Background: IL-7 can up-regulate CD95 expression on memory CD4+ T cells, but the mechanism remains unclear.
Results: IL-7 elevates miR-124 to decrease the expression of splicing regulator PTB and represses CD95 mRNA splicing.
Conclusion: IL-7 elevates CD95 expression by regulating its alternative splicing.
Significance: Our data demonstrate that the co-stimulation of IL-7 and CD95 ligand benefits HIV-1 reservoir maintenance.
Keywords: Alternative Splicing, Human Immunodeficiency Virus (HIV), Interleukin, MicroRNA (miRNA), Post-transcriptional Regulation, CD95, PTB
Abstract
Interleukin-7 (IL-7) has been used as an immunoregulatory and latency-reversing agent in human immunodeficiency virus type 1 (HIV-1) infection. Although IL-7 can restore circulating CD4+ T cell counts in HIV-1-infected patients, the anti-apoptotic and proliferative effects of IL-7 appear to benefit survival and expansion of HIV-1-latently infected memory CD4+ T lymphocytes. IL-7 has been shown to elevate CD95 on CD4+ T cells in HIV-1-infected individuals and prime CD4+ T lymphocytes to CD95-mediated proliferative or apoptotic signals. Here we observed that through increasing microRNA-124, IL-7 down-regulates the splicing regulator polypyrimidine tract binding protein (PTB), leading to inclusion of the transmembrane domain-encoding exon 6 of CD95 mRNA and, subsequently, elevation of CD95 on memory CD4+ T cells. Moreover, IL-7 up-regulates cellular FLICE-like inhibitory protein (c-FLIP) and stimulates c-Jun N-terminal kinase (JNK) phosphorylation, which switches CD95 signaling to survival mode in memory CD4+ T lymphocytes. As a result, co-stimulation through IL-7/IL-7R and FasL/CD95 signal pathways augments IL-7-mediated survival and expansion of HIV-1-latently infected memory CD4+ T lymphocytes. Collectively, we have demonstrated a novel mechanism for IL-7-mediated maintenance of HIV-1 reservoir.
Introduction
Interleukin-7 (IL-7) is a member of the γc cytokine family and binds to a cell-surface receptor (IL-7R)3 composed of an α chain (IL-7Rα, CD127) and a common γ-chain (CD132). It was originally identified as a pre-B cell growth factor and was also established to play a crucial role during early T cell development (1). Eventually, IL-7 was reported to have potent effects on the generation and maintenance of memory T cells (1). In vivo experiments in mice suggest that IL-7 promotes survival of effector CD4+ T cells and preserves IL-7R-positive effector T cells for further development into long-lived memory cells (2, 3). IL-7 also contributes to the maintenance of T lymphocytes and is a pivotal cytokine in T-cell homeostasis (4–6). In addition, IL-7 can increase T cell survival by maintaining a favorable balance of anti-apoptotic proteins over pro-apoptotic proteins (1).
However, the role played by IL-7 in lymphopenic conditions, such as HIV-1 infection, is quite complicated. Studies suggest that plasma IL-7 is increased during HIV-1 infection and is inversely correlated with CD127 expression (7–9). In addition, plasma IL-7 is negatively correlated with CD4+ T cell count and disease progression in HIV-1-infected patients (10–12). Although increased IL-7 results in elevated T cells and increased T cell reactivity in humans (13, 14), endogenous IL-7 is unable to rescue low CD4+ T cell counts when HIV-1-infected patients progress to AIDS, likely due to down-regulated CD127 expression and reduced IL-7 consumption (15). Moreover, IL-7 has been suggested to activate HIV-1 latency (16, 17). Clinical trials of IL-7 therapy in HIV-1-infected individuals receiving antiretroviral therapy (ART) reveal that despite the positive effect on circulating T cell populations, IL-7 can promote HIV-1 persistence by inducing survival and expansion of latently infected T cells (18, 19).
CD95 (Fas, APO-1, TNFRSF6) is a member of the tumor necrosis factor receptor superfamily and can transmit apoptotic signals when stimulated with its natural CD95 ligand (FasL, CD178) or anti-CD95 antibody. CD95 has multiple isoforms due to alternative pre-mRNA splicing. FasEX06Del (FasΔTM), which lacks the transmembrane domain due to the skipping of exon 6, can compete with the membrane-bound form of Fas as a soluble decoy (20, 21). The interaction of CD95 with its corresponding ligand, FasL, leads to the formation of death-inducing signaling complex (DISC) and subsequent apoptosis (22). It has been reported that CD95 mediates apoptosis of CD4+ T lymphocytes during HIV-1 infection (23). IL-7 has also been reported to enhance the expression of CD95 on the surface of CD4+ T cells isolated from HIV-1-infected individuals and increase sensitivity to CD95-mediated apoptosis in both naive and memory T cells (24, 25).
However, CD95 is now recognized as a dual function receptor. CD95 mediates not only apoptosis but also diverse non-apoptotic functions such as T cell proliferation, tumor growth, and injury-induced neurogenesis depending on the tissue and the conditions (26–29). The non-apoptotic pathways of CD95 possibly involve extracellular signal-regulated kinase (ERK), JNK, and nuclear factor-κB (NF-κB), although the detailed molecular mechanism of how this occurs is unclear (26–30). Although CD95 expression is high in several CD4+ T lymphocyte subsets including long-lived memory cells such as central memory T cells and stem cell-like memory T cells, CD95+ memory cells are most likely resistant to CD95-mediated apoptosis (31–33). Peripheral T cells primed with high IL-7 doses have been characterized by an increased sensitivity to CD95-mediated proliferation as well as apoptosis (34). The suboptimally-activated CD4+ T lymphocytes, primed by IL-7, could undergo compensatory expansion responding to a CD95-mediated proliferation signal during HIV-1 infection (34).
Currently, a major challenge of HIV-1 treatment is the existence of viral reservoir in HIV-1-infected patients receiving suppressive ART. Resting memory CD4+ T lymphocytes, including stem cell-like memory T cells and central memory T cells, function as the major HIV-1 reservoir (35, 36). Administration of IL-7 has been shown to benefit the survival and expansion of HIV-1-latently infected memory CD4+ T lymphocytes, and the molecular mechanism behind this phenomenon may involve co-factors that merit further investigation (18, 19). Because CD95 is up-regulated by IL-7 and is highly expressed on memory CD4+ T lymphocytes, the specific molecular mechanism of how IL-7 up-regulates CD95 expression is worth studying as is the possible effect of IL-7 upon non-apoptotic signal pathway of CD95 and the possible collaboration between IL-7 and CD95 in viral reservoir persistence. Here, using various approaches, we report that IL-7 up-regulates CD95 expression in resting memory CD4+ T cells via regulation of alternative splicing of CD95. Combination signaling of IL-7/IL-7R and FasL/CD95 promotes survival and expansion of memory CD4+ lymphocytes and, therefore, contributes to IL-7-mediated maintenance of the HIV-1 reservoir.
EXPERIMENTAL PROCEDURES
Ethics Statement
This research was approved by the Ethics Review Board of Sun Yat-Sen University. Written informed consent was provided by all study participants.
Blood Samples and Memory CD4+ T Lymphocyte Cultures
Peripheral blood mononuclear cells from HIV-negative controls or HIV-1-infected individuals enrolled under the criteria of suppressive viremia (<50/ml copies of HIV-1 RNA and CD4+ T cell count >400/ml) and taking suppressive ART for at least 2 years were isolated by Ficoll gradient centrifugation (TBD Science). CD4+ T cells were then purified with the human CD4+ T Lymphocyte Enrichment Set-DM (BD Biosciences IMagTM) according to the manufacturer's instructions. The selected cell populations contained >95% CD3+ CD4+ CD45RO+ CD45RA− T cells as measured by flow cytometry. Memory CD4+ T cells were separated by flow cytometry sorting (SORP FacsAria II, BD Biosciences) with anti-CD4-APC-conjugated mAbs (Pharmingen) and anti-CD45RA-FITC-conjugated mAbs (Pharmingen). CD4+ CD45RA− T cells (5 × 106 cells/ml) were then cultured in RPMI 1640 conditioned medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Human rhIL-7 (R&D) was added to cell cultures at the concentration of 25 ng/ml.
Flow Cytometric Analysis of Memory CD4+ T Cells
For flow cytometric measurement, anti-CD25-PE-conjugated mAbs, anti-CD69-PE-Cy5-conjugated mAbs, anti-CD95-APC-conjugated mAbs and the appropriate isotype control mAbs were used (all from Pharmingen). Cells were stained and washed followed by immediate flow cytometric analysis (FCM). Alternatively, cells were labeled after fixation and permeabilization using the Foxp3 staining buffer set (eBioscience) or BD Biosciences Cytofix/CytopermTM Fixation/Permeabilization kit (catalog no. 554714), and BD Biosciences GolgiPlugTM protein transport inhibitor (catalog no. 555028). Mean fluorescent intensities (MFI) were measured by FCM (LSRFsortessa, BD Biosciences), and data were analyzed using FlowJo software (Tree Star).
Detection of mRNA Expression in Memory CD4+ T Cells
Cellular RNA was isolated from memory CD4+ T cells using TRIzol reagent (Invitrogen) followed by DNase digestion (TURBO DNA-free, Ambion). Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase (Invitrogen). Reverse transcription PCR detection of CD95 mRNA variants was performed as described previously (37). Real-time PCR was performed using SYBR Ex-taq premix (TaKaRa) in a CFX96TM Real-time PCR Detection System (Bio-Rad). Human GAPDH and/or β-actin mRNA was measured as endogenous controls. The primer sets used were as follows: CD95 (forward primer, 5′-TTA TCT GAT GTT GAC TTG AG-3′; reverse primer, 5′-ATT ACG AAG CAG TTG AAC-3′); PTB (forward primer, 5′-TTA TAC CTG TTG TGA GAC C-3′; reverse primer, 5′-CAA TAC TGA GCC TGG AAT-3′); miR-124 (forward primer, 5′-TAA GGC ACG CGG TG-3′; reverse primer, 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACG GCA TT-3′) (38). Relative mRNA expression in IL-7-treated memory CD4+ T cells was compared with expression in non-treated cultured cells using the comparative C(T) method (39).
Western Blot
The Western blot procedure previously described by our group was followed (40). Immunoblots were developed with a chemiluminescence reagent kit (Thermo Pierce). Primary antibody sources used are as follows: Fas (4C3) mouse mAb (Cell Signaling Technology, 8023S), mouse anti-PTB (Invitrogen, catalog no. 325000), anti-Ago1 antibody (Abcam, ab98056), anti-Ago2/eIF2C2 antibody (Abcam, ab32381), FLIP (D16A8) rabbit mAb (Cell Signaling Technology, 8510S), anti-JNK1 antibody (Abcam, ab10664), phospho-SAPK/JNK (Thr-183/Tyr-185) antibody (Cell Signaling Technology, 9251S), β-actin (Cell Signaling Technology, 4970S), GAPDH Ab (Proteintech, 10494-1-AP).
Synthesis and Transfection of siRNA
The sequences of small interfering RNAs (siRNAs) used to specifically target human PTB mRNA have been described (41). The target sequences in human AGO1 and AGO2 for siRNAs were as follows: siGENOME SMARTpool siAGO1-1, 5′-GAG AAG AGG UGC UCA AGA A-3′; siGENOME SMARTpool siAGO1-2, 5′-GGA AAC AGU UCU ACA AUG G-3′; siGENOME SMARTpool siAGO1–3, 5′-GCU GUU ACC UCA CUG GAU A-3′; siGENOME SMARTpool siAGO1-4, 5′-GGA GUU ACU UUC AUA GCA U-3′; siGENOME SMARTpool siAGO2–1, 5′-GCA CGG AAG UCC AUC UGA A-3′; siGENOME SMARTpool siAGO2-2, 5′-GCA GGA CAA AGA UGU AUU A-3′; siGENOME SMARTpool siAGO2-3, 5′-GGG UCU GUG GUG AUA AAU A-3′; siGENOME SMARTpool siAGO2-4, 5′-GUA UGA GAA CCC AAU GUC A-3′. These siRNAs were synthesized by RIBOBIO (Guangzhou, China), and miR-124 mimics/inhibitors/negative control were purchased from GenePharma (Shanghai, China). Cells were transfected with small RNA oligonucleotides as previously described by our group (40).
Apoptosis Detection
Isolated primary memory CD4+ T cells were initially treated with or without rhIL-7 (25 ng/ml). Anti-CD95 or control IgG (Pharmingen) was added to cell cultures (1 μg/ml) 48 h later. After 24 h cells were labeled with FITC/phosphatidylethanolamine-conjugated annexin-V (Keygen) and analyzed by FCM.
Proliferation of CD4+ T Cells
Isolated primary CD4+ T cells were activated with 1 μg/ml anti-CD3 (Pharmingen). Cell divisions were detected using carboxyfluorescein diacetate succinimidyl ester labeling according to the manufacturer's instructions (Invitrogen). CD4+ T cells (0.5 × 106 cells/ml) were cultured in 96-well plates and treated with rhIL-7 for 48 h and then cultured with 1 μg/ml anti-CD95 or control IgG for 4 days as described previously (34). Cells were collected and analyzed by FCM.
Quantification of Integrated HIV-1 DNA
CD4+ T cells from HIV-1-infected individuals were isolated as described above and cultured in the presence of 10 μm azidothymidine (42). Cells were treated with or without 25 ng/ml rhIL-7 in combination with 1 μg/ml anti-CD95 or control IgG. Cells were lysed by TRIzol reagent (Invitrogen) followed by DNA extraction. Alu-gag PCR was performed to quantify integrated HIV-1 DNA copy numbers as previously described by others and our group (42, 43).
Measurement of HIV-1 Production
CD4+ T cells isolated from HIV-1-infected individuals receiving suppressive ART were cultured as described above. Cells were stimulated with 5 μg/ml anti-CD3, 25 ng/ml rhIL-7, and 1 μg/ml anti-CD95 or control IgG. Culture media was harvested after 7 days for viral RNA quantification. Viral RNA was extracted with TRIzol reagent (Invitrogen) and quantified using the Verso 1-step RT-qPCR kit (Thermo). Primers used for HIV-1 RNA quantification have been described previously by our group (44).
Statistical Analysis
Paired, two-tailed Student's t test was used to determine the significance of real-time PCR data or FCM data between differently treated sample groups. Wilcoxon matched-pairs signed rank test was used to determine the significance of data obtained from samples of HIV-1-infected individuals. Data were considered significant at p < 0.05.
RESULTS
IL-7 Increases CD95 Expression on Resting Memory CD4+ T Cells by Inducing Exon 6 Inclusion
IL-7 has been reported to promote survival of HIV-1-infected CD4+ cells (18). It can also elevate the expression of CD95 on CD4+ T cells during lymphopenic conditions such as HIV-1 infection (24). Therefore, the mechanism underlying this phenomenon warrants investigation in resting memory CD4+ T lymphocytes, the major reservoir for HIV-1. We first isolated CD4+ T cells from HIV-negative controls or HIV-1-infected individuals receiving suppressive ART and cultured them in the presence or absence of IL-7, and CD95 expression was analyzed. Cellular membrane-bound CD95 was significantly increased after treatment with IL-7 (Fig. 1A). However, the amount of CD95 mRNA did not change significantly (Fig. 1B), and Western blots revealed that CD95 protein in whole cell lysate did not change either (Fig. 1C). As transmembrane domain-encoded exon 6 inclusion of CD95 mRNA is reported to be increased during T cell activation (Fig. 1D) (45), it is possible that alternative splicing could also be involved in the induction of CD95 by IL-7. To validate this hypothesis, RT-PCR was then performed to identify possible changes in CD95 splicing pattern. Interestingly, after IL-7 treatment, alternative splicing of CD95 favored the cellular membrane-bound isoform over the soluble splice variant (FasΔTM) (Fig. 1E). Furthermore, CD95 levels in permeabilized or non-permeabilized T cells were analyzed. CD95 expression in IL-7-treated non-permeabilized memory CD4+ T cells was significantly elevated compared with non-treated cells (Fig. 1F, top panel). However, the expression of CD95 in the permeabilized cells (in the whole cell) remained relatively unchanged despite IL-7 treatment (Fig. 1F, bottom panel). These findings suggest that IL-7 increases CD95 expression on memory CD4+ T cells by inducing its exon 6 inclusion.
FIGURE 1.
IL-7 up-regulates CD95 expression on the surface of resting memory CD4+ T cells by promoting exon 6 inclusion. A, CD95 expression on memory CD4+ T cells from HIV-1-infected donors cultured for 2 days with or without IL-7 (25 ng/ml) was measured by FCM. MFI is shown with error bars representing S.D. calculated from six independent experiments. B, C, E, and F, memory CD4+ T cells from HIV-negative controls were isolated and cultured with or without 25 ng/ml IL-7 for 2 days. B, CD95 mRNA was measured by real-time PCR. Error bars represent S.D. calculated from three independent experiments. C, CD95 protein expression was measured by Western blot. A representative result out of eight independent experiments was shown. Values represent -fold changes of CD95 normalized against GAPDH and compared with control. D, a schematic map for two of splice variants of CD95 is shown. DD, death domain; TM, transmembrane domain. E, mRNAs of CD95 splice variants were measured by RT-PCR. Results of six independent experiments are shown. The column graph shows a statistical analysis of the percentage of mCD95/sCD95 (measured by relative density) after IL-7 treatment. mCD95, membrane-bound CD95; sCD95, soluble CD95. F, CD95 expression of permeabilized or non-permeabilized memory CD4+ T cells was measured by FCM. MFI is shown with error bars representing S.D. calculated from 10 independent experiments. A, C, and F, paired, two-tailed Student's t test: ***, p < 0.001. NS, non-significant.
IL-7 Modulates CD95 Splicing Regulator PTB
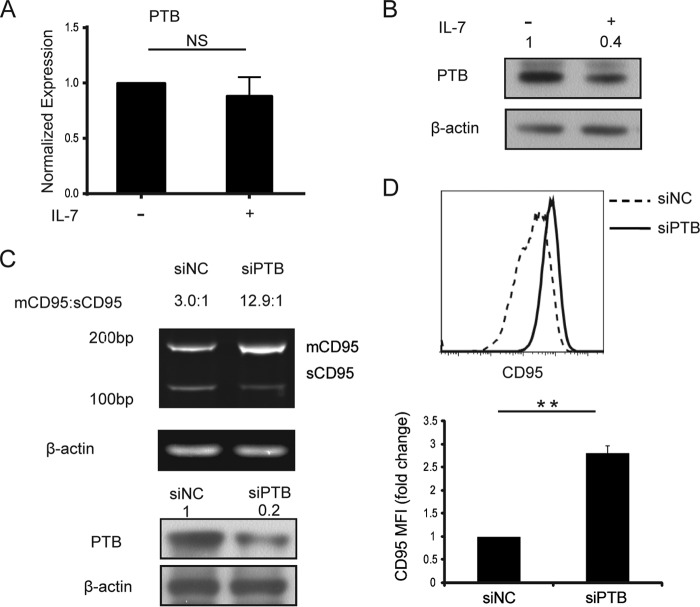
The polypyrimidine tract binding protein (PTB/PTBP1/hnRNP I) has been reported to promote CD95 exon 6 skipping by targeting the exon definition process (46). Therefore, we hypothesized that IL-7 could regulate PTB to increase expression of membrane-bound CD95. Real-time PCR was performed to measure PTB mRNA in IL-7-treated and non-treated memory CD4+ T cells, and PTB mRNA did not change significantly (Fig. 2A). However, Western blot analysis of PTB revealed a significant decrease after IL-7 treatment (Fig. 2B). To confirm the interaction between PTB and CD95, PTB gene silencing experiments were performed using the transfection of PTB-specific siRNAs (Fig. 2, C and D). CD95 in PTB-specific siRNA-transfected cells had similar splicing pattern changes as the IL-7-treated group (Figs. 1E and 2C). Flow cytometry analysis of membrane-bound CD95 expression in the PTB-silenced group also had significantly increased CD95 expression (Fig. 2D). Thus, these data support the hypothesis that IL-7 regulates CD95 splicing by depressing the expression of PTB.
FIGURE 2.
IL-7 down-regulates CD95 splicing regulator PTB. A and B, memory CD4+ T cells from HIV-negative controls were isolated and cultured with or without 25 ng/ml IL-7 for 2 days. A, cells were harvested for RNA extraction, and PTB mRNA was measured by real-time PCR. Error bars represent S.D. calculated from six independent experiments. B, cells were collected for Western blot analysis of PTB protein expression. A representative sample of five independent experiments is shown. Values represent -fold changes of PTB normalized against β-actin and compared with control. C and D, memory CD4+ T cells from HIV-negative controls transfected with PTB-specific siRNA or control siRNA were cultured with or without 25 ng/ml IL-7 for 2 days. C, top panel, mRNAs of CD95 splice variants were measured by RT-PCR. A representative sample of three independent experiments is shown. Bottom panel, silencing of PTB by PTB-specific siRNAs was confirmed by Western blot. mCD95, membrane-bound CD95; sCD95, soluble CD95. Values represent the ratios of the relative density of mCD95/sCD95. D, CD95 expression was measured by FCM. MFI is shown with error bars representing S.D. calculated from three independent experiments. NC, negative control. **, p < 0.01 in a paired t test. NS, non-significant.
IL-7 Up-regulates PTB-targeting miR-124
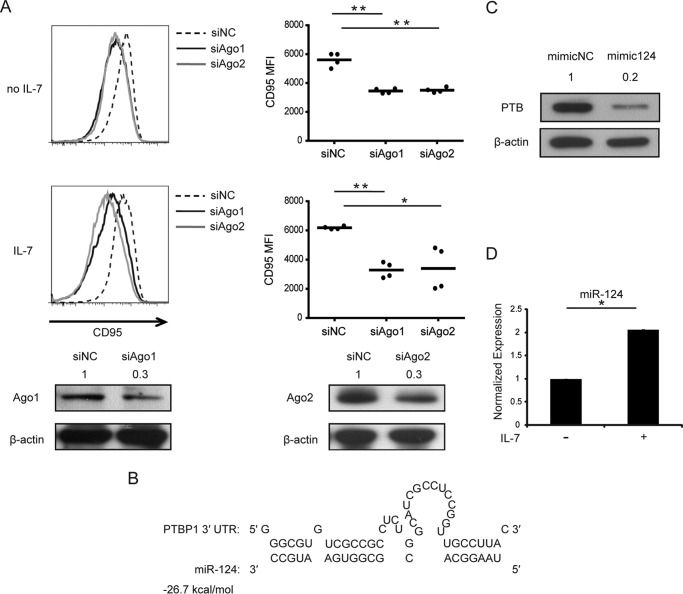
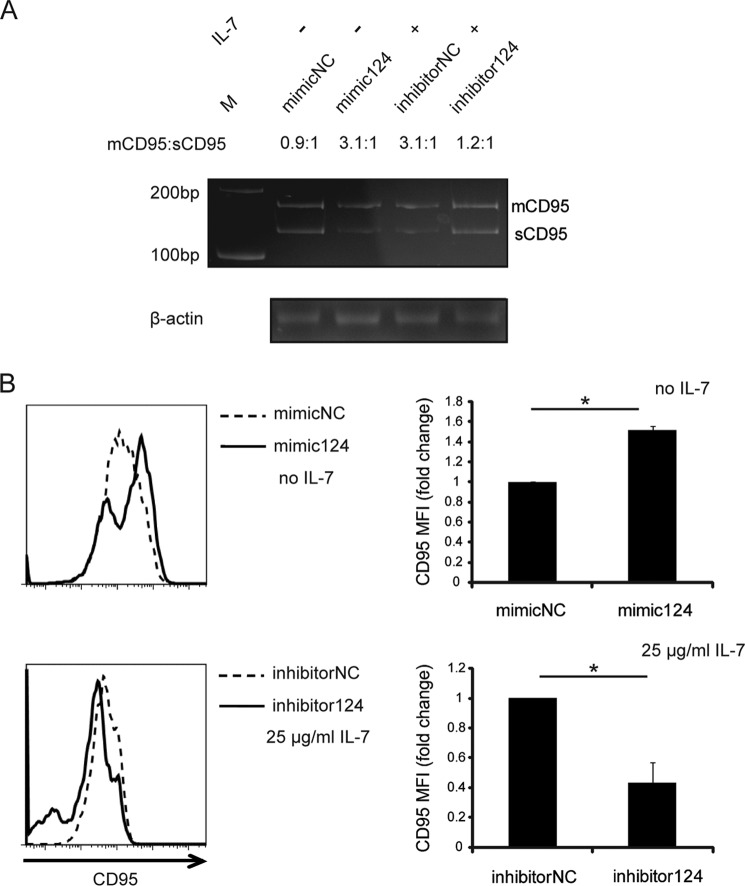
As noted above, PTB mRNA was only slightly decreased in IL-7-treated memory CD4+ T cells (Fig. 2A), but protein expression was significantly down-regulated, suggesting the possible involvement of miRNA. To confirm the participation of miRNA, AGO1 and AGO2 proteins, which play an important role in the miRNA pathway, were silenced using specific siRNAs before memory CD4+ T cells were stimulated with IL-7. Flow cytometric analysis indicated that CD95 expression decreased after silencing of AGO proteins, and the down-regulation of CD95 could not be rescued by IL-7 (Fig. 3A). Therefore, miRNAs, which target the 3′-UTR of PTB, were predicted using the RNAhybrid online tool (47). Several predicted miRNAs were analyzed (data not shown), and among them miR-124 was known to target PTB in the nervous system in mice (48). This was confirmed in human CD4+ T cells by Western blot (Fig. 3, B and C). After IL-7 treatment, miR-124 was significantly elevated (Fig. 3D). Transfection of miR-124 mimic increased CD95 exon 6 inclusion, and miR-124 inhibitor reversed the effect on the alternative splicing of CD95 (Fig. 4A). This phenomenon was also recapitulated by flow cytometric analysis of membrane CD95 expression on miR-124 mimic- or inhibitor-transfected memory CD4+ T cells (Fig. 4B). Collectively, these data suggested that IL-7 increases membrane-bound CD95 expression by up-regulating PTB-targeting miR-124.
FIGURE 3.
IL-7 up-regulates PTB-targeting miR-124. A, top panel, memory CD4+ T cells from HIV-negative controls were transfected with AGO1- and AGO2-specific siRNAs or control oligonucleotides and then cultured with or without the presence of 25 ng/ml IL-7 for 2 days. CD95 expression was measured by FCM. MFI is shown with error bars representing S.D. calculated from four independent experiments. Bottom panel, silencing of AGO1/2 by AGO1/2-specific siRNAs was confirmed by Western blot. B, predicted target site of miR-124 within the 3′-UTR of PTB is shown. C, memory CD4+ T cells from HIV-negative controls were transfected with miR-124 mimic or control oligonucleotides and harvested for Western blot of PTB protein expression. A representative sample of three independent experiments is shown. Values represent fold changes of PTB normalized against β-actin and values compared with control. D, memory CD4+ T cells from HIV-negative controls were isolated and cultured with or without 25 ng/ml IL-7 for 2 days. Cells were harvested for miR-124 quantification by stem-loop PCR. Error bars represent S.D. calculated from three independent experiments. NC, negative control. Paired, two-tailed Student's t test: *, p < 0.05; **, p < 0.01.
FIGURE 4.
miR-124 up-regulates CD95 expression on memory CD4+ T cells. A and B, memory CD4+ T cells from HIV-negative controls were transfected with miR-124 mimic, miR-124 inhibitor, or their respective control oligonucleotides. IL-7 was added to the cell cultures 12 h after miR-124 inhibitor transfection. A, the mRNAs of CD95 splice variants were measured by RT-PCR. A representative sample of three independent experiments is shown. mCD95, membrane-bound CD95; sCD95, soluble CD95. Values represent the ratios of the relative density of mCD95/sCD95. B, CD95 expression was measured by FCM. MFI is shown with error bars representing S.D. calculated from three independent experiments. NC, negative control. *, p < 0.05 in a paired t test.
IL-7 and CD95 Synergistically Promote Survival and Proliferation in Memory CD4+ T Cells
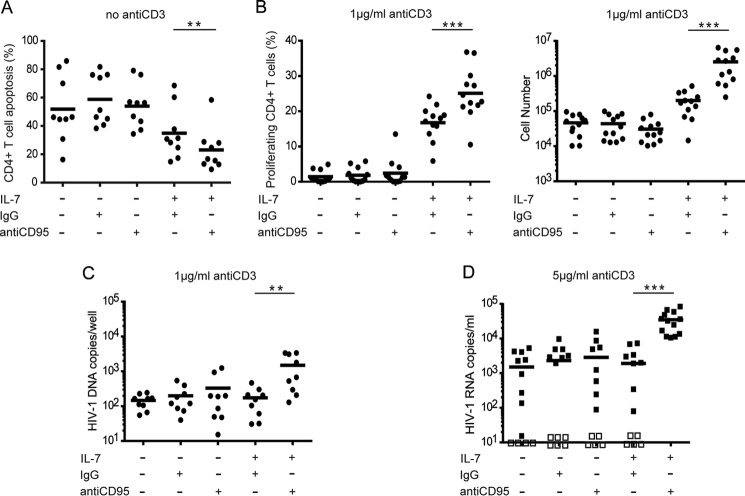
Peripheral T cells are reported to be primed for CD95-mediated apoptosis by high IL-7 doses and are also characterized by an increased sensitivity to CD95-mediated proliferation (34). To determine whether CD95 stimulation on resting CD4+ T cells leads to apoptosis or proliferation, we compared the sensitivity of resting or activated memory CD4+ T cells to CD95-mediated apoptosis or proliferation after IL-7 treatment. We found that IL-7 treatment results in different sensitivity to apoptotic or proliferative signals mediated by CD95 in memory CD4+ T cells. The percentage of apoptotic cells significantly decreased in resting memory CD4+ T cells in the presence of CD95 costimulating signals (Fig. 5A). However, after activation with a suboptimal dose of anti-CD3 mAb, the percentage of dividing cells significantly increased due to CD95 pathway activation (Fig. 5B). Thus, IL-7-treated memory CD4+ T cells have a decreased response to CD95-mediated apoptosis and an increased sensitivity to CD95-mediated proliferation.
FIGURE 5.
IL-7 and CD95 synergistically promote survival and proliferation in memory CD4+ T cells. A, B, C, and D, memory CD4+ T cells from HIV-negative controls were treated with or without 25 ng/ml IL-7 for 48 h. Cells were then stimulated with 1 μg/ml anti-CD95 or control IgG. Human anti-CD3 was added into culture along with anti-CD95 or control IgG in activated cultures (B). A, apoptosis of T cells was measured after 24 h of anti-CD95 activation using annexin-V staining. B. proliferation of T cells was measured at day 4 of anti-CD95 activation using carboxyfluorescein diacetate succinimidyl ester labeling. A and B consist of results from 12 independent experiments. C and D, protein levels of c-FLIP (C) and JNK/p-JNK (D) were detected by Western blot. A representative sample of three independent experiments is shown. Values in C and D represent -fold changes of c-FLIP (C) and JNK/p-JNK (D) normalized against GAPDH and compared with control. ***, p < 0.001 in a paired t test.
Possible Mechanism of IL-7-primed Proliferation with CD95 Signals
IL-7 is a pro-survival cytokine, and it can elevate anti-apoptotic proteins such as the inhibitor of apoptosis protein family as well as decrease the expression of pro-apoptotic proteins (1). Cellular FLICE-like inhibitory protein (c-FLIP/CFLAR) is well known to interact with the death domain of the CD95 receptor and prevent transduction of the apoptotic signal (49). IL-7 has been reported to not affect the expression of c-FLIP in recent thymic emigrants (50). Because IL-7R is low in recent thymic emigrants, the effect of IL-7 on c-FLIP in memory T cells warrants study. Western blots indicated that c-FLIPS (c-FLIPShort), splice variants of c-FLIP, was increased in memory CD4+ T cells treated with IL-7 and even more so in samples treated with both anti-CD3 antibody and IL-7 (Fig. 5C). The other splice variant of c-FLIP, c-FLIPL (c-FLIPLong) was only significantly enhanced in activated samples (Fig. 5C). These data showed that IL-7 inhibits CD95-mediated apoptosis by up-regulating anti-apoptotic protein c-FLIP.
Recently, the tumorigenic activity of CD95 has been suggested to be mediated by a pathway involving JNK (26, 51). To determine the possible signaling pathway of CD95-induced proliferation in IL-7-primed memory CD4+ T cells, the effect of IL-7 on JNK activation was measured. We did not observe any significant phosphorylation of JNK in memory CD4+ T cells treated with IL-7 alone, but a significant phosphorylation of JNK was observed in memory CD4+ T cells treated with both anti-CD3 antibody and IL-7 (Fig. 5D). These observations indicated that IL-7 can trigger CD95-mediated proliferation signal in activated CD4+ T cells but not resting T cells. This was consistent with the observation that resting T cells cannot be driven into cell replication cycle by IL-7 alone (50, 52).
IL-7 and CD95 Synergistically Promote Survival and Proliferation of HIV-1-latently Infected CD4+ T Cells in Vitro
To investigate the effect of IL-7 induced CD95 signaling on the HIV-1 reservoir, we measured apoptosis and proliferation of CD4+ T cells from HIV-1-infected individuals on suppressive ART. Data were consistent with those in HIV-negative controls and indicated that CD95 signaling could promote survival and proliferation in IL-7-treated CD4+ T cells from HIV-1-infected donors on ART (Fig. 6, A and B).
FIGURE 6.
IL-7 and CD95 synergistically promote the maintenance of HIV-1 reservoir in vitro. A, B, C, and D, CD4+ T cells from HIV-1-infected donors on suppressive ART were cultured in the presence of 10 μm azidothymidine (AZT) and treated with or without 25 ng/ml IL-7 for 48 h. Cells were then stimulated with 1 μg/ml anti-CD95 or control IgG. Human anti-CD3 were added into culture along with anti-CD95 or control IgG in activated cultures (B, C, and D). A, apoptosis of T cells was measured after 24 h of anti-CD95 activation using annexin-V staining. B, proliferation of T cells was measured at day 4 of anti-CD95 activation using carboxyfluorescein diacetate succinimidyl ester labeling (left) and cell counting (right). C. integrated HIV-1 DNA copy numbers per well after IL-7 and anti-CD95 treatment were quantified by Alu-gag PCR. D, HIV-1 RNA copy numbers per milliliter after IL-7 and anti-CD95 treatment were measured by real-time PCR. These data consist of results from different donors indicated in each graph. Empty squares in D represent negative results (<10 HIV-1 copies/ml). Wilcoxon matched-pairs signed rank test: **, p < 0.01; ***, p < 0.001.
We then measured the effect of IL-7 on the maintenance of the HIV-1 reservoir by quantifying copies of integrated HIV-1 DNA within CD4+ T cells before and after cytokine and anti-CD95 treatment in vitro (Fig. 6C). IL-7 and anti-CD95 treatment increased copy numbers of integrated HIV-1 DNA in CD4+ T cells, suggesting that CD4+ T cells harboring HIV-1-integrated DNA proliferated along with the whole CD4+ T cell population. HIV-1 RNA was also quantified to detect HIV-1 production after IL-7 and anti-CD95 treatment, and these proliferated CD4+ T cells could produce more HIV-1 particles into the medium (Fig. 6D). These results suggested that IL-7 and CD95 synergistically promote survival and proliferation of HIV-1-latently infected CD4+ T cells and, therefore, contribute to the maintenance of HIV-1 reservoir.
DISCUSSION
Here we confirm that IL-7 can elevate CD95 on the surface of memory CD4+ T cells by inducing the redistribution of CD95 from intracellular compartments to the cellular membrane (24). Our data indicate that IL-7 promotes CD95 elevation by regulating the alternative splicing process via suppression of splicing regulator PTB. Interestingly, our data reveal the involvement of miRNA in IL-7-mediated up-regulation of CD95. In the nervous system miR-124 has been reported to directly target PTB, which in turn leads to nonsense-mediated decay of its homolog nPTB (PTBP2/brPTB/PTBLP) and affects neuronal differentiation (48). We also found that miR-124 inhibits PTB expression and was up-regulated by IL-7 in memory CD4+ T cells (Fig. 3, C and D), leading to the inhibition of PTB and subsequently the inclusion of the transmembrane domain of CD95. In summary, we have described a signaling transduction pathway of IL-7-induced expression of CD95 molecule on the cellular membrane of memory CD4+ T lymphocytes.
We have also found that IL-7-primed resting memory CD4+ T cells were less susceptible to CD95-mediated apoptosis and that suboptimally activated cells were more sensitive to a CD95-mediated proliferation signal. These data are consistent with previous understandings that memory CD4+ T cells are CD95 type ΙΙ cells and are resistant to CD95-mediated apoptosis (31–33). Thus, memory CD4+ T cells can achieve homeostatic proliferation with the protection of IL-7 and acquire CD95 resistance toward bystander cell death during HIV-1 infection or antigen restimulation. Although IL-7 has been reported to induce CD95-mediated apoptosis in HIV-1-infected individuals, a recent study suggested that the pathway through which cells die during HIV-1 infection is decided by the permissivity status of the cell (53). Of CD4+ T cells encountered by HIV-1, 95% are non-permissive quiescent cells and undergo caspase-1-mediated pyroptosis instead of caspase-3-mediated apoptosis (53). Therefore, CD95-mediated apoptosis could only account for 5% of CD4+ T cell death during HIV-1 infection, and IL-7 induced CD95 expression may not play a major role in cell death.
Furthermore, IL-7 can up-regulate anti-apoptotic proteins to promote memory cell survival (1). As c-FLIP has been shown to play a role in regulating the outcome of CD95 stimulation (49, 54), investigating the effect of IL-7 on c-FLIP expression is a worthy pursuit. c-FLIP is composed of several alternatively spliced variants, among which are c-FLIPL (c-FLIPLong), c-FLIPS (c-FLIPShort), and c-FLIPR. All three variants can bind to the death domain, but only c-FLIPS and c-FLIPR could block apoptosis by inhibiting the recruitment and activation of procaspase-8 (55, 56). Our data showed that c-FLIPS was significantly increased in IL-7-treated memory CD4+ T cells regardless of cell activation status. However, investigations of c-FLIP regulation by interleukins did not generate consistent results (57). Ours is the first report to suggest that c-FLIPS is increased in IL-7-primed resting memory CD4+ T cells in vitro, suggesting that c-FLIP, which is not up-regulated in recent thymic emigrant T cells, might contribute to the mechanism of IL-7-mediated memory cell survival (50). Of course, IL-7 could also increase T cell survival by maintaining a favorable balance of anti-apoptotic proteins (e.g. Bcl-2, Mcl-1) over pro-apoptotic proteins (Bim, Bax, and Bad) (1). Therefore, the significance of c-FLIP in IL-7-mediated survival of lymphocytes still needs further investigation. In addition, previous work indicated that mitogen-activated protein kinase is involved in CD95-mediated proliferation pathway (26). Our data confirmed that phosphorylated JNK was increased in activated memory CD4+ T cells pretreated with IL-7. Thus, c-FLIP and JNK pathways may be the potential pathways for the non-apoptotic function of CD95 in memory CD4+ T lymphocytes. As a result, the function of FasL/CD95 up-regulated by IL-7 in memory CD4+ T lymphocytes is consistent with the canonical function of IL-7/IL-7R.
That IL-7 and CD95 could synergistically promote survival and proliferation of CD4+ T cells was observed in HIV-1-infected individuals on ART ex vivo. Furthermore, CD4+ T cells harboring integrated HIV-1 DNA could proliferate along with the entire CD4+ T cell subset, and these latently infected T cells could produce more HIV-1 particles when IL-7 and anti-CD95 were both present, suggesting that CD95 signaling could participate in the survival and expansion of HIV-1 reservoir after IL-7 priming (Fig. 6). Of note, our results suggest that proliferation stimulated by the CD95 pathway in IL-7-primed CD4+ T cells is a general effect, not specific for HIV-1-latently infected CD4+ T cells.
Our findings suggest a novel mechanism for immunopathogenesis of HIV-1. IL-7 maintains and expands the HIV-1 reservoir not only through its classical signal pathways but also through the anti-apoptotic and proliferation pathways mediated by CD95. Although IL-7 can contribute to restoring the CD4+ T cell pool in HIV-1-infected individuals, the therapeutic prospect of IL-7 has been questioned (58). The pro-survival and anti-apoptotic effects of IL-7 also benefit the preservation of HIV-1-latently infected cells and block the clearance of HIV-1 reservoir (18, 19). IL-7 cannot activate and eradicate the HIV-1 reservoir efficiently; rather, it elevates HIV-1-DNA in studied subjects, possibly due to IL-7-mediated T-cell stimulation and expansion (18, 19). HIV-1 DNA persistence is possibly achieved by homeostatic proliferation of latently infected cells (59). Our data support the argument that IL-7 is not efficient at reactivating HIV-1 latency. In contrast, given the abnormal increase of IL-7 during HIV-1 infection, the contribution of the FasL/CD95 signaling pathway to the maintenance of the HIV-1 reservoir warrants further investigation. Moreover, CD95 and other related molecules identified in our study may be possible therapeutic targets to eradicate the HIV-1 reservoir. Additional studies of these aspects may illuminate a novel mechanism(s) for maintaining and expanding the HIV-1 reservoir and identify a novel target(s) for activating HIV-1 latency and eradicating viral reservoirs.
Acknowledgments
We thank Dr. Yijun Zhang for advice during the preparation of this manuscript. We also appreciate the collaboration and help from the medical staff of Guangzhou Eighth People's Hospital and all the blood donors involved in this study.
This work was supported by grants from the National Special Research Program for Important Infectious Diseases (2013ZX10001004), Guangdong Innovative Research Team Program (2009010058), National Basic Research Program of China (973 Program) (2010CB912202), National Natural Science Foundation of China (30972620, 81101255, and 81471935), and Natural Science Foundation of Guangdong (9251008901000022).
- IL-7R
- IL-7 receptor
- PTB (PTBP1/hnRNP I)
- polypyrimidine tract binding protein
- ART
- antiretroviral therapy
- FasL
- CD95 ligand
- FCM
- flow cytometry
- MFI
- mean fluorescent intensity
- miRNA
- microRNA
- AGO
- Argonaute
- c-FLIP
- cellular FLICE-like inhibitory protein.
REFERENCES
- 1. Mackall C. L., Fry T. J., Gress R. E. (2011) Harnessing the biology of IL-7 for therapeutic application. Nat. Rev. Immunol. 11, 330–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kondrack R. M., Harbertson J., Tan J. T., McBreen M. E., Surh C. D., Bradley L. M. (2003) Interleukin 7 regulates the survival and generation of memory CD4 cells. J. Exp. Med. 198, 1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J., Huston G., Swain S. L. (2003) IL-7 promotes the transition of CD4 effectors to persistent memory cells. J. Exp. Med. 198, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schluns K. S., Kieper W. C., Jameson S. C., Lefrançois L. (2000) Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat. Immunol. 1, 426–432 [DOI] [PubMed] [Google Scholar]
- 5. Seddon B., Tomlinson P., Zamoyska R. (2003) Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat. Immunol. 4, 680–686 [DOI] [PubMed] [Google Scholar]
- 6. Tan J. T., Ernst B., Kieper W. C., LeRoy E., Sprent J., Surh C. D. (2002) Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J. Exp. Med. 195, 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swainson L., Verhoeyen E., Cosset F. L., Taylor N. (2006) IL-7R α gene expression is inversely correlated with cell cycle progression in IL-7-stimulated T lymphocytes. J. Immunol. 176, 6702–6708 [DOI] [PubMed] [Google Scholar]
- 8. Vranjkovic A., Crawley A. M., Gee K., Kumar A., Angel J. B. (2007) IL-7 decreases IL-7 receptor α (CD127) expression and induces the shedding of CD127 by human CD8+ T cells. Int. Immunol. 19, 1329–1339 [DOI] [PubMed] [Google Scholar]
- 9. Sasson S. C., Zaunders J. J., Zanetti G., King E. M., Merlin K. M., Smith D. E., Stanley K. K., Cooper D. A., Kelleher A. D. (2006) Increased plasma interleukin-7 level correlates with decreased CD127 and increased CD132 extracellular expression on T cell subsets in patients with HIV-1 infection. J. Infect Dis. 193, 505–514 [DOI] [PubMed] [Google Scholar]
- 10. Napolitano L. A., Grant R. M., Deeks S. G., Schmidt D., De Rosa S. C., Herzenberg L. A., Herndier B. G., Andersson J., McCune J. M. (2001) Increased production of IL-7 accompanies HIV-1-mediated T-cell depletion: implications for T-cell homeostasis. Nat. Med. 7, 73–79 [DOI] [PubMed] [Google Scholar]
- 11. Llano A., Barretina J., Gutiérrez A., Blanco J., Cabrera C., Clotet B., Esté J. A. (2001) Interleukin-7 in plasma correlates with CD4 T-cell depletion and may be associated with emergence of syncytium-inducing variants in human immunodeficiency virus type 1-positive individuals. J. Virol. 75, 10319–10325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koesters S. A., Alimonti J. B., Wachihi C., Matu L., Anzala O., Kimani J., Embree J. E., Plummer F. A., Fowke K. R. (2006) IL-7Rα expression on CD4+ T lymphocytes decreases with HIV disease progression and inversely correlates with immune activation. Eur. J. Immunol. 36, 336–344 [DOI] [PubMed] [Google Scholar]
- 13. Levy Y., Lacabaratz C., Weiss L., Viard J. P., Goujard C., Lelièvre J. D., Boué F., Molina J. M., Rouzioux C., Avettand-Fénoêl V., Croughs T., Beq S., Thiébaut R., Chêne G., Morre M., Delfraissy J. F. (2009) Enhanced T cell recovery in HIV-1-infected adults through IL-7 treatment. J. Clin. Invest. 119, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sereti I., Dunham R. M., Spritzler J., Aga E., Proschan M. A., Medvik K., Battaglia C. A., Landay A. L., Pahwa S., Fischl M. A., Asmuth D. M., Tenorio A. R., Altman J. D., Fox L., Moir S., Malaspina A., Morre M., Buffet R., Silvestri G., Lederman M. M. (2009) IL-7 administration drives T cell-cycle entry and expansion in HIV-1 infection. Blood 113, 6304–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rethi B., Fluur C., Atlas A., Krzyzowska M., Mowafi F., Grützmeier S., De Milito A., Bellocco R., Falk K. I., Rajnavölgyi E., Chiodi F. (2005) Loss of IL-7Rα is associated with CD4 T-cell depletion, high interleukin-7 levels, and CD28 down-regulation in HIV-infected patients. AIDS 19, 2077–2086 [DOI] [PubMed] [Google Scholar]
- 16. Wang F. X., Xu Y., Sullivan J., Souder E., Argyris E. G., Acheampong E. A., Fisher J., Sierra M., Thomson M. M., Najera R., Frank I., Kulkosky J., Pomerantz R. J., Nunnari G. (2005) IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J. Clin. Invest. 115, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scripture-Adams D. D., Brooks D. G., Korin Y. D., Zack J. A. (2002) Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J. Virol. 76, 13077–13082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vandergeeten C., Fromentin R., DaFonseca S., Lawani M. B., Sereti I., Lederman M. M., Ramgopal M., Routy J. P., Sékaly R. P., Chomont N. (2013) Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 121, 4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katlama C., Lambert-Niclot S., Assoumou L., Papagno L., Lecardonnel F., Tambussi G., Clotet B., Youle M., Costagliola D., Autran B., Group t. E.-S. (2013) Impact of Interleukin 7 and raltegravir plus maraviroc intensification on total HIV DNA reservoir: results from ERAMUNE 01. in 20th Conference on Retroviruses and Opportunistic Infections (CROI), Atlanta, GA, March 3, 2013, World Health Organization and CREATE [Google Scholar]
- 20. Cascino I., Fiucci G., Papoff G., Ruberti G. (1995) Three functional soluble forms of the human apoptosis-inducing Fas molecule are produced by alternative splicing. J. Immunol. 154, 2706–2713 [PubMed] [Google Scholar]
- 21. Cheng J., Zhou T., Liu C., Shapiro J. P., Brauer M. J., Kiefer M. C., Barr P. J., Mountz J. D. (1994) Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science 263, 1759–1762 [DOI] [PubMed] [Google Scholar]
- 22. Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K. J., Debatin K.-M., Krammer P. H., Peter M. E. (1998) Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17, 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Westendorp M. O., Frank R., Ochsenbauer C., Stricker K., Dhein J., Walczak H., Debatin K. M., Krammer P. H. (1995) Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature 375, 497–500 [DOI] [PubMed] [Google Scholar]
- 24. Fluur C., De Milito A., Fry T. J., Vivar N., Eidsmo L., Atlas A., Federici C., Matarrese P., Logozzi M., Rajnavölgyi E., Mackall C. L., Fais S., Chiodi F., Rethi B. (2007) Potential role for IL-7 in Fas-mediated T cell apoptosis during HIV infection. J. Immunol. 178, 5340–5350 [DOI] [PubMed] [Google Scholar]
- 25. Lelièvre J. D., Petit F., Arnoult D., Ameisen J. C., Estaquier J. (2005) Interleukin 7 increases human immunodeficiency virus type 1 LAI-mediated Fas-induced T-cell death. J. Virol. 79, 3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen L., Park S. M., Tumanov A. V., Hau A., Sawada K., Feig C., Turner J. R., Fu Y. X., Romero I. L., Lengyel E., Peter M. E. (2010) CD95 promotes tumour growth. Nature 465, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Desbarats J., Birge R. B., Mimouni-Rongy M., Weinstein D. E., Palerme J. S., Newell M. K. (2003) Fas engagement induces neurite growth through ERK activation and p35 up-regulation. Nat. Cell Biol. 5, 118–125 [DOI] [PubMed] [Google Scholar]
- 28. Corsini N. S., Sancho-Martinez I., Laudenklos S., Glagow D., Kumar S., Letellier E., Koch P., Teodorczyk M., Kleber S., Klussmann S., Wiestler B., Brüstle O., Mueller W., Gieffers C., Hill O., Thiemann M., Seedorf M., Gretz N., Sprengel R., Celikel T., Martin-Villalba A. (2009) The death receptor CD95 activates adult neural stem cells for working memory formation and brain repair. Cell Stem Cell 5, 178–190 [DOI] [PubMed] [Google Scholar]
- 29. Alderson M. R., Armitage R. J., Maraskovsky E., Tough T. W., Roux E., Schooley K., Ramsdell F., Lynch D. H. (1993) Fas transduces activation signals in normal human T lymphocytes. J. Exp. Med. 178, 2231–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Legembre P., Barnhart B. C., Zheng L., Vijayan S., Straus S. E., Puck J., Dale J. K., Lenardo M., Peter M. E. (2004) Induction of apoptosis and activation of NF-κB by CD95 require different signalling thresholds. EMBO Rep. 5, 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fas S. C., Baumann S., Krueger A., Frey C. R., Schulze-Bergkamen H., Brenner D., Stumpf C., Kappes K., Krammer P. H. (2006) In vitro generated human memory-like T cells are CD95 type II cells and resistant towards CD95-mediated apoptosis. Eur. J. Immunol. 36, 2894–2903 [DOI] [PubMed] [Google Scholar]
- 32. Inaba M., Kurasawa K., Mamura M., Kumano K., Saito Y., Iwamoto I. (1999) Primed T cells are more resistant to Fas-mediated activation-induced cell death than naive T cells. J. Immunol. 163, 1315–1320 [PubMed] [Google Scholar]
- 33. Desbarats J., Wade T., Wade W. F., Newell M. K. (1999) Dichotomy between naive and memory CD4(+) T cell responses to Fas engagement. Proc. Natl. Acad. Sci. U.S.A. 96, 8104–8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rethi B., Vivar N., Sammicheli S., Fluur C., Ruffin N., Atlas A., Rajnavolgyi E., Chiodi F. (2008) Priming of T cells to Fas-mediated proliferative signals by interleukin-7. Blood 112, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 35. Buzon M. J., Sun H., Li C., Shaw A., Seiss K., Ouyang Z., Martin-Gayo E., Leng J., Henrich T. J., Li J. Z., Pereyra F., Zurakowski R., Walker B. D., Rosenberg E. S., Yu X. G., Lichterfeld M. (2014) HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 20, 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tabler C. O., Lucera M. B., Haqqani A. A., McDonald D. J., Migueles S. A., Connors M., Tilton J. C. (2014) CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J. Virol. 88, 4976–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferenbach D. A., Haydon G. H., Rae F., Malcomson R. D., Harrison D. J. (1997) Alteration in mRNA levels of Fas splice variants in hepatitis C-infected liver. J. Pathol. 183, 299–304 [DOI] [PubMed] [Google Scholar]
- 38. Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 40. Liu C., Zhang X., Huang F., Yang B., Li J., Liu B., Luo H., Zhang P., Zhang H. (2012) APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J. Biol. Chem. 287, 29373–29383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wollerton M. C., Gooding C., Wagner E. J., Garcia-Blanco M. A., Smith C. W. (2004) Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 [DOI] [PubMed] [Google Scholar]
- 42. Huang J., Wang F., Argyris E., Chen K., Liang Z., Tian H., Huang W., Squires K., Verlinghieri G., Zhang H. (2007) Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 43. Liszewski M. K., Yu J. J., O'Doherty U. (2009) Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47, 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang H., Bagasra O., Niikura M., Poiesz B. J., Pomerantz R. J. (1994) Intravirion reverse transcripts in the peripheral blood plasma on human immunodeficiency virus type 1-infected individuals. J. Virol. 68, 7591–7597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grigoryev Y. A., Kurian S. M., Nakorchevskiy A. A., Burke J. P., Campbell D., Head S. R., Deng J., Kantor A. B., Yates J. R., 3rd, Salomon D. R. (2009) Genome-wide analysis of immune activation in human T and B cells reveals distinct classes of alternatively spliced genes. PLoS ONE 4, e7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Izquierdo J. M., Majós N., Bonnal S., Martínez C., Castelo R., Guigó R., Bilbao D., Valcárcel J. (2005) Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 19, 475–484 [DOI] [PubMed] [Google Scholar]
- 47. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007) The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rasper D. M., Vaillancourt J. P., Hadano S., Houtzager V. M., Seiden I., Keen S. L., Tawa P., Xanthoudakis S., Nasir J., Martindale D., Koop B. F., Peterson E. P., Thornberry N. A., Huang J., MacPherson D. P., Black S. C., Hornung F., Lenardo M. J., Hayden M. R., Roy S., Nicholson D. W. (1998) Cell death attenuation by “Usurpin,” a mammalian DED-caspase homologue that precludes caspase-8 recruitment and activation by the CD-95 (Fas, APO-1) receptor complex. Cell Death Differ. 5, 271–288 [DOI] [PubMed] [Google Scholar]
- 50. O'Neill R. M., Hassan J., Reen D. J. (2003) IL-7-regulated homeostatic maintenance of recent thymic emigrants in association with caspase-mediated cell proliferation and apoptotic cell death. J. Immunol. 170, 4524–4531 [DOI] [PubMed] [Google Scholar]
- 51. Curtin J. F., Cotter T. G. (2004) JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU 145 prostate carcinoma cells. J. Biol. Chem. 279, 17090–17100 [DOI] [PubMed] [Google Scholar]
- 52. Bosque A., Famiglietti M., Weyrich A. S., Goulston C., Planelles V. (2011) Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 7, e1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doitsh G., Galloway N. L., Geng X., Yang Z., Monroe K. M., Zepeda O., Hunt P. W., Hatano H., Sowinski S., Muñoz-Arias I., Greene W. C. (2014) Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scaffidi C., Schmitz I., Krammer P. H., Peter M. E. (1999) The role of c-FLIP in modulation of CD95-induced apoptosis. J. Biol. Chem. 274, 1541–1548 [DOI] [PubMed] [Google Scholar]
- 55. Golks A., Brenner D., Fritsch C., Krammer P. H., Lavrik I. N. (2005) c-FLIPR, a new regulator of death receptor-induced apoptosis. J. Biol. Chem. 280, 14507–14513 [DOI] [PubMed] [Google Scholar]
- 56. Schmitz I., Weyd H., Krueger A., Baumann S., Fas S. C., Krammer P. H., Kirchhoff S. (2004) Resistance of short term activated T cells to CD95-mediated apoptosis correlates with de novo protein synthesis of c-FLIPshort. J. Immunol. 172, 2194–2200 [DOI] [PubMed] [Google Scholar]
- 57. Budd R. C., Yeh W.-C., Tschopp J. (2006) cFLIP regulation of lymphocyte activation and development. Nat. Rev. Immunol. 6, 196–204 [DOI] [PubMed] [Google Scholar]
- 58. Rethi B., Vivar N., Sammicheli S., Chiodi F. (2009) Limited efficiency of endogenous interleukin-7 levels in T cell reconstitution during HIV-1 infection: will exogenous interleukin-7 therapy work? AIDS 23, 745–755 [DOI] [PubMed] [Google Scholar]
- 59. Wagner T. A., McKernan J. L., Tobin N. H., Tapia K. A., Mullins J. I., Frenkel L. M. (2013) An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J. Virol. 87, 1770–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]