Abstract
Objectives
Plasmid-mediated AmpC β-lactamases (PMABLs) and carbapenemases are emerging groups of antimicrobial-resistance determinants. The aims of the study were to evaluate the occurrence of PMABLs and carbapenemases in clinical isolates of Klebsiella pneumoniae and compare the test performance of various phenotypic methods for detection of these enzymes in Iran.
Methods
A total of 100 K. pneumoniae isolates were collected from clinical specimens obtained in Valiasr Hospital. AmpC production in all isolates was determined using the AmpC disk test, the cephamycin Hodge test, the AmpC Etest, and the boronic acid combined-disk test. In addition, carbapenemase production was determined using the modified Hodge test, the EDTA disk synergy test, and the boronic acid combined-disk test. The performances of various phenotypic methods were evaluated by the comparison of their results with polymerase chain reaction (PCR) method as the gold standard.
Results
Of the 100 isolates, 19 (19%) were demonstrated to harbor the PMABL-resistance gene by the multiplex PCR method. The PCR result indicated the presence of carbapenemase genes in 12 isolates. The performance of various phenotypic tests carried out for detection of carbapenemase-producing isolates varied widely, ranging in sensitivity from 30% to 100% and in specificity from 90.8% to 100%.
Conclusion
This is the first report of MOX-type AmpC β-lactamase and blaGES in K. pneumoniae in Iran. A comparison of the phenotypic methods showed that a combination of cefoxitin plus boronic acid is optimal for detecting plasmid-mediated AmpC enzymes in K. pneumoniae, whereas the implementation of molecular methods is often complex, requires specially trained personnel, and is associated with higher costs.
Keywords: AmpC β-lactamase genes, carbapenemase, Iran, Klebsiella pneumoniae, phenotypic tests
1. Introduction
AmpC β-lactamases are clinically important cephalosporinases encoded on the chromosome of many organisms such as Citrobacter spp., Enterobacter spp., Morganella spp., Hafnia spp., and Serratia spp. in which they mediate resistance to cephalothin, cefazolin, cefoxitin, most penicillins, and all β-lactamase inhibitor antibiotics except cefepime and imipenem [1]. AmpC β-lactamase hydrolyzes cephamycins and its activity is not inhibited by clavulanic acid. These features distinguish AmpC β-lactamase from extended-spectrum β-lactamases. Since 1989, plasmid-mediated AmpC β-lactamases (PMABLs) have been detected worldwide in the strains of Escherichia coli, Klebsiella spp., Proteus mirabilis, and Salmonella spp. This is because transmissible plasmids have acquired genes for AmpC enzymes, which consequently can appear in bacteria lacking (e.g., Klebsiella pneumoniae) or poorly expressing a chromosomal ampC gene (e.g., E. coli).
Although many phenotypic tests are available for AmpC detection, none of these is validated by the Clinical and Laboratory Standards Institute (CLSI). Therefore, it is important to develop methods that can reliably detect PMABLs [2,3]. Significant spread of carbapenem-resistant K. pneumoniae was observed in the United States, Europe, and some Asian countries [4,5]. Spreading of different types of carbapenemases, such as K. pneumoniae carbapenemase and the New Delhi metallo-β-lactamase-1, was reported in Iran and elsewhere [6,7]. Therefore, we evaluated the prevalence of PMABLs and carbapenemases in Iranain clinical isolates of K. pneumoniae and compared the performance of phenotypic tests for detection of these enzymes.
2. Materials and methods
2.1. Bacterial strains and susceptibility testing
In this cross-sectional study, 100 nonduplicate K. pneumoniae isolates were collected from clinical specimens obtained in Valiasr Hospital in the central province of Iran (March–September 2011).
The identification of all isolates was confirmed by the API 20E system (bioMerieux, Marcy-l'Etoile, France). Susceptibilities to 21 different antibiotics (MAST, Bootle, Merseyside, UK) were defined by disk diffusion according to the CLSI guidelines (Figure 1) [8].
Figure 1.
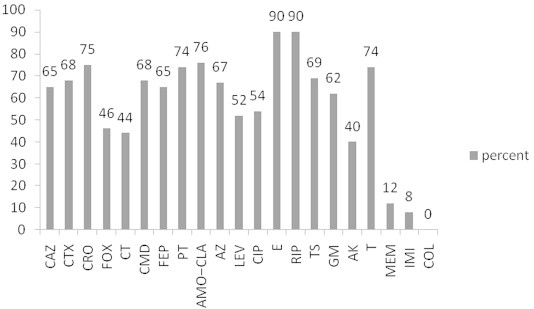
Resistance percent of isolates to the 21 antibiotics used in this study. AK = amikacin; AMO–CLA = amoxicillin–clavulanic acid; AZ = aztreonam; CAZ = ceftazidime; CIP = ciprofloxacin; CMD = cefamandole; COL = colistin; CRO = ceftriaxone; CT = cefotetan; CTX = cefotaxime; E = erythromycin; FEP = cefepime; FOX = cefoxitin; GM = gentamicin; IMI = imipenem; LEV = levofloxacin; MEM = meropenem; PT = piperacillin–tazobactam; RIP = rifampicin; T = tetracycline; TS = trimethoprim/sulfamethoxazole.
2.2. Polymerase chain reaction for detecting PMABLs and carbapenemases genes
DNA extraction was performed based on the method described by Pérez-Pérez and Hanson [9]. Polymerase chain reaction (PCR) was used for amplification of genes encoding PMABLs (blaMOX, blaCIT, blaDHA, blaACC, blaEBC, and blaFOX), Ambler class A (blaKPC and blaGES), metallo-β-lactamases (MBLs; blaIMP, blaVIM, blaNDM, blaSPM, blaSIM, and blaGIM), and class D carbapenemase (blaOXA48) [9–11]. PCR products were sequenced and analyzed by the Basic Local Alignment Search Tool algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. Phenotypic detection of PMABLs and carbapenemases
AmpC production in all isolates was determined by the following: AmpC Etest (Figure 2B), cephamycin Hodge test (CHT; Figure 2C), AmpC disk test (Figures 2E and 2F), and combination-disk test with boronic acid (Figure 2H) [12–14]. In addition, carbapenemase production was determined using the following: modified Hodge test (MHT; Figure 2D), MBL Etest (Figure 2A), EDTA disk synergy (EDS) testing (Figure 2I), and combination-disk testing with boronic acid (Figure 2G). The performances of these test methods were compared with PCR as the gold standard [15–17].
Figure 2.
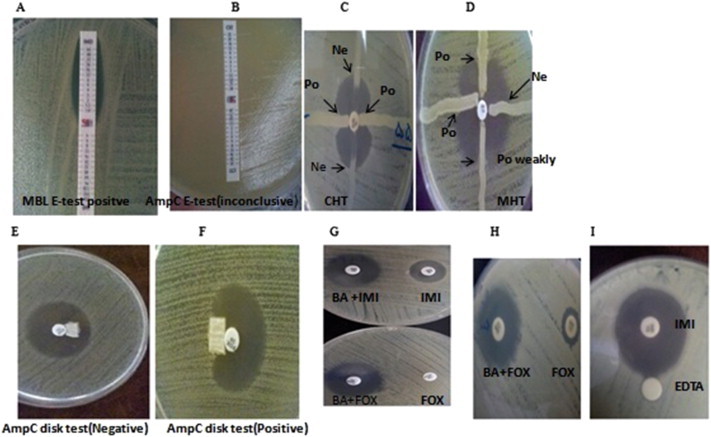
Results of the phenotypic methods for detection of plasmid-mediated AmpC and carbapenemase enzymes in Klebsiella pneumoniae isolates. (A) MBL-positive isolate, (B) AmpC Etest, inconclusive result in isolate, (C) cephamycin Hodge test (CHT), AmpC-negative and AmpC-positive isolates, (D) modified Hodge test (MHT), (E and F) AmpC-negative and AmpC-positive isolates, (G) a positive result obtained from blaGES using the imipenem + boronic acid disk test (top) and a positive result obtained from AmpC using the cefoxitin + boronic acid disk test (bottom), (H) cefoxitin + boronic acid disk test (positive isolate), (I) positive result obtained from MBL-producer isolate using the EDTA disk synergy (EDS) test. BA = boronic acid; CHT = cephamycin Hodge test; FOX = cefoxitin; IMI = imipenem; MBL = metallo-β-lactamase; MHT = modified Hodge test; Ne = negative; Po = positive.
3. Results
Clinical samples were from surgical wounds (n = 36; 36%), urines (n = 41; 41%), blood (n = 13; 13%), and respiratory secretions (n = 10; 10%) obtained in surgery and neurology and neurosurgery intensive care units. Antimicrobial resistances of the isolates are shown in Figure 1.
Of the 100 isolates tested, 19 (19%) harbored PMABLs as shown by multiplex PCR; of these 19 isolates, eight (42.2%) carried bla genes of the CIT group (CMY-2 genes), seven (36.8%) carried genes of the MOX group, whereas three (15.7%) and one (5.2%) carried genes belonging to the EBC and DHA groups (DHA-1; Table 1), respectively.
Table 1.
Characteristics of the 19 AmpC-positive isolates in this study.a
| No. of isolates | AmpC group | Cephalosporin phenotype |
AmpC disk test | Cephamycin Hodge test | AmpC Etest | Cefoxitin/cefoxitin+ boronic acid | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CTX | FOX | CT | FEP | ||||||
| 5 | CIT | S | S | S | S | S | − | − | − | + |
| 7 | CIT | S | S | S | S | S | − | − | − | + |
| 9 | MOX | R | R | R | R | R | − | − | IR | + |
| 11 | MOX | R | R | R | R | R | + | − | IR | + |
| 12 | MOX | R | R | R | R | R | − | − | IR | + |
| 17 | MOX | R | R | R | R | R | − | − | + | + |
| 1 | MOX | R | R | S | S | R | − | − | − | + |
| 21 | MOX | S | S | S | S | S | − | − | − | + |
| 22 | MOX | R | R | R | S | S | + | + | − | + |
| 3 | DHA | R | R | R | R | S | + | − | IR | + |
| 6 | EBC | S | S | S | S | S | − | − | − | + |
| 51 | EBC | R | R | S | S | R | − | − | − | + |
| 49 | CIT | R | R | S | S | R | − | − | − | + |
| 55 | CIT | R | R | R | R | R | + | + | IR | + |
| 60 | CIT | R | R | R | S | R | + | + | − | + |
| 68 | CIT | R | R | R | R | R | − | − | IR | + |
| 75 | CIT | R | R | R | R | S | + | + | IR | + |
| 89 | CIT | R | R | R | R | R | − | − | IR | + |
| 27 | EBC | R | R | R | R | S | + | + | + | + |
CAZ = ceftazidime; CT = cefotetan; CTX = cefotaxime; FEP = cefepime; FOX = cefoxitin; IMI = imipenem; IR = inconclusive result.
Results were rated inconclusive if the minimum inhibitory concentrations exceeded the scale of the Etest for cefotetan alone and/or cefotetan in combination with cloxacillin (Etest AmpC).
The PCR result showed that of the 100 isolates, 12 (12%) carried a carbapenemase gene, the most common being blaVIM (n = 10, 10%) and blaGES (n = 2; 2%). One isolate (1%) contained both blaVIM and blaAmpC (CIT group; Table 2), whereas blaFOX, blaACC, blaIMP, blaSIM, blaNDM, blaGIM, blaSPM, blaKPC, and blaOXA48 were not detected in our isolates.
Table 2.
Characteristics of 12 carbapenemase-positive isolates in this study.
| Isolates | Carbapenem phenotype |
β-Lactamases | IMI/IMI +boronic acid | MHT | IMI/IMI+ EDTA | CAZ/CAZ+ EDTA | MBL Etest | |
|---|---|---|---|---|---|---|---|---|
| IMI | MEM | |||||||
| 15 | R | R | VIM | + | − | + | + | + |
| 18 | R | R | VIM | + | + | + | + | + |
| 20 | R | R | VIM | − | − | + | + | + |
| 39 | S | R | VIM | + | +a | − | + | + |
| 43 | S | R | VIM | − | − | − | + | + |
| 47 | S | R | VIM | − | − | − | + | + |
| 55 | R | R | VIM and CIT | + | + | − | − | − |
| 69 | S | R | VIM | + | + | − | + | + |
| 70 | R | R | VIM | + | + | − | + | + |
| 74 | R | R | VIM | + | + | − | + | + |
| 23 | R | R | GES | + | + | + | + | − |
| 73 | R | R | GES | + | + | + | + | − |
CAZ = ceftazidime; IMI = imipenem; MBL = metallo-β-lactamases; MEM = meropenem; MHT = modified Hodge test.
Weakly positive.
The antimicrobial-susceptibility profiles of the 19 AmpC-producing isolates showed 100% (n = 19) resistance to rifampicin and erythromycin; 79% (n = 15) to cefamandole, ceftazidime, and cefotaxime; 73.6% (n = 14) to tetracycline, ceftriaxone, and aztreonam; 68.5% (n = 13) to piperacillin–tazobactam and amoxicillin–clavulanic acid; 63% (n = 12) to cefoxitin; 57.8% (n = 11) to cefepime; 52.6% (n = 10) to cefotetan, ciprofloxacin, levofloxacin, gentamicin, and trimethoprim/sulfamethoxazole; 31.5% (n = 6) to amikacin; 5.2% (n = 1) to imipenem and meropenem. None of the isolates was resistant to colistin in this study.
The results of phenotypic methods for detection of AmpC- and carbapenemase-producing isolates are shown in Tables 1 and 2, respectively.
All carbapenemase-producing isolates were resistant to rifampicin, erythromycin, ciprofloxacin, aztreonam, meropenem, piperacillin–tazobactam, amoxicillin–clavulanic acid, cefamandole, ceftazidime, cefoxitin, cefotaxime, and ceftriaxone. The frequency of resistance is as follows: 11/12 (91.6%) to tetracycline and amikacin; 10/12 (83.3%) to trimethoprim/sulfamethoxazole, gentamicin, and levofloxacin; 9/12 (75%) to cefotetan; and 8/12 (66.6%) to imipenem. The most effective antimicrobial agent against carbapenemase-producing K. pneumoniae in this study was colistin with 100% susceptibility. Eight of the 12 carbapenemase-producing isolates were AmpC positive in the MHT; AmpC was not detected in the other four isolates. Five noncarbapenemase-producing isolates were falsely positive in MHT. Of these five isolates, four were AmpC producers.
4. Discussion
The prevalence of PMABLs evaluated in this study (19%) is much higher than described in several other parts of the world. In an earlier study from Iran, the prevalence of PMABLs was 8.4% in K. pneumoniae. In Japan, the prevalence of PMABLs was 13% in K. pneumoniae [18]. In China, the rate was 10.1% in K. pneumoniae and the lowest rates of AmpC genes were reported from Switzerland (0.4%) [19]. High prevalence of AmpC genes was reported from Korea (77%) and Singapore (26%) [19]. In this study, similar to other reports, CIT- (CMY-2) and MOX-type genes were predominantly present in K. pneumoniae, followed by the EBC and DHA types, whereas the ACC and FOX genes were not detected. The CMY-2 is one of the most prevalent and most widely distributed PMABLs and has been previously found in several countries [1].
The rate of resistance to cefoxitin in our isolates was 46% (n = 46); only 12 isolates were AmpC producers and 34 isolates were negative. Cefoxitin resistance in these nonproducers could be due to some other resistance mechanism such as alterations in outer membrane permeability [2,20]. In this study, 63% (12/19 isolates) and 68.4% (13/19) of AmpC producers were found to be resistant to cefoxitin and amoxicillin–clavulanic acid, respectively.
Cefotetan has a lower sensitivity than cefoxitin concerning the detection of AmpC production (52.6% vs. 63%). These data indicate that although screening methods using cefoxitin and amoxicillin–clavulanic acid for detection of AmpC-harboring isolates are useful, they are not perfect [20,21]. Therefore, in contrast to Ingram et al [22], our study cannot recommend cefoxitin, cefotetan, and amoxicillin–clavulanic-acid-susceptibility testing for initial AmpC screening. Resistance to cefepime among 11/19 (58%) AmpC-positive K. pneumoniae isolates is another important finding. No synergy was found between expanded-spectrum cephalosporins and clavulanic acid using the double-disk synergy test (data not shown). Only 1/11 cefepime-resistant isolates in this study had carbapenemase genes (blaVIM) and thus, resistance to cefepime may be mediated by structurally modified AmpC β-lactamases [23].
We found that the cefoxitin ± boronic acid disk test detected 19/19 AmpC-positive K. pneumoniae isolates and gave one false-positive result. The AmpC disk test and CHT detected 7/19 and 5/19 AmpC-positive strains (12 and 14 false negatives), respectively. Possibly, inhibitory compounds exhibit different levels of inhibition among heterogeneous groups of AmpCs [24].
The AmpC disk test and CHT gave five false-positive results. Of these five isolates, three were MBL (blaVIM) producers and the two others were blaGES positive.
The AmpC Etest was easy to use, but was unsuitable for AmpC detection. Overall, 17/19 AmpC-producing isolates were not detected by this method. Test performance could be improved by developing an Etest strip with a broader minimum inhibitory concentration range. Inconclusive results using this test have been previously reported [21,22]. However, we indicate that combination-disk testing with boronic acid is potentially useful for clinical laboratories for the detection of PMABLs [13].
The prevalence of carbapenemases among our isolates was found to be 12%. The sensitivity and specificity of disk diffusion for their identification using imipenem disk were 66.6% and 100%, respectively, whereas meropenem disk demonstrated 100% sensitivity and specificity. Apparently, application of meropenem disk is superior for screening, which is in contrast to the study carried out by Benenson et al [25].
This is the first report of blaGES and blaMOX in Iran. Guiana extended-spectrum β-lactamase (GES β-lactamase) has been reported in Europe [26]. In our region, carbapenem resistance has emerged in K. pneumoniae, mostly as VIM-type MBLs. VIM-MBL producers have been reported in Greece, Kuwait, and Korea [27].
Four VIM-producing isolates were not detected by MHT (Table 2). It has been suggested that addition of zinc sulfate or the use of MacConkey agar may improve the sensitivity of the MHT for MBL producers [28]. In addition, the MHT gave five false positives. The overall sensitivity and specificity of the MHT were 66% and 94.3%, respectively. The sensitivity and specificity of the imipenem ± boronic acid disk test were 100% and 90.8%, respectively.
Although two GES-producing isolates were detected using the MHT, there were also nine false-positive results. Of these nine isolates, six were MBL (blaVIM) producers and three were AmpC producers. It is possible that other AmpC families, not detected by the multiplex PCR, were present in the six VIM-producer isolates in our study.
The MBL Etest was the most specific test (100%) for MBL detection in this study. In the EDS test, the EDTA–ceftazidime combination detected additional MBL producers, which were not identified by the EDTA–imipenem combination (90% sensitivity vs. 30% sensitivity). Therefore, ceftazidime appears to be the better substrate for the EDS test. Similar results were observed by Noyal et al [29].
In conclusion, this study confirmed the prevalence of AmpC-producing K. pneumoniae and carbapenemase (blaVIM and blaGES) in the central hospital of Arak (Iran) to be very high. By understanding the resistance pattern and prevalence of the β-lactamase-producing organisms, especially K. pneumoniae, we would be able to manage infection-control policy as well as proper and rational antibiotics prescription. The results indicate that a cefoxitin plus boronic acid is potential method for clinical laboratories to detect emerging AmpC-plasmid-mediated enzymes in K. pneumoniae, whereas the implementation of molecular methods is often complex, requires specially trained personnel, and is associated with higher costs.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors acknowledge the Vice Chancellor for Research at Arak University of Medical Sciences for financial support of the research.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Jacoby G.A. AmpC β-lactamases. Clin Microbiol Rev. 2009 Jan;22(1):161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan T.Y., Ng L.S., He J. Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrob Agents Chemother. 2009 Jan;53(1):146–149. doi: 10.1128/AAC.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasim K., Elsayed S., Pitout J.D. New method for laboratory detection of AmpC β-lactamases in Escherichia coli and Klebsiella pneumoniae. J Clin Microbiol. 2004 Oct;42(10):4799–4802. doi: 10.1128/JCM.42.10.4799-4802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantón R., Akóva M., Carmeli Y. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012 May;18(5):413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 5.Chung K.P., Tseng S.P., Huang Y.T. Arrival of Klebsiella pneumoniae carbapenemase (KPC)-2 in Taiwan. J Antimicrob Chemother. 2011 May;66(5):1182–1184. doi: 10.1093/jac/dkr025. [DOI] [PubMed] [Google Scholar]
- 6.Gaibani P., Ambretti S., Berlingeri A. Outbreak of NDM-1-producing Enterobacteriaceae in northern Italy, July to August 2011. Euro Surveill. 2011 Nov;16(47):20027. [PubMed] [Google Scholar]
- 7.Gupta N., Limbago B.M., Patel J.B. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011 Jul;53(1):60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 8.Wikler M.A. Clinical and Laboratory Standards Institute; Wayne, PA: 2010. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. [Google Scholar]
- 9.Pérez-Pérez F.J., Hanson N.D. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002 Jun;40(6):2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011 Oct;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellington M.J., Kistler J., Livermore D.M. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother. 2007 Feb;59(2):321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 12.Black J.A., Moland E.S., Thomson K.S. AmpC disk test for detection of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal AmpC β-lactamases. J Clin Microbiol. 2005 Jul;43(7):3110–3113. doi: 10.1128/JCM.43.7.3110-3113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coudron P.E. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J Clin Microbiol. 2005 Aug;43(8):4163–4167. doi: 10.1128/JCM.43.8.4163-4167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polsfuss S., Bloemberg G.V., Giger J. Evaluation of a diagnostic flow chart for detection and confirmation of extended spectrum β-lactamases (ESBL) in Enterobacteriaceae. Clin Microbiol Infect. 2012 Dec;18(12):1194–1204. doi: 10.1111/j.1469-0691.2011.03737.x. [DOI] [PubMed] [Google Scholar]
- 15.Cohen Stuart J., Leverstein-Van Hall M.A. Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int J Antimicrob Agents. 2010 Sep;36(3):205–210. doi: 10.1016/j.ijantimicag.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Lee W., Jung B., Hong S.G. Comparison of 3 phenotypic-detection methods for identifying plasmid-mediated AmpC beta-lactamase-producing Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis strains. Korean J Lab Med. 2009 Oct;29(5):448–454. doi: 10.3343/kjlm.2009.29.5.448. [DOI] [PubMed] [Google Scholar]
- 17.Segal H., Elisha B.G. Use of Etest MBL strips for the detection of carbapenemases in Acinetobacter baumannii. J Antimicrob Chemother. 2005 Sep;56(3):598. doi: 10.1093/jac/dki265. [DOI] [PubMed] [Google Scholar]
- 18.Yamasaki K., Komatsu M., Abe N. Laboratory surveillance for prospective plasmid-mediated AmpC beta-lactamases in the Kinki region of Japan. J Clin Microbiol. 2010 Sep;48(9):3267–3273. doi: 10.1128/JCM.02111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohamudha P.R., Harish B.N., Parija S.C. Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian J Med Res. 2012;135:114–119. doi: 10.4103/0971-5916.93433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manchanda V., Singh N.P. Occurrence and detection of AmpC β-lactamases among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003 Feb;51(2):415–418. doi: 10.1093/jac/dkg098. [DOI] [PubMed] [Google Scholar]
- 21.Polsfuss S., Bloemberg G.V., Giger J. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2011 Aug;49(8):2798–2803. doi: 10.1128/JCM.00404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingram P.R., Inglis T.J., Vanzetti T.R. Comparison of methods for AmpC β-lactamase detection in Enterobacteriaceae. J Med Microbiol. 2011 Jun;60(Pt 6):715–721. doi: 10.1099/jmm.0.029140-0. [DOI] [PubMed] [Google Scholar]
- 23.Mammeri H., Poirel L., Bemer P. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob Agents Chemother. 2004 Mar;48(3):716–720. doi: 10.1128/AAC.48.3.716-720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philippon A., Arlet G., Jacoby G.A. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother. 2002 Jan;46(1):1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benenson S., Temper V., Cohen M.J., Schwartz C., Hidalgo-Grass C., Block C. Imipenem disc for detection of KPC carbapenemase-producing Enterobacteriaceae in clinical practice. J Clin Microbiol. 2011 Apr;49(4):1617–1620. doi: 10.1128/JCM.02179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L., Le Thomas I., Naas T. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000 Mar;44(3):622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yong D., Choi Y.S., Roh K.H. Increasing prevalence and diversity of metallo-beta-lactamases in Pseudomonas spp., Acinetobacter spp., and Enterobacteriaceae from Korea. Antimicrob Agents Chemother. 2006 May;50(5):1884–1886. doi: 10.1128/AAC.50.5.1884-1886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miriagou V., Cornaglia G., Edelstein M. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clin Microbiol Infect. 2010 Feb;16(2):112–122. doi: 10.1111/j.1469-0691.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- 29.Noyal M.J., Menezes G.A., Harish B.N. Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res. 2009 Jun;129(6):707–712. [PubMed] [Google Scholar]


