The DEAD-box RNA helicase Prp5 is required for the formation of the prespliceosome through an ATP-dependent function to remodel U2 snRNPs and an ATP-independent function of unknown mechanism. Liang and Cheng show that Prp5 binds to the spliceosome in association with U2 by interacting with the branchpoint-interacting stem–loop and is released upon base-pairing of U2 with the branch site to allow the recruitment of the tri-snRNP.
Keywords: Prp5, splicing, branch site, U2, proofreading
Abstract
The DEAD-box RNA helicase Prp5 is required for the formation of the prespliceosome through an ATP-dependent function to remodel U2 small nuclear ribonucleoprotein particles (snRNPs) and an ATP-independent function of unknown mechanism. Prp5 has also been implicated in proofreading the branch site sequence, but the molecular mechanism has not been well characterized. Using actin precursor mRNA (pre-mRNA) carrying branch site mutations, we identified a Prp5-containing prespliceosome with Prp5 directly bound to U2 small nuclear RNA (snRNA). Prp5 is in contact with U2 in regions on and near the branchpoint-interacting stem–loop (BSL), suggesting that Prp5 may function in stabilizing the BSL. Regardless of its ATPase activity, Prp5 mutants that suppress branch site mutations associate with the spliceosome less tightly and allow more tri-snRNP binding for the reaction to proceed. Our results suggest a novel mechanism for how Prp5 functions in prespliceosome formation and proofreading of the branch site sequence. Prp5 binds to the spliceosome in association with U2 by interacting with the BSL and is released upon the base-pairing of U2 with the branch site to allow the recruitment of the tri-snRNP. Mutations impairing U2–branch site base-pairing retard Prp5 release and impede tri-snRNP association. Prp5 mutations that destabilize the Prp5–U2 interaction suppress branch site mutations by allowing progression of the pathway.
Introns of precursor mRNAs (pre-mRNAs) are removed via two steps of transesterification reaction, catalyzed by the spliceosome, which consists of five small nuclear RNAs (snRNAs; U1, U2, U4/U6, U5) and many protein factors. The snRNAs, each complexed with a number of proteins to form a small nuclear ribonucleoprotein particle (snRNP), bind to the pre-mRNA sequentially in an order of U1 and U2 and then U4/U6.U5 as a preformed tri-snRNP (Brow 2002; Wahl et al. 2009; Will and Lührmann 2011). Spliceosome assembly initiates with the binding of U1 to the 5′ splice, forming the commitment complex (CC), followed by binding of U2 to the branch site to form the prespliceosome. Respective recognition of the 5′ splice site and the branch site by U1 and U2 is mediated through RNA base-pairing interactions between the snRNAs and the intronic sequence. Prior to U2 addition, a Msl5–Mud2 heterodimer binds to the branch site and interacts with U1 snRNP, bridging the interaction between the 5′ splice site and the branch site (Berglund et al. 1997, 1998; Wang et al. 2008). Msl5–Mud2 is displaced from the branch site before U2 snRNA can base-pair with the branch site sequence in the formation of the prespliceosome. Following binding of the tri-snRNP, the spliceosome undergoes a major structural rearrangement to release U1 and U4 and forms new base pairs between U6 and the 5′ splice site and between U2 and U6. A protein complex associated with Prp19 (nineteen complex [NTC]) (Tarn et al. 1994) then binds to the spliceosome to stabilize the association of U5 and U6 in formation of the active spliceosome, which can catalyze the splicing reaction (Chan et al. 2003; Chan and Cheng 2005).
The spliceosome undergoes profound conformational changes during the splicing cycle to achieve splicing efficiency and accuracy (Wahl et al. 2009). DExD/H-box RNA helicases are important players in facilitating remodeling of the spliceosome (Staley and Guthrie 1998). They mediate rearrangements of RNA–RNA or RNA–protein interactions during progression of the splicing pathway, using the energy from ATP hydrolysis (Linder et al. 2001). Among the eight DExD/H-box ATPases required for the process, Sub2 and Prp5 are involved in the formation of the prespliceosome. The essential function of Sub2 can be bypassed in the absence of Mud2 when the association of Msl5 is weakened (Kistler and Guthrie 2001), suggesting that Sub2 may be required for destabilizing Msl5–Mud2 to allow the base pair interaction of U2 with the branch site sequence. Biochemical analysis revealed that Sub2 is also required for formation of CC2 but not CC1 (Kistler and Guthrie 2001).
Prp5 has been shown to mediate an ATP-dependent conformational change of U2 snRNP, and its ATPase activity is stimulated severalfold higher by U2 snRNA than any of the other snRNAs or nonspecific RNAs (O’Day et al. 1996). Prp5 has also been demonstrated to genetically interact with several components of the U2 snRNP, including Prp9, Prp11, Prp21, Cus1, Cus2, and the stem–loop IIa of U2 snRNA (Ruby et al. 1993; Wells and Ares 1994; Will et al. 2002; Xu et al. 2004). Prp5 was shown to interact with U1 and U2 snRNPs via the N-terminal domain to bridge the two snRNPs in the formation of the prespliceosome in the fission yeast Schizosaccharomyces pombe and humans (Xu et al. 2004). Such interactions were not found in Saccharomyces cerevisiae due to a lack of the corresponding U1-interacting domain in the Prp5 N terminus (Xu et al. 2004).
Genetic studies suggest that the helix II of U2 snRNA switches between two conformations—a functional IIa and a nonfunctional IIc form—throughout the splicing cycle (Hilliker et al. 2007; Perriman and Ares 2007). Mutations that disrupt U2 stem IIc bypass the ATP requirement for formation of the prespliceosome (Perriman and Ares 2000; Perriman et al. 2003). Prp5 was hypothesized to promote the formation of the IIa structure by displacing Cus2 from U2 in an ATP-dependent manner to form a functional U2 snRNP for its recruitment to the spliceosome (Perriman and Ares 2000; Perriman et al. 2003) and is further required for the formation of the prespliceosome independent of its ATPase function. Accordingly, the ATPase function of Prp5 is not required for splicing in cus2∆ extracts (Perriman et al. 2003). The underlying mechanism of the ATP-independent function is unknown. A U2 snRNA structure, the branchpoint-interacting stem–loop (BSL), was proposed to form prior to the interaction of U2 with the intron (Perriman and Ares 2010). The BSL is located between stem I and stem IIa, with the branch site-interacting sequence present in the loop region. The BSL is proposed to present the U2 nucleotides that will contact the intron and may be disrupted upon the engagement of U2 with the intron, likely mediated by Prp5.
Prp5 and several other DExD/H-box RNA helicases have been shown to play roles in splicing fidelity control (Burgess and Guthrie 1993a; Mayas et al. 2006; Xu and Query 2007; Yang et al. 2013). Prp16, the archetype of such proteins, was initially identified as a suppressor of pre-mRNA branchpoint A-to-C mutation (Burgess et al. 1990) but was later found to be required for the second catalytic step only after formation of lariat intron–exon 2 (Schwer and Guthrie 1992). Further studies have led to a proposal for a role of Prp16 in splicing fidelity control for the branch site by a kinetic proofreading mechanism (Burgess and Guthrie 1993a,b). Prp5, Prp22, and Prp28 were proposed to function in proofreading the branch site, the 3′ splice site, and the 5′ splice site, respectively, by a similar mechanism (Mayas et al. 2006; Xu and Query 2007; Koodathingal et al. 2010; Yang et al. 2013). Genetic analysis has revealed a set of prp5 SAT mutants to improve the splicing of suboptimal substrates with mutations in the branch site sequence (Xu and Query 2007). Prp5 was suggested to mediate splicing fidelity control of the branch site sequence by competing with base-pairings between U2 snRNA and the branch site sequence (Xu and Query 2007).
How Prp5 promotes prespliceosome formation remains elusive. It is also not clear whether Prp5 is further required for the spliceosome pathway after the formation of the prespliceosome. A GST-Prp5 fusion protein has been shown previously to associate with the spliceosome throughout the spliceosome pathway (Kosowski et al. 2009), but another study failed to detect Prp5 tightly associated with the spliceosome (Fabrizio et al. 2009). Although it is speculated that Prp5 may also mediate the IIc/IIa switch in U2 snRNP along the spliceosome pathway (Hilliker et al. 2007; Perriman and Ares 2007), no biochemical evidence for such an argument has been provided.
In this study, we first examined whether Prp5 also participates in the spliceosome pathway after prespliceosome formation by in vitro splicing assays. We found that Prp5 is not required for the progression of the spliceosome pathway after the formation of the prespliceosome, arguing against an essential role of Prp5 in mediating the U2 IIc/IIa switch after the prespliceosome step. When pre-mRNA carries mutations in the branch site sequence, the spliceosome is arrested after prespliceosome formation and before the addition of the tri-snRNP, with Prp5 tightly bound. Under such conditions, Prp5 could cross-link to U2 snRNA in regions on or near the BSL structure, suggesting a role of Prp5 in stabilizing the BSL. Interestingly, Prp5 mutants that suppress branch site mutations are less stably associated with the spliceosome, allowing more tri-snRNP to be recruited to the spliceosome, suggesting a biochemical mechanism for the suppression. The affinity of Prp5 with the spliceosome negatively correlates with the efficiency of suppression and is irrelevant to its ATPase activity. Our results suggest that Prp5, in association with U2, promotes the binding of U2 snRNP to the spliceosome and is then released from the spliceosome to allow the recruitment of the tri-snRNP. Mutations in the branch site sequence prohibit the release of Prp5 and block the splicing pathway, suggesting that proper base-pairing or interaction between U2 and the branch site may be important for Prp5 release. Our study provides novel mechanistic insights into the action of Prp5 in the splicing reaction.
Results
Prp5 is not required for splicing after prespliceosome formation
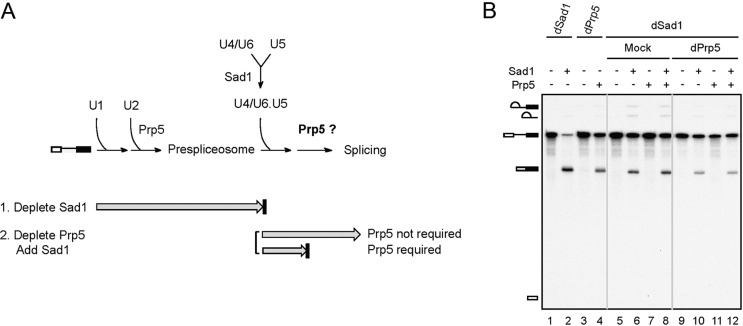
We first investigated whether Prp5 function is restricted to prespliceosome formation. We took advantage of the feature of an essential splicing factor, Sad1, that is required for maintaining the integrity of the tri-snRNP but is not tightly associated with any snRNP. In the absence of Sad1, the spliceosome pathway is blocked after formation of the prespliceosome due to a lack of functional tri-snRNP under normal splicing conditions (Fig. 1A; Huang et al. 2014). Splicing reactions were carried out in Sad1-depleted extracts (Fig. 1B, lanes 1,2,5–12), and Prp5 was then depleted from the reaction mixtures using anti-Prp5 antibody (Fig. 1B, lanes 9–12). Purified recombinant Sad1 was then added to the reaction mixtures with or without coaddition of recombinant Prp5 to see whether the splicing activity could be restored. We reasoned that if Prp5 were required after the prespliceosome step, the addition of Sad1 would not be sufficient to restore the splicing activity of Sad1-depleted extracts. On the other hand, if Prp5 were not required after the prespliceosome step, the splicing activity would be fully recovered by the addition of Sad1. The result shows that, as in mock-treated reactions (Fig. 1B, lane 6), Prp5 depletion had no effect on the splicing reaction after Sad1 was added to the reaction mixture (Fig. 1B, lane 10), and the addition of Prp5 did not improve the splicing efficiency (Fig. 1B, cf. lanes 10 and 12), suggesting that Prp5 is not required after the prespliceosome stage. Accordingly, the proposed U2 structural switch in the late steps of the spliceosome pathway is unlikely to be mediated by Prp5. Although GST-Prp5 has been shown previously to associate with the spliceosome throughout the splicing cycle (Kosowski et al. 2009), we did not see a stable association of Prp5 with the spliceosome at any stage of the pathway by immunoprecipitation studies using Prp5-V5-tagged extracts (Supplemental Fig. S1). An observation against the stable association of Prp5 with the spliceosome has also been reported (Fabrizio et al. 2009).
Figure 1.
Prp5 is not required for splicing after prespliceosome formation. (A) A schematic diagram showing early steps of spliceosome assembly and the involvement of Prp5 and Sad1. (B) Splicing reactions were carried out in Sad1-depleted extracts (lanes 1,2,5–12), and the reaction mixtures were then mock-treated (lanes 5–8) or depleted of Prp5 (lanes 9–12). Purified Sad1 and/or recombinant Prp5 proteins were added as indicated to the reaction mixtures, which were then further incubated for 20 min. The splicing reaction was carried out in Prp5-depleted extracts without (lane 3) or with (lane 4) the addition of Prp5.
Stable association of Prp5 with the spliceosome assembled on U257 mutant pre-mRNA
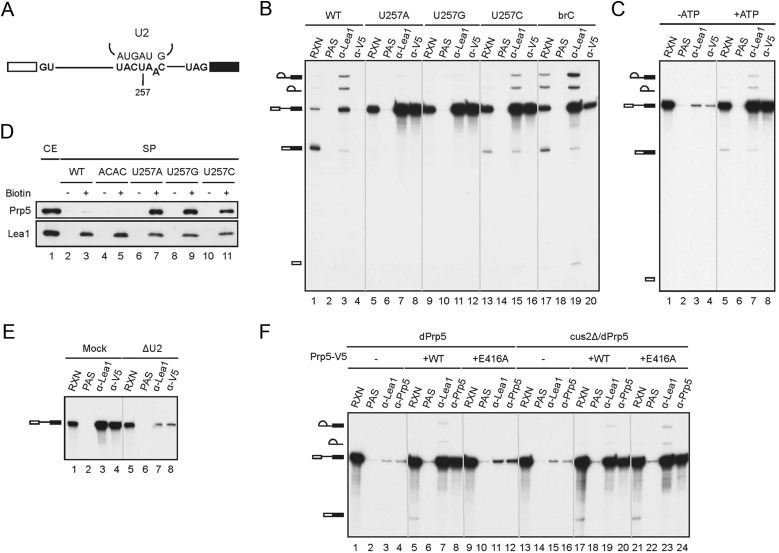
A group of SAT mutants of Prp5 were demonstrated in vivo to improve the splicing efficiency of pre-mRNA with branch site mutation, and, based on these studies, Prp5 was suggested to mediate fidelity control of spliceosome assembly at an early step (Xu and Query 2007). To explore the mechanism underlying Prp5-mediated splicing fidelity control, we set up in vitro assays using an actin pre-mRNA, with U257 in the conserved branchpoint sequence changed to A, G, or C (Fig. 2A). The U257 residue has been shown to base-pair with U2 snRNA, and such base-pairing is important for the splicing reaction (Parker et al. 1987). Several PRP5 alleles that suppress U257 mutations have been isolated (Xu and Query 2007). We first examined whether prespliceosome formation is affected by mutation in U257. Spliceosomes formed with wild-type and different U257 mutant pre-mRNAs were precipitated with antibodies against the U2 snRNP component Lea1 (Supplemental Fig. S2A) to see whether U2 could associate with mutant pre-mRNA (Fig. 2B). With wild-type pre-mRNA, while Lea1 coprecipitated precursor and lariat RNAs (Fig. 2B, lane 3), Prp5 did not coprecipitate any substrate RNA (Fig. 2B, lane 4), consistent with previous observations that Prp5 is not tightly associated with the spliceosome. Although splicing was completely (Fig. 2B, lanes 5,9) or severely (Fig. 2B, lane 13) inhibited with U257 mutant pre-mRNAs, large amounts of pre-mRNA were coprecipitated with Lea1 in all three mutants (Fig. 2B, lanes 7,11,15), indicative of the association of U2 with U257 mutant pre-mRNA. This result suggests that mutation in U257 does not prevent prespliceosome formation but blocks normal progression of the pathway at some point during or after prespliceosome formation. Interestingly, anti-V5 antibody also coprecipitated the mutant pre-mRNA (Fig. 2B, lanes 8,12,16) nearly as efficiently as anti-Lea1 antibody but did not coprecipitate wild-type pre-mRNA (Fig. 2B, lane 4). The branchpoint A-to-C mutant pre-mRNA brC was also coprecipitated with Prp5 but to a lesser extent (Fig. 2B, lane 20). Furthermore, ATP is required for the coprecipitation, at least for U257C (Fig. 2C, cf. lanes 4 and 8). ATP is known to be required for U2 snRNP binding to the spliceosome in the presence of Cus2, with Prp5 proposed to mediate Cus2 displacement to form functional U2 snRNP (Perriman et al. 2003). The fact that ATP is required for Prp5 binding to the spliceosome suggests that Prp5 may bind to the spliceosome only after U2 snRNP assumes a functional form. Association of Prp5 with the spliceosome was further confirmed by Western blotting of the spliceosome formed with biotinylated pre-mRNA and pulled down with streptavidin agarose (Fig. 2D). Prp5 was found to accumulate on the spliceosome formed on U257 mutant pre-mRNAs (Fig. 2D, lanes 7,9,11) but not on wild-type (Fig. 2D, lane 3) or 3′ splice site mutant ACAC (Fig. 2D, lane 5) pre-mRNA. These results indicate that mutation in U257 does not affect the association of U2 with pre-mRNA but results in retention of Prp5 on the spliceosome. The Prp5-containing spliceosomes accumulated on U257 mutant pre-mRNAs are called U257m spliceosomes, which may represent an intermediate in the formation of the mature prespliceosome.
Figure 2.
Stable association of Prp5 with the U257m spliceosome. (A) A diagram showing the sequence of the branch site and the position of the U257 mutation. (B) Splicing reactions were performed in Prp5-V5 extracts with wild-type and U257A, U257G, U257C, and brC mutant pre-mRNAs, and the reaction mixtures were immunoprecipitated with anti-Lea1 or anti-V5 antibody. (RXN) One-tenth of the reaction mixture; (PAS) protein A-Sepharose. (C) Splicing reactions were assembled with U257C mutant pre-mRNA in Prp5-V5 extracts in the presence of glucose (lanes 1–4) or ATP (lanes 5–8), and the reaction mixtures were immunoprecipitated with anti-Lea1 or anti-V5 antibody. (RXN) One-tenth of the reaction mixture; (PAS) protein A-Sepharose. (D) The spliceosome formed with biotinylated wild-type (lane 3), ACAC (lane 5), or U257m (lanes 7,9,11) pre-mRNA in Prp5-V5 extracts was isolated by precipitation with streptavidin-Sepharose, and the components were analyzed by Western blotting. (E) Splicing reactions were carried out in mock-depleted (lanes 1–4) or U2-depleted (lanes 5–8) Prp5-V5 extracts with U257G pre-mRNA. The reaction mixtures were immunoprecipitated with anti-Lea1 or anti-V5 antibody. (F) Splicing reactions were performed with U257G pre-mRNA in Prp5-depleted (lanes 1–12) or Prp5-depleted (lanes 13–24) cus2∆ extracts without (lanes 1–4,13–16) or with (lanes 5–8,17–20) the addition of wild-type Prp5 or the prp5-E416A mutant (lanes 9–12,21–24). The reaction mixtures were immunoprecipitated with anti-Lea1 or anti-V5 antibody. (RXN) One-tenth of the reaction mixture; (PAS) protein A-Sepharose.
Stable association of Prp5 with the spliceosome is U2-dependent
Prp5 has been shown to possess an ATP-independent function for the association of U2 with the spliceosome, but how Prp5 mediates U2 binding to the spliceosome is not known (Perriman et al. 2003). Prp5 may bind to the pre-mRNA first and then recruit U2 to the spliceosome or may restructure the spliceosome for binding of U2 to the branch site. We tested whether Prp5 could bind stably to the U257 spliceosome without U2. U2 snRNA was depleted from the extract by oligo-directed RNase H cleavage (Tarn et al. 1993). Despite incomplete cleavage, >70% of U2 snRNA was depleted after incubation with the oligo (Supplemental Fig. S2B). Splicing reactions were performed with U257G pre-mRNA in U2-depleted Prp5-V5 extracts, and the reaction mixtures were precipitated with anti-V5 antibody to determine the association of Prp5 (Fig. 2E). The result shows that the amount of pre-mRNA coprecipitated with Prp5 was greatly reduced (Fig. 2E, cf. lanes 4 and 8) congruent with that coprecipitated with Lea1 (Fig. 2E, cf. lanes 3 and 7), indicating that binding of Prp5 to the spliceosome requires the presence of U2. This result suggests that it is unlikely that Prp5 would first stably bind to the spliceosome to recruit U2 to the spliceosome and further suggests that Prp5 may act on the U2 snRNP to stabilize its association with pre-mRNA.
Previous studies have shown that Prp5 has an ATP-dependent and an ATP-independent function for prespliceosome formation. The ATPase-dependent activity of Prp5 is dispensable in cus2∆ cells, suggesting that the ATPase activity might function in displacing Cus2 for the formation of functional U2 snRNP. The association of U2 with the spliceosome is further promoted by an ATPase-independent activity of Prp5 (Perriman et al. 2003). To examine whether the ATPase activity of Prp5 is required for U2 association with the U257m spliceosome, we used the ATPase-defective Prp5 mutant E416A, with a mutation changing glutamine to alanine in the conserved DEAD motif, for complementation assays. Splicing reactions were carried out with U257G mutant pre-mRNA in Prp5-depleted extracts, and V5-tagged wild-type or E416A mutant Prp5 protein was then added (Fig. 2F, lanes 1–12). Immunoprecipitation analysis reveals that both Lea1 and Prp5 were associated with the spliceosome when wild-type Prp5 was added (Fig. 2F, lanes 7,8) but neither was associated with the spliceosome when prp5-E416A was added (Fig. 2F, lanes 11,12), suggesting that the ATPase activity of Prp5 is required for U2 association and Prp5 stabilization on the U257m spliceosome. To see whether the ATP-dependent activity is only necessary for counteracting Cus2 function (Perriman et al. 2003), we used extracts prepared from the cus2∆ strain to perform splicing reactions with U257G pre-mRNA after depletion of Prp5 (Fig. 2F, lanes 13–24). The result shows that, like wild-type pre-mRNA, Prp5 was required for the association of U2 with the U257G spliceosome in the absence of Cus2, as revealed by the association of Lea1 (Fig. 2F, cf. lanes 15 and 19), and the ATPase activity of Prp5 is not required for this function (Fig. 2F, lane 23). These results not only demonstrate the dependence on both U2 and Prp5 for the formation of the U257m spliceosome but also reveal a similar assembly process of the prespliceosome between U257m and wild-type pre-mRNA. This provided us with a tool for analyzing the mechanism of prespliceosome formation.
Prp5 directly interacts with U2 snRNA on the U257m spliceosome
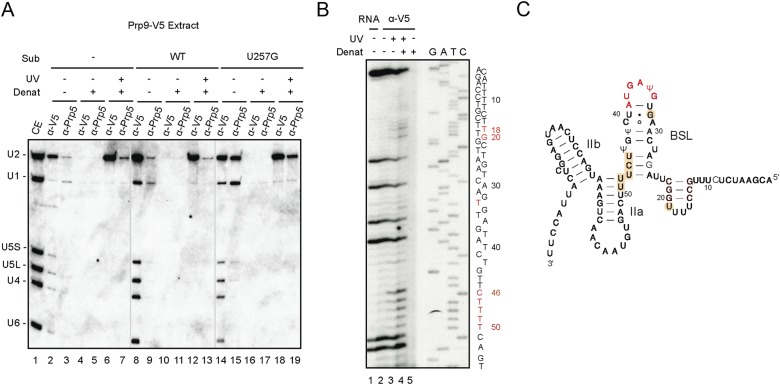
Prp5 has been demonstrated to physically interact with U1 and U2 snRNAs in S. pombe and humans, bridging the two snRNPs for prespliceosome formation (Xu et al. 2004), but direct interaction between Prp5 and U1 or U2 RNA has not been reported in S. cerevisiae. Stabilization of Prp5 on the U257m spliceosome allowed us to examine how Prp5 may interact with pre-mRNA or U1 or U2 snRNA on the prespliceosome. To examine the direct interaction between Prp5 and pre-mRNA, splicing reactions were carried out with U257C pre-mRNA in Hsh155-HA extracts, and the reaction mixtures were irradiated with UV254nm. Following denaturation, the reaction mixtures were immunoprecipitated with anti-Prp5 antibody and, as a control, anti-HA antibody for Hsh155, which has previously been shown to cross-link to pre-mRNA flanking the branchpoint (Gozani et al. 1998; McPheeters and Muhlenkamp 2003). As expected, Hsh155 was seen to cross-link to pre-mRNA, but Prp5 was not detected to cross-link to pre-mRNA (Supplemental Fig. S3), suggesting that Prp5 does not come into close contact with pre-mRNA. A similar experiment was performed and analyzed by Northern blotting to examine for cross-linking to snRNAs using Prp9-V5 extracts. In this experiment, Prp5 was found to directly interact with U2 snRNA (Fig. 3A, lane 19), although the interaction was slightly weaker than Prp9 as precipitated by anti-V5 antibody (Fig. 3A, lane 18). Weak cross-linking of Prp5 to U2 was also observed in the absence (Fig. 3A, lane 7) or presence (Fig. 3A, lane 13) of wild-type pre-mRNA, suggesting that Prp5 might be weakly associated with U2 snRNP even in the extract. These results provide definitive evidence for direct binding of Prp5 to U2 snRNA on the U257G prespliceosome. Prp5 cross-linking sites were mapped by primer extension using a primer spanning from position 62 to 82 of U2 snRNA (Fig. 3B). Strong stops were observed at U2 positions 46–50, corresponding to cross-linking at positions 45–49, a region encompassing the last three residues of the stem of the BSL and the first two residues of helix IIa of U2 snRNA. Two weaker cross-links at positions 17 and 19 in the loop region of U2 helix I and one at position 32 in the BSL stem were also observed (Fig. 3C). The fact that all cross-linking sites are either on or near the BSL suggests that Prp5 might play a role in stabilizing the BSL structure to direct the interaction of U2 with the branch site.
Figure 3.
Prp5 interacts directly with U2 snRNA on the U257G spliceosome. (A) Splicing reactions were carried out without (lanes 2–7) or with wild-type (lanes 8–13) or U257G (lanes 14–19) pre-mRNA in Prp9-V5 extracts, and the reaction mixtures were separated into three aliquots. One aliquot was not treated further (lanes 2,3,8,9,14,15), another was subjected to denaturation (lanes 4,5,10,11,16,17), and the third was irradiated with UV254nm followed by denaturation (lanes 6,7,12,13,18,19). The reaction mixtures were then precipitated with anti-V5 or anti-Prp5 antibody. Precipitated materials were analyzed by Northern blotting probed with five snRNAs. (B) Prp5-U2 cross-linked product was isolated as in A using Prp5-V5 extracts for precipitation of Prp5 cross-linked products with anti-V5 antibody and analyzed by primer extension to map cross-linking sites using primer U2-B. (C) The proposed structure of the U2–BSL form with the Prp5 cross-linked residues highlighted; branch site-interacting residues are indicated in red.
Splicing of U257 mutants is blocked for tri-snRNP association
To evaluate the progress of the blocked complexes in spliceosome assembly, we asked whether tri-snRNP is associated with the U257m spliceosome using antibodies against Prp8 (Fig. 4A). Splicing reactions were carried out with wild-type or U257 mutant pre-mRNA, and the reaction mixtures were subjected to immunoprecipitation with anti-Lea1 or anti-Prp8 antibody to examine the association of U2 snRNP and tri-snRNP, respectively, with pre-mRNA (Fig. 4A). While both Lea1 and Prp8 coprecipitated wild-type pre-mRNA, lariat intermediate, and product (Fig. 4A, lanes 3,5), Prp8 barely coprecipitated U257A (Fig. 4A, lane 10) or U257G (Fig. 4A, lane15) mutant pre-mRNA. Small amounts of U257C pre-mRNA and lariats were coprecipitated (Fig. 4A, lane 20) due to the presence of residual splicing activity. Western blotting of the purified spliceosomes assembled on biotinylated U257m pre-mRNAs also hardly detected tri-snRNP component Prp8 or NTC component Ntc20 on the spliceosome, except for a small amount of Ntc20 and Prp8 with U257C pre-mRNA (Fig. 4B, lanes 4–6). Northern blotting of the U257m spliceosomes further revealed the presence of U1 and U2 but only small amounts of U4, U5, and U6 snRNAs (Supplemental Fig. S4). All of these results indicate that U257 mutations prevent binding of the tri-snRNP to the spliceosome, leading to accumulation of a uniquely blocked prespliceosome-like complex that has failed to release Prp5.
Figure 4.
Binding of Prp5 and binding of the tri-snRNP on the spliceosome are mutually exclusive. (A) Splicing reactions were carried out with wild-type, U257A, U257G, and U257C pre-mRNAs in Prp5-V5 extracts, and the reaction mixtures were precipitated with anti-Lea1, anti-V5, or anti-Prp8 antibody. (B) Spliceosomes assembled on wild-type, ACAC, U257A, U257G, and U257C biotinylated pre-mRNAs using Prp5-V5 extracts were pulled down with streptavidin-Sepharose, and the components were analyzed by Western blotting probed with anti-V5, anti-Lea1, anti-Prp8, and anti-Ntc20 antibodies. (C,D) Splicing reactions were carried out in Prp5-V5 extracts with wild-type, U257A, U257G, and U257 pre-mRNAs (C) The spliceosome was precipitated with anti-V5 antibody, and the components were analyzed by Northern blotting probed for five snRNAs. (D) The spliceosome was precipitated with anti-Lea1, anti-V5, or anti-Prp8 antibody, and the components were analyzed by Western blotting probed with anti-Snu114, anti-V5, and anti-Lea1 antibodies.
Prp5 and tri-snRNPs are mutually exclusive on the spliceosome
Since U257 mutations result in retention of Prp5 on the prespliceosome and inhibition of tri-snRNP recruitment, we reasoned that retention of Prp5 might prevent binding of the tri-snRNP to the spliceosome. In this case, the association of Prp5 and the tri-snRNP with the spliceosome should be mutually exclusive. To test this hypothesis, we isolated Prp5-associated spliceosomes assembled on U257m pre-mRNAs in Prp5-V5 extracts by precipitation with anti-V5 antibody and examined the RNA and protein components. Northern blotting revealed no U4, U5, or U6 on the isolated spliceosome (Fig. 4C, lanes 5–7). Snu114 is a component of U5 snRNP and is also associated with U4/U6.U5 tri-snRNP (Supplemental Fig. S2). Western blotting also revealed no Snu114 on the spliceosome when precipitated with anti-Prp5 antibody (Fig. 4D, top panels, lanes 9–11) but detected small amounts of Snu114 when precipitated with anti-Lea1 antibody (Fig. 4D, top panels, lanes 4–6). In the reciprocal experiment, when the spliceosome was precipitated with anti-Prp8 antibody, Prp5 was not coprecipitated (Fig. 4D, middle panels, lanes 13–16). These results demonstrate that in the splicing of U257 mutant pre-mRNA, Prp5 is retained on the spliceosome, which inhibits the recruitment of the tri-snRNP. Only a small fraction of the spliceosome escapes from Prp5 retention and proceeds through the spliceosome pathway.
Prp5 mutants facilitate the association of the tri-snRNP
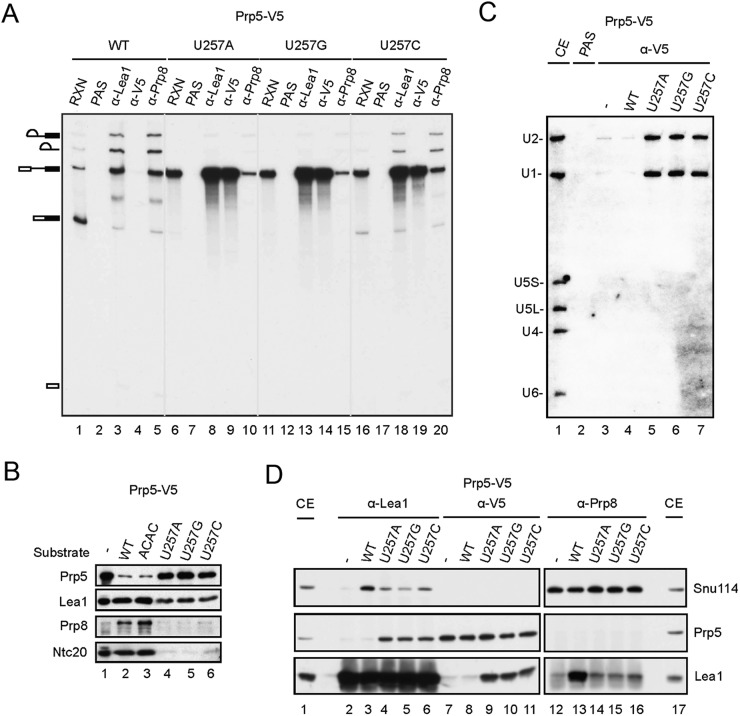
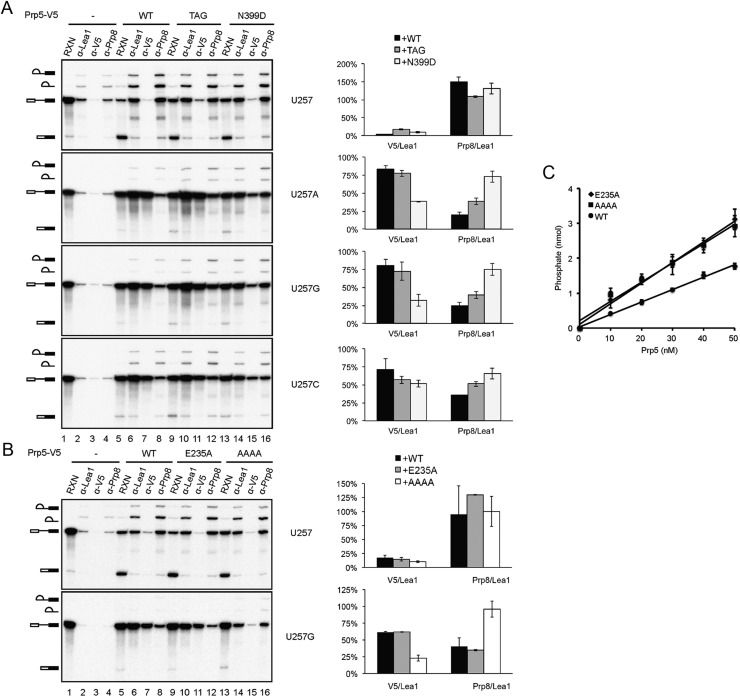
Specific alleles of PRP5 that improve splicing efficiency in vivo were identified in a genetic screen of suppressors to branch site mutations (Xu and Query 2007). Based on the results presented above, we anticipated these mutants to bind the spliceosome less tightly to allow progression of the splicing pathway. To see whether the identified mutants could improve the splicing efficiency in vitro, we purified two Prp5 branch site suppressor mutants—N399D and TAG proteins (Xu and Query 2007)—from Escherichia coli (Supplemental Fig. S5A) for complementation of Prp5-depleted extracts in splicing of U257 mutant pre-mRNA. All of these proteins are functional for complementation of Prp5-depleted cus2∆ extracts with slightly lower activities than wild-type Prp5 (Supplemental Fig. S5B), consistent with a previous report that the ATPase activity of Prp5 is not required for splicing of wild-type pre-mRNA in cus2∆ extracts (Perriman et al. 2003). To our disappointment, these mutant proteins did not significantly improve the splicing efficiency of U257m pre-mRNA in vitro (data not shown), possibly due to lower sensitivity in the in vitro assay system.
We then examined whether the Prp5 mutant proteins were altered to affect the association of itself or the tri-snRNP with the spliceosome in splicing of U257m pre-mRNA. In this experiment, extracts prepared from the cus2∆ strain were used to bypass the U2 activation step. Splicing reactions were carried out with U257m pre-mRNA in Prp5-depleted extracts supplemented with V5-tagged wild-type or mutant N399D or TAG prp5 protein, and the reaction mixtures were precipitated with anti-Lea1, anti-V5, or anti-Prp8 antibody. Figure 5A shows that in comparison with coprecipitation with Lea1, the amounts of the RNA coprecipitated with Prp5 were lower with N399D or TAG mutant protein than with wild-type Prp5 (lanes 7,11,15), indicating that these mutations may affect stable association of Prp5 on the U257m spliceosome. In contrast, the amounts of RNA coprecipitated with Prp8 were higher with mutant Prp5 proteins. Quantification of the precipitated RNA by PhosphorImager revealed an inverse correlation between the molar fraction of the Prp5-associated and Prp8-associated spliceosomes and Lea1-associated spliceosomes in U257 mutants. The N399D mutant protein showed less binding of Prp5 but more binding of Prp8 to the spliceosome than the TAG mutant. These results indicate that prp5 mutants may improve the splicing efficiency by destabilizing Prp5 association to allow the recruitment of the tri-snRNP in the splicing of pre-mRNA with branch site mutations.
Figure 5.
Prp5 mutants facilitate the association of tri-snRNP. (A) Splicing reactions were performed with wild-type, U257A, U257G, or U257C pre-mRNA in Prp5-depleted cus2∆ extracts without (lanes 1–4) or with the addition of wild-type Prp5 (lanes 5–8), prp5-N399D (lanes 9–12), or prp5-TAG (lanes 13–16) recombinant protein. The reaction mixtures were immunoprecipitated with anti-Lea1, anti-V5, or anti-Prp8 antibody. RNA was quantified by a PhosphorImager, and the molar ratios of total RNA precipitated by anti-V5 and anti-Prp8 antibodies to those precipitated by anti-Lea1 antibody are plotted in a bar graph. (B) The same as in A, with wild-type or U257G pre-mRNA for the splicing reaction using prp5-E235A (lanes 9–12) and prp5-AAAA (lanes 13–16) mutants. (C) Assays for the ATPase activity of wild-type and E235A and AAAA mutants of Prp5.
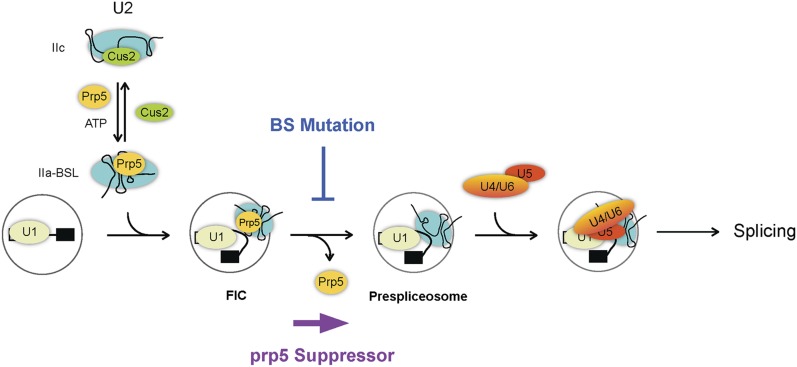
The structure of a Prp5 fragment containing the conserved helicase core domain and additional N-terminal and C-terminal flanking sequences has recently been determined (Zhang et al. 2013), revealing N399 located on the surface of the protein (Supplemental Fig. S5C). Although how Prp5 interacts with other proteins is not known, it is possible that this residue is involved in the interface of protein–protein interactions. Two residues, E235 and L238, located in the N-terminal region, were shown to possess higher ATPase activity and deteriorate the growth of branch site mutations U257C, A258C, and A258U when changed to alanine. These data support a previous hypothesis of the correlation between the ATPase activity of Prp5 and the growth defect of branch site mutations (Zhang et al. 2013). We examined whether the enhancer mutant of Prp5 would bind better to the spliceosome as opposed to loose binding for suppressor mutants. Residue 238 of Prp5 is polymorphic and codes for phenylalanine instead of leucine in the strain that we used. We therefore examined the binding property of the enhancer mutant E235A using U257G pre-mRNA. We observed only a very small increase in the level of Prp5 and a corresponding small decrease in that of Prp8 (Fig. 5B). Although such a small increase in the affinity of E235A for the spliceosome may not be significant to account for the enhancer phenotype, it contrasted with the suppressor mutants N399D and TAG in its affinity for the spliceosome relative to the wild-type protein. We further examined another mutant of Prp5 recently reported to be a strong suppressor of branch site mutations. A conserved DPLD motif present in the N-terminal domain of Prp5 has been shown to be important for interaction with U2 snRNP, and mutations in these residues suppress growth defects of branch site mutations (Shao et al. 2012). We purified mutant Prp5 with all four conserved residues changed to alanine (AAAA) and performed the same experiment using U257G pre-mRNA. Like the other branch site suppressors, N399D and TAG, the AAAA mutant showed lower affinity for the spliceosome and allowed more tri-snRNP binding (Fig. 5B). Moreover, the AAAA and E235A mutants gave comparable ATPase activities (both being higher than wild-type protein) (Fig. 5C) despite the fact that one suppresses and the other enhances the growth defect of branch site mutations. These results suggest that the level of ATPase activity in Prp5 is irrelevant to the suppression of branch site mutations. Our data support a mechanism in which suppression of branch site mutations is a consequence of weakening Prp5 association with the spliceosome to relieve a splicing block regardless of the ATPase function. A model for the functional roles of Prp5 in prespliceosome formation and proofreading the branch site sequence is shown in Figure 6. In this model, Prp5 activates U2 snRNP by displacing Cus2 and binding to U2 to stabilize the IIa–BSL structure. Prp5 then binds to the pre-mRNA in association with U2, forming a Prp5-associated intermediate complex (FIC, for five intermediate complex), and is subsequently released to form the prespliceosome, possibly upon base-pairing of U2 with the branch site sequence. The release of Prp5 allows binding of the tri-snRNP to the prespliceosome. Mutations at the branch site prevent Prp5 release, and prp5 mutations that destabilize the association of Prp5 with U2 relieve the block, allowing progression of the pathway.
Figure 6.
A model for the roles of Prp5 in the early steps of the spliceosome pathway. The newly identified Prp5-associated intermediate complex is marked as FIC.
Discussion
In this study, we elucidate how Prp5 functions in the splicing reaction by an in vitro reconstitution system using pre-mRNA branch site mutants. We demonstrate that Prp5 is required only for the formation of the prespliceosome but not for steps afterward in the spliceosome pathway. Unlike the GST-Prp5 fusion protein, which was previously reported to associate with the spliceosome throughout the splicing pathway (Kosowski et al. 2009), we did not see stable association of Prp5 with the spliceosome under normal splicing conditions by immunoprecipitation studies using authentic Prp5 or V5-tagged Prp5. We speculate that the GST-Prp5 fusion might alter structural properties of the protein so that it binds to the spliceosome less specifically.
The U2 helix II structure has been shown to switch between two different conformational states: IIa and IIc. Cus2 stabilizes the IIc conformation, whereas Prp5 promotes the IIa conformation (Perriman et al. 2003). Prp5 was proposed to mediate displacement of Cus2 from U2 snRNP in an ATP-dependent manner. In the absence of Cus2, the ATPase function of Prp5 is not required for prespliceosome formation, but Prp5 was known to be further needed by an unknown mechanism (Perriman et al. 2003). Using pre-mRNA branch site mutant U257m, we identified a Prp5-containing prespliceosome-like complex with Prp5 directly bound to U2, suggesting that Prp5 binds to the spliceosome in association with U2. Prp5 directly contacts U2 in several regions on or near the BSL structure (one in the stem of the BSL, another at the junction of the stem IIa and the BSL, and a third in the loop region of stem I), suggesting that Prp5 may function in promoting the formation of or stabilizing the BSL structure by binding to U2 snRNA. The BSL was proposed to present the U2 nucleotides that will contact the branch site and form prior to the interaction of U2 with the intron (Perriman and Ares 2010). It is likely that Prp5 binds to U2 snRNA after Cus2 is displaced to facilitate or stabilize the formation of IIa and the BSL to present U2 for base-pairing with the branch site. This unique role of Prp5 in BSL formation may also explain why Prp5 is unlikely to be involved in the IIc/IIa switch in later steps of the spliceosome pathway. The interaction of Prp5 with U2 in the extract may be too weak to sustain immunoprecipitation and was only barely detected by UV cross-linking (Fig. 3A).
Both human and S. pombe Prp5 proteins interact with U1 and U2 snRNPs via distinct domains located at the N terminus (Xu et al. 2004). SpPrp5 interacts with U1 snRNP via interaction with Rsd1, which interacts with U1A, and interacts with U2 snRNP via interaction with SF3b (Shao et al. 2012). The S. cerevisiae Prp5 protein lacks the U1-interacting domain but preserves the U2-interacting domain. Consistently, Prp5 was not seen to cross-link to U1 on the U257m spliceosome (Fig. 3A). The U2-interacting domain contains an evolutionarily conserved DPLD motif, but only the C-terminal half of the domain is conserved. The difference in the affinity for U2 may be attributed to the diverse sequence in the N-terminal half of the domain. Strong cross-linking was seen only in splicing with U257m but not with wild-type pre-mRNA (Supplemental Fig. S5). These results suggest that under normal splicing conditions, Prp5 is not stably associated with U2 or the spliceosome and is released immediately after engagement of U2 to the branch site. When pre-mRNA carries mutations in the branch site, Prp5 is unable to dissociate to allow tri-snRNP recruitment, thus preventing further progression of the splicing pathway to ensure the fidelity of branch site usage. Our results demonstrate that U257 mutations block splicing by preventing the release of Prp5 rather than inhibiting the binding of U2 snRNP to the spliceosome. This is the first direct demonstration of the association of Prp5 with the spliceosome as an intermediate complex in prespliceosome formation.
The cause of Prp5 retention on the branch site mutant spliceosome is not known. Conceivably, binding of U2 snRNP to the spliceosome induces a change in U2 conformation that stabilizes the interaction of Prp5 with U2, and Prp5 is immediately dissociated upon disruption of the BSL and base-pairing of U2 with the branch site. Mutations in U257 impair U2–branch site base-pairing and prevent Prp5 from dissociation. However, branchpoint mutant brC also retains Prp5 on the spliceosome but to a lesser extent (Fig. 2). Since the brC mutant does not affect the extent of U2–branch site base-pairing (Query and Konarska 2004; Tseng et al. 2011), full base-pairing is unlikely to be the only factor accounting for Prp5 dissociation. In this sense, the configuration of U2–branch site interaction may determine the stability of Prp5 on the spliceosome. It has been shown that the relative position of the branchpoint in the U2–branch site duplex would influence the efficiency of lariat formation (Smith et al. 2009). It would be interesting to see whether altering the position of the branchpoint also results in Prp5 retention. Alternatively, U257 mutations may affect the U2–branch site interaction by inhibiting the release of Msl5 and Mud2. Prior to U2 addition, the Msl5–Mud2 heterodimer binds to the branch site and is displaced from the branch site before U2 snRNA can base-pair with the branch site sequence. Whether the U257m spliceosome is arrested before or after the release of Msl5–Mud2 is not known. U257 mutations may impede Msl5–Mud2 release and, as a consequence, prevent Prp5 from dissociation. In this case, the identified prespliceosome intermediate would provide a tool for analyzing how Msl5–Mud2 and Sub2 act in concert with U2 and Prp5 in the formation of the prespliceosome.
The homeostatic property of U2 snRNP is highly similar to that of the U4/U6.U5 tri-snRNP. We previously demonstrated homeostasis of the tri-snRNP with dynamic association and dissociation between the U4/U6 di-snRNP and U5 snRNP (Huang et al. 2014). Brr2 promotes tri-snRNP dissociation using the energy from ATP hydrolysis, and Sad1 promotes U4/U6 and U5 association despite not being tightly associated with the tri-snRNP. Depletion of Sad1 results in complete dissociation of the tri-snRNP in the presence of ATP, and this consequently blocks the spliceosome pathway after formation of the prespliceosome. It is not known whether homeostatic control of the tri-snRNP and U2 snRNP is functionally significant or simply a reflection of the structural dynamics of the snRNPs outside the spliceosome pathway. Although Prp5 promotes the switch of U2 from stem IIc to IIa conformation to allow the binding of U2 to the spliceosome, our data suggest that Prp5 is dispensable after the formation of the functional prespliceosome, arguing against a role of Prp5 in mediating the conformational change of U2 snRNA at post-prespliceosome stages. In accordance, Cus2 is not detected to associate with the spliceosome during the splicing reaction but can coprecipitate a small amount of U2 in the cell extract. Cus2 has previously been shown to associate with U2 only weakly and is significantly coprecipitated with U2 only at lower salt concentrations (Yan et al. 1998). These results suggest that Cus2 and Prp5 may regulate only the homeostasis of the free U2 snRNP but are not involved in structural changes of U2 after the prespliceosome stage. Factors that mediate conformational changes of U2 on the spliceosome remain unknown.
Prp5 has previously been shown to modulate the fidelity of spliceosome assembly by proofreading the branch site sequence (Xu and Query 2007). ATPase-defective prp5 mutant alleles can suppress branch site mutations, and the ability of the mutants to suppress branch site mutation in general negatively correlates with the level of its ATPase activity (Perriman and Ares 2007; Xu and Query 2007). Based on these results, it was proposed that spliceosomes assembled on the pre-mRNA containing mutations in the branch site are retarded for the formation of a functional prespliceosome due to poor pairing of U2 snRNA with the branch site sequence. The spliceosomes are then directed to a discard pathway mediated by Prp5 using the energy from ATP hydrolysis. Mutations that reduce the ATPase activity of Prp5 slow down Prp5 action, allowing more time for duplex formation between U2 and the branch site (Xu and Query 2007; Zhang et al. 2013). Supporting this hypothesis, mutations that increase the ATPase activity of Prp5 were shown to deteriorate the growth defect of branch site mutants (Zhang et al. 2013). Incongruent with this hypothesis, the N399D mutant possesses full ATPase activity but exhibits strong suppression of branch site mutations, suggesting that the ATPase activity is not exclusively accountable for suppression (Xu and Query 2007). We demonstrate that the binding of U2 to the pre-mRNA is not affected by branch site mutations. Interestingly, the N399D mutant is less well retained than wild-type Prp5 on the spliceosome assembled on U257m pre-mRNA and allows more tri-snRNP binding to the spliceosome. This explains how N399D may suppress the splicing defect of U257 mutants. Consistent with this scenario, the SAT mutant TAG is a weaker suppressor than N399D and is better retained on the spliceosome than N399D but less well than wild-type Prp5. Notably, N399D assumes full ATPase activity, whereas TAG is half as active as the wild-type protein (Supplemental Fig. S4; Xu and Query 2007). Intriguingly, the AAAA mutant possesses higher ATPase activity than wild-type Prp5 at a level comparable with that of the enhancer mutant E235A but is a strong suppressor of branch site mutations. AAAA also binds the spliceosome less tightly than wild-type Prp5. Together, these results suggest that the affinity of Prp5 for the spliceosome is not correlated with its ATPase activity, and the primary cause of these Prp5 mutations in relieving splicing blocks of U257m pre-mRNA is alteration of the protein structure rather than reduction of the ATPase activity. This also raises a question of whether mutations in the SAT motif would cause a subtle conformational change of Prp5, since many of the branch site suppressors are mutated in SAT (Xu and Query 2007). Considering that the ATPase function of Prp5 is not required in cus2∆ mutants, it is unlikely that Prp5 proofreads the branch site sequence through its ATPase activity. Conformational defects may account for the suppression phenotype of those SAT mutants.
It has also been shown that in the cus2∆ genetic background, the growth defect of the prp5∆494 mutant is suppressed by U2 mutations that cause destabilization of the BSL structure, suggesting that Prp5∆494 may be a tight-binding mutant defective in BSL destabilization (Perriman and Ares 2010). Congruent with this notion, our in vitro data suggest that the Prp5∆494 mutant protein binds the U257G spliceosome slightly more tightly than wild-type Prp5 (data not shown). Our results support a model in which Prp5 binds directly to U2 snRNA to stabilize the BSL structure and chaperones U2 snRNP to the spliceosome. Under normal splicing conditions, Prp5 is subsequently released from the spliceosome to allow the recruitment of the tri-snRNP. Mutations in the branch site result in retention of Prp5 on the spliceosome and inhibition of tri-snRNP recruitment. Prp5 mutants that allow the protein to dissociate more easily would relieve the block of the pathway, eliciting a suppression phenotype. This suggests that Prp5 mediates splicing fidelity control by restraining its release from the impaired spliceosome after U2 binding rather than by competing for U2–branch site base-pairing.
Materials and methods
Splicing extracts, substrates, and splicing assays
Yeast whole-cell extracts were prepared according to Cheng et al. (1990). Substrates were prepared by in vitro transcription with SP6 RNA polymerase using plasmid DNA pSPact64-88 linearized with EcoRI as template. Splicing assays were carried out according to Cheng and Abelson (1987) for 30 min at 25°C unless otherwise indicated. Biotinylated pre-mRNA was synthesized according to the procedure described by Chan et al. (2003).
To calculate the fraction of the spliceosome containing Prp5 and the tri-snRNP in Figure 5, the amount of each RNA species was measured using a PhosphorImager and normalized to the number of uridine residues for each RNA species. The amount of total spliceosomes was calculated as L+I+P for each immunoprecipitation (where L, I, and P represent molar amounts of lariat-IVS-E2, lariat-IVS, and pre-mRNA, respectively). The ratio of Prp5-associated spliceosome to Lea1-associated spliceosome represents the fraction of U2-associated spliceosome that contains Prp5, and the ratio of Prp8-associated spliceosome to Lea1-associated spliceosome represents the fraction of U2-containing spliceosome that contains the tri-snRNP.
Expression and purification of recombinant Prp5 and Sad1 proteins
The PRP5 gene tagged with V5 at the C terminus was cloned into a modified pSUMO vector. Mutations in the DEAD motif (Xu et al. 2004) or SAT motif; at positions 235 (Zhang et al. 2013), 399 (Xu and Query 2007); or in the DPLD motif (Shao et al. 2012) were introduced by site-directed mutagenesis using primers P5-11 and P5-12, P5-15 and P5-16, P5-13 and P5-14, P5-17 and P5-18, or P5-21 and P5-22, respectively. Plasmids pSUMO.PRP5-V5, pSUMO.PRP5-TAG-V5, pSUMO.PRP5-N399D-V5, pSUMO.PRP5-E416A-V5, pSUMO.PRP5-E235A-V5, and pSUMO.PRP5-AAAA-V5 were transformed into E. coli strain Rosetta for expression of Prp5 proteins. Overnight cultures were inoculated into 8 L of LB medium containing 10 μg/mL ampicillin to a final A600 of 0.05 and grown at 37°C until A600 reached ∼0.5. The culture was chilled for 30 min on ice, IPTG was added to a final concentration of 1 mM, and the culture was incubated for 3 h at 18°C with constant shaking. Cells were harvested by centrifugation at 4000 rpm for 10 min. Cell pellets were stored at −80°C.
The following operations were performed at 4°C: Cell pellets were resuspended in 300 mL of lysis buffer (50 mM NaPO4 at pH 7.0, 750 mM NaCl, 5 mM imidazole, 10% glycerol, 1 mM β-mercaptoethanol) and twice disrupted using a microfludizer at a setting of 14,000 psi. Lysates were centrifuged at 15,000 rpm for 40 min to remove cell debris. The supernatant was applied at 1.25 mL/min to a 5-mL HisTrap HP column (GE Healthcare) and equilibrated with lysis buffer using the Akta FPLC system (GE Healthcare). After washing with 10 column volumes of wash buffer (50 mM NaPO4 at pH 7.0, 750 mM NaCl, 40 mM imidazole, 10% glycerol, 1 mM β-mercaptoethanol), bound proteins were eluted with 20 column volumes of elution buffer (50 mM NaPO4 at pH 7.0, 750 mM NaCl, 250 mM imidazole, 10% glycerol, 1 mM β-mercaptoethanol).
Eluted protein fractions were dialyzed against HEPES-binding buffer (50 mM HEPES-NaOH at pH 7.5, 600 mM NaCl, 20 mM imidazole, 10% glycerol) and loaded onto a HisTrap HP column again to remove nucleic acid (Warkocki et al. 2009). The column was washed with 10 column volumes of binding buffer followed by two column volumes of wash buffer (50 mM HEPES-NaOH at pH 7.5, 2 M LiCl, 10% glycerol) and two column volumes of binding buffer again. Proteins were eluted with 10 column volumes of HEPES elution buffer (50 mM HEPES-NaOH at pH 7.5, 600 mM NaCl, 250 mM imidazole, 10% glycerol).
The eluent was dialyzed against 2 L of a buffer containing 20 mM MES (pH 6.5), 300 mM NaCl, and 10% glycerol and loaded onto a 5-mL HiTrap SP HP column (GE Healthcare) equilibrated with the same buffer. Proteins were eluted with a gradient of 0.3–1 M NaCl. Fractions containing Prp5 were collected and concentrated with Ultra-15 (Millipore). The Prp5 proteins were further purified by size exclusion chromatography Superdex 200 (GE Healthcare) using a buffer containing 50 mM Tris-HCl (pH 8.0) and 300 mM NaCl. Fractions containing Prp5 were concentrated, with glycerol added to a final concentration of 10%. The purified proteins were separated into aliquots, flash-frozen in liquid nitrogen, and stored at −80°C. The purity of the proteins was assessed by SDS-PAGE (Supplemental Fig. S4A), and the concentrations of the proteins were determined by Bradford assays. The Sad1 protein was purified according to Huang et al. (2014).
Immunodepletion, immunoprecipitation, and precipitation of the spliceosome by streptavidin Sepharose
Immunodepletion of Prp5 or Sad1 was performed by incubation of 100 μL of splicing extracts with 150 μL of anti-Prp5 antibody or 150 μL of anti-Sad1 antibody conjugated to 50 μL of protein A-Sepharose (PAS). Immunoprecipitation was performed as described by Tarn et al. (1993). For every 20 μL of the splicing reaction, 1 μL of anti-V5 antibody, 5 μL of anti-Prp5 antibody, 5 μL of anti-Lea1 antibody, 1.5 μL of anti-Prp8 antibody, and 1 μL of anti-Snu114 antibody were used. Precipitation of the spliceosome with streptavidin Sepharose was carried out according to the procedure of Chan et al. (2003).
In vitro depletion of U2 snRNA
Depletion of U2 snRNA with oligo-directed RNase H cleavage was performed using primer U2-C according to the procedure of Tarn et al. (1993).
ATPase assay
ATPase assays were carried out according to Tanaka et al. (2007) in a buffer containing 40 mM Tris-HCl (pH 8.0), 2 mM DTT, 1 mM ATP, 1 mM MgCl2, 0.1 mM poly(A), and various amounts of recombinant Prp5 proteins. The reaction mixtures were incubated for 30 min at 25°C. Reactions were quenched by adding 1 mL of Biomol Green reagent (Biomol Research Laboratories), and the absorbance at A620 was measured after 20 min. The amounts of free phosphate were calculated by intrapolation of the A620 values to a phosphate standard curve.
UV cross-linking of Prp5 to RNA
UV cross-linking and analyses of cross-linked products were carried out as described in Chiang and Cheng (2013) except that splicing reactions were carried out in Prp9-V5 extracts for 30 min. For UV cross-linking of Prp5 to pre-mRNA, the splicing reaction was carried out using pre-mRNA with 10-fold specific radioactivity. RNA was isolated after UV irradiation and fractionated on 8 M urea–5% (29:1) polyacrylamide gels. For analysis of Prp5 cross-linking to snRNAs, splicing was carried out with 2 nM substrate pre-mRNA, and RNA was isolated after UV irradiation for Northern blotting probed with five snRNAs or for primer extension analysis using primer U2-B.
Supplementary Material
Acknowledgments
We thank M. Ares for careful comments on the manuscript, A. Peña for English editing, C.-S. Chung for quantification of Figure 1B, and members of the Cheng laboratory for helpful discussions. This work was supported by a grant from Academia Sinica and the Minister of Science and Technology (Taiwan) (MoST102-2745-B-001-001-ASP).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.253708.114.
References
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Abovich N, Rosbash M. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev 13: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA. 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. 1993a. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1392. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. 1993b. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci 18: 381–384. [DOI] [PubMed] [Google Scholar]
- Burgess S, Couto JR, Guthrie C. 1990. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60: 705–717. [DOI] [PubMed] [Google Scholar]
- Chan S-P, Cheng S-C. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with Pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199. [DOI] [PubMed] [Google Scholar]
- Chan S-P, Kao D-I, Tsai W-Y, Cheng S-C. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Cheng S-C, Abelson J. 1987. Spliceosome assembly in yeast. Genes Dev 1: 1014–1027. [DOI] [PubMed] [Google Scholar]
- Cheng S-C, Newman A, Lin R-J, McFarland GD, Abelson JN. 1990. Preparation and fractionation of yeast splicing extract. Methods Enzymol 181: 89–96. [DOI] [PubMed] [Google Scholar]
- Chiang T-W, Cheng S-C. 2013. A weak spliceosome-binding domain of Yju2 functions in first step and bypasses Prp16 in second step of splicing. Mol Cell Biol 33: 1746–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Luhrmann R. 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608. [DOI] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. 1998. A potential role for U2AF SAP155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-H, Chung C-S, Kao D-I, Kao T-C, Cheng S-C. 2014. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol Cell Biol 34: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev 15: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Novak T, Piccirilli JA, Staley JP. 2010. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell 39: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosowski TR, Keys HR, Quan TK, Ruby SW. 2009. DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle. RNA 15: 1345–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Tanner NK, Banroques J. 2001. From RNA helicases to RNPases. Trends Biochem Sci 26: 339–341. [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Muhlenkamp P. 2003. Spatial organization of protein–RNA interactions in the branch site–3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol 23: 4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Day CL, Dalbadie-McFarland G, Abelson J. 1996. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem 271: 33261–33267. [DOI] [PubMed] [Google Scholar]
- Parker R, Siliciano P, Guthrie C. 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base-pairing to the U2-like snRNA. Cell 49: 229–239. [DOI] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr. 2000. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev 14: 97–107. [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr. 2010. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell 38: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Barta I, Voeltz GK, Abelson J, Ares M Jr. 2003. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci 100: 13857–13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM. 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–353. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang T-H, Abelson J. 1993. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev 7: 1909–1925. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 11: 5033–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Kim H-S, Cao Y, Xu Y-Z, Query CC. 2012. A U1–U2 snRNP interaction network during intron definition. Mol Cell Biol 32: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Konarska MM, Query CC. 2009. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell 34: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Aronova A, Schwer B. 2007. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 21: 2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W-Y, Lee K-R, Cheng S-C. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol 13: 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W-Y, Hsu C-H, Huang K-T, Chen H-R, Kao H-Y, Lee K-R, Cheng S-C. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J 13: 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C-K, Liu H-L, Cheng S-C. 2011. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann RL. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang L, Lynn B, Rymond BC. 2008. A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast. Nucleic Acids Res 36: 2787–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243. [DOI] [PubMed] [Google Scholar]
- Wells SE, Ares M Jr. 1994. Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol 14: 6337–6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harbor Perspect Biol doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Urlauh H, Achsel T, Gentzel M, Wilm M, Lührmann R. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J 21: 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-Z, Query CC. 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-Z, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J 23: 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Perriman R, Igel H, Howe KJ, Neville M, Ares M Jr. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol 18: 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang X-Y, Zhang Z-M, Pu J, Fan Y-J, Zhou J, Query CC, Xu Y-Z. 2013. Splicing proofreading at 5′ splice sites by ATPase Prp28p. Nucleic Acids Res 41: 4660–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZM, Yang F, Zhang J, Tang Q, Li J, Gu J, Zhou J, Xu YZ. 2013. Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly. Cell Rep 5: 1269–1278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.