Abstract
Autosomal recessive osteogenesis imperfecta (OI) accounts for 10% of all OI cases, and, currently, mutations in 10 genes (CRTAP, LEPRE1, PPIB, SERPINH1, FKBP10, SERPINF1, SP7, BMP1, TMEM38B, and WNT1) are known to be responsible for this form of the disease. PEDF is a secreted glycoprotein of the serpin superfamily that maintains bone homeostasis and regulates osteoid mineralization, and it is encoded by SERPINF1, currently associated with OI type VI (MIM 172860). Here, we report a consanguineous Brazilian family in which multiple individuals from at least 4 generations are affected with a severe form of OI, and we also report an unrelated individual from the same small city in Brazil with a similar but more severe phenotype. In both families the same homozygous SERPINF1 19-bp deletion was identified which is not known in the literature yet. We described intra- and interfamilial clinical and radiological phenotypic variability of OI type VI caused by the same homozygous SERPINF1 19-bp deletion and suggest a founder effect. Furthermore, the SERPINF1 genotypes/phenotypes reported so far in the literature are reviewed.
Key Words: Autosomal recessive, Osteogenesis imperfecta type VI, SERPINF1
Osteogenesis imperfecta (OI) is a heterogeneous genetic disorder of connective tissue, characterized by short stature and susceptibility to fractures. It affects 6-7/ 100,000 people at birth, with clinical findings varying from a subtle increase in fracture frequency to intrauterine fractures and perinatal lethality [Becker et al., 2011; van Dijk et al., 2011]. Extraskeletal manifestations are common (blue sclerae, dentinogenesis imperfecta, and hearing loss) [Rauch and Glorieux, 2004]. The majority of OI cases (90%) are due to dominant mutations in the genes that encode the α-chains of collagen type 1, COL1A1 and COL1A2 [Becker et al., 2011; van Dijk et al., 2011]. Most recently, Cho et al. [2012] and Semler et al. [2012] described mutations in IFITM5 as the cause of the autosomal dominant OI type V. Approximately 10% of OI cases are due to other yet unknown genetic causes [van Dijk et al., 2011].
In the last few years, the genetic causes of several forms of autosomal recessive OI have been characterized [Rohrbach and Giunta, 2012], and there are currently 10 known genes responsible for these forms of OI. CRTAP (MIM 605497), LEPRE1 (MIM 610339), and PPIB (MIM 123841) encode components of the prolyl 3-hydroxylation complex of P986 in the proalpha1 (I)-collagen chains [Becker et al., 2011; van Dijk et al., 2012]. SERPINH1 (MIM 600943) and FKBP10 (MIM 607063) encode chaperones involved in the intracellular folding and trafficking of the collagen triple helix [Alanay et al., 2010; Becker et al., 2011; van Dijk et al., 2012; Venturi et al., 2012a]. SP7 (MIM 606633) is thought to play a role in regulating the differentiation of preosteoblasts to osteoblasts [Lapunzina et al., 2010; van Dijk et al., 2012]. BMP1 (MIM 112264) is involved in the C-terminal processing of procollagen chains [Asharani et al., 2012]. SERPINF1 (MIM 172860) encodes pigment epithelium-derived factor (PEDF), a secreted glycoprotein with high affinity to collagens of the extracellular matrix, and it is speculated that a loss of PEDF causes OI independent of collagen type I biosynthesis [Becker et al., 2011; Rauch et al., 2012; Venturi et al., 2012b]. TMEM38B (MIM 611236) encodes TRIC-B (trimeric intracellular cation channel type b), a ubiquitous component of TRIC, a monovalent cation-specific channel involved in Ca2+ release from intracellular stores that has been shown to act in cell differentiation. Mutations in this gene were recently described in the Saudi population [Shaheen et al., 2012; Volodarsky et al., 2013]. The Wnt (WNT1; MIM 164820) signaling pathway is one of the key regulators of normal skeletal development, homeostasis, and osteoblast function, and WNT1 recessive mutations have been described as being responsible for moderately severe and progressive recessive OI. Furthermore, heterozygous mutations have been described as causing autosomal dominant early-onset osteoporosis [Fahiminiya et. al, 2013; Faqeih et al., 2013; Keupp et al., 2013; Laine et al., 2013; Pyott et al., 2013; Wilson, 2013].
OI has been historically classified into subtypes primarily based on clinical/radiological findings and, recently, molecular findings [Sillence et al., 1979; Alanay et al., 2010]. The traditional Sillence classification groups the disease into 4 types based on clinical and radiological characteristics and inheritance patterns. Its main purpose was to describe the clinical heterogeneity of OI [Sillence et al., 1979; Rauch and Glorieux, 2004]. Since the discovery of COL1A1 and COL1A2 mutations being responsible for OI types I-IV, the Sillence classification was used mainly to indicate severity of the disease [van Dijk et al., 2011]. OI type I is the mildest form, OI type II is lethal in the neonatal period, OI type III comprises the most severe bone deformities in children that survive the neonatal period, and patients with mild to moderate skeletal involvement are classified as OI type IV [Sillence et al., 1979; Rauch and Glorieux, 2004; Venturi et al., 2012a]. Recently, this classification has been expanded in order to accommodate the new molecular findings associated with OI, and autosomal recessive OI caused by mutations in SERPINF1 is classified as OI type VI [Homan et al., 2011; Rauch et al., 2012].
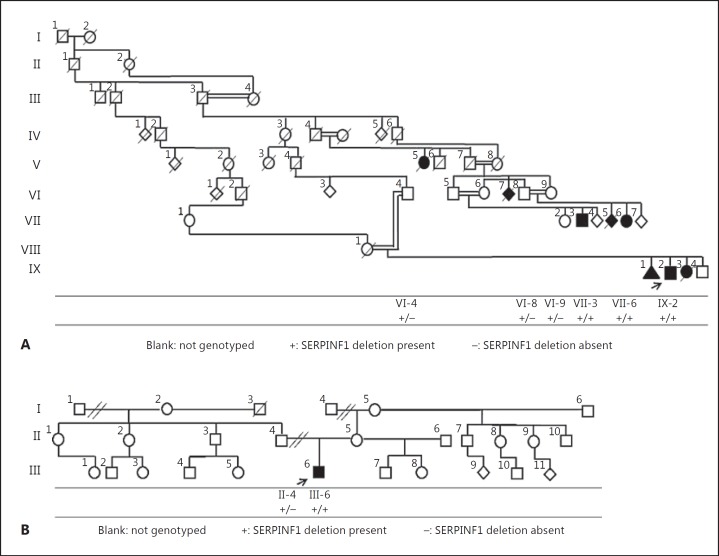
Here, we report a consanguineous Brazilian family from Bueno Brandao (family 1, fig. 1A) in which multiple individuals from at least 4 generations are affected with a severe form of OI. We also report a presumably unrelated individual (family 2, fig. 1B) from the same city with a similar phenotype. In both families, we identified a 19-bp homozygous SERPINF1 deletion not yet described in the literature which segregated with a severe form of autosomal recessive OI.
Fig. 1.
Pedigrees of families 1 (A) and 2 (B).
Clinical Reports
Patient IX-2
Patient IX-2 (family 1) (fig. 2A) was first evaluated at the age of 28 years. He has a history of 13 fractures since he was 1 year old (femur, humerus, and vertebrae) which prevented him from walking. Treatment with intravenous pamidronate started at the age of 32 years and led to mild improvement of pain and reduction of subsequential fractures. He has a moderate neurosensorial hearing loss that required hearing aids at the age of 33 years and sleep apnea. His physical examination revealed severe short stature (100 cm; >-2 SD), weight of 43 kg (>-2 SD), BMI of 43 (compatible with morbid obesity), lack of pigmentation in the right eyelashes and eyebrow, severe scoliosis that led to extreme chest deformity, rhizomelia of upper and lower limbs, no hypermobility of joints, and a high pitched voice. He does not have dentinogenesis imperfecta or blue sclerae. His skeletal survey showed wormian bones, severe osteopenia, severe thoracolumbar scoliosis, narrow pelvis, tapering ribs with depression of the right chest wall, rhizomelic shortening of upper and lower limbs, acetabular protrusion, bilateral femorotibial subluxation, and popcorn calcifications in the left femur (fig. 3A–F). His echocardiogram showed mild left ventricular diastolic dysfunction and minimum pulmonary reflux.
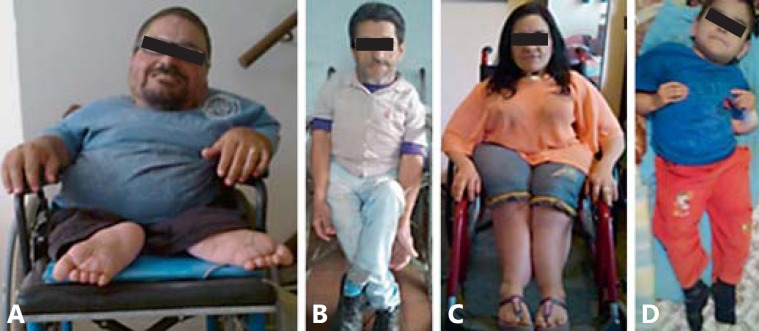
Fig. 2.
Patients 1-3 are from family 1 and patient 4 is from family 2. A Patient IX-2 had 13 sustained fractures and lack of pigmentation of the right eyelashes and eyebrow (feature not observed in any other patient). B Patient VII-3 had 20 sustained fractures. C Patient VII-6 had 12 sustained fractures. D Patient III-6 had 30 sustained fractures.
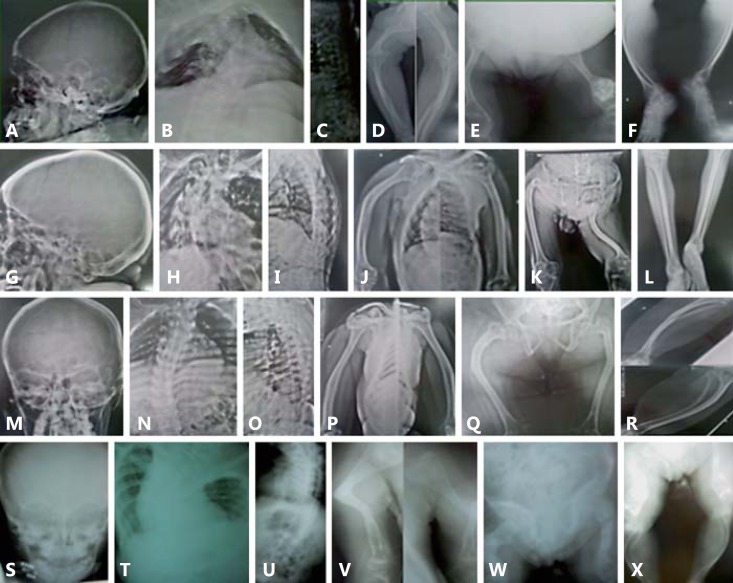
Fig. 3.
Radiological findings. A-F Patient IX-2. A Wormian bones. B Severe throracolumbar scoliosis and osteopenia, tapering of ribs with depression of the right chest wall. C Platyspondylia. D Rhizomelic shortening of upper limbs, flaring of proximal and distal metaphyses. E Rhizomelic shortening of lower limbs, popcorn calcification in left distal femur, bowing of femurs and bilateral femoro-tibial subluxation. F Tapering and bowing of tibiae and fibulae. G-L Patient VII-3. G Absence of wormian bones. H Severe thoracic scoliosis, no tapering of ribs. I Reduction of height of vertebral bodies and bone density, accentuation of kyphosis. J Bowing of humeri without rhizomelic shortening. K Acetabular protrusion, signs of old fractures of femurs, shortening and angulation of left femur. L Tapering of tibiae and fibulae. M-R Patient VII-6. M Absence of wormian bones. N Eleven pairs of ribs, no tapering of ribs, thoracolumbar scoliosis. O Reduction of height of vertebral bodies and osteopenia. P Bowing of humeri without rhizomelic shortening, osteopenia. Q Shortening and narrowing of iliac bones, hip subluxation, old fractures of femurs. R Medial bowing and tapering of tibiae and fibulae. S-X Patient III-6. S Absence of wormian bones. T, U Severe cervicothoracolumbar scoliosis with body deformity and osteopenia. V Signs of old fracture of left humerus with angulation and fragment deflexion, old fracture of right humerus, metaphyseal flaring of humeri. W Acetabular protrusion, right femur deformity characterized by multiple fractures at different consolidation stages, distal metaphyseal and epiphyseal flaring of left femur. X Severe bowing and tapering of tibiae and fibulae.
Patient VII-3
Patient VII-3 (family 1) (fig. 2B) was first evaluated at the age of 51 years. His first fracture occurred before 1 year of age (no congenital fractures were apparent) and the last at the age of 40 years (total of 20 fractures). He walks with crutches, and his physical examination showed slightly blue sclera, no dentinogenesis imperfecta, no hypermobility of joints, short stature (120 cm; >-2 SD), weight of 34.6 kg (>-2 SD), BMI of 24, asymmetry of chest and upper limbs because of scoliosis, and lower limb asymmetry because of left rhizomelic shortening. Formal audiologic exam was not performed, but he had no signs or symptoms of hearing loss. His skeletal survey revealed severe thoracic scoliosis and kyphosis, diffuse osteopenia, bowing of humeri, acetabular protrusion, shortening of left femur, and tapering of tibiae and fibulae (fig. 3G–L).
Patient VII-6
Patient VII-6 (family 1) (fig. 2C) was first examined at the age of 36 years. She had her first fracture at the age of 1 year and the last at the age of 11 years. She walks with crutches, and her physical examination showed that she had short stature (116 cm; >-2 SD), weight of 47.4 kg (>-2 SD), BMI of 35.2 (severe obesity), mild scoliosis, short chest, and no rhizomelic shortening of upper and lower limbs. She had no blue sclera, no dentinogenesis imperfecta, and no hypermobility of joints. Her skeletal survey showed 11 pairs of ribs, thoracolumbar scoliosis, bowing of humeri, shortening of iliac bones, medial bowing and tapering of tibiae and fibulae (fig. 3M–R).
Patient III-6
Patient III-6 (family 2) (fig. 2D) is an 11-year-old boy also from Bueno Brandao but unrelated to family 1. His first fracture was at 5 months of age. A hearing test was not performed, but there were no clinical signs or symptoms of hearing impairment. Treatment with intravenous pamidronate started 8 years ago, but there was no reduction of pain or in the number of fractures (he had 30 fractures). He is not able to sit, and at physical examination he had short stature (98 cm; >-2 SD), weight of 20 kg (>-2 SD), and a BMI of 20.8, rhizomelia of upper limbs, and bowing of upper and lower limbs. He did not have blue sclerae, dentinogenesis imperfecta, and hypermobility of joints. His skeletal survey showed severe osteopenia, acetabular protrusion, right femur deformity, distal metaphyseal and epiphyseal enlargement of the left femur, severe tapering of tibiae and fibulae, severe cervicothoracolumbar scoliosis with body deformity, and angulation and fragment deflexion of the left humerus (fig. 3S–X).
Material and Methods
DNA Extraction
Genomic DNA from affected individuals and their unaffected relatives was purified from fresh whole blood using the Gentra Puregene Kit (Qiagen Sciences, Germantown, Md., USA) according to the manufacturer's instructions.
SNP Genotyping
Whole genome SNP genotyping was performed on patients IX-2, VII-3, and VII-6 from family 1 using the Illumina HumanCytoSNP-12 DNA Analysis BeadChip Kit according to the manufacturer's instructions.
Targeted Capture and Whole Exome Sequencing
Whole exome sequencing (WES) was performed on patients IX-2, VII-3, and VII-6 from family 1 using the Illumina HiSeq2000 platform (Illumina, Inc., San Diego, Calif., USA). The target regions (consensus coding DNA sequence: exonic regions and flanking intronic regions totaling ∼50 Mb of genomic DNA) were captured using the Agilent SureSelect Human All Exon 50Mb Kit (Agilent Technologies, Santa Clara, Calif., USA) following the manufacturer's instructions. The samples were sequenced using paired 100 × 100 base pair reads. Each read was then aligned to the reference genome (NCBI human genome assembly build 36; Ensembl core database release 50_361) [Hubbard et al., 2009] using the Burrows-Wheeler Alignment tool [Marini et al., 2009], and single nucleotide variants (SNVs) and small insertion-deletions (indels) were identified using SAMtools [Li and Durbin, 2009]. PCR duplicates were removed using the Picard software (http://picard.sourceforge.net).
Identification of Potentially Causal Variants
We identified variants of interest using standard filtering criteria, SNV quality, SNV consensus score, indel consensus score ≥20, indel quality ≥50, number of reads supporting SNV or indel ≥3. Because the family was consanguineous, we first searched for functional variants (missense, nonsense, splice site variants, and indels) that were homozygous in 1 of the 7 known genes associated with autosomal recessive OI at the time (SERPINF1, CRTAP, LEPRE1, PPIB, SERPINH1, FKBP10, SP7).
Validation and Segregation of Candidate Variant Using PCR and Sanger Sequencing
Using Primer 3, we designed primers to amplify a 365-bp fragment surrounding the 19-bp deletion in SERPINF1. We performed PCR using standard methods, and agarose electrophoresis of the product revealed specific fragments of the predicted size. We sequenced these directly on an ABI 3730 (Applied Biosystems, Life Technologies, Carlsbad, Calif., USA) using Life Technologies Big Dye Terminator V 1.1 cycle sequencing kit and analyzed the data using CodonCode Aligner 3.6.1 by comparing the sequences to the reference (GRCh37/hg19).
Results
Identification of Potentially Causal Variants
In all 3 affected individuals sequenced, we identified a homozygous 19-bp deletion in exon 8 of SERPINF1 (c.1152_1170del; p.384_390del). The deletion was not found in dbSNP 131, Exome Variant Server, the 1000 Genomes Project, or in 50 in-house controls.
Validation and Segregation of the Candidate Variant Using PCR and Sanger Sequencing
To analyze the segregation of the c.1152_1170del (p.384_390del) in family 1, we genotyped 5 other individuals in that family for the deletion. Individuals VII-3 and VII-6, affected members of family 1, were homozygous and the unaffected individuals VI-4, VI-8, and VI-9 were heterozygous for the same deletion. Next, we genotyped patient III-6 and his unaffected father (II-4) from family 2 and found that the patient was homozygous and his unaffected father was heterozygous for the same deletion.
SNP Genotyping
The whole genome SNP genotyping detected no CNVs that met our criteria for pathogenicity. Using 202,949 independent SNPs from the Illumina HumanCytoSNP-12 DNA Analysis BeadChip Kit, we estimated that the inbreeding coefficient of patients IX-2, VII-3, and VII-6 from family 1 were 0.1312, 0.004468, and 0.1245, respectively, consistent with the consanguineous pedigree. The IBD sharing analysis showed that the homozygous 19-bp deletion in exon 8 of SERPINF1 belongs to a 3-Mb haplotype (chr17: 18,901-3,045,626) shared by these 3 individuals.
Discussion
OI type VI is a moderate to severe form of OI and was identified as a separate disease entity about a decade ago with an estimated incidence of 4% [Glorieux et al., 2002; Rauch and Glorieux, 2004; Homan et al., 2011; Rauch et al., 2012]. Sclerae and teeth do not appear to be affected, patients are healthy at birth, and fractures usually occur after 6 months of age [Glorieux et al., 2002; Basel and Steiner, 2009; Homan et al., 2011; Rohrbach et al., 2012]. The pathognomonic histological finding of OI type VI is the large amount of unmineralized osteoid tissue, osteomalacia, and disorganization of the bone matrix where the lamellar pattern is replaced by a fish scale appearance [Homan et al., 2011; Rohrbach et al., 2012]. Radiological findings are non-distinctive in this type of OI and include bowing of long bones, vertebral compression fractures, generalized osteopenia, hyperplastic callus formation, and absence of wormian bones in the majority of cases [Becker et al., 2011]. Patients with OI type VI apparently do not respond to bisphosphonate treatment as well as patients with other types of OI [Venturi et al., 2012b].
Here, we report 2 Brazilian families from a small city called Bueno Brandao in Southeast Brazil with a severe deforming form of autosomal recessive OI, in which a novel 19-bp homozygous deletion in exon 8 of SERPINF1 was identified by WES.
There is inter- and intrafamilial variability among individuals with the same OI type and also among those presenting the same mutation [Basel and Steiner et al., 2009]. All affected individuals in this study had their first fracture approximately at the age of 1 year (except for patient III-6 who had his first fracture at 5 months of age), none of them have dentinogenesis imperfecta, and only patient VII-3 presented slight blue sclerae. Patients VII-3 and VII-6 can walk with crutches, but patient IX-2 uses a wheelchair, and patient III-6 is not able to sit at the age of 11 years; only 1 patient has hearing loss (patient IX-2). The radiological features among our patients are also variable: patient IX-2 has wormian bones, slightened ribs, and popcorn calcifications in the right distal femur, patient VII-3 has tapering of tibia and fibulae, while the other patients have bowing and tapering of these bones (table 1), being patient VII-6 the one with the mildest deformities and patient VII-3 with the most severely deforming phenotype; all patients presented severe scoliosis and osteopenia.
Table 1.
Clinical, radiological, and molecular findings of our patients
| Patient IX-2 | Patient VII-3 | Patient VII-6 | Patient III-6 | |
|---|---|---|---|---|
| OI type | VI | VI | VI | VI |
| Birth weight/birth length | – | – | – | – |
| Confirmed prenatal fractures | no | no | no | no |
| Age at first fracture, months | 12 | 11 | 12 | 5 |
| Color of sclerae | white | slightly blue | white | white |
| Dentinogenesis imperfecta | no | no | no | no |
| Retarded gross motor functions | yes | no | no | yes |
| Normal intelligence | yes | yes | yes | yes |
| Hearing impairment | yes | no | no | no |
| Hypermobility of joints | no | no | no | no |
| Age at start of treatment with pamidronate, years | 32 | – | – | 3 |
| Ability to walk (with crutches) | No | yes | yes | no |
| Weight, kg/BMI of last visit | 43/43 | 34.6/24 | 47.4/35.2 | 20/20.8 |
| Length, cm/SD of last visit | 100/>–2 | 120/>–2 | 116/>–2 | 98/>–2 |
| Wormian bones | yes | no | no | no |
| Vertebral fractures | yes | no | no | no |
| Old fractures of extremities | yes | yes | yes | yes |
| Tapering of ribs | yes | no | no | no |
| Popcorn calcification | yes | no | no | no |
| Rhizomelic shortening of limbs | severe | moderate (left femur) | no | moderate (upper limbs) |
| Bowing of upper extremities | severe | moderate | mild | severe |
| Bowing and tapering of femur, tibiae, and fibulae | severe | mild tapering of tibiae and fibulae | moderate | severe |
All patients showed the same 19-bp deletion in exon 8 of SERPINF1 (c.1152_1170del, p.384_390del) and were of Portuguese/Dutch/native-born origin.
Families 1 and 2 are from a small city in Southeast of Brazil which has approximately 10,000 inhabitants. Since the deletion we found is hitherto not described in the literature and is responsible for OI type VI with phenotypic variability in these 2 unrelated families, we hypothesize that there is a founder effect in this city.
Mutations in SERPINF1 associated with OI type VI were first described by Becker et al. [2011] (online suppl. table 1, www.karger.com/doi/10.1159/000369108) who applied next-generation sequencing and identification of homozygous regions to analyze the exome of a single male individual who had a severe form of OI and a similarly affected older brother. Their parents were from the United Arab Emirates and were second cousins. A single homozygous truncating mutation affecting SERPINF1 on chromosome 17p13.3, that was embedded into a homozygous stretch of 2.99 Mb, was identified. The mutation was also homozygous in the affected brother, whereas both parents and 2 unaffected sisters were heterozygous carriers. Becker et al. [2011] also identified homozygosity for 2 different truncating SERPINF1 mutations in 2 unrelated Turkish patients with OI and parental consanguinity. All 4 individuals with SERPINF1 mutations had severe OI compatible with OI type III. Fractures of long bones and severe vertebral compression fractures with resulting deformities were observed as early as the first year of life in these individuals. Collagen analyses with cultured dermal fibroblasts displayed no evidence for impaired collagen folding, posttranslational modification, or secretion.
In a large consanguineous family, Homan et al. [2011] performed homozygosity mapping and next-generation sequencing of the candidate gene region and identified loss-of-function mutations in SERPINF1 in 2 affected members of this family (g.4130C>T, p.R99X) and in an additional unrelated 14-year-old boy with OI type VI (g.10440_10443dupATCA, p.H389fsX392). In 3 siblings born to consanguineous parents, who show an initially mild and then progressively worsening form of OI with severe deformities of the long bones, Venturi et al. [2012b] sequenced the SERPINF1 gene and identified a homozygous deletion in exon 4 (c.423delG) which leads to a loss-of-function phenotype due to nonsense-mediated decay of mRNA. In 3 Saudi families with OI, Shaheen et al. [2012] identified homozygous mutations in the SERPINF1 gene (c.1118_1119del, c.1-4796dupT, c.653delT). All affected individuals had early-childhood-onset fractures, and in 2 families the affected individuals had blue sclera. There was no apparent involvement of the teeth or other organs. All responded well to bisphosphonate therapy. In a boy with cystinosis and OI type VI, Tucker et al. [2012] sequenced SERPINF1 and identified an in-frame duplication of 9 bp in exon 3 [c.271_279dupGCCCTCTCG (p.Ala91_Ser93dup)] which adds 3 amino acids to the PEDF protein sequence. Recently, Cho et al. [2013] reported a compound heterozygous mutation in SERPINF1 in an 8-year-old Korean girl [c.77dupC (p.Glu27Glyfs*38) in exon 2 and c.421dupC (p.Arg141Profs*5) in exon 4] that had severe progressive deforming OI, poor response to bisphosphonate treatment, absence of teeth, blue sclerae, and hearing manifestations. Caparrós-Martin et al. [2013] described 4 patients from 3 families with Egyptian origin presenting the SERPINF1 mutations c.752_753insKC847088.1:g.51_393 and c.651G>A (p.Trp217) with a history of onset of fractures in infancy, severe progressive bone deformities, inability to walk independently, slightly opalescent teeth in 1 patient, and no hearing impairment.
PEDF is encoded by SERPINF1 (8 exons) and is a 50-kDa secreted glycoprotein of 418 amino acid residues with high affinity to collagens of the extracellular matrix. It is the most potent antiangiogenic factor and is expressed mainly in bone cells, but its function is still not well characterized [Becker et al., 2011]. Yet, it has other functions, including immunomodulation, protection against oxidative stress, and expansion of the neural stem cell niche [Becker et al., 2011; Venturi et al., 2012b].
The fact that loss of function of PEDF in human cells causes osteogenesis imperfecta is somehow unexpected, since the knock-out murine model did not present a skeletal phenotype, but PEDF was capable of inhibiting osteoclast differentiation and hence bone reabsorption via osteoprotegerin and RANKL (receptor activator of NF-κB ligand) in a dose-dependent manner [Becker et al., 2011]. It is thus speculated that OI caused by SERPINF1 mutations is due to a pathomechanism primarily independent of alterations in type I collagen synthesis or intracellular processing [Becker et al., 2011].
Supplementary Material
Supplementary Table
Acknowledgements
We are grateful to our patients and their families for participating in this study. Our work was supported in part by grants from the National Institutes of Health/National Human Genome Research Institute, number 1U54HG006542.
References
- 1.Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asharani PV, Keupp K, Semler O, Wang W, Li Y, et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am J Hum Genet. 2012;90:661–674. doi: 10.1016/j.ajhg.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basel D, Steiner RD. Osteogenesis imperfecta: recent findings shed new light on this once well-understood condition. Genet Med. 2009;11:375–385. doi: 10.1097/GIM.0b013e3181a1ff7b. [DOI] [PubMed] [Google Scholar]
- 4.Becker J, Semler O, Gilissen C, Li Y, Bolz HJ, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88:362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparrós-Martin JA, Valencia M, Pulido V, Martinez-Glez V, Rueda-Arenas I, et al. Clinical and molecular analysis in families with autosomal recessive osteogenesis imperfecta identifies mutations in five genes and suggests genotype-phenotype correlations. Am J Med Genet A. 2013;161A:1354–1369. doi: 10.1002/ajmg.a.35938. [DOI] [PubMed] [Google Scholar]
- 6.Cho TJ, Lee KE, Lee SK, Song SJ, Kim KJ, et al. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91:343–348. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SY, Ki CS, Sohn YB, Kim SJ, Maeng SH, Jin DK. Osteogenesis imperfecta type VI with severe bony deformities caused by novel compound heterozygous mutations in SERPINF1. J Korean Med Sci. 2013;28:1107–1110. doi: 10.3346/jkms.2013.28.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahiminiya S, Majewski J, Mort J, Moffatt P, Glorieux FH, Rauch F. Mutations in WNT1 are a cause of osteogenesis imperfecta. J Med Genet. 2013;50:345–348. doi: 10.1136/jmedgenet-2013-101567. [DOI] [PubMed] [Google Scholar]
- 9.Faqeih E, Shaheen R, Alkuraya FS. WNT1 mutation with recessive osteogenesis imperfecta and profound neurological phenotype. J Med Genet. 2013;50:491–492. doi: 10.1136/jmedgenet-2013-101750. [DOI] [PubMed] [Google Scholar]
- 10.Glorieux FH, Ward LM, Rauch F, Lalic L, Roughley PJ, Travers R. Osteogenesis imperfecta type VI: a form of brittle bone disease with a mineralization defect. J Bone Miner Res. 2002;17:723–728. doi: 10.1359/jbmr.2002.17.1.30. [DOI] [PubMed] [Google Scholar]
- 11.Homan EP, Rauch F, Grafe I, Lietman C, Doll JA, et al. Mutations in SERPINF1cause osteogenesis imperfecta type VI. J Bone Miner Res. 2011;26:2798–2803. doi: 10.1002/jbmr.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard TJ, Aken BL, Ballester B, Beal K, Bragin E, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keupp K, Beleggia F, Kayserili H, Barnes AM, Steiner M, et al. Mutations in WNT1 cause different forms of bone fragility. Am J Hum Genet. 2013;92:565–574. doi: 10.1016/j.ajhg.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med. 2013;368:1809–1816. doi: 10.1056/NEJMoa1215458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapunzina P, Aglan M, Temtamy S, Caparrós-Martin JA, Valencia M, et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87:110–114. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marini C, Scheffer LE, Nabbout R, Mei D, Cox K, et al. SCN1A duplications and deletions detected in Dravet syndrome: implications for molecular diagnosis. Epilepsia. 2009;50:1670–1678. doi: 10.1111/j.1528-1167.2009.02013.x. [DOI] [PubMed] [Google Scholar]
- 18.Pyott SM, Tran TT, Leistritz DF, Pepin MG, Mendelsohn NJ, et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet. 2013;92:590–597. doi: 10.1016/j.ajhg.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004;363:1377–1385. doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 20.Rauch F, Husseini A, Roughley P, Glorieux FH, Moffatt P. Lack of circulating pigment epithelium-derived factor is a marker of osteogenesis imperfecta type VI. J Clin Endocrin Metab. 2012;97:1–7. doi: 10.1210/jc.2012-1827. [DOI] [PubMed] [Google Scholar]
- 21.Rohrbach M, Giunta C. Recessive osteogenesis imperfecta: clinical, radiological and molecular findings. Am J Med Genet C Semin Med Genet. 2012;160C:175–189. doi: 10.1002/ajmg.c.31334. [DOI] [PubMed] [Google Scholar]
- 22.Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, et al. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012;91:349–357. doi: 10.1016/j.ajhg.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaheen R, Alazami AM, Alshammari MJ, Fageih E, Alhashmi N, et al. Study of autosomal recessive osteogenesis imperfect in Arabia reveals a novel locus defined by TMEM38B mutation. J Med Genet. 2012;49:630–635. doi: 10.1136/jmedgenet-2012-101142. [DOI] [PubMed] [Google Scholar]
- 24.Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tucker T, Nelson T, Sirrs S, Roughley P, Glorieux FH, et al. A co-occurrence of osteogenesis imperfecta type VI and cystinosis. Am J Med Genet A. 2012;158A:1422–1426. doi: 10.1002/ajmg.a.35319. [DOI] [PubMed] [Google Scholar]
- 26.Van Dijk FS, Cobben JM, Kariminejad A, Maugeri A, Nikkels PG, et al. Osteogenesis imperfecta: a review with clinical examples. Mol Syndromol. 2011;2:1–20. doi: 10.1159/000332228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Dijk FS, Byers PH, Dalgleish R, Malfait F, Maugeri A, et al. EMQN best practice guidelines for the laboratory diagnosis of osteogenesis imperfecta. Eur J Hum Genet. 2012;20:11–19. doi: 10.1038/ejhg.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venturi G, Monti E, Dalle Carbonare L, Corradi M, Gandini A, et al. A novel splicing mutation in FKBP10 causing osteogenesis imperfecta with a possible mineralization defect. Bone. 50:343–349. doi: 10.1016/j.bone.2011.10.023. (2012a). [DOI] [PubMed] [Google Scholar]
- 29.Venturi G, Gandini A, Monti E, Dalle Carbonare L, Corradi M, et al. Lack of expression of SERPINF1 the gene coding for pigment epithelium-derived factor, causes progressively deforming osteogenesis imperfecta with normal type I collagen. J Bone Miner Res. 27:723–728. doi: 10.1002/jbmr.1480. (2012b). [DOI] [PubMed] [Google Scholar]
- 30.Volodarsky M, Markus B, Cohen I, Staretz-Chacham O, Flusser H, et al. A deletion mutation in TMEM38B associated with autosomal recessive osteogenesis imperfecta. Hum Mutat. 2013;34:582–586. doi: 10.1002/humu.22274. [DOI] [PubMed] [Google Scholar]
- 31.Wilson C. Wnt-1-a key player in the regulation of human bone mass? Nat Rev Endocrinol. 2013;9:377. doi: 10.1038/nrendo.2013.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table