Abstract
Williams-Beuren syndrome (WBS) is a rare neurodevelopmental condition caused by a recurrent chromosomal microdeletion involving about 28 contiguous genes at 7q11.23. Most patients display a specific congenital heart defect, characteristic facial features, a particular behavior, and intellectual disability. Cases from sub-Saharan Africa have been seldom reported. The present study describes 3 Cameroonian patients affected by WBS, aged 19 months, 13 and 14 years, in whom the diagnosis was confirmed by fluorescent in situ hybridization (FISH) and comparative genomic hybridization (CGH). The first patient presented with a congenital heart defect, the second and third with learning difficulties as well as developmental and behavioral issues. In the latter 2 cases, the facial phenotypes were similar to those of the unaffected population with the same ethnic background. However, the cardiovascular anomalies and friendly behavioral attitudes led to suspicion of WBS. FISH revealed the deletion of the WBS critical region in the first patient, and array-CGH detected a heterozygous ∼1.4-Mb deletion in the 7q11.23 region in the second and third patient. This preliminary report suggests that for sub-Saharan Africans clinical suspicion of WBS could be mostly based on behavioral phenotype and structural heart defects, and less on the classical facial dysmorphic signs.
Key Words: Cameroon, Clinical diagnosis, Sub-Saharan Africans, Williams-Beuren syndrome
Williams-Beuren syndrome (WBS; OMIM 194050) is a well-known genetic disorder caused by a heterozygous deletion within chromosome region 7q11.23. It is a rare condition with a prevalence of 1/7,500 to 1/20,000 live births [Strømme et al., 2002]. It occurs sporadically in almost all of the cases, although some familial cases have been reported with parent-to-child transmission [Scherer et al., 2005]. Paracentric inversion of the WBS chromosomal region (7q11.23) is known as a genomic polymorphism in about 5% of the general population. This inversion is found in up to 30% of unaffected transmitting parents and appears to be a major predisposing factor for the WBS-related deletion [Osborne et al., 2001; Bayés et al., 2003; Hobart et al., 2010]. The causative deletion in chromosome 7q11.23 spans ∼28 contiguous genes [Schubert, 2009], including the elastin gene (ELN). Elastin haploinsufficiency is the established cause of cardiovascular and connective tissue anomalies in WBS [Ewart et al., 1993; Metcalfe et al., 2000]. The specific contribution of each of the remaining genes in the clinical picture of the disease is not yet conclusive. Nevertheless, CLIP2 (CYLN2), LIMK1, GTF2I, and GTF2IRD1 are strongly suspected as being the most important contributors in the cognitive and neurobehavioral phenotypes of the disease [Frangiskakis et al., 1996; Hirota et al., 2003; Morris et al., 2003; van Hagen et al., 2007; Schubert, 2009]. The typical deletion affects segments of 1.5 or 1.84 Mb, with variable frequencies of 95 and 5%, respectively [Schubert, 2009]. The clinical features observed in most patients include specific facial dysmorphism, a congenital heart defect with supravalvular aortic stenosis (SVAS) in about 75% of the cases, connective tissue disorders, mild to severe intellectual disability, endocrine anomalies, and a specific behavior characterized by overfriendliness, empathy and anxiety [Hobart et al., 2010; Morris, 2010a; Pober, 2010]. It has been found that co-morbidity of WBS and autistic disorder is frequent, resulting in a social communication impairment ranging from absence of verbal language to poor social relationships, in opposition to the classical behavioral phenotype of WBS [Tordjman et al., 2012]. Genetic testing for WBS is routinely performed using either fluorescence in situ hybridization (FISH) or deletion/duplication testing. The latter includes quantitative PCR, multiplex ligation-dependent probe amplification and comparative genomic hybridization on microarray (array-CGH). In this study, we report the clinical phenotype and genetic investigations of 3 patients of African descent with WBS and discuss the practical implication of the clinical suspicion of WBS in sub-Saharan Africans.
Clinical Reports
Patient 1
The child is a 19-month-old female, the third out of 3 children of non-consanguineous parents, with no relevant family history. She was born to a 32-year-old woman by spontaneous vaginal delivery at 38 weeks of an uncomplicated pregnancy (birth weight 2,900 g, 10th centile; head circumference 35 cm, 50th centile; height 49 cm, 25th centile). Following several episodes of respiratory distress from the age of 2 weeks, she was noted to have a congenital heart defect by echocardiography, especially SVAS, aortic isthmus stenosis and stenosis on different levels of the pulmonary artery branches. The serum calcium was in the normal range following multiple checks. In terms of developmental milestones, she smiled at 3 months, sat at 7 months, crawled at 9 months, walked without support at 16 months, and was able to pronounce 2 consecutive words from 18 months. On clinical examination, weight was 11 kg (50th centile), head circumference 49 cm (25-50th centile) and height 85 cm (75-90th centile). The patient displayed mild facial dysmorphism (fig. 1a), and cardiac examination revealed a heart murmur. Owing to her young age, evidence for diagnosis of attention-deficit/hyperactivity disorder was not applicable (all clinical data reported in table 1). WBS was suspected and FISH was performed.
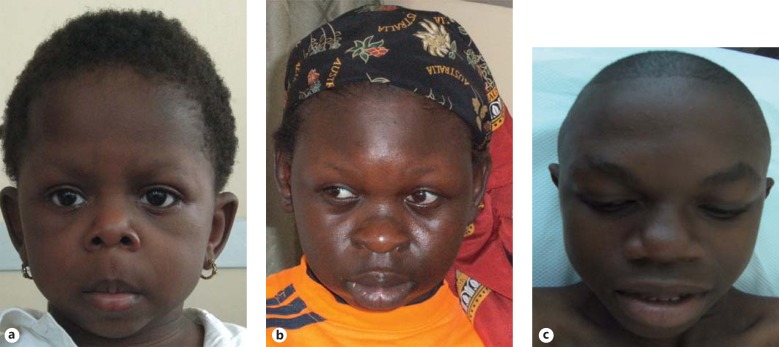
Fig. 1.
Facial features of the 3 patients. a Front view of patient 1 at 19 months of age showing anteverted nares, long philtrum, bulbous nasal tip, and mild lower-lip fullness. b Front view of patient 2 at 13 years of age. Note broad forehead, flat nasal bridge, supraorbital fullness, bulbous nasal tip, downturned mouth, and full lips. c Front view of patient 3 at 14 years of age. Note supraorbital fullness, flat nasal bridge, bulbous nasal tip, wide mouth, and mild lower-lip fullness.
Table 1.
Clinical findings in the 3 patients with WBS
| Clinical featuresa | Patient 1 | Patient 2 | Patient 3 |
|---|---|---|---|
| Age | 19 months | 13 years | 14 years |
| Sex | F | F | M |
| Facial features | |||
| Bitemporal narrowing | – | – | – |
| Epicanthal folds or flat nasal bridge | – | + | + |
| Short nose or anteversion of nares | + | – | – |
| Full cheeks | – | – | – |
| Long philtrum | + | – | – |
| Small, widely spaced teeth | – | – | – |
| Wide mouth | – | – | + |
| Prominent lobe | – | – | – |
| Broad brow | – | – | – |
| Periorbital fullness | – | + | + |
| Stellate lacy iris pattern | – | – | – |
| Bulbous or full nasal tip | + | – | + |
| Malar hypoplasia (flat cheek bone) | – | – | – |
| Full prominent lips | + (mild) | + | + |
| Small jaw | – | – | – |
| Cardiovascular | |||
| Supravalvular aortic stenosis | + | NS | NS |
| Cardiac murmur | + | – | – |
| Other | – | NS | NS |
| Development and cognition | |||
| Developmental delay | – | + | + |
| Delayed speech acquisition, followed by excessive talking | – | + | + |
| Weakness in visuospacial skills | – | – | + |
| Global cognitive impairment | – | + | + |
| Genitourinary | |||
| Delayed toilet training | NA | + | + |
| Voiding frequency, urgency, enuresis | NA | + | – |
| Other | – | – | – |
| Personality, behavior and emotional well-being | |||
| Friendly personality | NA | + | + |
| Anxiety and phobias, excessive-compulsive trait | – | + | – |
| Attention-deficit/hyperactivity disorder | NA | + | – |
| Skin and integument | |||
| Soft skin with mild premature aging | – | + | – |
NA = Not applicable; NS = not studied; + = present; – = absent.
Adapted from the AAP [Committee on Genetics, 2001] and Pober [2010].
Classical clinical features of WBS (auditory and ear, nose, and throat; dental; endocrine; gastrointestinal and weight-related; musculoskeletal; neurologic; and ophthalmologic) completely absent in the 3 patients of this study are not listed.
Patient 2
The patient is a 13-year-old female (fig. 1b). She is her mother's last child out of 10 children, with 4 living and 5 deceased siblings. Four of the 5 siblings died a few hours after birth and the fifth sibling died at the age of 16 years in a context of seizures and intellectual disability. The cause of death was unknown in all the cases. There was a familial history of intellectual disability: a half-sister from the father's side and 3 paternal uncles. None of these patients were available for clinical assessment. The patient was born by normal vertex delivery at 41 weeks of gestation to a 41-year-old mother (birth weight 2,850 g, 10th centile; head circumference 34 cm, 25th centile; height 48 cm, 25th centile). Developmentally, she walked independently from 24 months and said her first sentence, although with some incomprehension, at 3 years of age. She had been noted to have learning difficulties, anxiety, overfriendliness, and enuresis. At clinical examination, her weight was 42 kg (25-50th centile), head circumference 53 cm (25-50th centile) and height 150 cm (10-25th centile). Her milestones were globally delayed, and she displayed mild facial dysmorphism with a normal calcium study. The clinical cardiac examination was unremarkable; no echocardiography was performed (table 1). An unbalanced familial chromosomal translocation was suspected in first line and WBS in second line. Standard karyotype and array-CGH were performed.
Patient 3
The patient is a 14-year-old male and is the third out of 3 children of unrelated Cameroonian parents with no relevant family history. He was born spontaneously at 39 weeks of an uncomplicated pregnancy to a 33-year-old mother (birth weight 3,200 g, 50th centile; head circumference 35.5 cm, 50th centile; height 47 cm, 10th centile). Following a few years of education in an ordinary school, he was reported to have learning difficulties with an inability to read and write. His milestones were delayed, especially with regard to language. He spoke his first word at 17 months and first intelligible sentences at 3 years. He sat at 9 months and started to walk independently at 17 months. At clinical examination, he was highly interactive with good visual contact and remarkably friendly behavior. He weighed 42 kg (10-25th centile), head circumference was 54 cm (25th centile) and height 153 cm (10-25th centile). He displayed some minor facial dysmorphism in relation to the general population standard (fig. 1c) and had no heart murmurs; no echocardiography was performed. The calcium study was normal (table 1). A microdeletion syndrome was considered in first line and WBS suspected secondarily. Array-CGH was performed.
Methods
FISH
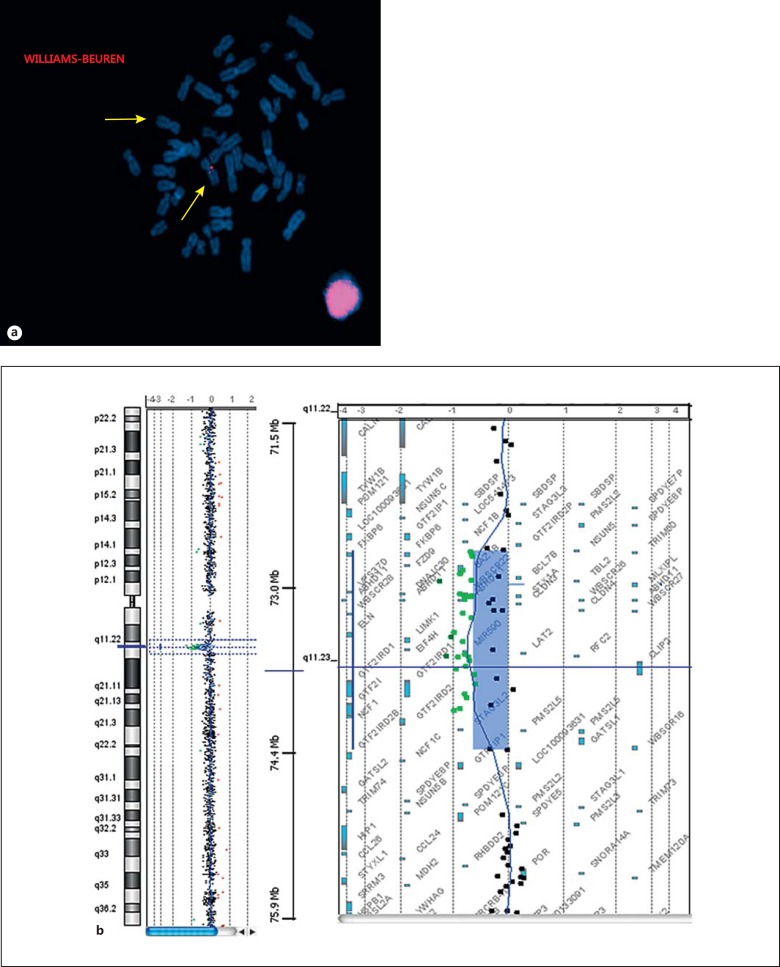
FISH analysis was performed on metaphase chromosome spreads from peripheral blood lymphocytes of patient 1 at the Cytogenetic Laboratory of the Geneva University Hospitals, Switzerland, according to the protocol of the manufacturer (Cytocell®, Cambridge, UK). The FISH clones WBSCR9, LIMK1 and CYLN2 (Cytocell) are located in the WBS critical region. In patient 1, the FISH analysis using these probes revealed a heterozygous deletion of the corresponding sequences, confirming the deletion of the WBS critical region in 7q11.23: 46,XX.ish del(7)(q11.23q11.23)(WBSCR9-, LIMK1-, CYLN2-) (fig. 2a). The exact size of the deletion was not defined by FISH analysis, but the minimum size encompasses the probes WBSCR9 (chr7: 72,854,744-72,856,458 bp) and CYLN2 (chr7: 73,703,805-73,820,273), spanning a region of ∼1 Mb.
Fig. 2.
Molecular cytogenetic results. a FISH result for patient 1. Metaphase FISH analysis with probes WBSCR9, LIMK1, CYLN2 labeled in red (Texas Red Spectrum). The arrows indicate both chromosomes 7, one showing no signal, confirming the WBS deletion in patient 1. b Array-CGH result in patients 2 and 3. Note a deletion of 1.4 Mb encompassing the region 7q11.23. Breakpoints were located between positions 72,726,578 and 74,139,331 bp (hg19) for both patient 2 and patient 3.
GTG-Banding
Cytogeneticanalysis, initially performed on GTG-banded metaphases from peripheral blood lymphocyte cultures of patient 2 with a resolution of 400-550 bands, showed a 46,XX karyotype.
Array-CGH
DNA samples of patients 2 and 3 as well as their respective mothers were extracted from peripheral blood following the instructions of the commercial kit (Puregene blood kit, Qiagen®, USA) at the Molecular Diagnostic Laboratory of Yaoundé Gynaeco-Obstetric and Pediatric Hospital, Cameroon. Array-CGH was performed at the Cytogenetic Laboratory of the Geneva University Hospitals, Switzerland, following the Agilent Oligonucleotide 180K Array-Based CGH for Genomic DNA Analysis Protocol with sex-matched DNA control from Promega (ref. G152A). Data were obtained by Agilent Feature Extraction software v10.5 and analyzed by CGH Analytics 3.5.14 software. In both patients, an identical minimum deletion spanning 1.4 Mb at band 7q11.23 between positions 72,726,578 bp and 74,139,331 bp confirmed the diagnosis of WBS (fig. 2b). The minimum deleted segment was mapped by the first probe A_14_P117491 and the last one A_14_P200801. The maximum size of the deletion is ∼1.9 Mb according to the design of the 180K Agilent array, showing the first non-deleted probe A_16_P01729704 (chr7: 72,401,086-72,401,145) and the last one A_16_P17983225 (chr7: 74,338,985-74,339,044).
For patients 2 and 3, the de novo status of the deletion was difficult to evaluate as no deletion was found by array-CGH testing in their mothers' DNA, and samples from fathers were not available. However, all 4 of the parents were apparently unaffected.
Discussion
Hundreds of WBS cases have been reported in the literature. However, few reports involving African-Americans and sub-Saharan Africans are available [Morris, 2010b]. This discrepancy may be explained by the limited facilities and access to health care, and most probably by the absence of medical genetics services in the majority of sub-Saharan African countries. The diagnosis of the present 3 cases is due to the recent establishment of a medical genetics service in Cameroon [Wonkam et al., 2011]. The present study discloses some difficulties in the clinical diagnosis of WBS, even when performed by experienced medical geneticists. Indeed, the classical dysmorphic features from the current international diagnostic criteria [Committee on Genetics, 2001], which do not take specific account of the phenotype of African populations, could also lead to under-diagnosis based on dysmorphic features alone that are relatively highly scored. According to the American Academy of Pediatrics (APP) [Committee on Genetics, 2001], the clinical diagnosis of WBS is established on the basis of 7 groups of parameters, each corresponding to a specific score. These include growth disorders (1 point), behavior and development (1 point), facial features or dysmorphism (3 points), echocardiogram problems with SVAS or peripheral pulmonary artery stenosis (5 points) and other cardiovascular problems (1 point), connective tissue abnormality (2 points), and calcium studies (2 points). The clinical diagnosis of WBS is unlikely if the total score for a patient is <3. For a total score of ≥3, genetic investigations for WBS are recommended.
The facial phenotype can be extremely variable from one patient to another. This feature is critical in most clinical settings, as it represents the first clinical feature attested by the physician at patient arrival. Although some mild dysmorphic features observed could independently raise our attention to the diagnosis of WBS without the behavioral phenotype and heart defect (fig. 1), all 3 patients described here display a facial appearance similar to many unaffected sub-Saharan African individuals. With a normal clinical cardiac examination in patient 2 and 3, one could suspect that they might not meet the diagnosis criteria of the APP; unfortunately, echocardiograms were not performed in both patients.
FISH analysis with a specific WBS probe was performed for patient 1, confirming the clinical diagnosis. For patient 2, at first a normal karyotype was obtained, ruling out an unbalanced familial chromosomal translocation; array-CGH analysis was performed secondarily, considering the behavioral phenotype associated with WBS. In patient 3, array-CGH analysis was performed first, screening for a microdeletion syndrome, since the WBS score evaluation was weak.
Could patient 1 possibly also have a 1.41-Mb deletion? This cannot be excluded. However, patients 2 and 3 were neither from the same subpopulation group, nor from the same geographical area. Patient 2 was originally from Chad, a neighboring country north of Cameroon, and patient 3 was originally from the Southern part of Cameroon. Since FISH was performed in patient 1 as a part of our routine work-up with the only purpose of diagnosis, our resources were limited to carry out extra experiments like microarray analysis. Nevertheless, with the possibility of a similar atypical deletion as reported in patients 2 and 3, we believe that future studies addressing the question of the deletion size in patient 1 would be of great value in understanding the genomic mechanism underlying WBS and its phenotypic correlation in this subpopulation group.
In conclusion, this preliminary study suggests that for the clinical diagnosis approach of WBS in patients of African descent, behavioral phenotype and structural heart defects should be scored more and dysmorphic features less. The present report raises the hypothesis that the AAP score might not be fully applicable in WBS patients of African descent, and complementary studies on a larger sample number are needed to address this question.
Acknowledgements
The authors would like to thank the families for their participation.
References
- 1.Bayés M, Megano LF, Rivera N, Flores R, Pérez Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Committee on Genetics. American Academy of Pediatrics: Health care supervision for children with Williams syndrome. Pediatrics. 2001;107:1192–1204. [PubMed] [Google Scholar]
- 3.Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- 4.Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, et al. LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- 5.Hirota H, Matsuoka R, Chen XN, Salandanan LS, Lincoln A, et al. Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genet Med. 2003;5:311–321. doi: 10.1097/01.GIM.0000076975.10224.67. [DOI] [PubMed] [Google Scholar]
- 6.Hobart HH, Morris CA, Mervis CB, Pani AM, Kistler DJ, et al. Inversion of the Williams syndrome region is a common polymorphism found more frequently in parents of children with Williams syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:220–228. doi: 10.1002/ajmg.c.30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metcalfe K, Rucka AK, Smoot L, Hofstadler G, Tuzler G, et al. Elastin: mutational spectrum in supravalvular aortic stenosis. Eur J Hum Genet. 2000;8:955–963. doi: 10.1038/sj.ejhg.5200564. [DOI] [PubMed] [Google Scholar]
- 8.Morris CA. The behavioral phenotype of Williams syndrome: a recognizable pattern of neurodevelopment. Am J Med Genet C Semin Med Genet. 154C:427–431. doi: 10.1002/ajmg.c.30286. (2010a). [DOI] [PubMed] [Google Scholar]
- 9.Morris CA. Introduction: Williams syndrome. Am J Med Genet C Semin Med Genet. 154C:203–208. doi: 10.1002/ajmg.c.30266. (2010b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris CA, Mervis CB, Hobart HH, Gregg RG, Bertrand J, et al. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet A. 2003;123:45–59. doi: 10.1002/ajmg.a.20496. [DOI] [PubMed] [Google Scholar]
- 11.Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pober BR. Williams-Beuren syndrome. N Engl J Med. 2010;362:239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 13.Scherer SW, Gripp KW, Lucena J, Nicholson L, Bonnefont JP, et al. Observation of a parental inversion variant in a rare Williams-Beuren syndrome family with two affected children. Hum Genet. 2005;117:383–388. doi: 10.1007/s00439-005-1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert C. The genomic basis of the Williams-Beuren syndrome. Cell Mol Life Sci. 2009;66:1178–1197. doi: 10.1007/s00018-008-8401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strømme P, Bjørnstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17:269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- 16.Tordjman S, Anderson GM, Botbol M, Toutain A, Sarda P, et al. Autistic disorder in patients with Williams-Beuren syndrome: a reconsideration of the Williams-Beuren syndrome phenotype. PLoS One. 2012;7:e30778. doi: 10.1371/journal.pone.0030778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Hagen JM, van der Geest JN, van der Giessen RS, Lagers-van Haselen GC, Eussen HJ, et al. Contribution of CYLN2 and GTF2IRD1 to neurological and cognitive symptoms in Williams syndrome. Neurobiol Dis. 2007;26:112–124. doi: 10.1016/j.nbd.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Wonkam A, Tekendo NC, Sama DJ, Zambo H, Dahoun S, et al. Initiation of a medical genetics service in sub-Saharan Africa: experience of prenatal diagnosis in Cameroon. Eur J Med Genet. 2011;54:e399–e404. doi: 10.1016/j.ejmg.2011.03.013. [DOI] [PubMed] [Google Scholar]