS, NMD, or DI-containing are plotted for each gene after 2 h of treatment with DMSO (blue bars) or CB19 (orange bars). Error bars indicate ±95% confidence intervals. (D) RT–PCR products from mESCs treated with DMSO or CB19 for 2 h. Splice products and intermediates are indicated by the schematics at the right. Srsf10 shows no response to CB19, and Sf1 served as a loading control. (E) All genes containing CB19-responsive DIs were divided into functional categories, and the distribution of the log2 fold change in splicing is indicated. Categories in blue have a median direction of “spliced less,” while those in orange have a median direction “spliced more.” Whiskers indicate 1.5× interquartile range. See also Supplemental Figures S4 and S5.
If changes in transcription rates in response to CB19 treatment were responsible for the observed differences in DI levels due to higher or lower proportions of nascent transcripts in the poly(A)-selected population, we would expect to see a corresponding change in reads mapping to all introns within the set of CB19-responsive genes and not just the DIs. We identified >500 genes showing this transcriptional signature when all nondetained intron reads within the gene were summed together, which probably reflects increased or decreased transcription. However, the majority of DI-containing genes showed no evidence of transcriptional differences, and, for those that did, there was no correlation between the direction of transcriptional change and that of intron detention levels (Supplemental Fig. S5). The DIs in Clk1 were previously shown to be spliced post-transcriptionally when TG003 treatment was performed following transcriptional blocking with α-amanitin. To determine directly whether the effect of CB19 treatment on DI splicing occurs post-transcriptionally, we tested whether CB19 treatment increased splicing of DIs in cells with no ongoing nascent transcription. In order to avoid detecting partial transcripts associated with blocked polymerase, we treated cells with flavopiridol for 30 min to allow completion of elongation and processing of nascent transcripts. Cells were then treated with CB19 or DMSO, and RNA was collected 60 min later. Using qRT–PCR, DIs in Clk1 and Nasp were significantly reduced in CB19-treated cells compared with DMSO treatment, while the normally spliced introns were not, indicating that the increased splicing upon Clk inhibition occurs post-transcriptionally (Supplemental Fig. S4C). Together, these data indicate that the mechanism by which Clk inhibition affects intron detention/splicing is highly specific, can modulate intron levels in both directions, and can act post-transcriptionally.
Several of the most highly responsive DIs are in SR protein genes. Srsf5 and Srsf7 showed increased splicing of DIs that was confirmed by RT–PCR; splicing of Srsf10 was unaltered in both the RNA-seq and RT–PCR data (Fig. 5D). In addition to Clk1/4 and multiple SR proteins, many genes containing DIs that were altered by CB19 treatment encode proteins involved in splicing or RNA processing. A number of these factors are RNA-binding proteins, and half of them contain an arginine–serine-rich (RS) domain, which is the domain phosphorylated by the Clk kinases (Supplemental Fig. S4D). We assigned the genes containing CB19-altered DIs to functional categories and plotted the distribution of changes in intron levels between control and CB19-treated cells for each category (Fig. 5E). Although every category contains DIs changing in both directions, splicing, RNA processing, and subcellular trafficking-related genes were the only functional categories for which the median change in DI levels were positive; that is, spliced more. Among these genes with increased DI splicing, many have a known or predicted NMD–CDS switch exon with flanking DIs, and the increased splicing leads to different outcomes depending on the gene. Clk1 and Clk4, for example, show increased inclusion of the exon producing more coding mRNA, while Srsf3 and Srsf5 had increased inclusion of poison exons that lead to NMD (Fig. 5B,C). Consequently, even after only 2 h of CB19 treatment, total coding transcript levels of the Clk kinases are elevated, while those of the SR proteins are decreased. Inhibition of Clk kinase activity thus leads to alterations in the splicing of many DIs that in turn lead to rapid changes in transcript levels in both directions, and, as a result, the transcripts encoding many splicing and RNA processing factors as well as other functional classes of protein are shifted in abundance.
Srsf4 is a Clk kinase target and is associated with CB19-activated splicing events
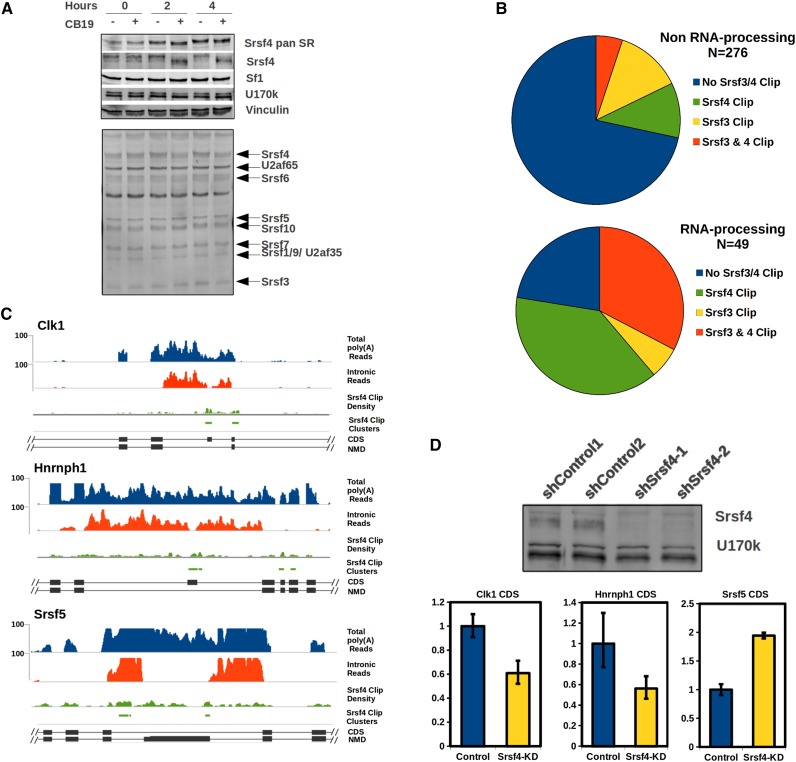
The Clk family of kinases phosphorylate RS domains on splicing factors, including SR proteins, in the nucleus (Colwill et al. 1996; Aubol et al. 2013). Because both hypophosphorylation and hyperphosphorylation of SR proteins have been shown to inhibit splicing in vitro and in vivo (Prasad et al. 1999; Shi and Manley 2007), a plausible mechanism by which Clk kinase activity could promote intron detention is through the hyperphosphorylation of SR proteins. To identify RS domain-containing proteins in mESCs whose phosphorylation is rapidly shifted in response to CB19 treatment, we used a phospho-RS-specific antibody (1H4) to perform Western blots on CB19-treated versus DMSO-control-treated mESCs (Fig. 6A, bottom panel). The predominant change observed was the decreased phosphorylation of Srsf4. This was validated by blotting with a Srsf4-specific antibody with a prominent shift to higher mobility (Fig. 6A, top panels). This result is not unexpected, as Srsf4 phosphorylation has been observed to be particularly sensitive to Clk kinase inhibitors, likely because it contains the largest RS domain of the classical SR protein family (Graveley 2000; Yomoda et al. 2008). The strong decrease in Srsf4 phosphorylation upon CB19 treatment suggested that it may play a role in regulating DIs whose splicing is sensitive to Clk inhibition.
Figure 6.
Srsf4 is a major Clk kinase substrate, and Srsf3/4 Clip tags are prevalent among CB19-sensitive RNA processing gene DIs. (A) Srsf4 is the predominant SR protein dephosphorylated in mESCs upon Clk kinase inhibition. Western blots of mESC total cell lysate (+phosphatase inhibitors) treated with DMSO or CB19 for 0, 2, or 4 h. Antibodies used to blot (from the top down) were α-pan SR protein, α-Srsf4, α-Sf1, α-U170k, and α-vinculin. (Bottom panel) α-Phospho-SR protein (1H4). (B) Srsf3 and Srsf4 Clip clusters were overlapped with DIs that respond to CB19, and the corresponding genes were divided into non-RNA processing (top pie chart) and RNA processing (bottom chart). (C) Total read density (top) and intron read density (bottom) are shown for 2-h DMSO-treated (blue) and CB19-treated (red) cells; Srsf4 Clip density and Clip clusters are shown in green across the NMD switch exon regions flanked by DIs in the Clk1, Hnrnph1, and Srsf5 genes. (D) Western blot showing Srsf4 knockdowns blotted with α-Srsf4; α-U170k was used as a loading control. RNA from lentivirus-infected mESCs was assayed for splicing of Clk1, Hnrnph1, and Srsf5 coding variants. Combined control shRNA mean (n = 4–6 ± SEM) are shown in blue, and that of the combined shRNAs targeting Srsf4 are shown in yellow for each transcript.
To identify DIs that are potentially regulated by Srsf4 or other SR proteins, we reprocessed data from a high-resolution Srsf3 and Srsf4 iClip (individual nucleotide-resolution cross-linking immunoprecipitation) data set performed in P19 mouse embryonal carcinoma cells (Anko et al. 2012). Although these iClip assays were performed in a different cell line, we reasoned that many of the same targets were likely bound by the same SR protein in mESCs. Because SR proteins typically bind to exonic sequences, we looked for iClip clusters overlapping not only introns but also the flanking upstream and downstream 100 nt. Thirty-six percent (115 of 325) of all DIs whose splicing was sensitive to CB19 treatment were bound by Srsf3, Srsf4, or both; this is highly enriched compared with DIs that do not respond to CB19 treatment (P ≤ 2.2 × 10−16). Strikingly, when we subdivided CB19-responsive DIs between those encoding RNA processing proteins versus other functional categories, 78% (38 of 49) of the RNA processing DIs were bound by one or both SR proteins, compared with 29% of the DIs in other gene categories (Fig. 6B). Among the regions that contained Srsf4 Clip clusters and showed increased splicing in response to CB19 treatment were DIs in Clk1, Srsf5, and Hnrnph1 (Fig. 6C). In all three genes, the Srsf4 Clip clusters are within NMD switch exons flanked by DIs—coding exons in Clk1 and Hnrnph1 and a poison exon in Srsf5. Upon Srsf4 knockdown, the coding isoforms of Clk1 and Hnrnph1 decreased by 50%–60% due to skipping of the coding exons, and the skipping of the poison exon in Srsf5 increased approximately twofold. This indicates that Srsf4 is required for efficient inclusion of all three exons (Fig. 6D). Importantly, in all three cases, the direction of splicing in response to CB19 inhibition is consistent with Clk kinase activity activating the splicing pattern enforced by Srsf4. Together, these data indicate that DIs found in a specific set of splicing regulators, among other genes, are direct targets of Clk kinase activity and that many of these DIs are likely regulated by Srsf4.
DNA damage triggers changes in splicing of a subset of DIs
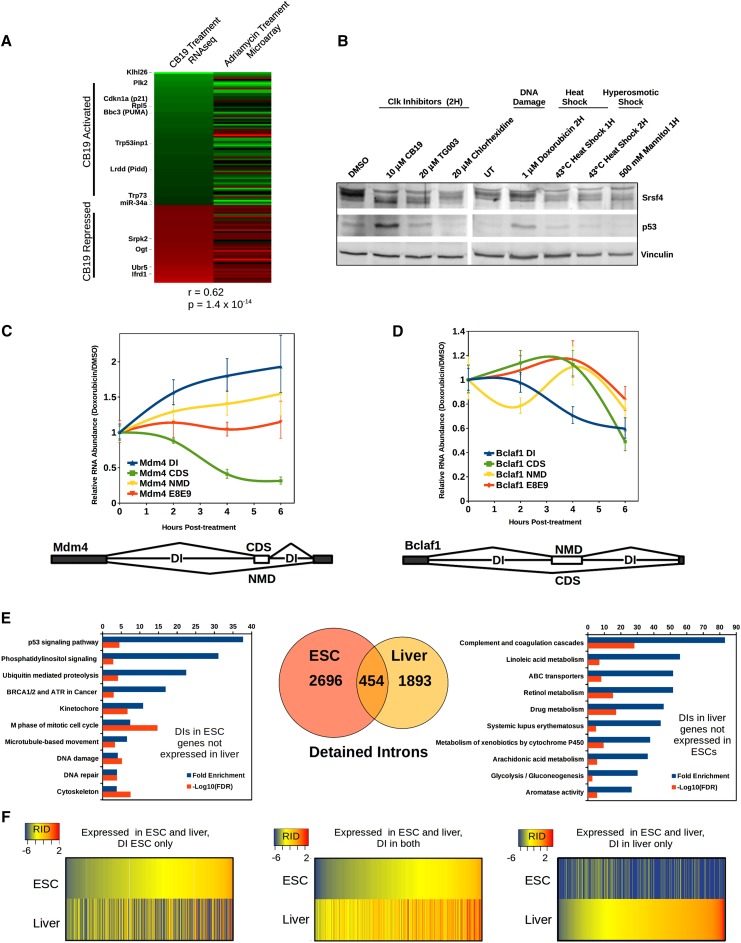
To look at later effects on gene expression, we quantified transcript abundance at the gene level after 4 h of CB19 treatment. We identified 408 genes with significant gene-level expression differences in CB19-treated cells compared with DMSO-treated cells. Strikingly, 30% (121) of these genes were previously identified as direct transcriptional targets of p53 in mESCs; that is, genes with p53 CHIP signal in their promoter that undergo a p53-dependent change in expression upon doxorubicin treatment (Li et al. 2012). This enrichment is highly significant (P = 2.2 × 10−15), and when the direction and magnitude of gene expression changes in our CB19 data or the previously published doxorubicin treatment were compared with each other, the correlation was also highly significant (P = 1.4 × 10−14), indicating that both p53-activated and p53-repressed genes change in similar directions and magnitudes in CB19-treated cells and doxorubicin-treated cells (Fig. 7A). Consistent with the presence of a p53 transcriptional signature, we observed that within 2 h of either doxorubicin or CB19 treatment, p53 protein levels are significantly elevated (Fig. 7B). Up-regulation of p53 protein in response to Clk inhibition has been previously observed where it was assumed to be the result of stress induced by general splicing inhibition (Allende-Vega et al. 2013). Thus, both DNA damage and Clk1 inhibition specifically regulate partially overlapping subsets of genes enriched in the DI class. In addition, this indicates that rather than acting only as a splicing modulator, CB19 also induces changes indirectly through a transcriptional response that is dominated by p53.
Figure 7.
DIs are differentially spliced in response to DNA damage and show cell type-dependent abundance. (A) Genes showing significant change in expression at 4-h after CB19 treatment were compared with mESC p53 targets responding to Adriamycin (doxorubicin) treatment (Li et al. 2012). Genes going up in expression are colored green, and those going down are in red. Correlation was determined by Wilcoxon rank sum. (B) Western blot of Srsf4 (top panels) to indicate Clk kinase activity and of p53 protein (middle panel) and vinculin loading control (bottom panel) with various Clk inhibitors or stress conditions as indicated. (C,D) Real-time qRT–PCR was performed on RNA from doxorubicin-treated cells or DMSO-treated cells. All transcript-specific signals were first normalized to Mylpf transcript levels and then normalized to the signal at time 0. The plot shows the ratio of DIs relative to the total transcript level of each gene in doxorubicin compared with DMSO and the relative change in spliced isoform levels of each gene of the NMD substrate (NMD), CDS, and total transcript in Mdm4 (C) and Bclaf1 (D). Bars represent the average, and error bars are SEM; n = 3. Schematics indicating the structure of the NMD switch exons and flanking DIs are shown below. (E, middle) DIs in genes expressed in either mESCs or adult mouse liver samples or both (Venn diagram). DAVID GO analysis of genes expressed only in mESCs (left) or livers (right) that contain DIs are shown with fold enrichment and the −log10 of the FDR for each category graphed. (F) Relative intron detention (RID) levels for all introns expressed in both mESCs and livers. (Left) RID for introns found in genes expressed in both cell types but only detained in mESCs and not livers. (Right) Introns detained only in livers but not mESCs. (Middle) Introns detained in both cell types. See also Supplemental Figs. S6 and S7.
Consistent with the up-regulation of p53 protein, Clk inhibition was shown previously to induce an increase in the NMD isoform of Mdm4, a negative regulator of p53 (Allende-Vega et al. 2013; Bezzi et al. 2013). We identified the introns flanking the Mdm4 NMD switch exon as DIs and therefore reasoned that their splicing might be regulated under physiological conditions that induce p53, such as DNA damage. To determine whether DIs in Mdm4 are directly altered by DNA damage, we treated mESCs with 1 μM doxorubicin and quantified the effect on DIs in Mdm4 as well as several other genes that encode proteins with known roles in the DNA damage response. Indeed, the amount of Mdm4 transcript containing a DI was increased almost twofold by 4–6 h after doxorubicin treatment (Fig. 7C). In addition to the change in the amount of DI-containing transcripts, Mdm4 spliced isoforms switched from coding to NMD isoforms. Both the increase in the DI pool and the increased skipping of the NMD switch exon resulted in a decrease in the cellular pool of coding mRNA, although the total amount of Mdm4 transcript, as assayed by primers spanning exons 8 to 9, remained essentially the same throughout the treatment. While CB19 and doxorubicin treatment had a similar magnitude of effect on Mdm4 splicing, both drugs together had an additive effect, indicating overlapping but distinct effects (Supplemental Fig. S6A). DIs in Clk1, Srsf5, and Morc3 also increased following DNA damage (Supplemental Fig. S6B,C). In contrast, Bclaf1, which is an RS domain-containing RNA-binding protein that has been shown to associate with Brca1 upon DNA damage (Savage et al. 2014), showed the opposite switch upon doxorubicin treatment. The DI in Bclaf1 was reduced by almost half, and the total transcript level as well as the coding isoform fraction were up-regulated by 4 h, while the NMD isoform was down-regulated, consistent with the DI pool contributing to increased coding message (Fig. 7D). The DI in a fourth gene, Baz1b, which is also involved in DNA damage response, showed an initial increase in splicing of the DI at 2 h that leveled out through 6 h as total mRNA levels dropped (Supplemental Fig. S6C). The bidirectional changes in the abundance of multiple DIs in response to DNA damage indicates that this subset of DIs is dynamically regulated by physiological stress signals.
Adult livers contain overlapping and unique sets of DIs
The homogeneity of cell lines enables robust statistical identification of DIs. To determine whether DIs are present and quantifiable in differentiated cells within the tissue of an animal, we used poly(A)-selected RNA-seq data from two individual adult mouse livers produced by ENCODE (The ENCODE Project Consortium 2012). The liver was chosen due to the higher degree of cell homogeneity (60%–80% hepatocytes) compared with other organs. With our computational pipeline, we identified a set of 2347 DIs in adult mouse livers, representing 6.8% of introns in expressed genes. When compared with the set of ESC-specific DIs, we found a set of 454 introns that were detained in both ESCs and the liver, a highly significant overlap (P < 2.2 × 10−16). In addition to this set of shared DIs, among genes that were expressed in both cell types, 2154 DIs were present only in ESCs, and 1261 DIs were present only in the liver. Finally, 632 DIs were found in genes expressed only in the liver, and 538 DIs were found in genes expressed only in ESCs (Fig. 7E,F). To determine whether DIs present in both or only in one tissue type fall within genes with particular functions, we performed GO analysis on the different subsets of DI-containing genes. Among the genes containing DIs that are expressed in ESCs but not the liver, there was a strong enrichment for DNA damage response as well as DNA replication and cell cycle. Of the DI-containing genes expressed exclusively in the liver, complement and coagulation factors, lipid metabolism, and drug metabolism—all related to stress and homeostatic response—were highly overrepresented. Among the functional categories overrepresented by genes with DIs in both ESCs and the liver were the spliceosome, glycerolipid metabolism, and ubiquitin-mediated proteolysis (Supplemental Fig. S7A). Thus, DIs are present in a set of common genes expressed in both ESCs and differentiated adult tissue, but each cell type also contained a set of DIs in genes with specific functional classes relevant to the function and activity of the cell in which they are found. Importantly, the presence of DIs in genes expressed in both tissues but with a DI in only one is strong evidence that intron detention is the result of regulated splicing rather than simply slow processing due to poor recognition of the splice sites.
Discussion
Surprisingly, we found a large number of introns in polyadenylated RNA from human and mouse cell lines. This observation is unexpected in light of several recent transcriptome-wide analyses suggesting that completion of most splicing events occurs cotranscriptionally, prior to transcription termination and polyadenylation (Carrillo Oesterreich et al. 2010; Ameur et al. 2011; Khodor et al. 2011; Tilgner et al. 2012). On the other hand, there is extensive literature proposing post-transcriptional splicing of individual introns (Bauren and Wieslander 1994; Smith et al. 1999; Melcak et al. 2001; Vargas et al. 2011; Hao and Baltimore 2013; Pandya-Jones et al. 2013). In addition, other RNA-seq-based studies examining biochemically fractionated cells, metabolically labeled RNA, or lipid A-induced transcriptional activation and tracking the maturation of nascent transcripts from chromatin to cytoplasm have indicated varying degrees of post-transcriptional splicing (Rabani et al. 2011; Bhatt et al. 2012; Khodor et al. 2012; Windhager et al. 2012; Pandya-Jones et al. 2013). All of these genome-wide studies used population-based quantification of introns. Our approach differed from these previous studies by measuring, genome-wide, the abundance of each individual intron rather than aggregate populations. First, by applying the proper statistical constraints, we were able to use all reads mapping to an entire intron rather than, for example, only exon–intron and exon–exon junction-spanning reads, which are much less abundant; this provided robust quantification despite the “shot noise” inherent to a sampling-based technique like RNA-seq (Anders and Huber 2010). Second, using biological replicates, we could confidently compare the observed abundance of each intron with its expected abundance, assuming all introns within a gene are equally present in the population. This intron-centric approach uncovered a large set of individual introns that remain unspliced in nuclear-retained poly(A) transcripts in which other introns are spliced. These DIs are concentrated in the nucleus, and either they are processed at a slow rate or the intron-containing precursor is degraded or both. In either case, regulation of the processing of DIs can limit the synthesis of particular isoforms and thus regulate gene expression. As direct evidence that DIs can contribute to gene regulation, we showed that inhibition of Clk kinase activity as well as DNA damage can modulate the rate of splicing for particular subsets of DIs, enabling coordinated control of specific genes.
We found that DIs are present in not only cell lines but also tissue from adult animals. A common set of DIs is found in genes expressed in both ESCs and adult livers, alongside sets of tissue-specific DIs that are enriched for genes with particular functions related to the cell type in which they occur. Many of the functional gene categories in which DIs are overrepresented encode products whose physiological concentrations must be tightly controlled; for example, the complement and coagulation cascade factors produced in the liver. Regulation of DI splicing kinetics may contribute an additional control point for fine-tuning of gene expression in these cases. The identification of DIs in an adult tissue underscores the robustness of our computational method, since we were able to identify DIs despite the presence of cell type variation within the organ, differences between individual animals, and differences in environmental conditions at the time of sample collection. Most importantly, we found that a substantial number of DIs are present in the transcripts of only one of two cell types, even though the gene is expressed in both cases. This indicates that these DIs are not simply poor splicing substrates—since, in that case, they would appear with equal abundance wherever the gene is expressed—but rather are under cell type-specific or environmental regulation.
A significant subset of DIs is evolutionarily conserved, suggesting that this property serves an important regulatory function. DIs are statistically enriched adjacent to alternatively spliced exons, but the majority of DIs are categorized as constitutive introns, thus separating this property from processes that regulate alternative splicing. Indeed, constitutive DIs are also statistically evolutionarily conserved, further indicating that they have important regulatory functions. As identification of orthologous introns is complicated by a large amount of evolutionarily neutral intron sequences, we consider the conserved set of DIs to be a stringent estimate that likely underestimates the number of functionally conserved DIs. The slow rate of splicing of some nonconserved DIs relative to their flanking introns may simply be due to less efficient engagement with spliceosomal processes resulting from variation in splice sites, making their excision rate limiting for transport. However, as discussed above, we showed that subsets of DIs undergo changes in splicing rates in response to cellular signals and/or under cell type-specific conditions, indicating that the slower splicing of these DIs is a regulated process rather than the result of suboptimal splice site variation.
DIs constitute a uniquely regulated subset that spans all types of splicing events and are not simply a manifestation of processes associated with alternative splicing. Although the vast majority of DIs contain PTCs, these transcripts are not NMD substrates, as they do not increase in abundance when NMD is inhibited and are confined to the nuclear fraction in a distinct pattern from known NMD substrates. This suggests that DIs are present in incompletely processed transcripts rather than transcripts that are exported and translated. The slow decay rate of DIs upon inhibition of transcription is consistent with a fate of ultimately being either spliced or degraded as part of the precursor RNA within the nucleus. The latter may be related to a recently discovered developmental regulation mechanism in which inhibition of splicing of a 3′-terminal intron leads to destruction of the transcript through nuclear degradation (Yap et al. 2012). The possibility that many DIs represent a rate-limiting intermediate is suggestive of a process observed in recent studies using lipid A-induced macrophages (Bhatt et al. 2012; Pandya-Jones et al. 2013) in which there is a delay of tens of minutes between transcription of a gene and the removal of all introns, which occurs in a chromatin-associated fraction, thus demonstrating that splicing rates can determine the kinetics of gene expression. However, in these cases, all or most introns in the genes appeared to be similarly delayed, whereas DIs are distinctly present in transcripts that are otherwise fully spliced. Similarly, a large number of transcripts containing all of their introns unspliced accumulated in nuclear fractions from heat-shocked fibroblasts, while cotranscriptionally spliced genes were unaffected (Shalgi et al. 2014). Finally, a large group of introns enriched in polyadenylated transcripts that may partially overlap DIs were recently characterized (Braunschweig et al. 2014). However, these introns were enriched in untranslated regions (UTRs), were negatively correlated with gene expression, and were frequently targeted by NMD, which indicates that many of this set of introns are distinct from the ones that we identify here and further underscores the complexity of regulatory processes involving regulation of splicing kinetics.
The Hagiwara group (Ninomiya et al. 2011) previously identified introns in the Clk1 and Clk4 kinase genes that we now assign to a much larger class of DIs. We confirmed their previous observations that the introns in Clk1 undergo rapid post-transcriptional splicing upon drug inhibition of Clk kinase activity and performed the first genome-wide analysis of the effects of Clk inhibitor. We found that, beyond the autoregulation of DIs in their own transcripts, the Clk kinases likely modulate splicing rates of >300 other DIs directly, since the response to Clk inhibition occurs at a very early time (2 h) after treatment. Assuming a constant rate of transcription, the magnitude by which the mRNA pool increases in response to CB19 treatment is dependent on the amount of precursor RNA that is degraded under steady-state conditions. For example, we observed a twofold increase in Clk1 coding mRNA and a 90% decrease in Clk1 DI-containing transcripts 2 h after CB19 treatment. The half-life of Clk1 mRNA was previously estimated at 39 min (Friedel et al. 2009), and since this is much shorter than 2 h, with no detectable increase in transcription, the increase in mRNA levels must reflect an increase in the efficiency of splicing. The increase in spliced mRNA could result from either enhanced splicing efficiency in general or, more likely, redirection of DI-containing precursors fluxing through a nuclear degradation pathway to an efficient splicing pathway or both (see the model in Supplemental Fig. S7B).
An important clue into a physiological role of the Clk kinases is that heat shock and osmotic stress activate the same induction of DI splicing in the Clk precursor RNA as inhibition of the kinase activity (Ninomiya et al. 2011). Three other Clk family members have interesting roles in specific contexts, including insulin response, splicing of exons involved in neuronal tauopathies, and the replication cycle of HIV (Duncan et al. 1998; Hartmann et al. 2001; Jiang et al. 2009; Rodgers et al. 2010; Wong et al. 2011). These studies suggest that Clk kinases, through regulation of splicing, are positioned to channel multiple cellular signaling pathways in different cell types and disease states, including global stress responses. It has previously been observed that the Clk inhibitor TG003 triggered an increase in Mdm4 NMD isoform splicing, a corresponding decrease in Mdm4 protein, and up-regulation of p53 protein levels (Allende-Vega et al. 2013; Bezzi et al. 2013). This effect was attributed to induction of stress due to a general inhibition of splicing, since knockdown or genetic perturbation of core spliceosome components had a similar effect on Mdm4 and p53. Here we performed the first genome-wide analysis of the effect of Clk inhibitors, and our data clearly indicate that, far from being a general inhibitor of splicing, CB19 has a very specific effect on a subset of splicing events. Both Clk inhibition and DNA damage affect the splicing of subsets of DIs, and the additive effect on Mdm4 splicing when both drugs are present simultaneously indicates that they may act through overlapping but distinct pathways. The rapid and profound effect on Mdm4 splicing seen upon treatment with CB19, doxorubicin, or both (70% coding down to <20% at 4 h after treatment), accompanied by an approximately twofold increase in the amount of DI-containing transcripts, is strong evidence for the physiological role of regulated DI splicing, and the observed shift in splicing of Mdm4 transcripts is likely responsible at least in part for the up-regulation of p53 protein and the corresponding p53 transcriptional response.
While the earliest steps in spliceosome assembly require SR protein phosphorylation to enhance specific RNA binding as well as interaction with the U1-70k RS domain (Tacke et al. 1997; Xiao and Manley 1997; Cho et al. 2011), progression from early spliceosome to completion of splicing requires the dephosphorylation of SRs by PP1/2A phosphatases, likely related to their involvement in the recruitment of the U4/U5/U6 tri-snRNP and other snRNP-associated proteins (Crispino et al. 1994; Mermoud et al. 1994; Tarn and Steitz 1994; Roscigno and Garcia-Blanco 1995; Cao et al. 1997; Kanopka et al. 1998; Xiao and Manley 1998; Shi et al. 2006). Both hyperphosphorylation of SR proteins by overexpression of Clk1 and, in some contexts, hypophosphorylation upon overexpression of a dominant-negative kinase-dead mutant suppress splicing (Colwill et al. 1996; Prasad et al. 1999). These data suggest that Clk kinases may function in the context of DIs by maintaining the hyperphosphorylation of nearby SR proteins, presumably maintaining a prespliceosome-type state and counteracting the dephosphorylation step that is required for splicing completion. CB19 treatment causes splicing changes in a specific subset of ∼10% of all DIs, indicating that, if similar mechanisms regulate other DIs, further layers of specificity exist. In addition to combinatorial control by different SR proteins and/or Clk kinases, since many spliceosomal proteins—including Snrnp70 (U170k), Sf3b1, and Eftud2 (U5-116k)—undergo phosphorylation/dephosphorylation cycles during completion of splicing (Tazi et al. 1993; Shi et al. 2006), it is easy to speculate that other specific kinases may affect different subsets of DIs.
Materials and methods
RNA-seq data are accessible through Gene Expression Omnibus under accession number GSE57231.
Cell culture, small-molecule inhibitors, and RNAi
V6.5 cells were cultured on 0.2% gelatin-coated plates in HEPES-buffered DMEM (Life Technologies) supplemented with 15% FBS (HyClone) and 1000 U/mL LIF (ESGRO). Flavopiridol (1 μM in DMSO; Sigma) and KH-CB19 (10 μM in DMSO; CalBiochem) were added to culture medium for the indicated times. siRNAs targeting Upf1 and control siRNAs (Mm_Rent1_1, Mm_Rent1_6, and AllStars negative control siRNA; Qiagen) were transfected using HiPerFect (Qiagen) according to the manufacturer’s instructions. shRNAs targeting Srsf4, GFP, or luciferase were cloned into pLKO.1. Knockdowns were performed by infecting V6.5 cells with lentivirus followed by selection with 1 μg/mL puromycin (Sigma) for 48 h to enrich for infected cells.
RNA-seq
Total RNA from V6.5 mESCs was treated with Turbo DNase (Life Technologies) and poly(A)-selected [Poly(A)Purist MAG, Life Technologies]. Poly(A)+ RNA was then digested with 5′ exonuclease (Terminator, Epicentre) to further enrich for capped mRNAs. Two micrograms of poly(A)+ RNA was shattered under alkaline conditions in the presence of Mg++ (Mortazavi et al. 2008). A 3′ linker containing both priming sites and a 6-nt random 3′ overhang was ligated by splint ligation using T4 DNA ligase (New England Biolabs), reverse-transcribed at 60°C (SuperScript III, Life Technologies), and treated with RNaseH/A (Life Technologies, New England Biolabs) to remove RNA, and the cDNAs were size-selected on a 10% PAGE gel under denaturing conditions. The extracted DNA was circularized (CircLigase II, Epicentre) and amplified using 18 cycles (Phusion Polymerase, New England Biolabs). The resulting product was again purified on a 2% agarose gel followed by standard paired-end sequencing (2 × 36 nt, Illumina Genome Analyzer IIx). Libraries for the CB19 experiment were prepared using the Illumina TruSeq stranded library kit with two passes over oligo-dT beads following the manufacturer’s instructions with minor modifications. Two independent biological replicates for each condition (DMSO and CB19) at time 0, 2, and 4 h after drug treatment were made into separate barcoded libraries. Input for each library was 4 μg of total RNA per sample, fragmentation time was decreased to 5 min to produce longer insert length, and samples were size-selected on a 2% agarose gel following PCR amplification. Barcoded samples were paired-end sequenced (2 × 100 nt) in multiplex on an Illumina HiSeq 2000 instrument.
Read mapping
Each lane of raw reads was first mapped, all together, using Bowtie 1.0.1 (Langmead et al. 2009) with standard parameters requiring at most two mismatches to mm9, and unique mapping was enforced using -best -strata parameters. TopHat 2.0.9 (Trapnell et al. 2009) with standard mapping parameters was also used to map junction-spanning reads against a custom junctions file and allowing novel junctions. The aligned reads for each lane were then split into individual samples using barcodes specific to each sample. See the Supplemental Material for further details.
RT–PCR
Total RNA was harvested directly from tissue culture plates using Trizol (Life Technologies) according to the manufacturer’s directions. In all experiments, total RNA was treated with Turbo DNase (Life Technologies) to remove any residual DNA contamination, extracted in acid-buffered phenol/chloroform, and ethanol-precipitated. Reverse transcription was performed using 5 μg of DNase-treated total RNA in a 20-μL reaction following the manufacturer’s protocol (SuperScript III, Life Technologies) for 1 h at 50°C. cDNA reactions were digested with RNase H and diluted 1:10, and 1 μL was used in each PCR reaction. RT–PCR was performed using AmpliTaq (Perkin-Elmer). Real-time qRT–PCR was performed using Power SYBR Green (ABI) on an ABI 7500 machine. Normalization to the highly stable mRNA Mylpf was used to compare samples using a ΔΔCt calculation.
Western Blots and antibodies
Cells were harvested in cold PBS/2 mM EDTA, pelleted, and lysed in RIPA buffer (0.1% SDS, 0.5% sodium deoxycholate, 1% Igepal [NP-40], 150 mM NaCl, 1× Complete protease inhibitors [Roche], 1× phosphatase inhibitor [Pierce]). Lysates were run on Laemmli PAGE (10% acrylamide) gels followed by transfer to Hybond LFP PVDF membranes (GE) and blotted using standard protocols. The antibodies used were α-phospho-SR (1H4, Millipore), α-SR (16H3, Life Technologies), α-Srsf4 (06-1367, Millipore), α-U170K (H111, SynapticSystems), α-Vinculin (Abcam), and α-Sf1 (SAB2102119, Sigma). Antibody detection was performed using ECL Plex Cy5-conjugated secondary antibodies (GE), and blots were imaged and quantified on a Typhoon Trio PhosphorImager (GE).
Subcellular fractionation
Subcellular fractionations were performed as in Pandya-Jones and Black (2009) with minor modifications. Briefly,V6.5 cells grown in 100-mm dishes were trypsinized, pelleted by gentle centrifugation, washed in cold PBS, and lysed for 4 min in ice-cold buffer (0.15% Igepal [Sigma], 10 mM Tris at pH 7.5, 150 mM NaCl). Lysates were immediately transferred onto a sucrose cushion (15% sucrose, 10 mM Tris at pH 7.5, 150 mM NaCl) and spun for 10 min at 4°C. Cytoplasmic fractions were carefully removed, and the nuclear pellets were washed twice with PBS. Nuclear pellets and cytoplasmic fractions were extracted with Trizol (Life Technologies). Total RNA extracted per fraction was quantified and used to calculate cell equivalents for the qRT–PCR quantification.
Bioinformatics
Open source software and R modules were used to perform statistics, and data processing was performed in R, Python, and OpenOffice/LibreOffice using custom scripts. See the Supplemental Material for details.
Relative intron detention (RID) was determined by taking the log2 of the ratio between the read count within an intron normalized to its length and the total number of mapped reads per sample and the Cufflinks-estimated FPKM of the same gene. Intron read counts were the average across all relevant biological replicates.
Supplementary Material
Acknowledgments
We acknowledge the ENCODE project and the Gingeras laboratory for RNA-seq data. This work was supported by National Institutes of Health grant R01 GM34277-23 (P.A.S.), by an American Cancer Society/Massachusetts Biotechnology Council Challenge/Novartis Institutes for Biomedical Research Postdoctoral Research Fellowship (P.L.B.), and in part by Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute. We acknowledge the Massachusetts Institute of Technology BioMicro Center for RNA-seq services.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.247361.114.
References
- Allende-Vega N, Dayal S, Agarwala U, Sparks A, Bourdon JC, Saville MK. 2013. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene 32: 1–14. [DOI] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. 2011. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol 18: 1435–1440. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. 2012. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD, Adams JA. 2013. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J Mol Biol 425: 2894–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Wieslander L. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192. [DOI] [PubMed] [Google Scholar]
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. 2013. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev 27: 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. 2012. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell 150: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond U. 1988. Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J 7: 3509–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL. 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24: 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA. 1997. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3: 1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM. 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581. [DOI] [PubMed] [Google Scholar]
- Chang DD, Sharp PA. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789–795. [DOI] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. 2011. Interaction between the RNA binding domains of Ser–Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci 108: 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J 15: 265–275. [PMC free article] [PubMed] [Google Scholar]
- Crispino JD, Blencowe BJ, Sharp PA. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265: 1866–1869. [DOI] [PubMed] [Google Scholar]
- Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. 2005. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell 122: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Dufu K, Lei H, Reed R. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 1: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC. 1998. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res 241: 300–308. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O, Huber K, Eisenreich A, Filippakopoulos P, King O, Bullock AN, Szklarczyk D, Jensen LJ, Fabbro D, Trappe J, et al. 2011. Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing. Chem Biol 18: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R. 2009. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res 37: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, Urlaub H, Hartmuth K, Luhrmann R. 2012. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun 3: 994. [DOI] [PubMed] [Google Scholar]
- Graveley BR. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu XD. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369: 678–682. [DOI] [PubMed] [Google Scholar]
- Hao S, Baltimore D. 2013. RNA splicing regulates the temporal order of TNF-induced gene expression. Proc Natl Acad Sci 110: 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann AM, Rujescu D, Giannakouros T, Nikolakaki E, Goedert M, Mandelkow EM, Gao QS, Andreadis A, Stamm S. 2001. Regulation of alternative splicing of human τ exon 10 by phosphorylation of splicing factors. Mol Cell Neurosci 18: 80–90. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, zur Hausen A, Bettendorf H, Jager M, Stickeler E. 2009. Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Cancer Res 69: 2082–2090. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. 2004. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci 101: 9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveals expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Boutz PL, Paul P, Sharp PA. 2014. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev 28: 637–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, Cooper DR. 2009. Akt2 regulation of Cdc2-like kinases Clk/Sty serine/arginine-rich SR protein phosphorylation and insulin-induced alternative splicing of PKCβII messenger ribonucleic acid. Endocrinology 150: 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393: 185–187. [DOI] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, Rosbash M. 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M. 2012. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA 18: 2174–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929. [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583. [DOI] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. 2012. distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 46: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcak I, Melcakova S, Kopsky V, Vecerova J, Raska I. 2001. Prespliceosomal assembly on microinjected precursor mRNA takes place in nuclear speckles. Mol Biol Cell 12: 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen P, Lamond AI. 1992. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res 20: 5263–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Cohen PT, Lamond AI. 1994. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J 13: 5679–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr. 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev 21: 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya K, Kataoka N, Hagiwara M. 2011. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J Cell Biol 195: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A. 2011. Pre-mRNA splicing during transcription in the mammalian system. Wiley Interdiscip Rev RNA 2: 700–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Bhatt DM, Lin CH, Tong AJ, Smale ST, Black DL. 2013. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA 19: 811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. 2009. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad J, Colwill K, Pawson T, Manley JL. 1999. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol 19: 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29: 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Haas W, Gygi SP, Puigserver P. 2010. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab 11: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscigno RF, Garcia-Blanco MA. 1995. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA 1: 692–706. [PMC free article] [PubMed] [Google Scholar]
- Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, Pellagatti A, Lukashchuk N, McCance DJ, McCluggage WG, et al. 2014. Identification of a BRCA1–mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell 54: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertz H, Tolley ND, Foulks JM, Denis MM, Risenmay BW, Buerke M, Tilley RE, Rondina MT, Harris EM, Kraiss LW, et al. 2006. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J Exp Med 203: 2433–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Lindquist S, Burge CB. 2014. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Reports 7: 1362–1370. [DOI] [PubMed] [Google Scholar]
- Shi Y, Manley JL. 2007. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell 28: 79–90. [DOI] [PubMed] [Google Scholar]
- Shi Y, Reddy B, Manley JL. 2006. PP1/PP2A phosphatases are required for the second step of pre-mRNA splicing and target specific snRNP proteins. Mol Cell 23: 819–829. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111: 407–417. [DOI] [PubMed] [Google Scholar]
- Shin C, Feng Y, Manley JL. 2004. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427: 553–558. [DOI] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol 144: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. 1997. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci 94: 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura R, Takeiwa T, Taniguchi I, McCloskey A, Ohno M. 2011. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells 16: 1035–1049. [DOI] [PubMed] [Google Scholar]
- Taniguchi I, Masuyama K, Ohno M. 2007. Role of purine-rich exonic splicing enhancers in nuclear retention of pre-mRNAs. Proc Natl Acad Sci 104: 13684–13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes & Dev 8: 2704–2717. [DOI] [PubMed] [Google Scholar]
- Tazi J, Kornstadt U, Rossi F, Jeanteur P, Cathala G, Brunel C, Luhrmann R. 1993. Thiophosphorylation of U1-70K protein inhibits pre-mRNA splicing. Nature 363: 283–286. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. 2012. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 22: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. 2011. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147: 1054–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhager L, Bonfert T, Burger K, Ruzsics Z, Krebs S, Kaufmann S, Malterer G, L’hernault A, Schilhabel M, Schreiber S, et al. 2012. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res 22: 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW. 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13: 91–100. [DOI] [PubMed] [Google Scholar]
- Wong R, Balachandran A, Mao AY, Dobson W, Gray-Owen S, Cochrane A. 2011. Differential effect of CLK SR kinases on HIV-1 gene expression: potential novel targets for therapy. Retrovirology 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. 1997. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes & Dev 11: 334–344. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 17: 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Walker D, Bernardo A, Brodbeck J, Balestra ME, Huang Y. 2008. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J Neurosci 28: 1452–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. 2012. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev 26: 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Yomoda J, Muraki M, Kataoka N, Hosoya T, Suzuki M, Hagiwara M, Kimura H. 2008. Combination of Clk family kinase and SRp75 modulates alternative splicing of adenovirus E1A. Genes Cells 13: 233–244. [DOI] [PubMed] [Google Scholar]
- Yost HJ, Lindquist S. 1986. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 45: 185–193. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Fu XD. 2013. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122: 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.