Background: CHD2 is a conserved ATPase and deletions of CHD2 have been linked to developmental and neurological disorders.
Results: The regions flanking the ATPase domain of CHD2 confer substrate specificity and couple ATP hydrolysis to remodeling.
Conclusion: CHD2 possesses nucleosome assembly activity regulated by its accessory domains.
Significance: Understanding the mechanisms of chromatin remodeling is crucial for delineating how remodeling defects contribute to human diseases.
Keywords: ATPase, Chromatin Remodeling, DNA-binding Protein, Epilepsy, Nucleosome, CHD2, Chromodomains
Abstract
Chromodomain helicase DNA-binding protein 2 (CHD2) is an ATPase and a member of the SNF2-like family of helicase-related enzymes. Although deletions of CHD2 have been linked to developmental defects in mice and epileptic disorders in humans, little is known about its biochemical and cellular activities. In this study, we investigate the ATP-dependent activity of CHD2 and show that CHD2 catalyzes the assembly of chromatin into periodic arrays. We also show that the N-terminal region of CHD2, which contains tandem chromodomains, serves an auto-inhibitory role in both the DNA-binding and ATPase activities of CHD2. While loss of the N-terminal region leads to enhanced chromatin-stimulated ATPase activity, the N-terminal region is required for ATP-dependent chromatin remodeling by CHD2. In contrast, the C-terminal region, which contains a putative DNA-binding domain, selectively senses double-stranded DNA of at least 40 base pairs in length and enhances the ATPase and chromatin remodeling activities of CHD2. Our study shows that the accessory domains of CHD2 play central roles in both regulating the ATPase domain and conferring selectivity to chromatin substrates.
Introduction
Chromodomain helicase DNA-binding protein 2 (CHD2)2 is a member of the SNF2-like family of helicase-related enzymes, which includes all known ATP-dependent chromatin remodeling factors (1–3). In general, chromatin remodeling enzymes catalyze non-covalent changes in histone:DNA contacts leading to alterations in the structure of nucleosomes. Distinct groups of chromatin remodeling enzymes have been shown to act on chromatin in different ways (reviewed in Ref. 4). For example, ISWI and CHD1 can catalyze the assembly of periodic nucleosome arrays, whereas the SWI/SNF factors BRG1 and BRM have been linked to the disruption and disassembly of nucleosomes. Finally, the INO80 family of chromatin remodelers has been shown to exchange histones into, or out of, nucleosomes.
The mechanisms through which chromatin remodelers couple ATP hydrolysis to a particular remodeling activity are not well understood. While all remodeling enzymes contain a conserved ATPase domain, they also possess accessory domains that likely regulate the activity of the ATPase domain and confer distinct remodeling activities (reviewed in Ref. 4). CHD2 is part of the CHD family of chromatin remodelers that possess tandem chromodomains (CDs) on the N-terminal side of the core ATPase domain (5). While only one CHD protein (CHD1) is expressed in yeast, flies express four (CHD1, 3, 4, and 7), and vertebrates express nine (CHD1–9). Members of the CHD family have been further divided into subgroups, based on the presence of additional accessory domains. For example, CHD2 is grouped with CHD1 because both contain a putative SANT-SLIDE-like DNA-binding domain (DBD) near their C termini that is not as well conserved in the other seven CHD proteins (5).
While little is known about the biochemical and cellular activities of CHD2, several studies have shown that CHD2 is required for proper development. Homozygous mice containing a C-terminal truncation of the CHD2 protein are not viable, while the heterozygous mice exhibit decreased survival rates (6). In humans, a variety of developmental defects were observed in a heterozygous patient with one CHD2 allele disrupted by a translocation (7). More recently, a number of chromosome deletions and somatic nonsense, frameshift, splice-site, and missense mutations that prematurely truncate or mutate CHD2 have been linked to epileptic encephalopathies in humans (8–12). Similarly, heterozygous knockdown of CHD2 in zebrafish leads to increased epileptic seizures and stunted growth and development when compared with controls (12). A study of human CHD2 in myoblasts reported that loss of the chromodomains from CHD2 leads to decreased expression of muscle-specific genes and lower rates of differentiation of the myoblasts into muscle cells (13). Thus, CHD2 may be required for the expression of tissue-specific genes. Together, these findings suggest that CHD2 plays an integral role in specific developmental pathways. However, our understanding of how CHD2 functions in development is limited by the lack of insight into its biochemical activity.
In the present study, we purified human CHD2 and characterized its remodeling activity. We also dissected the roles of its accessory domains to determine how the ATPase and chromatin remodeling activities of CHD2 are regulated. Our study, which is the first to investigate the biochemical activity of CHD2, helps shed light on the potential role of this protein in cells.
EXPERIMENTAL PROCEDURES
Purification of Recombinant CHD2
In this study, we cloned and purified wild-type CHD2 (WT) and a mutant version of CHD2 (Mut), which contains a two-amino acid alanine substitution of the residues Asp-617 and Glu-618 in the Walker B motif that were introduced by site-directed mutagenesis. We also cloned and purified three CHD2 proteins with systematic truncations: central core ATPase consisting of amino acid residues 450–1129 (Core); DNA-binding truncation consisting of amino acid residues 1–1129 (Core+CD); and chromodomain truncation consisting of amino acid residues 450–1828 (Core+DBD). The human CHD2 proteins were cloned and expressed as N-terminal FLAG- fusion proteins in Sf9 cells using the Bac-to-Bac baculovirus expression system (Invitrogen). Of note, the Core+DBD protein also contained a 6× His-tag at the C terminus that helped stabilize the protein against proteolysis. All plasmid constructs were verified by sequencing. The CHD2 proteins were purified from infected Sf9 cells by immunoaffinity purification, as previously described for other ATPases (14). Briefly, infected Sf9 cells were suspended in cold Lysis Buffer (20 mm Tris-Cl, pH 7.6, 500 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 20% glycerol, 0.01% Nonidet P-40, 1 mm DTT supplemented with the following protease inhibitors: 0.5 mm benzamidine, 1 μg/ml of leupeptin, aprotinin, pepstatin A, bestatin, 5 μg/ml caspase-1 inhibitor I (EMD Biochemicals), and 10 μg/ml E-64). The cells were lysed by 30 strokes of a Dounce homogenizer on ice and immediately centrifuged at 48,384 × g for 10 min at 4 °C. The supernatant was centrifuged a second time for an additional 5 min at 4 °C to remove any residual insoluble material. The cleared cell lysate was then incubated with anti-FLAG resin (Sigma; pre-washed with Lysis Buffer) for 2–4 h at 4 °C, with rotation. The resin was pelleted by centrifugation, transferred to a 1.7 ml microcentrifuge tube, and washed two times with Lysis Buffer, and two times with Wash Buffer (Lysis Buffer containing only 100 mm NaCl). The CHD2 proteins were eluted with 8 sequential incubations of the resin with an equal volume of Elution Buffer (Wash Buffer containing 0.2 mg/ml FLAG peptide; Sigma). For the purification of the Core+DBD protein, all of the wash and elution steps were performed using Lysis Buffer, as the Core+DBD protein had a tendency to precipitate in the Wash Buffer containing less salt. The concentration and quality of the purified CHD2 proteins were assessed by Bradford assay and SDS-PAGE/Coomassie staining. The purest fractions were pooled, flash-frozen with liquid nitrogen, and stored at −80 °C for subsequent biochemical experiments.
In Vitro Chromatin Assembly
The in vitro chromatin assembly assay was performed essentially as described (15) with minor modifications. Native Drosophila core histones (used throughout this study), the catalytic domain of topoisomerase I (Topo I), NAP-1, and ACF were purified as described (15).
Micrococcal Nuclease (MNase) Analysis
A standard 70 μl assembly reaction contained purified core histones (1.4 μg), plasmid DNA (1.4 μg; 3.2 kilobases (kb); pGIE-0), Topo I, NAP-1 (13 μg), recombinant human CHD2 (100 nm final concentration), and an ATP regeneration system (3 mm ATP, 5 mm MgCl2, 30 mm phosphocreatine, and 5 ng/μl creatine phosphokinase). Briefly, the histone cores were incubated first with the histone chaperone NAP-1 for 20 min on ice. In parallel, the plasmid DNA/Topo I reaction was set up for 10 min at 30 °C and kept at room temperature until use. After 20 min, the ATP regeneration system, relaxed plasmid DNA (with Topo I still present), and CHD2 were added to the histone/chaperone mixture. For minus ATP reactions, only ATP was left out; all other components of the regeneration system were included. The reaction was allowed to proceed for 1 h at room temperature and then partially digested by adding 2 mm CaCl2 and MNase (Worthington; low concentration = 7 mU μl−1, high concentration = 28 mU μl−1). After 4 min at room temperature, the reactions were stopped with the addition of 125 μl of Stop Buffer (1% (w/v) SDS, 200 mm NaCl, 250 μg/ml glycogen, 20 mm EDTA pH 8.0) and proteinase K (Worthington) at a final concentration of 50 μg/ml. The DNA was then extracted with phenol:chloroform, EtOH precipitated, resolved by agarose gel eletrophoresis, and visualized by ethidium bromide (EtBr) staining. The marker is a 123-bp repeat ladder (Invitrogen).
Supercoiling Analysis
A standard 70 μl assembly reaction contained core histones (0.35 μg), plasmid DNA (0.35 μg; 3.2 kb; pGIE-0), Topo I, NAP-1 (2 μg), recombinant, purified ACF, or human CHD2 (20 nm final concentration), and an ATP regeneration system (3 mm ATP, 5 mm MgCl2, 30 mm phosphocreatine, and 5 ng/μl creatine phosphokinase). Briefly, the histone cores were incubated first with the histone chaperone NAP-1 for 30 min on ice. In parallel, the plasmid DNA/Topo I reaction was set up for 10 min at 30 °C and kept at room temperature until use. After 30 min, the ATP regeneration system, relaxed plasmid DNA (with Topo I still present), and CHD2 were added to the histone/chaperone mixture. For minus ATP reactions, AMP-PNP was used instead of ATP. The reaction was allowed to proceed for 1 h at room temperature and then the reactions were stopped with the addition of 125 μl of Stop Buffer and proteinase K at a final concentration of 50 μg/ml. The DNA was then extracted with phenol:chloroform, EtOH precipitated, resolved by agarose gel electrophoresis, and visualized by ethidium bromide (EtBr) staining. Plasmid DNA or DNA extracted from the plasmid DNA/Topo I mixture was used to assess where supercoiled (sc) and relaxed (rel) DNA run for comparison in agarose gel electrophoresis.
Radiometric ATPase Assay
Each ATPase reaction (10 μl) contained 10 mm HEPES-K+, pH 7.6, 50 mm KCl, 5 mm MgCl2, 0.1 μg/μl BSA, 500 nm ATP, and 5 μCi of [γ-32P]ATP as a tracer. Where indicated, reactions also contained 50 ng/μl of short, double-stranded DNAs (dsDNAs) 15, 20, 30, 40, 50, or 60 base pairs (bp) in length, plasmid DNA (3.2 kb), or plasmid that had been pre-assembled into chromatin by salt dialysis (final concentration 50 ng/μl). CHD2 was then added to a final concentration of 100 nm to start the ATPase reaction. Aliquots (1 μl) of the reaction were removed and added to 4 μl of 125 mm EDTA, pH 8 at the selected time points (0, 0.5, 1, 5, 15, 30, 60, and 90 min). From each stopped reaction, 0.5 μl was spotted onto a PEI-cellulose plate (Sigma), air-dried, and resolved by thin-layer chromatography (TLC) with 1 m acetic acid, 0.25 m lithium chloride. The TLC plate was then air-dried and exposed to a storage phosphor screen.
Quantification of the [γ-32P]ATP and the released inorganic phosphate (32Pi) was performed using ImageLab software. After background subtraction, the fraction of hydrolyzed ATP was calculated for each time point and plotted versus time (min). Curve fitting was done with the Prism software (GraphPad) using the Michaelis-Menten model. Error bars representing standard deviations from the mean (S.D.) were calculated from at least three experiments.
Restriction Endonuclease Accessibility (REA) Assay
The REA assay was performed essentially as described (16). A standard 20 μl REA reaction contained 1 μg of plasmid DNA (3.2 kb; pGIE-0) or plasmid pre-assembled into chromatin by salt dialysis, 5 units of HaeIII (NEB) in a buffer consisting of 20 mm Tris acetate, 10 mm magnesium acetate, 50 mm potassium acetate, 1 mm DTT, pH 7.9, and either 3 mm AMP-PNP (as a minus ATP control) or ATP. CHD2 (100 nm) or the chromatin-remodeling and assembly factor ACF (100 nm) was included, where indicated. After 2 h in a 30 °C water bath, the reactions were stopped by the addition of 125 μl of Stop Buffer and proteinase K at a final concentration of 50 μg/ml. The DNA was then phenol:chloroform extracted, EtOH precipitated, resolved by agarose gel electrophoresis, and visualized by EtBr staining.
The DNA signal in each lane was quantified using the QuantityOne (Bio-Rad) software. To facilitate the comparison of the amount of DNA digested in each lane, which indirectly measures chromatin remodeling, we created a Digestion Index (DI). The DI is calculated from the difference between the amount of DNA fragments larger than 1 kb (X) and the amount of DNA fragments smaller than 1 kb (Y), divided by the total amount of DNA in each lane [DI = (Y-X)/(Y+X)]. Bar graphs of the calculated DI values for each condition represent mean and S.D. (n = 3). The Student's t test was used to obtain the p value for the Core+CD protein.
Electrophoretic Mobility Gel Shift Assay (EMSA)
Each 10 μl gel shift reaction included CHD2 (0, 10, 20, 50, 100, 200, or 400 nm) and a dsDNA probe in a buffer consisting of 20 mm HEPES-K+, pH 7.6, 50 mm KCl, 5 mm magnesium acetate, 0.1 μg/μl BSA, 5% glycerol, 1 mm DTT, 0.2 mm EDTA, and 0.01% Nonidet P-40. The dsDNA probes were generated by annealing an oligonucleotide that was 5′ fluorescently-labeled with IRDye-700 (IDT) to its complement. The short, dsDNA probes used are the same as those used in the radiometric ATPase assays and were 15, 20, 30, 40, 50, and 60 bp in length at a final concentration of 5 nm. The samples were incubated for 1 h on ice and resolved on a 5% polyacrylamide/0.5× TBE gel at 4 °C. The gel was then scanned using a fluorescent imager (Li-Cor).
Using the QuantityOne software (Bio-Rad), we quantified the intensity of the free and bound DNA bands. We then adjusted for background and used these values to calculate the fraction of DNA bound for each lane. Curve fitting was done with the Prism software (GraphPad) using the Allosteric Sigmoidal model and an estimated Kd calculated. The graphed data represent mean and S.D. (n = 3).
RESULTS
CHD2 is an ATP-dependent Chromatin Assembly Factor
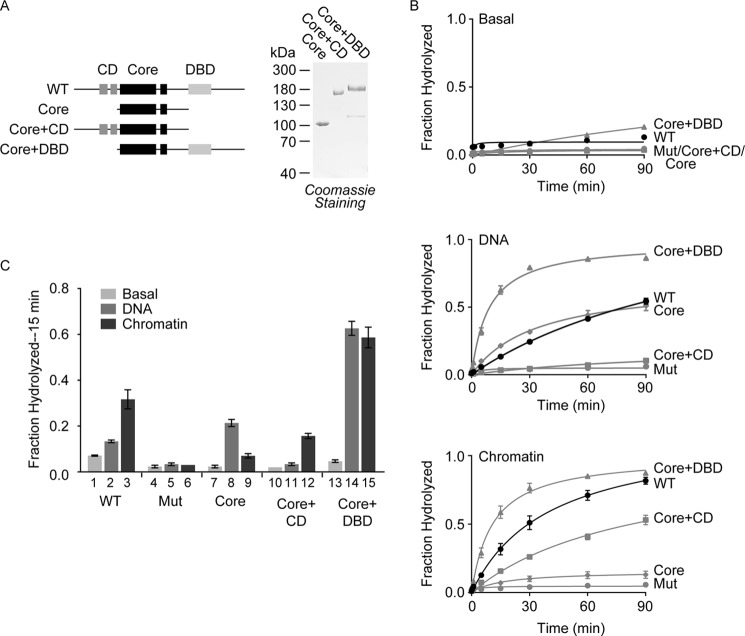
Despite the knowledge that CHD2 targets chromatin in vivo (13), little is known about the ATP-dependent activity of CHD2 and whether CHD2 possesses chromatin remodeling activity. Sequence alignments with other remodeling factors (5) have highlighted conserved domains within human CHD2 (Fig. 1A): a central core SNF2-like ATPase domain (Core) flanked by tandem chromodomains (CDs) on the N-terminal side and a DNA-binding domain (DBD) on the C-terminal side.
FIGURE 1.
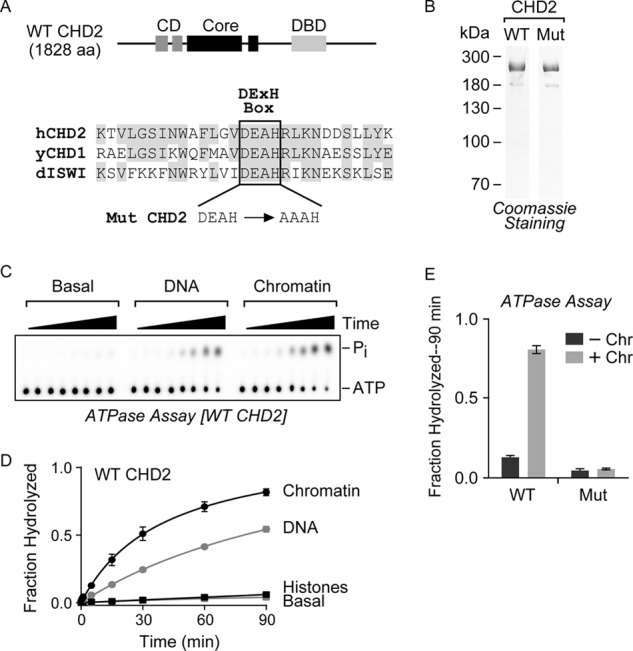
WT CHD2 is a chromatin-stimulated ATPase. A, top, the WT human CHD2 protein contains a central SNF2-like ATPase domain (Core) that is flanked by tandem CDs and a putative DBD. Bottom, a partial alignment of human CHD2 (hCHD2), yeast CHD1 (yCHD1), and Drosophila ISWI (dISWI) highlights the conserved DEXH sequence of the Walker B box. We used site-directed mutagenesis to clone a mutant version of CHD2 (Mut) that contains a two-amino acid alanine substitution of the Asp-617 and Glu-618 residues. B, WT and Mut CHD2 were purified from baculovirus-infected cells and analyzed by SDS-PAGE and Coomassie staining. C, a representative radiometric ATPase assay used to measure the ability of CHD2 to hydrolyze ATP over time. ATPase reactions with WT CHD2 alone (Basal) or containing DNA or chromatin were incubated for 0, 0.5, 1, 5, 15, 30, 60, or 90 min, stopped by the addition of EDTA, and resolved by TLC on PEI-cellulose plates. The positions of the ATP and released phosphate on the TLC plate are indicated. D, quantification of the ATPase assays with WT CHD2 stimulated by chromatin, DNA, or core histones. E, quantification of the radiometric ATPase assays using purified WT or Mut CHD2 protein in the presence or absence of chromatin. The fraction of ATP hydrolyzed was calculated and the values shown are mean and S.D.; n = 3.
To investigate the ATP-dependent activity of CHD2, we first cloned and purified the full-length wild-type human CHD2 protein (WT CHD2) and a mutant version of CHD2 (Mut CHD2), which carries a two-amino acid substitution (D617A and E618A) in the Walker B motif, using a baculovirus expression system (Fig. 1B). We then evaluated whether purified WT CHD2 is able to hydrolyze ATP. For this analysis, we used a radiometric ATPase assay in which we resolved the amount of unhydrolyzed [γ-32P]ATP and released inorganic phosphate (32Pi) on PEI-cellulose TLC plates (a representative TLC plate is shown in Fig. 1C). Because CHD2 contains tandem chromodomains, which are predicted to interact with histones (17), and a putative DNA-binding domain (6), we measured the effects of adding core histones, plasmid DNA, or pre-assembled, salt-dialyzed chromatin on the ATPase activity of CHD2. The fraction of ATP hydrolyzed was measured over a time course spanning 90 min. Using this assay, we found that WT CHD2 exhibits nearly undetectable ATPase activity alone (Basal) or in the presence of core histones (Histones; Fig. 1D). In contrast, both plasmid DNA (DNA) and plasmid that had been pre-assembled into chromatin by salt dialysis (Chromatin) stimulate the ATPase activity of CHD2, with chromatin providing the most stimulation (Fig. 1D). To confirm that Mut CHD2 is catalytically inactive, we measured the fraction of ATP hydrolyzed in the absence or presence of chromatin. In the absence of chromatin, both WT and Mut CHD2 exhibit low levels of ATP hydrolysis (Fig. 1E). In contrast, under conditions where WT CHD2 is active in the presence of chromatin, Mut CHD2 remains inactive (Fig. 1E). There was no detectable ATPase activity with the addition of plasmid or chromatin to the ATPase reaction in the absence of WT CHD2 (data not shown).
The sequence similarity of CHD2 to other remodeling enzymes and the fact that CHD2 is preferentially stimulated by chromatin suggest that CHD2 is also capable of remodeling chromatin (Fig. 1E). Chromatin remodeling factors have been shown to exhibit distinct remodeling activities (reviewed in Ref. 4); these include nucleosome assembly, disassembly, sliding, unwrapping (18), and histone exchange. So far, only the remodeling factors ISWI (the ATPase in ACF) and CHD1 from Saccharomyces cerevisiae and Drosophila melanogaster have been shown to catalyze the ATP-dependent assembly of periodic nucleosomes (19–21). Because CHD2 is ∼60% identical and 80% similar to CHD1 we considered the possibility that CHD2 can also catalyze nucleosome assembly. To test this, we used an established in vitro chromatin assembly assay, which relies on purified components to generate regularly spaced nucleosome arrays (15, 19). We pre-incubated purified core histones with the histone chaperone NAP-1, and then added naked plasmid DNA, Topo I, and an ATP regeneration system (Fig. 2A, left). The assembly of the naked plasmid DNA into periodic nucleosomal arrays was first analyzed by partial MNase digestion followed by agarose gel electrophoresis. In the absence of an ATP-dependent assembly factor and ATP, a limited amount of mono- and di-nucleosomes were observed, as previously reported (Fig. 2A, right; 19, 22). In contrast, the reactions that contain CHD2 and ATP yield periodic arrays of nucleosomes, as indicated by the extended DNA ladder formed following MNase digestion (Fig. 2A, right). The assembly of nucleosomes by CHD2 does not occur in reactions that lack ATP or when Mut CHD2 is used, demonstrating that chromatin assembly by CHD2 depends both on the presence of ATP and on a functional ATPase domain.
FIGURE 2.
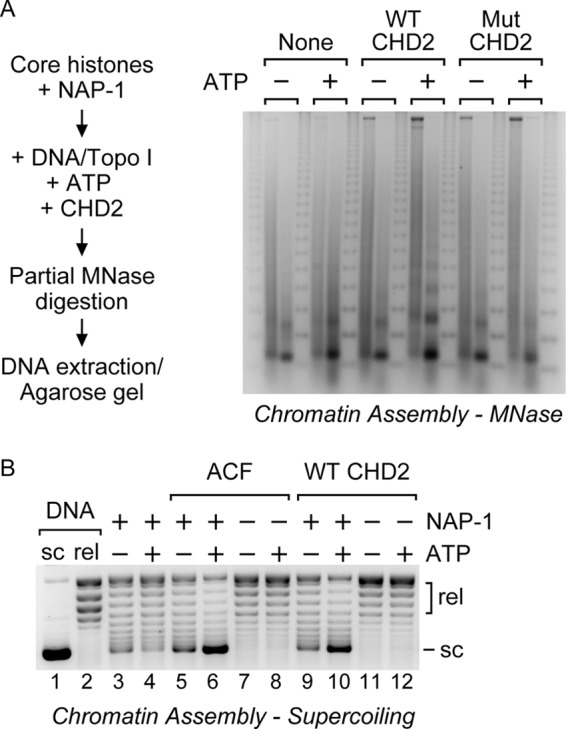
CHD2 catalyzes the assembly of periodic nucleosome arrays. A, left, nucleosome assembly reactions were performed to determine whether CHD2 assembles periodic nucleosome arrays. Core histones (1.4 μg) were pre-incubated with the histone chaperone NAP-1 (13 μg). Naked plasmid DNA (1.4 μg), Topo I, and WT or Mut CHD2 (100 nm) was then added in the presence or absence of ATP. The reactions were then partially digested with a low (left-hand lane) or a high (right-hand lane) concentration of MNase. The digested DNA was precipitated and analyzed by agarose gel electrophoresis and EtBr-staining. A 123-bp repeat marker was run between each pair of samples. Right, the assembly of periodic nucleosome arrays was analyzed by partial MNase digestion. A representative agarose gel showing the presence of a DNA ladder formed upon assembly of nucleosomes by CHD2 and ATP. B, supercoiling analysis of chromatin assembly reactions. Core histones (0.35 μg) were pre-incubated with the histone chaperone NAP-1 (2 μg). Naked plasmid DNA (0.35 μg), Topo I, and ACF (20 nm) or WT CHD2 (20 nm) was then added in the presence or absence of ATP (AMP-PNP used as negative ATP control). The plasmid DNA was then precipitated and analyzed by agarose gel electrophoresis and EtBr-staining. sc, supercoiled; rel, relaxed.
We next used DNA supercoiling to monitor the assembly of plasmid DNA into chromatin. In the presence of Topo I, plasmid DNA that assembles into chromatin will supercoil once the reactions are stopped (15). In contrast, unassembled plasmid DNA will be relaxed by Topo I. In Fig. 2B, a comparison of the amount of supercoiled DNA in lane 10 with that in lane 9 indicates CHD2 catalyzes the efficient ATP-dependent assembly of plasmid DNA into chromatin at levels comparable to that of ACF, a factor previously shown to assemble nucleosomes (Fig. 2B, compare lane 6 with lane 5; 19, 21). Both ACF and CHD2 require ATP and NAP-1 for efficient chromatin assembly, as significantly less supercoiling was detected in reactions that contain the non-hydrolyzable ATP analog AMP-PNP (Fig. 2B, lanes 5 and 9), or in reactions that lack the histone chaperone NAP-1 (Fig. 2B, lanes 7 and 8 and 11 and 12).
Previous reports have shown that chromatin assembly likely occurs in at least two steps (19, 21–23). The first step involves the deposition of histones onto DNA by NAP-1 and leads to the formation of an intermediate species termed pre-nucleosomes (22). The second step involves the rapid conversion of the pre-nucleosomes into canonical nucleosomes by an ATP-dependent motor protein (22). In the absence of a motor protein, a fraction of the histones deposited by NAP-1 may spontaneously fold into canonical nucleosomes (22–23). In our reactions lacking a motor protein or ATP, we also observe the formation of a limited amount of canonical mono- and di-nucleosomes that can be converted into extensive nucleosome arrays by CHD2 and ATP (Fig. 2A) and see a low, but detectable, amount of supercoiling (Fig. 2B, lanes 3–5 and 9), consistent with the formation of pre-nucleosomes and a limited amount of nucleosomes. Conversion of the pre-nucleosomes generated by NAP-1 into an extended array of canonical nucleosomes only occurs with the addition of an ATP-utilizing motor protein such as ACF or CHD2 and ATP to the reactions containing NAP-1, histones, and DNA (Fig. 2A and lanes 6 and 10 of Fig. 2B). Together, these results show that at least three distinct ATP-dependent chromatin assembly enzymes exist: ISWI (ACF), CHD1, and CHD2.
The CDs and DBD Regulate the Activity and Substrate Specificity of Human CHD2
Recent studies of other chromatin remodeling factors have shown that accessory domains outside of the core ATPase domain often play important roles in regulating the biochemical activity of the factor (24–28). To understand the role that the CDs and DBD play in the regulation of CHD2 activity, we cloned, expressed, and purified a series of CHD2 constructs missing either the DBD-containing C-terminal region (Core+CD) or the CD-containing N-terminal region (Core+DBD), or both, leaving the conserved ATPase domain (Core; Fig. 3A).
FIGURE 3.
The accessory domains of CHD2 regulate the core ATPase domain. A, left, a series of CHD2 deletion proteins was generated that include removal of the C-terminal region containing the putative DNA-binding domain (Core+CD), removal of the N-terminal region containing the tandem chromodomains (Core+DBD), or removal of both regions, leaving the central ATPase domain (Core). Right, as done with WT CHD2, the deletion proteins were purified from baculovirus-infected cells and analyzed by SDS-PAGE and Coomassie staining. B, ATPase reactions consisting of CHD2 (100 nm) and 50 ng/μl of DNA or in vitro salt-dialyzed, pre-assembled chromatin were performed to measure the ability of DNA and chromatin to stimulate of the ATPase activity of WT CHD2 and the deletion proteins. The reactions were stopped at various timepoints (0, 0.5, 1, 5, 15, 30, 60, and 90 min) and resolved by TLC. The fraction of ATP hydrolyzed was measured for each time point. C, for comparison, we took the experiments shown in B and graphed the fraction of ATP hydrolyzed at the 15-min time point. All values are mean and S.D.; n = 3.
To study how the accessory domains regulate the activity of CHD2, we first measured the ability of the CHD2 deletion proteins to hydrolyze ATP in the absence of DNA or chromatin. As observed with WT CHD2, all of the deletion proteins exhibit little to no activity in the absence of DNA or chromatin (Fig. 3, B and C, Basal). We then examined the ability of the deletion proteins to hydrolyze ATP in the presence of DNA or chromatin. We found that the core ATPase domain of CHD2 is stimulated by DNA to levels similar to WT CHD2, but is poorly stimulated by chromatin (Fig. 3, B and C). This finding is consistent with the prediction that SNF2-like ATPases are DNA-dependent (2, 29), suggesting that other regions of CHD2 are responsible for conferring its selectivity for chromatin.
We next examined the Core+CD protein, which contains the core ATPase domain and the tandem CDs but lacks the DBD (Fig. 3A). The Core+CD protein shows modest chromatin-stimulated ATPase activity but almost no DNA-stimulated ATPase activity (Fig. 3, B and C). Thus, the presence of the N-terminal region and CDs confers chromatin selectivity to the ATPase domain, which alone is preferentially stimulated by DNA. This finding also suggests that the N terminus negatively regulates the ATPase domain, possibly by limiting access of the ATPase domain to DNA.
We then examined the Core+DBD protein, which contains the core ATPase domain and the C-terminal DNA-binding domain (Fig. 3A). Unlike WT CHD2 and the Core+CD, which are preferentially stimulated by chromatin, the Core+DBD is stimulated almost equally by both DNA and chromatin, and the levels of stimulation are significantly higher than those observed with WT CHD2 (Fig. 3, B and C). This finding further implicates the N terminus and CDs in both providing the selectivity of human CHD2 for chromatin substrates and in repressing the ATPase activity of the core ATPase domain. The inhibition of ATPase activity by regions such as the CDs, which are able to sense unique features of chromatin such as specific histone modification patterns in the histone tails (17), is consistent with models proposed for other remodeling enzymes (24–27). Our results also suggest that the DBD acts to increase the affinity of CHD2 to both naked DNA and DNA present in the context of chromatin, leading to an overall increase in ATPase activity when compared with the core ATPase domain.
The Chromodomain-containing N Terminus of CHD2 Couples ATP Hydrolysis to Chromatin Remodeling
After examining the differences in substrate-stimulated ATPase activity between WT CHD2 and the CHD2 deletion proteins, we assessed how removal of the accessory domains affects the chromatin remodeling activity of CHD2. Because it is difficult to quantify the assembly of nucleosomal arrays, we used a restriction endonuclease accessibility (REA) assay to measure the extent of chromatin remodeling by CHD2. REA assays have been used successfully to investigate the remodeling activity of other enzymes including the chromatin assembly factors Drosophila ACF (a heterodimer that contains the ISWI ATPase) and Drosophila CHD1 (16, 30). This assay relies on the principle that when a plasmid is reconstituted into chromatin via salt dialysis, specific restriction sites are occluded by the presence of nucleosomes, but can be made accessible by remodeling (16, 31–34). For this analysis, we used a 3.2 kb plasmid (pGIE-0) that contains 15 HaeIII restriction sites. The plasmid alone is completely digested by HaeIII, regardless of the presence of AMP-PNP (minus ATP control) or ATP (Fig. 4A, compare lane 2 with lane 1). Reactions that contain the plasmid-assembled chromatin are not affected by the presence of AMP-PNP or ATP either but are only partially digested due to occlusion of some HaeIII sites (Fig. 4A, compare lanes 3 and 4 with lanes 1 and 2). To facilitate the comparison of the remodeling activities exhibited by the different proteins, we established a Digestion Index (DI) that takes into account the difference between the amount of DNA fragments larger than 1 kb (i.e. less digested) and the amount of DNA smaller than 1 kb (i.e. more digested) relative to the total amount of DNA in each lane (see Fig. 4B and “Experimental Procedures”).
FIGURE 4.
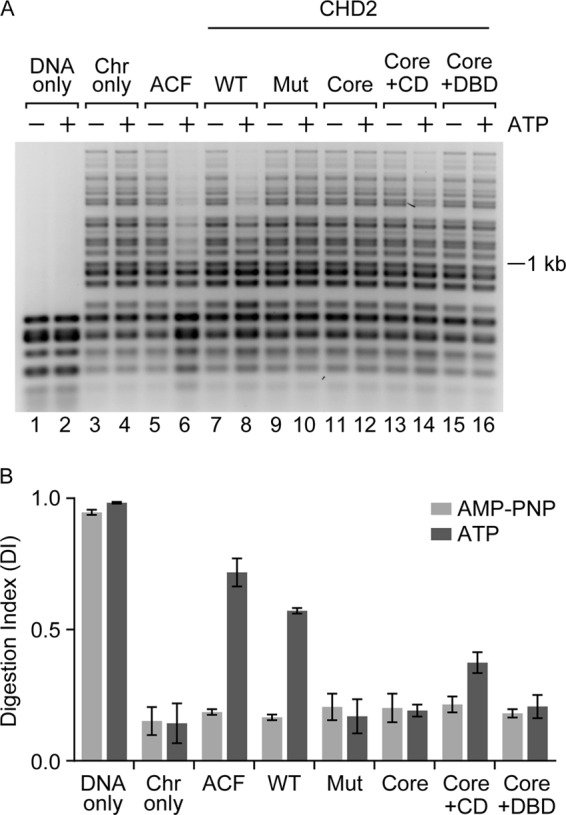
The chromodomains of CHD2 couple ATP-hydrolysis to chromatin remodeling. A, a restriction endonuclease accessibility (REA) assay was performed to measure the extent of chromatin remodeling by CHD2. The indicated proteins (100 nm) were incubated with the restriction enzyme HaeIII and salt-dialyzed, pre-assembled chromatin (1 μg), which contains 15 HaeIII restriction sites. The reactions also included AMP-PNP (as a minus ATP control) or ATP. Following digestion with HaeIII, the DNA was deproteinized, purified, and resolved by agarose gel electrophoresis. B, quantification of replicate REA assays. A Digestion Index (DI) was calculated as described in the “Experimental Procedures.” The graphed data represent mean and S.D.; n = 3.
Using the REA assay, we observed an ATP-dependent increase in HaeIII accessibility in reactions containing WT CHD2 (Fig. 4A, compare lanes 7 and 8) or our positive control, ACF (Fig. 4A, compare lanes 5 and 6). The catalytically-inactive Mut CHD2 does not exhibit any detectable remodeling activity, consistent with its lack of ATPase and chromatin assembly activities (Fig. 4A, compare lanes 9 and 10). Similarly, the core ATPase domain did not show any detectable remodeling activity during the 2-h incubation with chromatin and HaeIII (Fig. 4A, compare lanes 11 and 12). The Core+DBD deletion did not show any detectable remodeling activity during this 2-h incubation as well (Fig. 4A, compare lanes 15 and 16), despite having robust chromatin-stimulated ATPase activity (Fig. 3D, bar 15). This result suggests that the N terminus helps couple ATP hydrolysis with efficient chromatin remodeling and implies that the sustained stimulation of the ATPase domain by chromatin does not guarantee that chromatin remodeling will occur.
In contrast to the Core+DBD protein, the Core+CD protein shows low, but significant (p = 0.0024), remodeling activity after the 2-h incubation with chromatin and HaeIII (Fig. 4A, compare lanes 13 and 14). This finding is consistent with the results from the ATPase assays, which show that the Core+CD protein exhibits chromatin-stimulated ATPase activity, albeit at lower levels compared with WT CHD2 (Fig. 3C, compare bar 12 to bar 3). We extended the incubation time of the REA assay to 4 h and were still unable to detect any remodeling activity for the core ATPase domain or the Core+DBD protein (data not shown). Our results show that the N-terminal region containing the CDs with the core ATPase is sufficient for chromatin remodeling (Fig. 4B, DI of Core+CD in the presence of ATP). While the C-terminal region containing the DBD is not needed for chromatin remodeling in our assays, its presence enhances the remodeling activity (Fig. 4B, compare the DI of the Core+CD to that of WT CHD2 in the presence of ATP).
CHD2 Uses the DBD to Bind DNA in a Length-dependent Manner
Thus far, our results have supported the prediction that the C-terminal region of CHD2 contains a DBD. Loss of the DBD (Core+CD) reduces the ability of CHD2 to be stimulated by DNA, whereas addition of the DBD to the core ATPase domain (Core+DBD) greatly increases the ability of DNA and chromatin to stimulate the ATPase domain (Fig. 3, B and C). To examine the overall DNA-binding properties of CHD2, we performed electrophoretic mobility gel shift assays (EMSAs) using a series of short, 15–60 bp, dsDNA substrates as probes (Fig. 5A). We found that WT CHD2 shows weak association with 15, 20, or 30 bp probes (Fig. 5, A and B). In contrast, WT CHD2 shows clear preference for probes that are 40 bp or longer in length (Fig. 5, A and B). This correlates well with the DNA-stimulated ATPase activity of CHD2, which increases in a dsDNA length-dependent manner (Fig. 5C). We did observe low but detectable stimulation of the ATPase activity of CHD2 with dsDNA substrates less than 40 bp, and the effect is likely due to direct stimulation of the core ATPase domain. To determine whether the C-terminal region of CHD2 plays a role in binding dsDNA, we performed EMSA with the Core+CD protein and the 40 bp probe and were unable to detect significant binding even at high concentrations of the protein (Fig. 6A). This implies that the C-terminal region of CHD2 contains a DNA-binding domain.
FIGURE 5.
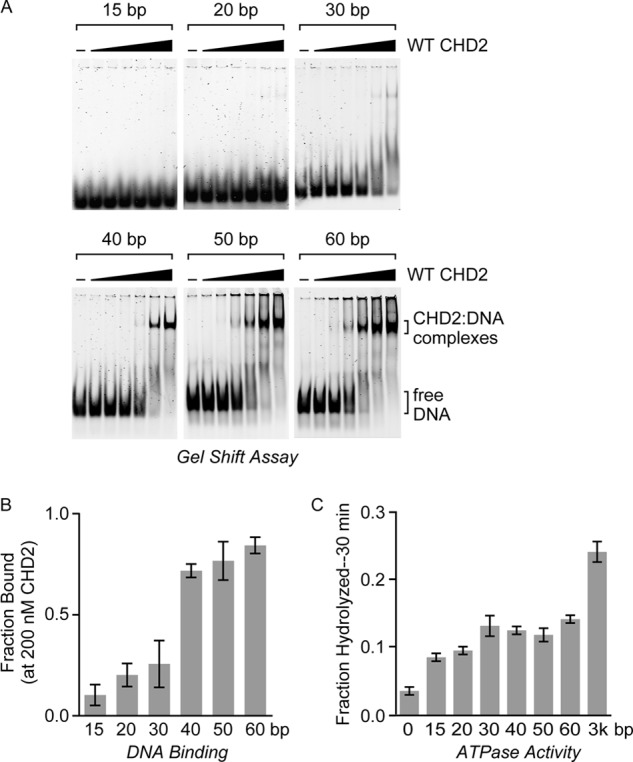
CHD2 binds dsDNA substrates that are at least 40 bp in length. A, electrophoretic mobility gel shift assays (EMSAs) were performed using WT CHD2 and a series of fluorescently-labeled dsDNA probes. Increasing amounts of CHD2 (0, 10, 20, 50, 100, 200, or 400 nm) were incubated with a dsDNA probe (5 nm) of the indicated length. The samples were then resolved by native PAGE and imaged with a fluorescent laser scanner. B, for comparison, the fraction of DNA bound by CHD2 at 200 nm was calculated and graphed. All values are mean and S.D.; n = 3. C, quantification of the fraction of ATP hydrolyzed by CHD2 (100 nm) in the presence of the same dsDNA probes (15, 20, 30, 40, 50, 60 bp) used in gel shift assays after 30 min as compared with fraction of ATP hydrolyzed in the presence of plasmid DNA (∼3 kb). All values are mean and S.D., n = 3.
FIGURE 6.
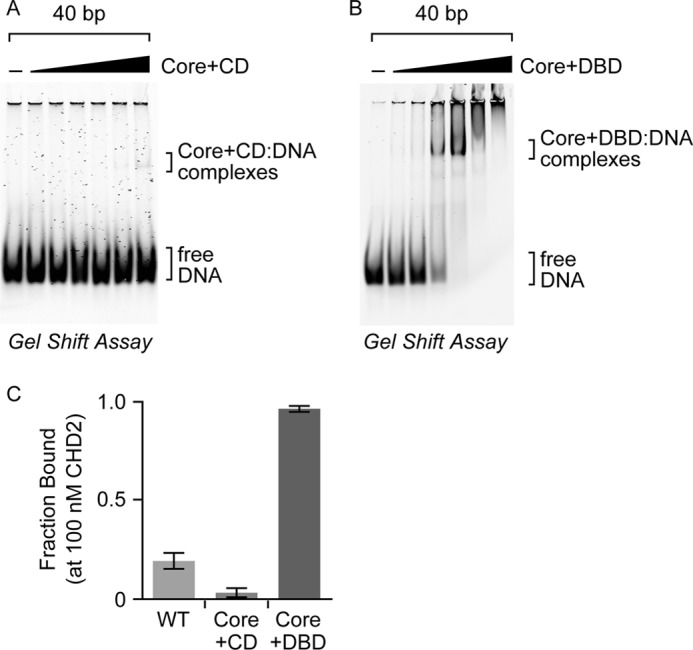
The accessory domains of CHD2 regulate its DNA-binding activities. EMSAs were performed using increasing amounts (0, 10, 20, 50, 100, 200, or 400 nm) of (A) the Core+CD, or (B) the Core+DBD on a 40 bp dsDNA probe. C, for comparison, the fraction of DNA bound by WT CHD2 and the two deletion proteins at 100 nm was calculated and graphed. All values are mean and S.D.; n = 3. Estimated Kd for WT CHD2 is ∼160 nm and the estimated Kd for Core+DBD is ∼50 nm.
We also examined how the loss of the CDs affects the binding of CHD2 to DNA. Our ATPase results show that the Core+DBD protein exhibits robust DNA- and chromatin-stimulated ATPase activity (Fig. 3, B and C), suggesting that the Core+DBD protein is capable of interacting with DNA. We then performed EMSA with the Core+DBD and the 40 bp probe (Fig. 6B) and found that the Core+DBD binds with significantly higher affinity than that observed for the WT protein (Fig. 6C). Based on our analyses, the Core+DBD exhibits ∼3-fold higher affinity for dsDNA than WT CHD2 (Kd for WT CHD2 is ∼160 nm; the Kd for Core+DBD is ∼50 nm). The fact that the Core+DBD protein shows enhanced DNA binding likely explains its higher DNA-dependent ATPase activity, as compared with WT CHD2 (Fig. 3C, compare bar 14 with bar 2). It further reinforces the model that the N terminus plays an inhibitory role in cis by reducing the affinity of the core ATPase domain and DBD for DNA.
DISCUSSION
In this study, we identified human CHD2 as a new ATP-dependent chromatin assembly factor. The identification of a new assembly factor suggests that vertebrate cells have at least three distinct ATP-dependent chromatin assembly factors: ISWI, CHD1, and CHD2. The reason vertebrate cells would need three assembly activities is not known, although they all appear to be required for proper development (6, 35, 36). This suggests ISWI, CHD1, and CHD2 play essential, non-overlapping roles in vivo.
The Core ATPase
In addition to characterizing the remodeling activity of human CHD2, we dissected the role that the accessory domains play in regulating both substrate specificity and chromatin remodeling activity (see model, Fig. 7). We show that the core ATPase domain alone exhibits DNA-stimulated ATPase activity, and that the level of stimulation is similar to that observed for WT CHD2 (Fig. 3C). In contrast, the core ATPase domain is poorly stimulated by chromatin (Fig. 3C) and is unable to remodel chromatin (Fig. 4B), which has less free DNA than the plasmid, suggesting that the core ATPase domain requires the accessory domains to be stimulated by chromatin.
FIGURE 7.
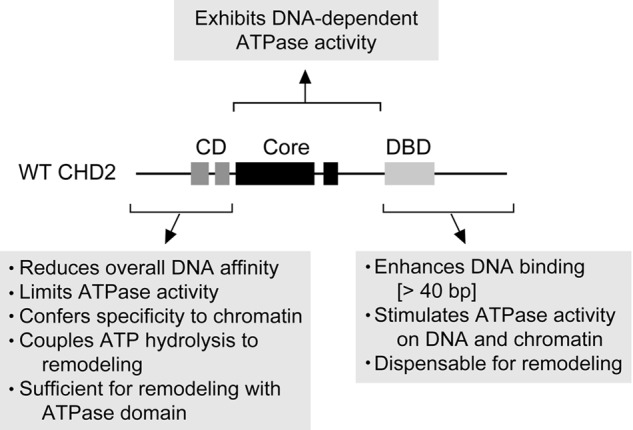
A schematic summarizing the findings of this study. The CD-containing N-terminal region plays an inhibitory role, reducing the overall DNA affinity of CHD2, limiting the DNA-stimulated ATPase activity of the Core, and thereby conferring chromatin specificity. This region is also needed to couple ATP hydrolysis to efficient chromatin remodeling. In contrast, the DBD-containing C-terminal region is not necessary for chromatin remodeling, but positively stimulates ATPase activity on DNA and chromatin and enhances both remodeling and binding to dsDNA greater than 40 bp in length.
The CD-containing N-terminal Region
Our findings show that the addition of the CD-containing N-terminal region to the ATPase domain (Core+CD) abrogates the ability of the core ATPase domain to be stimulated by DNA but not chromatin (Fig. 3C), supporting the idea that chromatin relieves the CD-dependent inhibition of the ATPase domain. This finding also shows that the DNA present in the chromatin is not sufficient to stimulate the ATPase activity when the CDs are present. It is possible that the CDs act to provide specificity for chromatin by restricting the ability of naked DNA to stimulate the ATPase activity and by increasing the affinity of CHD2 for the nucleosome, which would otherwise be refractory to its core ATPase domain alone.
Furthermore, the core ATPase domain with the CD-containing N-terminal region appears to be sufficient for chromatin-stimulated ATPase activity and chromatin remodeling activity (Figs. 3C and 4B). Though this N-terminal region provides selectivity for chromatin, its inhibitory role may limit the overall activity of the protein and explain why the ATPase and remodeling activities of the Core+CD protein are lower than that of WT CHD2.
In addition to providing specificity to chromatin substrates, the CDs are required for chromatin remodeling. The fact that the Core+DBD protein does not show detectable remodeling activity (Fig. 4B) suggests that the CDs have a crucial role in coupling ATP hydrolysis to efficient remodeling. The question still remains as to how the N terminus couples ATP hydrolysis to chromatin remodeling. If the only role of this region is to provide selectivity of CHD2 for chromatin over free DNA, then we would expect the Core+DBD protein, which exhibits robust chromatin-stimulated ATPase activity, to also possess remodeling activity; however, this is not the case.
There are several ways that the N-terminal region of CHD2 could act to couple ATP hydrolysis with remodeling. The CDs may act to auto-inhibit the ATPase activity until the CDs bind histones, after which the auto-inhibition is relieved. Sustained interaction with the histones could be required for subsequent remodeling steps. Since remodeling factors are thought to disrupt histone:DNA contacts, the interaction with both the histones and DNA could be necessary for this disruption to occur efficiently. Another possibility is that the remodeling action of CHD2 may involve cycling through auto-inhibited and activated states that are dependent on transient binding of the CDs to the histone tails. That cycling would require the continued presence of the CDs, may be regulated by ATP hydrolysis, and could ultimately contribute to the translocation force needed for remodeling.
The DBD-containing C-terminal Region
Our results show that the while the N-terminal region containing the tandem CDs provides selectivity for chromatin substrates, it also represses the activity of the ATPase domain (Fig. 3C). In contrast, the presence of the C-terminal region containing the DBD appears to counteract this inhibitory effect by stimulating the ATPase activity. Our EMSA studies suggest that the stimulation likely occurs through an overall increase in the affinity of CHD2 for DNA.
We also show that CHD2 requires at least 40 bp of dsDNA for high-affinity binding (Fig. 5, A and B) and that this correlates with increasing DNA-dependent ATPase activity (Fig. 5C), suggesting that the SANT-SLIDE-like DNA-binding domain of CHD2 might serve to bind the linker region between nucleosomes. This would be consistent with CHD1 and ISWI, which have SANT-SLIDE-like DNA-binding domains that are needed to bind extranucleosomal DNA (37–41). Moreover, the 40 bp requirement for high-affinity binding is within the estimated range for the linker lengths in humans (42–43).
Future studies will help shed light on the direct contributions of the CDs and DBD to chromatin remodeling and how the assembly activity of CHD2 complements those of the other factors, ISWI and CHD1. While we do not rule out the possibility that other motifs of CHD2 also contribute to the regulation of the ATPase domain, it is apparent that the CDs and DBD co-evolved to balance the strong auto-inhibition from the N terminus with the robust DNA-binding activity provided by the C terminus. Our studies here have helped elucidate the biochemical properties of CHD2 and set the foundation for delineating the mechanisms by which loss of normal CHD2 function leads to diseases such as epilepsy.
Acknowledgments
We thank J. Kadonaga, V. Nguyen, and R. Ceccaldi for critical reading of the manuscript. We would also like to thank A. Leschziner, R. Gaudet, S. Mango, G. Verdine, D. Nijhawan, and members of the Yusufzai Laboratory for their helpful discussions. Sincere appreciation is extended to the Kadonaga Laboratory for providing us with the Drosophila embryos.
Footnotes
- CHD2
- chromodomain helicase DNA-binding protein 2
- CD
- chromodomain
- DBD
- DNA-binding domain
- DI
- digestion index
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- dsDNA
- double-stranded DNA
- EMSA
- electrophoretic mobility gel shift assay.
REFERENCES
- 1. Gorbalenya A. E., Koonin E. V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3, 419–429 [Google Scholar]
- 2. Eisen J. A., Sweder K. S., Hanawalt P. C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Flaus A., Martin D. M., Barton G. J., Owen-Hughes T. (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clapier C. R., Cairns B. R. (2009) The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 5. Woodage T., Basrai M. A., Baxevanis A. D., Hieter P., Collins F. S. (1997) Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 11472–11477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marfella C. G. A., Ohkawa Y., Coles A. H., Garlick D. S., Jones S. N., Imbalzano A. N. (2006) Mutation of the SNF2 Family Member Chd2 Affects Mouse Development and Survival. J. Cell. Physiol. 209, 162–171 [DOI] [PubMed] [Google Scholar]
- 7. Kulkarni S., Nagarajan P., Wall J., Donovan D. J., Donell R. L., Ligon A. H., Venkatachalam S, Quade B. J. (2008) Disruption of Chromodomain Helicase DNA Binding Protein 2 (CHD2) Causes Scoliosis. Am. J. Med. Genet. 146, 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carvill G. L., Heavin S. B., Yendle S. C., McMahon J. M., O'Roak B. J., Cook J., Khan A., Dorschner M. O., Weaver M., Calvert S., Malone S., Wallace G., Stanley T., Bye A. M., Bleasel A., Howell K. B., Kivity S., Mackay M. T., Rodriguez-Casero V., Webster R., Korczyn A., Afawi Z., Zelnick N., Lerman-Sagie T., Lev D., Møller R. S., Gill D., Andrade D. M., Freeman J. L., Sadleir L. G., Shendure J., Berkovic S. F., Scheffer I. E., Mefford H. C. (2013) Targeted resequencing in epileptic encephalopathis identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 45, 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lund C., Brodtkorb E., Øye A.-M., Røsby O., Selmer K. K. (2014) CHD2 mutations in Lennox-Gastaut Syndrome. Epilepsy Behav. 33, 18–21 [DOI] [PubMed] [Google Scholar]
- 10. Courage C., Houge G., Gallati S., Schjelderup J., Rieubland C. (2014) 15q26.1 microdeletion encompassing only CHD2 and RGMA in two adults with moderate intellectual disability, epilepsy and truncal obesity. Eur. J. Med. Genet. 10.1016/j.ejmg.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 11. Chénier S., Yoon G., Argiropoulos B., Lauzon J., Laframboise R., Ahn J. W., Ogilvie C. M., Lionel A. C., Marshall C. R., Vaags A. K., et al. (2014) CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy an neurobehavioural problems. J. Neurodev. Disord. 10.1186/1866-1955-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suls A., Jaehn J. A., Kecskés A., Weber Y., Weckhuysen S., Craiu D. C., Siekierska A., Djémié T., Afrikanova T., Gormley P., von Spiczak S, Kluger G., Iliescu C. M., Talvik T., Talvik I., Meral C., Caglayan H. S., Giraldez B. G., Serratosa J., Lemke J. R., Hoffman-Zacharska D., Szczepanik E., Barisic N., Komarek V., Hjalgrim H., Møller R. S., Linnankivi T., Dimova P., Striano P., Zara F., Marini C., Guerrini R., Depienne C., Baulac S., Kuhlenbäumer G., Crawford A. D., Lehesjoki A. E., de Witte P. A., Palotie A., Lerche H., Esguerra C. V., De Jonghe P., Helbig I., EuroEPINOMICS RES Consortium (2013) De Novo Loss-of-Function Mutations in CHD2 Cause a Fever-Sensitive Myoclonic Epileptic Encephalopathy Sharing Features with Dravet Syndrome. Am. J. Hum. Genet. 93, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harada A., Okada S., Konno D., Odawara J., Yoshimi T., Yoshimura S., Kumamaru H., Saiwai H., Tsubota T, Kurumizaka H., Akashi K., Tachibana T., Imbalzano A. N., Ohkawa Y. (2012) Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 31, 2994–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yusufzai T., Kadonaga J. T. (2008) HARP is an ATP-driven annealing helicase. Science 322, 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fyodorov D. V., Kadonaga J. T. (2003) Chromatin Assembly In Vitro with Purified Recombinant ACF and NAP-1. Methods Enzymol. 371, 499–515 [DOI] [PubMed] [Google Scholar]
- 16. Alexiadis V., Kadonaga J. T. (2002) Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16, 2767–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brehm A., Tufteland K. R., Aasland R., Becker P. B. (2004) The many colours of chromodomains. Bioessays. 26, 133–140 [DOI] [PubMed] [Google Scholar]
- 18. Quan J., Yusufzai T. (2014) The Tumor Suppressor Chromodomain Helicase DNA-binding Protein 5 (CHD5) Remodels Nucleosomes by Unwrapping. J. Biol. Chem. 289, 20717–20726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito T., Bulger M., Pazin M. J., Kobayashi R., Kadonaga J. T. (1997) ACF, an ISWI-Containing and ATP-Utilizing Chromatin Assembly and Remodeling Factor. Cell 90, 145–155 [DOI] [PubMed] [Google Scholar]
- 20. Robinson K. M., Schultz M. C. (2003) Replication-Independent Assembly of Nucleosome Arrays in a Novel Yeast Chromatin Reconstitution System Involves Antisilencing Factor Asf1p and Chromodomain Protein Chd1p. Mol. Cell Biol. 23, 7937–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lusser A., Urwin D. L., Kadonaga J. T. (2005) Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12, 160–166 [DOI] [PubMed] [Google Scholar]
- 22. Torigoe S. E., Urwin D. L., Ishii H., Smith D. E., Kadonaga J. T. (2011) Identification of a Rapidly Formed Nonnucleosomal Histone-DNA Intermediate that Is Converted into Chromatin by ACF. Mol. Cell 43, 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakagawa T., Bulger M., Muramatsu M., Ito T. (2001) Multistep Chromatin Assembly on Supercoiled Plasmid DNA by Nucleosome Assembly Protein-1 and ATP-utilizing Chromatin Assembly and Remodeling Factor. J. Biol. Chem. 276, 27384–27391 [DOI] [PubMed] [Google Scholar]
- 24. Alexiadis V., Lusser A., Kadonaga J. T. (2004) A Conserved N-terminal Motif in Rad54 Is Important for Chromatin Remodeling and Homologous Strand Pairing. J. Biol. Chem. 279, 27824–27829 [DOI] [PubMed] [Google Scholar]
- 25. Lake R. J., Geyko A., Hemashettar G., Zhao Y., Fan H-Y. (2010) UV-Induced Association of the CSB Remodeling Protein with Chromatin Requires ATP-Dependent Relief of N-Terminal Autorepression. Mol. Cell 37, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hauk G., McKnight J. N., Nodelman I. M., Bowman G. D. (2010) The Chromodomains of theChd1 Chromatin Remodeler Regulate DNA Access to the ATPase Motor. Mol. Cell 39, 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clapier C. R., Cairns B. R. (2012) Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492, 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mueller-Planitz F., Klinker H., Ludwigsen J., Becker P. B. (2013) The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 20, 82–89 [DOI] [PubMed] [Google Scholar]
- 29. Henikoff S. (1993) Transcriptional activator components and poxvirus DNA-dependent ATPases comprise a single family. Trends Biochem. Sci. 18, 291–292 [DOI] [PubMed] [Google Scholar]
- 30. Torigoe S. E., Patel A., Khuong M. T., Bowman G. D., Kadonaga J. T. (2013) ATP-dependent chromatin assembly is functionally distinct from chromatin remodeling. Elife. 0.7554/eLife. 00863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Almer A., Rudolph H., Hinnen A., Hörz W. (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Varga-Weisz P. D., Wilm M., Bonte E., Dumas K., Mann M., Becker P. B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388, 598–602 [DOI] [PubMed] [Google Scholar]
- 33. Boyer L. A., Shao X., Ebright R. H., Peterson C. L. (2000) Roles of the Histone H2A-H2B Dimers and the (H3-H4)2 Tetramer in Nucleosome Remodeling by the SWI-SNF Complex. J. Biol. Chem. 275, 11545–11552 [DOI] [PubMed] [Google Scholar]
- 34. Fan H. Y., He X., Kingston R. E., Narlikar G. J. (2003) Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 11, 1311–1322 [DOI] [PubMed] [Google Scholar]
- 35. Stopka T., Skoultchi A. I. (2003) The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. U.S.A. 100, 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M. J., Heidersbach A., Ramalho-Santos J., McManus M. T., Plath K., Meshorer E., Ramalho-Santos M. (2009) Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stockdale C., Flaus A., Ferreira H., Owen-Hughes T. (2006) Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 281, 16279–26288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J. G., Madrid T. S., Sevastopoulos E., Narlikar G. J. (2006) The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 13, 1078–1083 [DOI] [PubMed] [Google Scholar]
- 39. Gangaraju V. K., Bartholomew B. (2007) Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 618, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryan D. P., Sundaramoorthy R., Martin D., Singh V., Owen-Hughes T. (2011) The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J. 30, 2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Racki L. R., Yang J. G., Naber N., Partensky P. D., Acevedo A., Purcell T. J., Cooke R., Cheng Y., Narlikar G. J. (2009) The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature 462, 1016–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Widom J. (1992) A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 89, 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaffney D. J., McVicker G., Pai A. A., Fondufe-Mittendorf Y. N., Lewellen N., Michelini K., Widom J., Gilad Y., Pritchard J. K. (2012) Controls of nucleosome positioning in the human genome. PLoS Genet. 10.1371/journal.pgen.1003036 [DOI] [PMC free article] [PubMed] [Google Scholar]