Background: SCCRO/DCUN1D1 is commonly amplified in squamous cell carcinomas. Although the SCCRO PONY domain is required for its function, the contribution of its UBA domain is undefined.
Results: Binding of polyubiquitin chains to UBA domain inhibits SCCRO-promoted neddylation and oncogenic activity.
Conclusion: UBA domain serves as a sensor and regulator of SCCRO.
Significance: Targeting the UBA domain may inhibit SCCRO function in human cancers.
Keywords: Head and Neck Cancer, Head and Neck Squamous Cell Carcinoma (HNSCC), Oncogene, Ubiquitin, Ubiquitination, DCUN1D1, SCCRO, Neddylation, Ubiquitin Binding Domain, Ubiquitin-associated Domain
Abstract
Amplification of squamous cell carcinoma-related oncogene (SCCRO) activates its function as an oncogene in a wide range of human cancers. The oncogenic activity of SCCRO requires its potentiating neddylation domain, which regulates its E3 activity for neddylation. The contribution of the N-terminal ubiquitin-associated (UBA) domain to SCCRO function remains to be defined. We found that the UBA domain of SCCRO preferentially binds to polyubiquitin chains in a linkage-independent manner. Binding of polyubiquitin chains to the UBA domain inhibits the neddylation activity of SCCRO in vivo by inhibiting SCCRO-promoted nuclear translocation of neddylation components and results in a corresponding decrease in cullin-RING-ligase-promoted ubiquitination. The results of colony formation and xenograft assays showed a mutation in the UBA domain of SCCRO that reduces binding to polyubiquitin chains, significantly enhancing its oncogenic activity. Analysis of 47 lung and head and neck squamous cell carcinomas identified a case with a frameshift mutation in SCCRO that putatively codes for a protein that lacks a UBA domain. Analysis of data from The Cancer Genome Atlas showed that recurrent mutations cluster in the UBA domains of SCCRO, lose the ability to bind to polyubiquitinated proteins, and have increased neddylation and transformation activities. Combined, these data suggest that the UBA domain functions as a negative regulator of SCCRO function. Mutations in the UBA domain lead to loss of inhibitory control, which results in increased biochemical and oncogenic activity. The clustering of mutations in the UBA domain of SCCRO suggests that mutations may be a mechanism of oncogene activation in human cancers.
Introduction
Posttranslational modification by ubiquitin regulates the activity of proteins involved in a wide range of essential cellular processes. Although ubiquitin modifications primarily target proteins for degradation by the 26 S proteasome, they can also signal for nonproteolytic effects on protein function. The pleiotropic effects of the ubiquitin signal are encoded for by variations in length and linkage of the ubiquitin chain. In general, Lys48-linked polyubiquitin chains signal for protein degradation, whereas monoubiquitin and non-Lys48-linked polyubiquitin chains signal for nonproteolytic effects (Lys6, Lys11, Lys27, Lys29, Lys33, and Lys63) (1–5). However, the same ubiquitin linkage can have varying consequences: for example, Lys11-linked chains can promote degradation of the targeted proteins by either the proteasome or the endoplasmic reticulum as well as promote endocytosis and nondegradative activities that lead to NF-κB activation (6). The diversity of effects imparted by the ubiquitin signal are mediated by motifs that bind to ubiquitin and/or ubiquitin chains and transduce their effects, analogous to the role played by binding of phosphorylated proteins to phospho-binding domains (7–10). Collectively known as ubiquitin-binding domains (UBDs),2 these motifs comprise more than 20 different families. The ubiquitin-associated (UBA) domain, the prototypic and most common type of UBD, is composed of a motif of 40 residues that are arranged in three tightly packed α-helices that share structural homology with coupling of ubiquitin conjugation to endoplasmic reticulum degradation (CUE)-type UBDs (9, 11–14). It is estimated that more than 79 proteins contain UBA domains. The majority of UBA domains recognize a canonical Ile44/Val70-binding patch on ubiquitin and preferentially bind to polyubiquitin chains (15, 16). However, the affinities of individual UBA domains vary considerably, binding to ubiquitin chains of different lengths and linkages (10). UBA domains are thought to limit ubiquitin chain elongation and to help with localization of the bound ubiquitinated proteins to the 26 S proteasome for degradation. Recent studies show that UBA domains can also transduce their effects for a variety of other consequences, including modulation of protein interactions and subcellular localization as well as nonproteasomal turnover of the ubiquitinated proteins and/or the UBA domain-containing proteins (9, 17, 18).
The presence of a UBA domain in SCCRO (also known as DCUN1D1), a protein that functions as an E3 in neddylation (conjugation of the ubiquitin-like protein NEDD8), raises several intriguing possibilities about its function. In contrast to ubiquitination, only a limited number of proteins are subject to neddylation with the cullin protein family (CUL1, CUL2, CUL3, CUL4, CUL5, and CUL7) being the best characterized targets (19). Cullins serve as the scaffold for assembly of cullin-RING-ligase (CRL)-type E3 ligases, the most common type of ubiquitination E3s (20, 21). Neddylation of cullins promotes assembly of the CRL complex and optimizes its conformation to allow efficient transfer of ubiquitin from the E2 to the substrate protein. In its role in promoting neddylation, SCCRO activity regulates levels of ubiquitination activity in cells: an increase in SCCRO activity increases levels of ubiquitinated proteins by promoting CRL activity, whereas a decrease in SCCRO activity has the opposite effect (22–27).
So what is the role of the UBA domain in SCCRO? The high degree of conservation and the existence of splice forms of the SCCRO orthologue DCN1 in lower organisms and SCCRO paralogues in higher organisms that lack a UBA domain suggest that, although it is not required, the UBA domain plays an important role in the function of SCCRO. In vitro studies confirm that the UBA domain is not required for the neddylation activity of SCCRO (26). Our finding that SCCRO regulates neddylation in vivo by promoting nuclear translocation of the neddylation components (27) combined with recent reports that the UBA domain promotes monoubiquitination of SCCRO leading to its nuclear export (28) suggests that the UBA domain may regulate the neddylation activity of SCCRO in vivo. In this study, we show that binding of polyubiquitin chains to the UBA domain inhibits the activity of SCCRO, a core protein in the ubiquitin-proteasome pathway. In this role, the UBA domain serves as a feedback regulator of the in vivo neddylation and oncogenic activity of SCCRO. Moreover, clustering of mutations in the UBA domain of SCCRO that mitigate binding to polyubiquitinated proteins suggests that it is a mechanism for oncogene activation. Furthermore, the presence of mutations in the UBA domain of many other proteins suggests that this may be an underappreciated mechanism of oncogene activation in human cancers. Given the conserved binding characteristics of the UBA domain, targeting it may have therapeutic benefit in the treatment of human cancer.
EXPERIMENTAL PROCEDURES
Alignment and Sequence Analyses
Database and BLAST searches were performed at the National Center for Biotechnology Information. Sequence alignments were performed using the ClustalW program.
Reagents
All mutants were generated by PCR mutagenesis and verified by sequencing. Proteins were expressed as glutathione S-transferase (GST) fusions in Escherichia coli using the pGEX4T3 vector. The proteins were purified using glutathione-Sepharose 4B beads (Amersham Biosciences) followed by thrombin cleavage where required. CUL1-ROC1 was expressed and purified from E. coli as described previously (29). APPBP1/UBA3, UBC12, NEDD8, and all ubiquitin chains were obtained from a commercial source (Boston Biochem, Cambridge, MA). The proteasome inhibitor Z-Leu-Leu-Leu-al (here referred to as MG132) was purchased from Sigma-Aldrich. The following antibodies were used in this study: anti-CUL1 (Zymed Laboratories Inc., South San Francisco, CA); anti-CUL3 (BD Biosciences); anti-UBC12 (Rockland Immunochemicals, Gilbertsville, PA); anti-NEDD8 (Invitrogen); anti-CAND1 (BD Biosciences); anti-α-tubulin (Calbiochem); anti-HA, anti-His6, and anti-ROC1 (Abcam, Cambridge, MA); and anti-MYC and anti-ubiquitin (Santa Cruz Biotechnology, Dallas, TX). Anti-SCCRO (rabbit polyclonal) antibody was produced and used as described previously (30). Anti-SCCRO monoclonal antibody was raised against an N-terminal region of the protein in SCCRO−/− mice. This antibody was found to be highly sensitive and specific (data not shown). All cell lines used in this study were obtained from the American Type Culture Collection (Manassas, VA) and maintained as recommended.
cDNAs for mammalian transfection were cloned into the pBABE-puro vector (Addgene, Cambridge, MA) or pUSE-amp (Clontech). NIH 3T3 cells were doubly exposed to virus in the presence of 7 μg/ml Polybrene (Sigma-Aldrich) for 24 h each. Infected cells were selected for by use of puromycin (Sigma-Aldrich). pUSE-amp transfection was performed using Lipofectamine 2000 reagent (Invitrogen) in accordance with the manufacturer's specifications.
In Vitro Neddylation
Cullin-ROC1 substrate for in vitro neddylation reactions was derived from either HeLa cell lysates or bacteria (see above). For reactions using HeLa-derived cullin-ROC1 complexes, 50 μg of lysate (CUL1 concentration, ∼20 fmol) was added to reactions containing 10 nm APPBP1/UBA3, 4 nm UBC12, and 0.9 μm NEDD8 in neddylation buffer (50 mm Tris-HCl (pH 7.5), 55 mm NaCl, and 5 mm MgCl2). Reactions were incubated at 30 °C and stopped with the addition of 6× Laemmli buffer. Reactions with bacterially derived substrate were performed at 30 °C in neddylation buffer containing 27 nm CUL1-ROC1, 4 mm ATP, 10 nm APPBP1/UBA3, 20 nm UBC12, and 0.9 μm NEDD8. Proteins were resolved by SDS-PAGE and subjected to Western blot analysis. For quantification, Western blots were imaged using NIH ImageJ software.
GST Pulldown Assay and Immunoprecipitation
GST-tagged proteins were bound to glutathione-Sepharose beads by gentle rotation at 4 °C for 45 min. The beads were washed five times with EBC buffer (50 mm Tris-HCl (pH 7.5), 2.5 mm MgCl2, 150 mm NaCl, and 0.5% Nonidet P-40) at 20× bead volume. The beads were incubated with HeLa cell lysate or purified proteins as indicated at 4 °C for 1 h followed by five washes with EBC buffer at 20× bead volume. Bound proteins were eluted by the addition of 6× Laemmli buffer, resolved by SDS-PAGE, and analyzed by Western blotting. GST-tagged proteins were visualized by use of Coomassie R-250 staining (Sigma-Aldrich). Immunoprecipitations were performed essentially as described previously (30). In brief, 1 mg of whole-cell lysate was incubated with antibody bound to agarose beads by gentle rocking at 4 °C for 2 h. The wash and detection were the same as described above for the GST pulldown assay.
Immunofluorescence
Rhodamine-conjugated anti-MYC and FITC-conjugated anti-HA antibodies were obtained from a commercial source (Jackson ImmunoResearch Laboratories, West Grove, PA). Cells transfected with plasmid(s) were seeded in 6-well plates with cover glass. Forty-eight hours after transfection, cells were washed with PBS and fixed in 4% formaldehyde for 10 min. The fixed cells were permeabilized with 0.5% Triton X-100 for 5 min, incubated in blocking buffer (PBS containing 10% FCS) for 30 min, and stained overnight at 4 °C with fluorochrome-conjugated antibodies. The cells were washed three times with PBS, counterstained with DAPI, covered with ProLong Gold antifade reagent (Invitrogen), and examined with a Leica inverted confocal microscope fitted with appropriate fluorescence filters.
Colony Formation and Xenograft Assays
A soft agar assay was performed essentially as described previously (30). Xenograft assays were performed as described previously and in accordance with Institutional Animal Care and Use Committee guidelines. In brief, NIH 3T3 cells (5 × 106) were implanted into the flanks of 8-week-old female BALB/c severe combined immunodeficiency (SCID) mice (Taconic, Hudson, NY). Tumor size was measured every 3 days by a single observer and analyzed using Microsoft Excel and GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). According to institutional guidelines, mice were sacrificed when tumor burden impeded normal feeding and movement.
UBA Mutation Analysis
Sixty-four human UBA domain-containing proteins from Simple Modular Architecture Research Tool (SMART) and 15 UBA domain-containing proteins from the literature were analyzed. Of these proteins, 28 were included in the TCGA data set. These 28 proteins were assessed for the presence and location of mutations against 29 cancer types by use of the cBioPortal for Cancer Genomics.
Statistical Analysis
Paired t tests were used to compare the means ± S.D. of soft agar colony formation counts by use of SPSS 19 software (IBM, Armonk, NY). Survival curves were generated using the Kaplan-Meier method and compared using the log rank test by use of GraphPad Prism. Disease-free survival was defined as the time from primary treatment of cancer to development of recurrence. Survival outcomes were censored for patients who died of intercurrent disease or who survived to the end of the study. A two-tailed p value ≤0.05 was considered to indicate statistical significance.
RESULTS
The UBA Domain of SCCRO Binds Polyubiquitin Chains in a Linkage-independent Manner
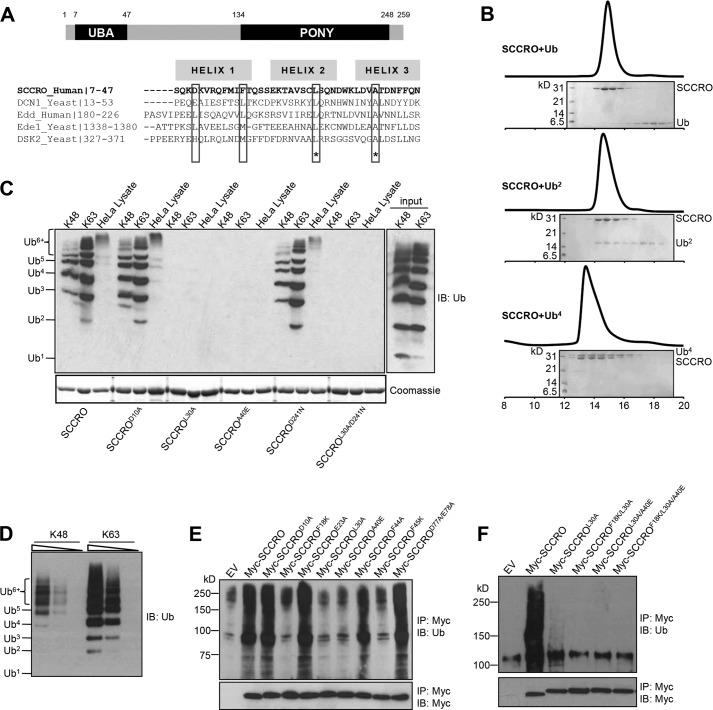
Results of sequence and structural analyses show that the UBA domain of SCCRO is composed of a three-helical bundle architecture that shares homology with several other UBA and CUE domains (26, 31). Hydrophobic residues that are critical for interactions with ubiquitin chains in structurally related domains are highly conserved in the UBA domain of SCCRO (Fig. 1A). Whereas previous studies have shown that the UBA domain of the yeast orthologue of SCCRO (DCN1) binds to monoubiquitin, structurally related domains preferentially bind to polyubiquitin chains and have a low affinity for monoubiquitin (16, 22). To define the binding preferences, we used in vitro synthesized Lys48-linked ubiquitin chains (Ub2 and Ub4) and assessed their binding to SCCRO by size exclusion chromatography. The results showed a preferential 1:1 co-elution of the Ub4-SCCRO complex even in the presence of excess Ub or Ub2 (Fig. 1B). To validate these binding preferences and precisely map the residues involved, GST-SCCRO and selected UBA and PONY domain mutants were used in a pulldown assay containing ubiquitin chains of varying lengths and linkages (Lys48 or Lys63). The results showed that SCCRO bound to both Lys48- and Lys63-linked Ub4 or greater length chains with higher affinity than Ub or Ub2 chains (Fig. 1C). Pulldown assays repeated using serial dilutions of equal concentrations of Lys48- and Lys63-linked polyubiquitin chains showed a higher affinity for binding to Lys63-linked chains (Fig. 1D). Mutations in conserved hydrophobic residues in the UBA domain of SCCRO (L30A and A40E) reduced binding, whereas mutations of nonconserved residues in the UBA domain or of residues in the PONY domain did not affect binding to polyubiquitin chains (Fig. 1C). Immunoprecipitation for MYC in lysates from U2OS cells expressing MYC-tagged SCCRO or SCCRO mutants followed by Western blot analysis for ubiquitin showed that SCCRO binds to polyubiquitinated proteins in vivo and that this interaction requires conserved hydrophobic residues (Phe18, Leu30, Ala40, and Phe45) but not other residues in the UBA domain (Fig. 1E). Mutations in two (SCCROF18K/L30A and SCCROL30A/A40E) or three conserved residues (SCCROF18K/L30A/A40E) in the UBA domain of SCCRO resulted in complete loss of binding to polyubiquitinated proteins (Fig. 1F). These findings confirm the requirement of the UBA domain in binding to polyubiquitinated proteins. Previous studies have suggested that UBA domain-containing proteins can be subclassified on the basis of the length and linkage of the ubiquitin chains to which they bind (32). Our results show that the UBA domain of SCCRO is best included in the class 4 subgroup, although it does not bind to monoubiquitin.
FIGURE 1.
The UBA domain of SCCRO binds polyubiquitin chains in a linkage-independent manner. A, schematic depiction of the domain structure of SCCRO (top panel) and sequence analysis of the UBA domain of SCCRO with several other UBA and CUE domains (bottom panel). Highlighted are conserved hydrophobic residues critical for binding to ubiquitinated proteins. B, SCCRO preferentially binds Ub4 over Ub or Ub2. Lys48-linked ubiquitin chains (Ub2 and Ub4) were synthesized in vitro. Pictured is a size exclusion chromatogram showing that SCCRO was co-eluted with Ub4 but not with Ub or Ub2. C, pulldown assay using GST-SCCRO and its mutants on Lys48 or Lys63 ubiquitin chains or HeLa cell lysates followed by Western blot analysis for ubiquitin showing that SCCRO binds to the ubiquitin chain in a linkage-independent manner. D, pulldown assay using GST-SCCRO on serially diluted Lys48 or Lys63 ubiquitin chains showing that SCCRO has preferential binding toward Lys63-linked ubiquitin chains. E and F, immunoblot (IB) analysis of lysates from U2OS cells transfected with MYC-SCCRO and selected mutants probed with antibody against ubiquitinated proteins following immunoprecipitation (IP) for MYC showing that conserved hydrophobic residues in the UBA domain of SCCRO are critical for binding to ubiquitin chains. EV, empty vector.
The UBA Domain of SCCRO Binds to Polyubiquitinated CRL Substrates
To determine whether SCCRO binds to proteins that are ubiquitinated by CRL complexes activated by neddylation, we performed tandem affinity purification to assess binding to known CUL1- and CUL3-anchored CRL substrates. GST-SCCRO pulldowns on whole-cell lysate from HeLa cells pretreated with MG132 were performed to enrich for SCCRO-binding proteins. After thrombin cleavage to release these proteins from the glutathione beads, immunoprecipitation was performed using anti-ubiquitin antibody to enrich for ubiquitinated proteins. Immunoblotting showed that SCCRO bound to polyubiquitinated Aurora B and RHOA, established substrates of CUL3-anchored CRL complexes, but not to cyclin E or p27, substrates of CUL1-anchored CRL complexes (Fig. 2A). However, as there is a limited amount of ubiquitinated Aurora B and RHOA in cellular lysates, extended exposure of the Western blot was required to detect binding. To verify these results, we repeated binding experiments on HeLa cells transfected with Aurora B or RHOA. Transfected cells were treated with MG132 and subjected to tandem affinity purification following the approach described above. As an additional control, we also included pulldowns with GST-SCCROL30A on the same lysates. We found clear binding of polyubiquitinated Aurora B and RHOA to GST-SCCRO but not to GST or GST-SCCROL30A (Fig. 2B). Combined, these findings suggest that SCCRO binds to CUL3-anchored CRL targets.
FIGURE 2.
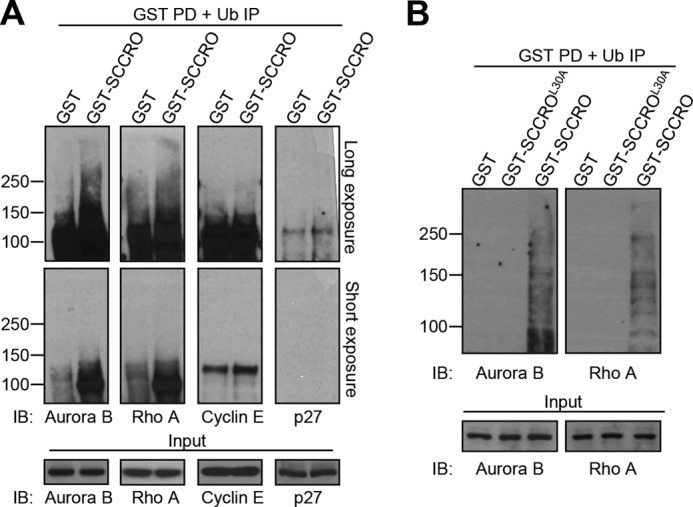
The UBA domain of SCCRO binds to polyubiquitinated CRL substrates. A, GST-SCCRO pulldown products from HeLa cell lysate pretreated with MG132 were released by thrombin cleavage. SCCRO and SCCRO-interacting proteins were further immunoprecipitated with antibody against polyubiquitinated proteins and analyzed by Western blotting with antibodies against Aurora B and RHOA (CUL3 substrates) and cyclin E and p27 (CUL1 substrates), which showed that SCCRO selectively bound to polyubiquitinated Aurora B and RHOA. B, experiments were performed as described for A except that HeLa cells were transfected with Aurora B or RHOA before MG132 treatment. Note that GST-SCCRO, but not GST or GST-SCCROL30A, bound to polyubiquitinated Aurora B and RHOA. IP, immunoprecipitation; IB, immunoblotting; PD, pulldown.
The UBA Domain Does Not Affect Neddylation Activity of SCCRO in Vitro
To determine whether the UBA domain modulates the neddylation activity of SCCRO, we performed structure-function studies to investigate its effect in in vitro assays. In vitro reactions contained recombinant NEDD8, APPBP1/UBA3 (E1), UBC12 (E2), ATP, and whole-cell lysate from HeLa cells (as a source of CUL-ROC1 substrate) in the presence of varying concentrations of SCCRO or selected SCCRO mutants. As expected, the addition of SCCRO increased cullin neddylation in a dose-dependent manner. Deletion or mutation of the UBA domain (SCCROΔN or SCCROL30A) did not alter the neddylation-promoting activity of SCCRO, whereas deletion or mutation of the PONY domain (SCCROΔC or SCCROD241N) abrogated its activity (Fig. 3A) (23). Moreover, the addition of a 20-fold molar excess of Lys48-linked Ub4 did not affect the efficiency with which SCCRO and SCCROL30A increased levels of neddylated CUL1 or CUL3 (Fig. 3B). These findings are consistent with those reported previously for DCN1 in Caenorhabditis elegans and yeast (26), confirming that binding of ubiquitin chains to the UBA domain does not affect the neddylation activity of SCCRO in vitro.
FIGURE 3.
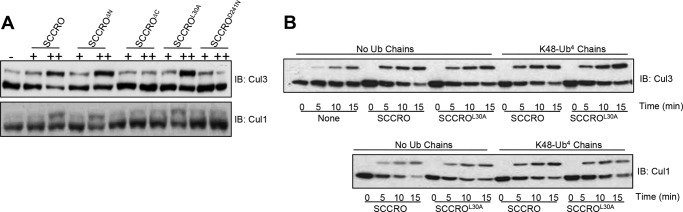
The UBA domain of SCCRO does not affect neddylation activity in vitro. A, in vitro neddylation reaction with concentration gradients of SCCRO or SCCRO mutants. Mutations in the PONY domain, but not the UBA domain, blocked the neddylation activity of SCCRO (ΔN, SCCRO with N-terminal 1–45 amino acids deleted; ΔC, SCCRO with C-terminal 151–259 amino acids deleted). B, in vitro neddylation reaction with purified recombinant SCCRO or SCCROL30A followed by Western blot analysis for CUL1 and CUL3 showing that the presence of a 20-fold molar excess of Lys48-linked tetraubiquitin failed to inhibit the effect of SCCRO or SCCROL30A on neddylation. IB, immunoblotting.
The UBA Domain Affects Neddylation Activity of SCCRO in Vivo
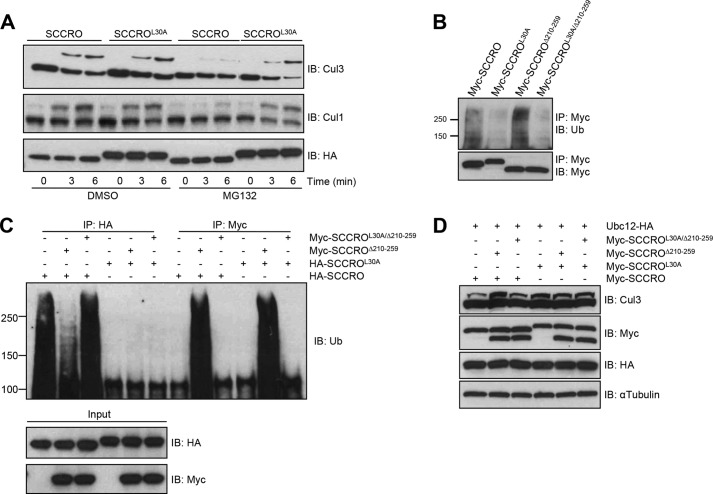
Given our recent findings showing differential requirements for SCCRO in neddylation in vitro and in vivo (27), we next assessed the requirement of the UBA domain for the neddylation activity of SCCRO in vivo. To determine whether these effects were caused by binding of polyubiquitinated proteins to the UBA domain, HeLa cells were treated with sublethal doses of proteasome inhibitor (MG132), which resulted in accumulation of polyubiquitinated proteins relative to DMSO-treated controls. Whereas transfection of either SCCRO or SCCROL30A increased levels of neddylated CUL1 and CUL3 in DMSO-treated cells, levels of neddylated CUL1 and CUL3 increased only in SCCROL30A-transfected HeLa cells treated with MG132 (Fig. 4A).
FIGURE 4.
Mutations in the UBA domain augment the neddylation activity of SCCRO in vivo. A, neddylation reaction on HeLa cell lysates with and without pretreatment with MG132 showing that the level of neddylated CUL1 and CUL3 was reduced in cells transfected with HA-SCCRO but not with HA-SCCROL30A when pretreated with MG132. B, immunoblot (IB) analysis with antibody against polyubiquitinated proteins following immunoprecipitation (IP) for MYC on lysates from U2OS cells transfected with MYC-SCCRO mutants showing that SCCROΔ210–259, but not SCCROL30A or SCCROL30A/Δ210–259, retains binding to ubiquitinated proteins. C, immunoblot analysis with antibody against polyubiquitinated proteins following immunoprecipitation for either HA or MYC on lysates from U2OS cells transfected as indicated showing that SCCROΔ210–259, but not SCCROL30A/Δ210–259, competes with SCCRO for polyubiquitinated proteins. D, Western blot analysis of neddylated CUL3 in U2OS cells transfected as indicated showing that coexpression of SCCRO with SCCROΔ210–259, but not with SCCROL30A/Δ210–259, resulted in increased levels of neddylated CUL3 (lanes 1–3). In contrast, neither of the SCCRO mutants affected neddylation of CUL3 promoted by SCCROL30A (lanes 4–6).
To validate these findings, we expressed MYC-tagged SCCRO, SCCROL30A, SCCROΔ210–259, a neddylation-deficient mutant that retains binding to polyubiquitinated proteins, or SCCROL30A/Δ210–259, a neddylation-deficient mutant with reduced binding to polyubiquitinated proteins in U2OS cells, and confirmed effects of UBA mutations on binding to polyubiquitinated proteins by immunoprecipitation and Western blotting for Ub (Fig. 4B). Immunoprecipitation using anti-HA antibody on lysates co-transfected with HA-SCCRO and MYC-SCCROΔ210–259 or MYC-SCCROL30A/Δ210–259 followed by Western blotting for Ub showed reduced binding of polyubiquitinated proteins to HA-SCCRO when it was co-transfected with MYC-SCCROΔ210–259 but not with MYC-SCCROL30A/Δ210–259 (Fig. 4C, compare lanes 1–3) even though HA-SCCRO was expressed at equal levels (Fig. 4C, see IB: HA). This is likely due to sequestration of polyubiquitinated proteins by MYC-SCCROΔ210–259, reducing the pool available to bind HA-SCCRO. Co-expression of MYC-SCCRO with MYC-SCCROΔ210–259 increased the levels of neddylated CUL3 relative to cells transfected with MYC-SCCRO alone (Fig. 4D, lanes 1 and 2). Co-transfection of either MYC-SCCROΔ210–259 or MYC-SCCROL30A/Δ210–259 did not affect levels neddylated CUL3 in MYC-SCCROL30A-transfected U2OS cells (Fig. 4D, lanes 4–6). Combined, our observations suggest that binding of polyubiquitinated proteins to the UBA domain of SCCRO inhibits its neddylation activity in vivo. These findings suggest that the UBA domain may serve as a sensor and inhibitor of the neddylation activity of SCCRO in response to increased levels of polyubiquitinated proteins.
The UBA Domain Affects Compartmentalization Activity of SCCRO
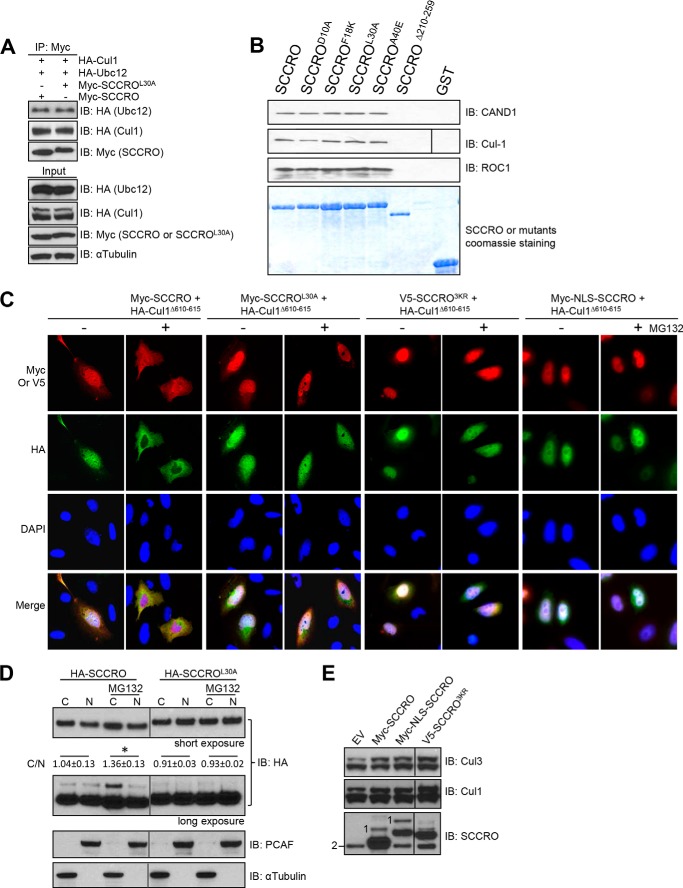
To determine the mechanisms by which the UBA domain modulates the neddylation activity of SCCRO, we first investigated whether binding of polyubiquitinated proteins to the UBA domain affects interactions between SCCRO and other neddylation components (27). Lysates from HeLa cells transfected with HA-UBC12, HA-CUL1, and MYC-SCCRO or MYC-SCCROL30A were subjected to immunoprecipitation using anti-MYC antibody followed by Western blotting for neddylation components. We found no differences between the binding of MYC-SCCRO and MYC-SCCROL30A to neddylation components (HA-CUL1 or HA-UBC12) (Fig. 5A). Similar results were obtained from GST pulldown assays with no detectable differences in binding to neddylation E3 components between GST-SCCRO and GST-SCCROL30A in HeLa cell lysates treated with DMSO or MG132 (Fig. 5B).
FIGURE 5.
Binding of ubiquitinated proteins affects SCCRO-promoted nuclear translocation of cullins. A, immunoblot (IB) analysis of lysates from U2OS cells transfected with the indicated constructs following immunoprecipitation (IP) for MYC showing no differences between the binding of SCCRO and SCCROL30A to neddylation components. B, Western blot analysis of the pulldown products of GST-SCCRO and SCCRO mutants from HeLa cell extracts probed with the indicated antibodies showing that SCCRO binds to CAND1, CUL1, and ROC1. The cullin-binding mutant SCCROΔ210–259 loses its ability to bind to CAND1, CUL1, and ROC1; in contrast, UBA mutants do not. C, immunofluorescence analysis using rhodamine-conjugated anti-MYC or anti-V5 and FITC-conjugated anti-HA on U2OS cells transfected with MYC-SCCRO, MYC-SCCROL30A, V5-SCCRO3KR, or MYC-NLS-SCCRO and HA-CUL1Δ610–615 in the presence or absence of MG132. Pretreatment with MG132 caused translocation of a significant proportion of MYC-SCCRO, but not MYC-SCCROL30A, V5-SCCRO3KR, or MYC-NLS-SCCRO, from the nucleus to the cytoplasm (first row). In addition, pretreatment with MG132 blocked nuclear translocation of HA-CUL1Δ610–615 promoted by SCCRO but not by SCCROL30A, V5-SCCRO3KR, or MYC-NLS-SCCRO (second row). D, fractionation analysis of U2OS cells transfected with HA-SCCRO showing increased levels of cytosolic SCCRO and monoubiquitinated SCCRO but not SCCROL30A after treatment with MG132. Results from densitometry measurement of Western blots from three independent experiments are shown (C, cytoplasm; N, nucleus; C/N stands for ratios of the levels of cytosolic SCCRO or SCCROL30A versus the levels of nuclear SCCRO or SCCROL30A). *, p = 0.043. E, Western blot analysis of neddylated CUL1 and CUL3 in U2OS cells transfected as indicated showing that expression of NLS-SCCRO or SCCRO3KR resulted in increased levels of neddylated CUL1 and CUL3 to an extent similar to expression of SCCRO. Western blotting for SCCRO on the same lysates showed that although both MYC-SCCRO and MYC-NLS-SCCRO underwent monoubiquitination V5-SCCRO3KR did not under identical conditions (1, monoubiquitinated SCCRO; 2, endogenous SCCRO). PCAF, p300/CBP-associated factor; EV, empty vector.
Given the importance of nuclear localization to the function of SCCRO (27), we next investigated the effects of MG132 pretreatment on the subcellular localization of MYC-SCCRO or MYC-SCCROL30A in U2OS cells. Treatment of U2OS cells with MG132 resulted in increased cytoplasmic localization of MYC-SCCRO compared with treatment with DMSO cells (Fig. 5C). In contrast, treatment with MG132 had no effect on localization of MYC-SCCROL30A, which remained primarily nuclear. Next, we cotransfected U2OS cells with MYC-SCCRO or MYC-SCCROL30A and HA-CUL1Δ610–615, a mutant that depends on SCCRO for nuclear translocation and neddylation in vivo (27). Both MYC-SCCRO and MYC-SCCROL30A promoted nuclear translocation of HA-CUL1Δ610–615 in U2OS cells treated with DMSO. However, treatment with MG132 blocked MYC-SCCRO- but not MYC-SCCROL30A-promoted nuclear translocation of HA-CUL1Δ610–615 (Fig. 5C). These observations suggest that the subcellular localization of SCCRO is regulated by binding of polyubiquitinated proteins to the UBA domain. Combined with our previous work showing the importance of SCCRO-promoted nuclear translocation of CRL complexes for the neddylation of cullins, these results provide an explanation for why the UBA domain of SCCRO differentially affects cullin neddylation in vitro and in vivo.
Binding of Polyubiquitinated Proteins to the UBA Domain Promotes Monoubiquitination and Nuclear Export of SCCRO
Previous studies have shown that the UBA domain is required for monoubiquitination of SCCRO, which promotes its nuclear export (28). We therefore sought to determine whether cytoplasmic localization of SCCRO in MG132-treated cells is due to increased monoubiquitination of SCCRO. We expressed HA-SCCRO or HA-SCCROL30A in U2OS cells. Whole-cell lysates from cells treated with either MG132 or DMSO were fractionated and immunoblotted for HA. HA-SCCRO, but not HA-SCCROL30A, was monoubiquitinated and was present only in the cytoplasmic fraction of MG132-treated cells (Fig. 5D). Consistent with previous findings (28), SCCROL30A was not monoubiquitinated under any conditions tested (Fig. 5D).
To determine whether monoubiquitination is required for nuclear export promoted by binding of polyubiquitinated proteins to the UBA domain of SCCRO, we cotransfected V5-SCCRO3KR (a monoubiquitination-deficient mutant; Fig. 5E) and HA-CUL1Δ610–615 into U2OS cells. In contrast to cells expressing SCCRO, treatment with MG132 had no effect on localization of V5-SCCRO3KR- or V5-SCCRO3KR-promoted nuclear translocation of HA-CUL1Δ610–615 (Fig. 5C), suggesting that monoubiquitination is important for nuclear export. We next questioned whether the nuclear export of SCCRO is important for the inhibitory effects exerted by binding of polyubiquitinated proteins to the UBA of SCCRO. MYC-NLS-SCCRO and HA-CUL1Δ610–615 when cotransfected into U2OS cells did not translocate to the cytoplasm upon treatment with MG132 (Fig. 5C). Combined with the observed changes in localization and neddylation activity of SCCRO that resulted from treatment with MG132, these data suggest that binding of polyubiquitinated proteins to the UBA domain promotes monoubiquitination and nuclear export of SCCRO, thereby inhibiting its neddylation activity.
Mutation of the UBA Domain Increases Oncogenic Activity of SCCRO
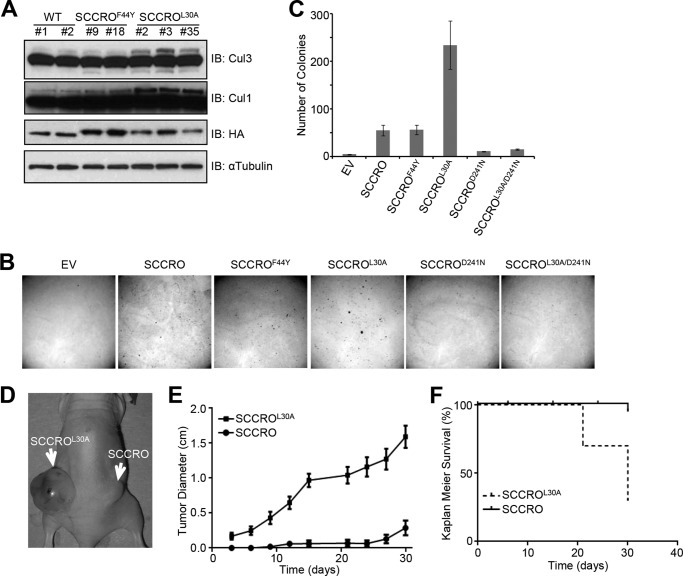
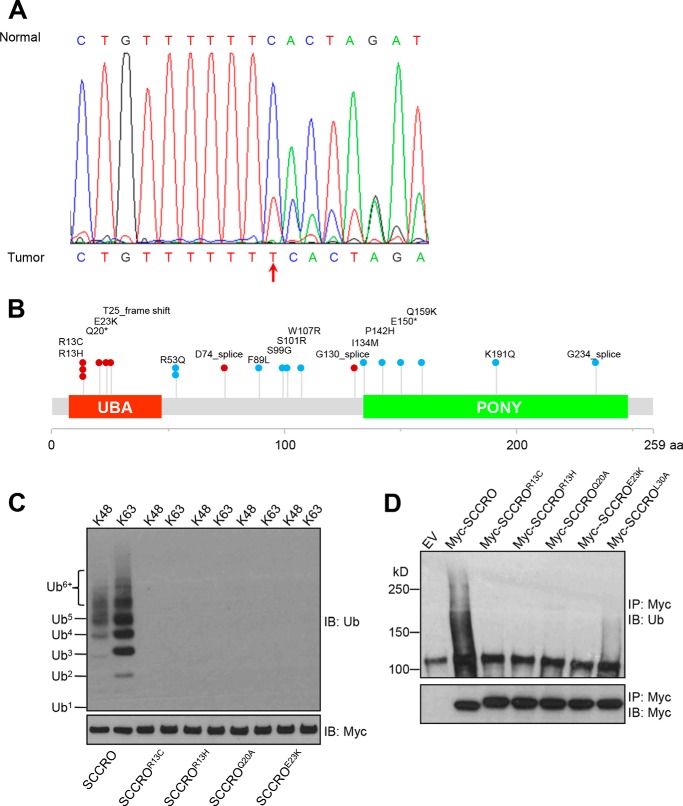
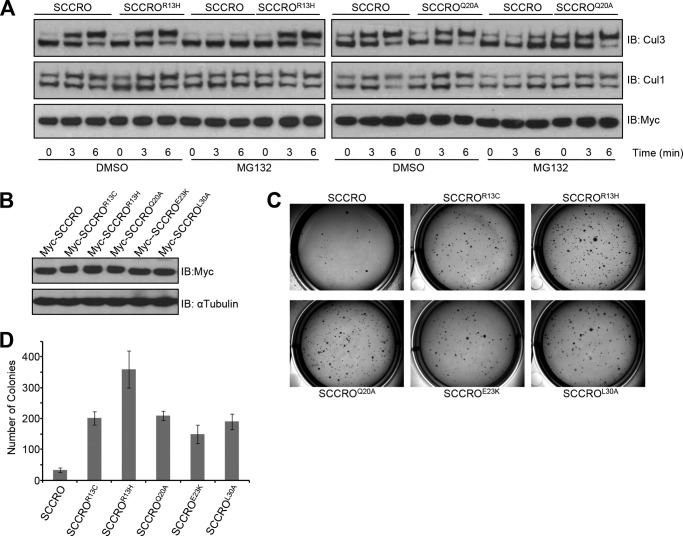
We reported previously that overexpression of SCCRO increases cullin neddylation, which promotes malignant transformation in vivo. To determine whether the UBA domain affects the function of SCCRO in vivo, we assessed the effects of stable expression of HA-SCCRO, HA-SCCROL30A, or HA-SCCROF44Y on transformation of NIH 3T3 cells. Independent stable clones with relatively equal levels of transgene expression were selected for each construct. We first confirmed the functional effects of SCCRO and SCCRO UBA mutants. Levels of neddylated CUL1 or CUL3 were assessed by immunoblot analysis on lysates from at least two independent stable clones for each construct. We found that levels of neddylated CUL1 and CUL3 were higher in cells transfected with SCCROL30A than in cells transfected with SCCRO or SCCROF44Y (Fig. 6A, compare lanes 5, 6, and 7 with the other lanes). These observations suggest that the UBA domain modulate the neddylation activity of SCCRO in these cells. Next we assessed oncogenicity in stably transfected NIH 3T3 clones by soft agar assay, which showed that clones expressing SCCROL30A formed significantly more colonies than NIH 3T3 clones expressing SCCRO (Fig. 6, B and C). In contrast, no differences in colony formation were seen between clones expressing SCCROF44Y (which retains binding to polyubiquitinated proteins) and those expressing SCCRO. To confirm that the increase in anchorage-independent growth in UBA domain mutants depends on the neddylation activity of SCCRO, we investigated the transforming activity of the neddylation-dead mutant (SCCROD241N) and a double mutant in both the UBA domain and the PONY domain (SCCROL30A/D241N). Neither the SCCROD241N mutant nor the SCCROL30A/D241N double mutant was able to form colonies in soft agar, which suggests that the neddylation activity of SCCRO plays an essential role in its oncogenic activity (Fig. 6, B and C). To validate these findings, we performed in vivo xenograft assays in BALB/c nude mice using SCCRO- or SCCROL30A-transfected NIH 3T3 clones. We found that there was shortened latency and significantly higher growth for SCCROL30A-transfected than SCCRO-transfected NIH 3T3 cells (Fig. 6, D–F). Necropsy of mice and histopathologic analyses showed that tumor xenografts resulting from injection of SCCROL30A-expressing NIH 3T3 cells were poorly differentiated and metastasized to pelvic lymph nodes in all mice (data not shown). Conversely, tumor xenografts resulting from injection of SCCRO-expressing NIH 3T3 cells were more differentiated and rarely metastasized to lymph nodes. These results suggest that the oncogenic activity of SCCRO is modulated by its UBA domain. To determine the relevance of these findings to human tumors, we screened 47 lung and head and neck squamous cell carcinomas for mutations using the Sequenom method and identified one with a frameshift mutation that putatively produces a truncated SCCRO protein with an alternate start site downstream of the mutation and without a UBA domain (Fig. 7A). However, because of a lack of tumor tissue, we could not confirm the effect of this mutation on the production of SCCRO protein. We further screened available whole-genome sequencing results from TCGA projects for different human cancers. This analysis showed that, although they are rare, mutations in SCCRO cluster in the UBA domain (Fig. 7B). To assess whether TCGA-identified UBA domain mutants bind to ubiquitin chains, we carried out an immunoprecipitation assay to assess their binding to ubiquitin chains of varying lengths and linkages (Lys48 or Lys63). In contrast to wild-type SCCRO, we found that none of the tumor-derived UBA mutants bound to ubiquitin chains (Fig. 7C). Moreover, immunoprecipitation for MYC in lysates from U2OS cells expressing MYC-tagged SCCRO or UBA mutants identified in TCGA followed by Western blot analysis for ubiquitin showed that all TCGA mutants lost the ability to bind to polyubiquitinated proteins in vivo (Fig. 7D).
FIGURE 6.
Disruption of ubiquitin binding increases transforming activity of SCCRO. A, Western blot analysis showing that levels of neddylated CUL1 and CUL3 are higher in NIH 3T3 cells transfected with HA-SCCROL30A than in those transfected with HA-SCCRO (WT) or HA-SCCROF44Y. B, representative results from soft agar assay showing that in NIH 3T3 cells stable expression of SCCROL30A resulted in increased colony formation compared with expression of SCCRO. Note that NIH 3T3 cells expressing PONY domain mutants (SCCROD241N and SCCROL30A/D241N) had a similar number of colonies as empty vector (EV) controls did. C, graph quantification of colonies for B. D and E, in vivo xenograft assays in nude mice with NIH 3T3 clones expressing SCCRO or SCCROL30A showing more aggressive tumor growth (D) with a larger tumor diameter (E) for SCCROL30A than for SCCRO. F, Kaplan-Meier survival curves showing that mice transplanted with NIH 3T3 clones expressing SCCROL30A had shorter survival than those transplanted with NIH 3T3 clones expressing SCCRO. IB, immunoblotting.
FIGURE 7.
Mutations in the UBA domain of SCCRO are present in human cancers. A, frameshift mutation in the UBA domain identified in one case of 47 lung and head and neck squamous cell carcinomas assessed by use of the Sequenom method. A truncated SCCRO mutant without a UBA domain is predicted. B, mutations in SCCRO identified in TCGA projects. Red dots represent UBA domain point mutations or splice mutations putatively producing truncated SCCRO without a UBA domain; light blue dots represent non-UBA domain mutations. Each dot represents a separate cancer. The asterisk (*) indicates a nonsense mutation. C, immunoprecipitation (IP) assay using MYC-SCCRO and its TCGA UBA mutants on Lys48- or Lys63-linked ubiquitin chains followed by Western blot analysis for ubiquitin showing that TCGA-identified UBA domain mutants do not bind ubiquitin chains. D, immunoblot (IB) analysis of lysates from U2OS cells transfected with MYC-SCCRO or TCGA UBA mutants probed with antibody against ubiquitinated proteins following immunoprecipitation for MYC showing that all mutants lose the ability to bind to polyubiquitinated proteins. EV, empty vector.
To determine whether UBA mutants identified in the TCGA data set affect the biochemical and oncogenic functions of SCCRO, we assessed their effect on cullin neddylation and transformation. Whereas transfection of SCCRO, SCCROR13H, or SCCROQ20A increased levels of neddylated CUL1 and CUL3 in DMSO-treated HeLa cells, neddylated CUL1 and CUL3 only increased in SCCROR13H- or SCCROQ20A-transfected cells after MG132 treatment (Fig. 8A). Next, to assess the transforming activities of tumor-derived SCCRO UBA mutants, we stably transfected NIH 3T3 cells and selected stable clones with similar transgene expression levels (Fig. 8B). These cells were subjected to soft agar assays, which showed significantly higher colony formation in NIH 3T3 cells expressing TCGA-identified SCCRO UBA mutants relative to those expressing wild-type SCCRO (Fig. 8, C and D). Combined, these findings suggest that the UBA domain of SCCRO modulates its neddylation activity and that this has functional relevance to tumorigenesis.
FIGURE 8.
Tumor-derived UBA mutations increase transforming activity of SCCRO. A, TCGA-identified mutations in the UBA domain augment the neddylation activity of SCCRO in vivo. A neddylation reaction on lysates derived from HeLa cells with and without pretreatment with MG132 shows that the levels of neddylated CUL1 and CUL3 are reduced by pretreatment with MG132 in MYC-SCCRO-transfected but not MYC-SCCROR13H- or MYC-SCCROQ20A-transfected cells. Results were confirmed by densitometry measurement. B, Western blot analysis of lysates from NIH 3T3 cells stably transfected with the indicated SCCRO constructs and probed with anti-MYC antibody showing equivalent expression. C, representative results from a soft agar assay showing that stable expression of TCGA-identified UBA domain mutants in NIH 3T3 cells results in increased colony formation in soft agar relative to expression of SCCRO. D, graph showing the average number of colonies for each NIH 3T3 clone from B. IB, immunoblotting.
DISCUSSION
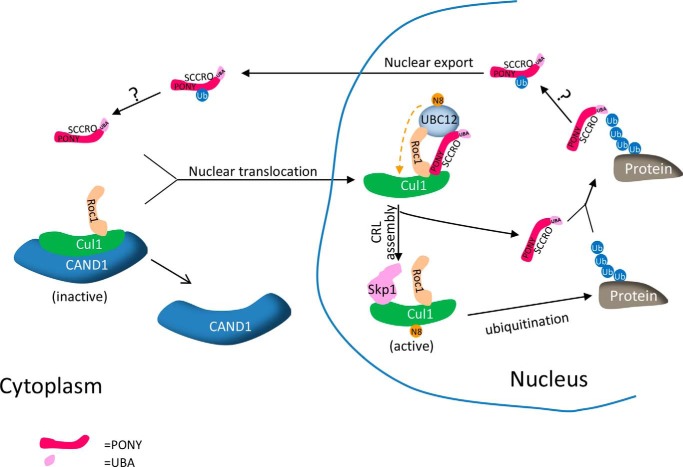
Through its role as an E3 in neddylation, SCCRO promotes CRL activity, which results in an increase in cellular levels of polyubiquitinated proteins (27). We found that binding of polyubiquitinated proteins to the N-terminal UBA domain of SCCRO inhibits its neddylation activity, suggesting the presence of a classical negative feedback loop that regulates CRL-promoted ubiquitination activity (Fig. 9). In this model, the N-terminal UBA domain of SCCRO serves as a sensor of polyubiquitinated proteins as well as an effector by promoting the autoubiquitination of SCCRO. The effects of the UBA domain do not appear to be allosteric as the addition of polyubiquitin chains or mutation of the UBA domain had no effect on the neddylation function of SCCRO in vitro, whereas accumulation of polyubiquitinated proteins showed classical negative feedback dynamics in vivo. Interestingly, the requirement for SCCRO in neddylation was also different in vitro and in vivo with its essential effects in vivo involving compartmentalization of neddylation components. As UBC12, the E2 for neddylation, is primarily located in the nucleus, SCCRO-promoted nuclear translocation of cullin-ROC1 complexes is required for neddylation. Consistent with this, we found that the effect that binding of polyubiquitinated proteins to the UBA domain had on the function of SCCRO was imparted by cytoplasmic translocation of SCCRO: an increase in levels of polyubiquitinated proteins increased cytoplasmic localization of SCCRO and inhibited nuclear localization of neddylation components, whereas mutation in the UBA domain abrogated these effects.
FIGURE 9.
Proposed model of UBA domain-mediated regulation of SCCRO.
These findings raise obvious questions about how binding of polyubiquitinated proteins to the UBA domain affects the subcellular localization and function of SCCRO. Recent work by Wu et al. (28) shows that the UBA domain of SCCRO is required for its monoubiquitination and that monoubiquitination promotes nuclear export of SCCRO. Conversely, mutations in the UBA domain blocked monoubiquitination of SCCRO and consequently its nuclear export (Fig. 5, C and D) (28). These findings suggest that the binding of polyubiquitinated proteins to the UBA domain serves as a signal for the monoubiquitination of SCCRO. A limitation of this work is the reliance on SCCRO mutants to assess structure-function relationships. It remains possible that the generated mutants affect protein function in unanticipated ways. Mitigating this issue at least in part, we found that blocking the monoubiquitination of SCCRO by mutation of ubiquitination residues in SCCRO or restricting the nuclear export of SCCRO by NLS tag had identical effects on SCCRO localization and function as UBA domain mutants expressed in cells exposed to MG132.
UBD-coupled autoubiquitination has been reported for several other proteins, including STS1, STS2, EPS15, EPS15R, Vps27p/HRS, Vps9p, and RABEX-5 (9). Although the precise underlying mechanisms remain to be defined, a range of functional consequences are known to accompany UBD-coupled autoubiquitination, including effects on inter- and intraprotein interactions and subcellular localization. Various UBDs have been implicated in coupled autoubiquitination, including ubiquitin-interacting motif, CUE, motif interacting with ubiquitin, and A20 zinc finger domains; however, this has not been reported previously for UBA domain-containing proteins. In the case of SCCRO, coupled autoubiquitination leads to its cytoplasmic translocation and functional inhibition in vivo. What remains to be defined is how monoubiquitinated SCCRO is translocated and sequestered in the cytoplasm. Given the relatively low affinity of the UBA domain in SCCRO for monoubiquitin, it is unlikely that intraprotein interaction is the cause of changes in the localization of SCCRO. These effects are also unlikely to be caused by changes in interactions with neddylation components as the presence of polyubiquitin chains and/or mutations in the UBA domain did not affect binding between SCCRO and CUL1 or UBC12. Given that SCCRO and other neddylation E3 components lack a canonical nuclear localization sequence, one possibility for nuclear export is that monoubiquitination of SCCRO inhibits interactions with NLS-containing protein partners involved in nuclear import.
In the broader context, the principal presence of the UBA domain in E2s and E3s (33) raises the question of whether UBA domains serve as general feedback regulators of the ubiquitination pathway. In a related model, autoubiquitination and ubiquitin binding of the UBD in RABEX-5 affect its ubiquitin ligase activity (34). It remains to be determined whether UBA domains play similar roles in other proteins.
Although specific mechanisms require definition, it is quite clear that mutations in the UBA domain that inhibit interactions with ubiquitinated proteins increase the neddylation activity of SCCRO in vivo. We have shown previously that increased neddylation activity resulting from overexpression of SCCRO promotes oncogenesis in vitro and in vivo (30). In primary tumors, the oncogenic activity of SCCRO is promoted by amplification that is present in many types of cancers, including lung, ovarian, head and neck, and esophageal cancers (35–37). Interestingly, we found that transgenic expression of UBA domain mutants of SCCRO was more oncogenic than that of wild-type SCCRO. Moreover, we found that, although they are rare, somatic mutations cluster in the UBA domain of SCCRO. As the spontaneously occurring somatic UBA domain mutations lose the ability to bind to polyubiquitinated proteins and have increased neddylation and transforming activity, they represent an alternate mechanism for activation of the oncogenic activity of SCCRO in human cancers.
The role of the UBA domain in cancer pathogenesis has been well established. For example, germ line mutations in the UBA domain of SQSTM1 are associated with the development of Paget disease of bone. These mutations impair the ability of SQSTM1 to bind to ubiquitin chains, altering proteasome- and/or autophagy-based turnover of the bound ubiquitinated proteins (38–42). Although the specific polyubiquitinated proteins involved remain to be identified, the carcinogenic effects that result from loss of binding due to mutation in the UBA domain of SQSTM1 are achieved through activation of NF-κB signaling (39, 40). Moreover, the identification of mutation clusters in the UBA domains of other proteins suggests that these domains may play a larger and more direct role in oncogenesis than appreciated previously (supplemental Table 1). In the broader context, functional domains typically have similar activities even when present in different proteins. As such, functionally related mutations in cancers may cluster in domains rather than in individual genes or gene families. Domain-based analysis of genome-wide sequencing data may identify novel targets for anticancer therapies. For example, the activity of an oncogene may be mitigated by agents that bind to or mimic the UBA domain of the oncoprotein. In one such instance, overexpression of the UBA domain of hFAF1 significantly promotes cell death by increasing the degradation of polyubiquitinated proteins (43).
Acknowledgments
We thank David B. Sewell for constructive editing and Dr. Zhen-Qiang Pan for generously providing SCCRO3KR construct. We also thank Molecular Cytology Core Facility (sponsored by Core Grant P30 CA008748) for great help.
This work was supported, in whole or in part, by National Institutes of Health Grant P30 CA008748, and NCI Cancer Center Support grant. This work was also supported by a Hackers for Hope grant (to B. S.) and The Clinical Innovator Award from the Flight Attendant Medical Research Institute (to B. S.).

This article contains supplemental Table 1.
- UBD
- ubiquitin-binding domain
- UBA
- ubiquitin-associated
- SCCRO
- squamous cell carcinoma-related oncogene
- DCUN1D1
- defective in cullin neddylation 1 domain-containing 1
- PONY
- potentiating neddylation domain
- TCGA
- The Cancer Genome Atlas
- CRL
- cullin-RING-ligase
- CUE
- coupling of ubiquitin conjugation to endoplasmic reticulum degradation
- NLS
- nuclear localization signal
- Ub
- ubiquitin.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 3. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 4. Mukhopadhyay D., Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 5. Aguilar R. C., Wendland B. (2003) Ubiquitin: not just for proteasomes anymore. Curr. Opin. Cell Biol. 15, 184–190 [DOI] [PubMed] [Google Scholar]
- 6. Kulathu Y., Komander D. (2012) Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 [DOI] [PubMed] [Google Scholar]
- 7. Madura K. (2002) The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle 1, 235–244 [PubMed] [Google Scholar]
- 8. Buchberger A. (2002) From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol. 12, 216–221 [DOI] [PubMed] [Google Scholar]
- 9. Hurley J. H., Lee S., Prag G. (2006) Ubiquitin-binding domains. Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su V., Lau A. F. (2009) Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell. Mol. Life Sci. 66, 2819–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trempe J. F., Brown N. R., Lowe E. D., Gordon C., Campbell I. D., Noble M. E., Endicott J. A. (2005) Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 24, 3178–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohno A., Jee J., Fujiwara K., Tenno T., Goda N., Tochio H., Kobayashi H., Hiroaki H., Shirakawa M. (2005) Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure 13, 521–532 [DOI] [PubMed] [Google Scholar]
- 13. Swanson K. A., Hicke L., Radhakrishnan I. (2006) Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J. Mol. Biol. 358, 713–724 [DOI] [PubMed] [Google Scholar]
- 14. Lowe E. D., Hasan N., Trempe J. F., Fonso L., Noble M. E., Endicott J. A., Johnson L. N., Brown N. R. (2006) Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. D Biol. Crystallogr. 62, 177–188 [DOI] [PubMed] [Google Scholar]
- 15. Searle M. S., Garner T. P., Strachan J., Long J., Adlington J., Cavey J. R., Shaw B., Layfield R. (2012) Structural insights into specificity and diversity in mechanisms of ubiquitin recognition by ubiquitin-binding domains. Biochem. Soc. Trans. 40, 404–408 [DOI] [PubMed] [Google Scholar]
- 16. Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3, 939–943 [DOI] [PubMed] [Google Scholar]
- 17. Husnjak K., Dikic I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 [DOI] [PubMed] [Google Scholar]
- 18. Trempe J. F. (2011) Reading the ubiquitin postal code. Curr. Opin. Struct. Biol. 21, 792–801 [DOI] [PubMed] [Google Scholar]
- 19. Xirodimas D. P. (2008) Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 36, 802–806 [DOI] [PubMed] [Google Scholar]
- 20. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 21. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 22. Kurz T., Ozlü N., Rudolf F., O'Rourke S. M., Luke B., Hofmann K., Hyman A. A., Bowerman B., Peter M. (2005) The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435, 1257–1261 [DOI] [PubMed] [Google Scholar]
- 23. Kim A. Y., Bommeljé C. C., Lee B. E., Yonekawa Y., Choi L., Morris L. G., Huang G., Kaufman A., Ryan R. J., Hao B., Ramanathan Y., Singh B. (2008) SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J. Biol. Chem. 283, 33211–33220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Scott D. C., Monda J. K., Grace C. R., Duda D. M., Kriwacki R. W., Kurz T., Schulman B. A. (2010) A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell 39, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heir P., Sufan R. I., Greer S. N., Poon B. P., Lee J. E., Ohh M. (2013) DCNL1 functions as a substrate sensor and activator of cullin 2-RING ligase. Mol. Cell. Biol. 33, 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kurz T., Chou Y. C., Willems A. R., Meyer-Schaller N., Hecht M. L., Tyers M., Peter M., Sicheri F. (2008) Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29, 23–35 [DOI] [PubMed] [Google Scholar]
- 27. Huang G., Kaufman A. J., Ramanathan Y., Singh B. (2011) SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J. Biol. Chem. 286, 10297–10304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu K., Yan H., Fang L., Wang X., Pfleger C., Jiang X., Huang L., Pan Z. Q. (2011) Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. J. Biol. Chem. 286, 34060–34070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 30. Sarkaria I., O-charoenrat P., Talbot S. G., Reddy P. G., Ngai I., Maghami E., Patel K. N., Lee B., Yonekawa Y., Dudas M., Kaufman A., Ryan R., Ghossein R., Rao P. H., Stoffel A., Ramanathan Y., Singh B. (2006) Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 66, 9437–9444 [DOI] [PubMed] [Google Scholar]
- 31. Yang X., Zhou J., Sun L., Wei Z., Gao J., Gong W., Xu R. M., Rao Z., Liu Y. (2007) Structural basis for the function of DCN-1 in protein neddylation. J. Biol. Chem. 282, 24490–24494 [DOI] [PubMed] [Google Scholar]
- 32. Raasi S., Varadan R., Fushman D., Pickart C. M. (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12, 708–714 [DOI] [PubMed] [Google Scholar]
- 33. Hofmann K., Bucher P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21, 172–173 [PubMed] [Google Scholar]
- 34. Mattera R., Tsai Y. C., Weissman A. M., Bonifacino J. S. (2006) The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J. Biol. Chem. 281, 6874–6883 [DOI] [PubMed] [Google Scholar]
- 35. Huang G., Singh B. (2013) Coamplification and cooperation: toward identifying biologically relevant oncogenes. Clin. Cancer Res. 19, 5549–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J., Qian J., Hoeksema M. D., Zou Y., Espinosa A. V., Rahman S. M., Zhang B., Massion P. P. (2013) Integrative genomics analysis identifies candidate drivers at 3q26–29 amplicon in squamous cell carcinoma of the lung. Clin. Cancer Res. 19, 5580–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Y., McGee J., Chen X., Doman T. N., Gong X., Zhang Y., Hamm N., Ma X., Higgs R. E., Bhagwat S. V., Buchanan S., Peng S. B., Staschke K. A., Yadav V., Yue Y., Kouros-Mehr H. (2014) Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS One 9, e98293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seibenhener M. L., Geetha T., Wooten M. W. (2007) Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Layfield R., Searle M. S. (2008) Disruption of ubiquitin-mediated processes in diseases of the brain and bone. Biochem. Soc. Trans. 36, 469–471 [DOI] [PubMed] [Google Scholar]
- 40. Lucas G. J., Daroszewska A., Ralston S. H. (2006) Contribution of genetic factors to the pathogenesis of Paget's disease of bone and related disorders. J. Bone Miner. Res. 21, Suppl. 2, P31–P37 [DOI] [PubMed] [Google Scholar]
- 41. Daroszewska A., Ralston S. H. (2005) Genetics of Paget's disease of bone. Clin. Sci. 109, 257–263 [DOI] [PubMed] [Google Scholar]
- 42. Layfield R., Ciani B., Ralston S. H., Hocking L. J., Sheppard P. W., Searle M. S., Cavey J. R. (2004) Structural and functional studies of mutations affecting the UBA domain of SQSTM1 (p62) which cause Paget's disease of bone. Biochem. Soc. Trans. 32, 728–730 [DOI] [PubMed] [Google Scholar]
- 43. Lee J. J., Kim Y. M., Jeong J., Bae D. S., Lee K. J. (2012) Ubiquitin-associated (UBA) domain in human Fas associated factor 1 inhibits tumor formation by promoting Hsp70 degradation. PLoS One 7, e40361. [DOI] [PMC free article] [PubMed] [Google Scholar]