Background: MITF is paramount for melanocyte development and melanoma pathogenesis.
Results: CRD-BP restricts the action of miR-340 by preventing its access to the mRNA of MITF, thereby establishing a novel mode for the regulation of MITF.
Conclusion: Regulation of MITF by CRD-BP contributes to the effects of CRD-BP on melanoma survival and progression.
Significance: CRD-BP is a potential target for the prevention and treatment of melanoma.
Keywords: Melanogenesis, Melanoma, MicroRNA (miRNA), mRNA, mRNA Decay, Ribonuclear Protein (RNP), MITF, CRD-BP, IGF2BP1, Melanoma, miRNA, Alternative Cleavage and Polyadenylation
Abstract
Alternative cleavage and polyadenylation generates multiple transcript variants producing mRNA isoforms with different length 3′-UTRs. Alternative cleavage and polyadenylation enables differential post-transcriptional regulation via the availability of different cis-acting elements in 3′-UTRs. Microphthalmia-associated transcription factor (MITF) is a master regulator of melanocyte development and melanogenesis. This central transcription factor is also implicated in melanoma development. Here, we show that melanoma cells favor the expression of MITF mRNA with a shorter 3′-UTR. We also establish that this isoform is regulated by a micro RNA (miRNA/miR), miR-340. miR-340 interacts with two of its target sites on the MITF 3′-UTR, causing mRNA degradation as well as decreased expression and activity of MITF. Conversely, the RNA-binding protein, coding region determinant-binding protein, was shown to be highly expressed in melanoma, directly binds to the 3′-UTR of MITF mRNA, and prevents the binding of miR-340 to its target sites, resulting in the stabilization of MITF transcripts, elevated expression, and transcriptional activity of MITF. This regulatory interplay between RNA-binding protein and miRNA highlights an important mechanism for the regulation of MITF in melanocytes and malignant melanomas.
Introduction
MITF2 is a basic helix-loop-helix leucine zipper dimeric transcription factor belonging to the MYC superfamily of proteins (1). MITF forms dimers and binds to specific sequence motifs present in the promoter region of its target genes to activate their transcription. Various studies documented the role of MITF in the induction of genes required for normal melanocyte development as well as melanin formation. MITF regulates the transcription of three major pigmentation enzymes, TYR (tyrosinase), TYRP-1 (tyrosine-related protein-1), and DCT (dopachrome-tautomerase) (2–4). It is also an amplified oncogene in a subset of melanomas and was reported to regulate a distinct set of target genes, which in turn are responsible for a neoplasia-related phenotype in these cancers. Primarily, this action of MITF in melanoma is mediated by increasing cell proliferation and triggering cell cycle progression. TBX-2 (T-box transcription factor 2) is one such target that suppresses senescence via down-regulation of p21 (5, 6). CDK2 is another target, regulation of which modulates cell cycle progression required for melanoma clonogenic growth (7). MITF was also reported to inhibit apoptosis and thus promote oncogenesis by increasing the expression of its direct target, the anti-apoptotic factor BCL2 (8). Furthermore, MITF contributes toward the metastasis of melanoma through transcriptional activation of the c-MET proto-oncogene (9). Therefore, understanding the mechanisms controlling the expression and function of MITF is extremely important because it will identify key components for melanocyte development and may uncover novel targets for the treatment and/or prevention of melanoma.
Previous research demonstrated that regulation of MITF expression occurs at multiple levels. Transcriptional regulation of MITF expression takes place through the MITF-M promoter via Wnt (10–13) and cAMP-CREB (14) pathways and also PAX3 (15). There is also evidence demonstrating that post-translational regulation of MITF affects the availability of functional protein. Specifically, MAPK components phosphorylate MITF protein at Ser-73, resulting in ubiquitination and subsequent degradation of the protein (16, 17). On the other hand, phosphorylation of the Ser-298 residue by GSK3β is believed to increase the DNA binding activity of MITF (18). However, apart from transcriptional and post-translational regulation, another important mode of regulating gene expression is to manipulate the stability of mature mRNA. Recent reports indicate MITF is a target of several miRNAs, including the miR-96/183/182 cluster (19) and miR-137 (20), suggesting the importance of a post-transcriptional regulatory step in the expression of MITF.
miRNAs are small ∼22-nucleotide non-coding RNAs that are an important class of gene regulators. They are present in both plant and animal genomes, and after being transcribed and processed, the final form of mature miRNAs bind to sites on target mRNAs typically in the 3′-UTR and may also bind to sites in the coding region (21). Additional protein components participate in the process of forming the miRNA-induced silencing complex, ultimately leading to translational repression and/or degradation of the target mRNA.
APA generates multiple transcript variants of a particular gene, creating mRNA isoforms with different 3′-UTR lengths, depending on the position of APA signals. The exclusion of large 3′-UTR regions allows those mRNA isoforms to escape miRNA-dependent regulation. Recently, it was observed in fast proliferating cells that mRNA isoforms with shorter 3′-UTRs are favored (22). This preference helps oncogenes to avoid regulation that would otherwise interfere with cell cycle progression and, ultimately, cellular transformation (22, 23). There are several different isoforms of MITF-M mRNA with varying 3′-UTR lengths reported (19, 20). In this study, we investigated the expression pattern of three such mRNA isoforms in melanoma cell lines and normal human melanocytes. We found that MITF mRNAs with a short 3′-UTR are more abundant in melanoma cell lines, and, additionally, this isoform undergoes miR-340-mediated regulation, leading to the destabilization of MITF mRNA. We also elucidated the role of an RBP, CRD-BP (IMP-1, IGF2BP1), in the regulation of MITF mRNA. This protein was previously found to attenuate miRNA-dependent degradation of several mRNAs (21) and is also overexpressed in melanoma (24). Further, CRD-BP is involved in the regulation of metastatic melanoma cell proliferation and invasion by hypoxia (25), and its inhibition sensitizes melanoma cells to chemotherapeutic agents (26). Our results here show that CRD-BP restricts the action of miR-340 by preventing its access to MITF mRNA, thereby proposing a novel mode of regulation for MITF.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture, and Transfection Conditions
Normal human melanocytes (NHMs) were maintained in Ham's F-10 medium (Mediatech Inc., Manassas, VA) supplemented with human melanocyte growth supplement (Cascade Biologics Inc., Portland, OR), 5% fetal bovine serum (FBS), and 1% antibiotic-antimycotic solution, containing penicillin, streptomycin, and amphotericin B (PSM). 451Lu cells were maintained in minimal essential medium (Invitrogen) supplemented with 1% sodium pyruvate, 1% non-essential amino acids, 5% FBS, and 1% PSM. The melanoma cell lines 928 mel, 1011 mel, and 1242 mel were kindly provided by Dr. P. Robbins (Center for Cancer Research, NCI, National Institutes of Health, Bethesda, MD) and were maintained in DMEM (Invitrogen) supplemented with 10% bovine calf serum and 1% PSM. Colorectal cancer cell lines HCT116, RKO, and DLD1 along with corresponding exon 5-disrupted Dicer cell lines were kindly provided by Drs. K. Kinzler and B. Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD) (27). 293T cells were obtained from ATCC (Manassas, VA). Colorectal cancer and 293T cell lines were cultured in DMEM (Invitrogen) supplemented with 10% FBS and 1% PSM. NHMs were electroporated using the AMAXA NucleofectorTM (Lonza, Switzerland) according to the manufacturer's protocol. Transfections of all other cells were performed using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen).
Expression Vectors
Full-length MITF cDNA was purchased from Open Biosystems Inc. (Huntsville, AL; clone ID 6066096/accession number BC065243/cloned in pCMV-SPORT6) and amplified using fragment 1 forward and 3′-UTR reverse primers (Table 1). The PCR product was then cloned into the pBI-G vector (Clontech), digested with NotI and SalI, and end-filled with Klenow enzyme (New England Biolabs, Ipswich, MA) and the clone with the correct orientation was selected. Three fragments of the MITF coding region (nt 1–421, 422–841, and 842–1260), the full-length cDNA (nt 1–1822), and 3′-UTR (nt 1261–1822) were subcloned into pcDNA3.1. The 3′-UTR of MITF cDNA was PCR-amplified and cloned into the pBI-GL vector (Clontech) just after the stop codon. To obtain the deletion mutants for miR-340 sites in pBI-G or pBI-GL constructs, we performed blunt end ligation of PCR products and treated them with DpnI as recommended in the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
TABLE 1.
oligonucleotide sequences for plasmid contruction

Plasmids for CRD-BP expression or the production CRD-BP shRNA were characterized previously (28). The pcDNA5-CMV-d2eGFP vector and control sponge-CXCR4 construct were kindly provided by Dr. P. Sharp (MIT, Cambridge, MA). The constructs of sponge-340 and sponge-584c-3p were designed as described (29). Separate oligonucleotides with seven bulged binding sites for miR-340 and miR-584c-3p containing 4-nt spacer sequences between them (Table 1) were annealed, gel-purified, and cloned into the pcDNA5-CMV-d2eGFP vector and linearized with XhoI and ApaI (New England Biolabs). Plasmids for miR-340 expression, pCMV-MIR and pCMV-MIR340, were purchased from Origene (Rockville, MD).
RNA Isolation and Quantitative Real-time PCR
Total RNA from cells was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH). For quantitative RT-PCRs (qRT-PCRs), total RNA was treated with 2 units of DNase I (Promega, Madison, WI). Reverse transcription was performed using the Advantage RT-for-PCR kit (Clontech), and qRT-PCR was performed with Power SYBR Green PCR master mix (Invitrogen), according to the manufacturer's recommendations. Reactions were performed on an ABI Prism 7000 machine using ABI Prism 7000 SDS software (Invitrogen). Sequences of primers used for qRT-PCR are presented in Table 2.
TABLE 2.
Primer sequences for qRT-PCR
| Name of the oligonucleotide | Sequence (5′ → 3′) |
|---|---|
| GAPDH FWD | ATGGTTGCCACTGGGGATCT |
| GAPDH REV | TGCCAAAGCCTAGGGGAAGA |
| MITF FWD | CCAGGCATGAACACACATTC |
| MITF REV | GCAGACCTTGGTTTCCATAAAG |
| Long 3′-UTR Detection FWD | CCAAGAGGCAGTGGTTTGGG |
| Long 3′-UTR Detection REV | AACCAAATGCTTTAATGAGGCTATC |
| Medium 3′-UTR Detection FWD | GAAAACCGAACTGGGCATATTTC |
| Medium 3′-UTR Detection REV | GGTTATCAATCTCCAAGAATATTGC |
| Short 3′-UTR Detection FWD | CGAATCCTCCCTGCACTGC |
| Short 3′-UTR Detection REV | ACAGAATACATATTTCTTTAAATAG |
| Stem-loop RT primer for miR-340 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAATCAGTCT |
| Forward primer for miR-340: | GCGGCGGTTATAAAGCAATG |
| Universal reverse primer | GCGGGTGCAGGGTCCGAGGT |
Stem-Loop Quantitative Real-time PCR
The expression of mature miR-340 was detected using a two-step process. First, using total RNA isolated by TRI reagent, the stem-loop reverse transcription (RT) primer for miR-340 (designed according to Ref. 30) was hybridized to the miRNA molecule by incubating at 16 °C for 30 min and then reverse transcribed for 30 min at 42 °C using an Advantage RT-for-PCR kit (Clontech). After heat-inactivating the reverse transcriptase at 95 °C for 5 min, one-tenth of the RT product was used for either end point PCR or qRT-PCR with Power SYBR Green PCR master mix (Invitrogen) using the miRNA-340-specific forward and universal reverse primers (Table 2).
Droplet Digital PCR
To quantify miR-340 RNAs at high resolution, droplet digital PCR (ddPCR) was performed as described elsewhere (31) on the QX100 ddPCR system (Bio-Rad), using TaqMan FAM amidite-labeled probes for hsa-miR-340-5p and RNU6B as an endogenous control (Invitrogen). 20 μl reactions were assembled with cDNA, probes, and ddPCR supermix for probes (no dUTP) (Bio-Rad). Droplets were generated using the manufacturer's suggested protocol using the QX100 droplet generator machine, droplet generator cartridges, gaskets, and droplet generation oil for probes (Bio-Rad). Generated droplets were placed into a twin.tec PCR 96-well plate (Eppendorf, Hauppauge, NY). Plates were sealed using pierceable foil heat seal and a PX1 PCR plate sealer (Bio-Rad). A PCR under the following conditions was performed on a Mastercycler gradient thermal cycler (Eppendorf): 5 min at 95 °C, 30 s at 95 °C, 1 min at 60 °C for 40 cycles with a ramp speed of 2 °C/s, 98 °C for 10 min, and hold at 12 °C overnight. FAM amidite-generated fluorescence in the droplets was evaluated using the QX100 droplet reader machine and QuantaSoft software version 1.3.2.0 (Bio-Rad). ddPCR data presented are representative of absolute copies of transcripts in reaction samples.
mRNA Degradation in Vivo
To investigate the stability of mRNA transcripts, we used the Tet-Off gene expression system (Clontech) as described elsewhere (28). Melanoma, colorectal cell lines, or 293T cells were transfected with 4–6 μg of Tet-Off and 2–3 μg of pBI-G response plasmid expressing the mRNA transcripts of interest. Transcription was stopped by adding doxycycline (1 mg/ml) into media 48 h after co-transfection. Cells were harvested at various time points after treatment, and total RNA was extracted as described above. mRNA levels were analyzed using qRT-PCR.
Antibodies and Immunoprecipitation Techniques
Mouse anti-FLAG M2 antibody (Sigma-Aldrich), anti-β-actin, and anti-MITF (Santa Cruz Biotechnology, Inc., Dallas, TX) primary antibodies and secondary antibodies conjugated with horseradish peroxidase (Chemicon, Billerica, MA) were all purchased. Immunoblotting procedures are described elsewhere (32).
Protein Purification
To obtain whole-cell lysates for Western blot analysis, cells were lysed using denaturing radioimmune precipitation buffer containing PBS (pH 7.4), 0.5% sodium deoxycholate, 0.1% SDS, 1% (v/v) Nonidet P-40, 100 mm sodium orthovanadate, and protease inhibitor mixture (Sigma-Aldrich). Protein extracts for UV cross-linking reactions were made in non-denaturing lysis buffer (10 mm HEPES (pH 7.6), 3 mm MgCl2, 40 mm KCl, 2 mm DTT, 5% (v/v) glycerol, 0.5% (v/v) IGEPAL, and protease inhibitor mixture).
Preparation of RNA Substrates for UV Cross-link Analysis
Plasmids containing full-length MITF mRNA or fragments of its coding region under the control of the T7 promoter were linearized with XbaI (New England Biolabs), purified with phenol-chloroform, and used for in vitro transcription using [32P]UTP RNA and the Riboprobe in vitro transcription system (Promega). Radiolabeled RNA substrates were gel-purified before use in UV cross-link reactions.
UV Cross-link Reaction
Analysis of RNA-protein complexes was performed as described before (28). Briefly, protein extracts (20 μg) and internally labeled RNA (1.5–2 × 106 cpm) were incubated in 20 μl of reaction buffer (non-denaturing lysis buffer without protease inhibitors) for 30 min at room temperature. After incubation, RNA-protein complexes were cross-linked with a 30 min exposure to 254-nm UV light, treated with RNase A and RNase T1, incubated with anti-FLAG antibodies for 6 h, and, last, incubated with M2-agarose beads overnight. Immunoprecipitates were washed six times in lysis buffer, boiled, and separated using a 10% SDS-polyacrylamide gel. Last, the gel was dried and exposed to x-ray film for 3–14 days.
Luciferase Reporter Assay
Melanoma cells were transfected with the previously described shRNA constructs (28) or miR-sponge constructs, pSV40 β-gal and pHTRPL4 plasmid (33, 34), or p-BI-GL-MITFwt or p-BI-GL-MITFδmiR-340 plasmids. 48 h after transfection, luciferase activity was measured using the luciferase reporter assay according to the manufacturer's protocol (Promega).
Senescence-associated β-Galactosidase Staining
Cells were fixed for 20 min using a solution of 20% formaldehyde and 2% glutaraldehyde, washed with PBS, and then stained with X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution overnight at 37 °C. Cells stained blue were counted under a microscope (20×), and the percentage of stained cells was estimated.
RESULTS
Melanoma Cell Lines Preferentially Express MITF mRNA with a Short 3′-UTR
As discussed earlier, several full-length cDNA isoforms of MITF-M with the same coding sequence exist, but the isoforms vary in their 3′-UTR length. Separate previous reports demonstrate that MITF mRNA is regulated by two different miRNAs; however, the studies used two different MITF mRNA isoforms. The study by Bemis LT et al. (20) showed a functional miR-137 binding site in the 3′-UTR using a full-length cDNA containing 1,143 bp of the 3′-UTR sequence (termed hereafter as the medium 3′-UTR) (Fig. 1A). In a different study, Segura et al. (19) used the full-length cDNA with a 3′-UTR length of ∼3 kb (termed the long 3′-UTR) in order to find a functional miR-182 site (Fig. 1A). We obtained the full-length MITF cDNA clone having a 3′-UTR ∼0.57 kb in length, which we termed the MITF mRNA with a short 3′-UTR (Fig. 1A). The finding of one mRNA with different 3′-UTRs being regulated by different miRNAs led us to investigate the abundance of the various MITF mRNA isoforms in several melanoma cell lines. We designed primers unique to the short, medium, and long 3′-UTRs and, using qRT-PCR, evaluated the relative abundance of the distinct mRNA isoforms in four different melanoma cell lines versus NHMs (Fig. 1B). The results showed that in NHMs, the levels of all three isoforms were comparable. But in all four melanoma cell lines tested, the relative proportion of MITF mRNA with a short 3′-UTR predominated. This suggested that fast proliferating melanoma cells preferentially express MITF mRNA with a shorter 3′-UTR.
FIGURE 1.
MITF mRNA with a short 3′-UTR is more abundant in melanoma cell lines. A, graphical representation of different MITF mRNA isoforms with varying 3′-UTR lengths. The approximate positions of reported miRNA target sites are shown on long and medium 3′-UTRs. B, expression of MITF mRNA isoforms containing various 3′-UTR lengths evaluated by qRT-PCR using primers specific for the long, medium, and short 3′-UTR.
miRNA Regulates the Short 3′-UTR MITF mRNA Isoform
As discussed before, miRNAs act on MITF mRNAs with medium and long 3′-UTRs. Because we found the mRNA isoform with the short 3′-UTR most prevalent in melanomas, we investigated whether this isoform is also being regulated by miRNAs. We measured the half-life of the short 3′-UTR MITF mRNA in cells defective in Dicer1 (DicerEx5/Ex5) function and, thus, deficient in miRNA maturation (27). In all three Dicer1Ex5/Ex5 hypomorphic cell lines examined, the half-life of short 3′-UTR MITF mRNA was 2–3-fold higher compared with their normal counterpart (DicerWT) where functional miRNAs were present (Fig. 2 (A–C) and Table 3). This indicated that miRNAs also regulate the MITF isoform with a short 3′-UTR. Next, we used a bioinformatics approach to determine possible miRNA binding sites in this region. Two miRNAs, miR-340 and miR-548c-3p, with sites located in the short 3′-UTR of MITF mRNA were selected for further investigation (Fig. 2D). To inhibit the function of these miRNAs in melanoma cells, we used miR-sponge constructs as described previously (29) for both miR-340 and miR-548c-3p. Our results show that the amount of short 3′-UTR MITF mRNA increased significantly only when the function of miR-340 was inhibited (Fig. 2E). Moreover, inhibition of miR-340 function in 451Lu cells also increased MITF protein levels (Fig. 2F). These data demonstrate that MITF mRNA with a short 3′-UTR is regulated by miR-340.
FIGURE 2.
The abundant short 3′-UTR MITF mRNA is also regulated by miRNA. A, Dicerwt and DicerEx5/Ex5 HCT116 cells were co-transfected with Tet-Off plasmid and p-BI-G-MITF plasmid with a short 3′-UTR. Transcription was stopped by treatment with doxycycline for the indicated durations. The stability of MITF transcripts was analyzed by measuring MITF mRNA levels with qRT-PCR and normalized to GAPDH expression. B, p-BI-G-MITF plasmid with short 3′-UTR was expressed in DLD1 cells, Dicerwt and DicerEx5/Ex5, under the control of the Tet-Off system. The stability of MITF mRNA was analyzed as in A. C, the stability of MITF transcripts with short 3′-UTR expressed in RKOwt and RKO DicerEx5/Ex5 cells was analyzed as in A. All results are representative of three separate experiments and expressed as the mean ± S.D. (error bars). The average half-lives of mRNAs are presented in Table 3. D, sequence of the short 3′-UTR of MITF showing binding sites for miR-340 (in boldface type) and miR-548c-3p (underlined). E, the levels of endogenous MITF mRNA in 451Lu cells, transfected with the indicated miR-sponge constructs, were estimated by qRT-PCR after normalization to GAPDH expression. Results are representative of three separate experiments and presented as a percentage of control (SP-CXCR4) as the mean ± S.D. (error bars). F, immunoblot analysis of MITF expression in 451Lu cells transfected with indicated miR-sponge constructs (top panel). The lower panel shows β-actin expression.
TABLE 3.
The average half-lives of mRNA
The half-life of mRNA (t½) was calculated with the equation, t½ = (t × log 2)/(log AmtE/AmtE), where t½ is the half-life of mRNA, t is elapsed time after the addition of doxycycline, AmtB is the average beginning amount of mRNA, and AmtE is the average ending amount of mRNA.
| Figure | Group | Average half-life |
|---|---|---|
| h | ||
| 2A | WT | 2.5 |
| 2A | Ex5 | 7.1 |
| 2B | WT | 2.7 |
| 2B | Ex5 | 5.6 |
| 2C | WT | 2.6 |
| 2C | Ex5 | 5.6 |
| 3A | Sponge CXCR4 | 3.2 |
| 3A | Sponge-340 | 6.6 |
| 3B | Sponge CXCR4 | 2.4 |
| 3B | Sponge 340 | 6.8 |
| 3D | WT | 2.8 |
| 3D | 340 mutant | 5.1 |
| 4E | pcDNA | 2.8 |
| 4E | CRD-BP | 14 |
| 5A | Control | 2.5 |
| 5A | Sponge-340 | 5.4 |
| 5A | CRD-BP | 5.4 |
| 5A | CRD-BP + Sponge-340 | 5.3 |
| 5E | Control | 6.4 |
| 5E | Sponge-340 | 7.8 |
| 5E | CRD-BP | 6.8 |
| 5E | CRD-BP + Sponge-340 | 7.6 |
miR-340 Destabilizes MITF mRNA
After the initial observations regarding miR-340 involvement in the regulation of MITF mRNA, we set out to investigate whether miR-340 regulates the turnover of MITF mRNA. The result shows a 2-fold increase in mRNA half-life upon miR-340 inhibition (Fig. 3A and Table 3), indicating that miR-340 is responsible for the destabilization of MITF mRNA. We further tested the efficacy of miR-340 action using a reporter construct. We cloned the short 3′-UTR of MITF mRNA just after the luciferase coding region into pBI-GL (Clontech). Inhibition of miR-340 function in melanoma cell lines significantly increased the half-life of the chimeric RNA as well as luciferase enzyme activity (Fig. 3, B and C and Table 3). This finding suggested that the short 3′-UTR of MITF mRNA was sufficient for miR-340-mediated inhibition. Next, both sites of miR-340 were deleted from the full-length MITF mRNA as well as from the reporter construct to generate corresponding mutant versions. The half-life of the deleted full-length MITF mRNA increased significantly (Fig. 3D and Table 3). The luciferase activity of the mutated chimeric mRNA was also increased about 2.5-fold, comparable with the outcome when miR-340 function was inhibited (Fig. 3C). On the other hand, miR-340 inhibition had no additional effect on the luciferase activity of mutant mRNA (Fig. 3C). Together these data suggest that miR-340 acts through its target sites on the 3′-UTR of MITF mRNA, leading to its destabilization.
FIGURE 3.
MITF mRNA is a target of miR-340. A, 451Lu cells were co-transfected with Tet-Off plasmid, p-BI-G-MITF plasmid with short 3′-UTR, and the indicated miR-sponge construct. Transcription was stopped by treatment with doxycycline for the indicated durations. The stability of MITF transcripts was analyzed by measuring MITF mRNA levels with qRT-PCR, normalized to GAPDH expression. B, 451Lu cells were co-transfected with Tet-Off plasmid, p-BI-GL-MITF-Short 3′-UTR, and the indicated miR-sponge construct. The turnover of chimeric luciferase-MITF-3′-UTR transcripts was analyzed as in A. C, 451Lu cells were co-transfected with β-gal-expressing plasmid, Tet-Off plasmid, p-BI-GL-MITFwt, or p-BI-GL-MITFδmiR-340 and the indicated miR-sponge construct. After 24 h, luciferase activity was measured. Values represent luciferase activity normalized to β-gal. D, 451Lu cells were co-transfected with Tet-Off plasmid, p-BI-G-MITFwt, or p-BI-G-MITFδmiR-340. The stability of MITF transcripts was analyzed as in A. All results are representative of three separate experiments and are expressed as the mean ± S.D. (error bars). The average half-lives of mRNAs are presented in Table 3.
CRD-BP Stabilizes MITF mRNA
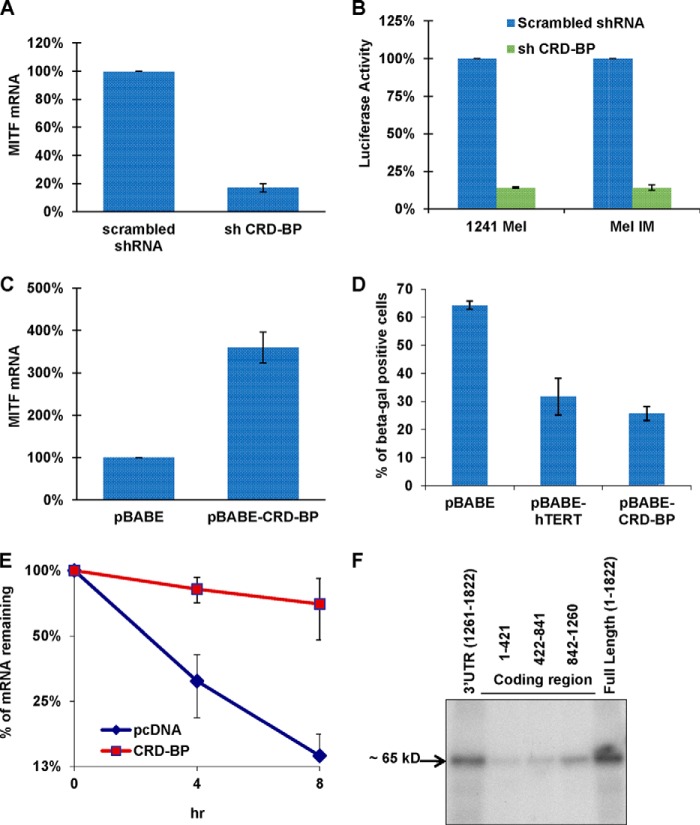
CRD-BP is a multifunctional mRNA-binding protein that modulates the stability, localization, and translation of several RNAs (c-myc, IGF-II, H19, CD4, MDR-1, Gli1, etc.) (35–42). As discussed earlier, previous research shows that CRD-BP protects another mRNA, βTrCP1, from miRNA-mediated destabilization (21). Moreover, CRD-BP is overexpressed in melanoma cell lines and human melanoma samples, thus contributing toward oncogenesis (24). Here we show that knockdown of CRD-BP in 451Lu cells decreased the level of endogenous MITF mRNA by more than 5-fold (Fig. 4A). Further, when CRD-BP was knocked down, MITF-dependent luciferase activity was drastically reduced (Fig. 4B). On the other hand, the levels of endogenous MITF mRNA were increased 3-fold when CRD-BP was overexpressed in NHMs (Fig. 4C). Overexpression of CRD-BP in NHMs also resulted in the significant inhibition of senescence, consistent with the function of MITF during cell cycle progression in melanocytes (Fig. 4D). Interestingly, the effect of CRD-BP on melanocyte senescence was similar to the effect of a powerful senescence inhibitor, hTERT (Fig. 4D). Together, these data indicate that CRD-BP regulates MITF expression as well as its function. CRD-BP overexpression also dramatically increased the half-life of MITF mRNA (Fig. 4E and Table 3), indicating that MITF expression is regulated post-transcriptionally by CRD-BP. This result underscores previous reports of how CRD-BP regulates its other target mRNAs. Last, to test whether this regulation is mediated through a direct interaction of CRD-BP protein with MITF mRNA, we performed a UV cross-linking and immunoprecipitation experiment using radiolabeled fragments or full-length MITF mRNA with protein extracts from FLAG-CRD-BP-expressing cells. The results show that CRD-BP directly interacts with the short 3′-UTR of MITF mRNA (Fig. 4F). Altogether, these experiments demonstrate CRD-BP directly interacts with MITF mRNA and stabilizes it.
FIGURE 4.
CRD-BP is a positive regulator of MITF expression. A, 451Lu cells were transfected with either control shRNA or shRNA against CRD-BP. 48 h after transfection, cells were collected and assayed for levels of MITF mRNA by qRT-PCR, normalized to GAPDH expression, and presented as a percentage of control (scrambled shRNA). B, 1241 mel and mel IM cells were co-transfected with the indicated shRNA-expressing plasmids, β-gal-expressing plasmid, and pHTRPL4, where the luciferase gene is expressed under an MITF-dependent promoter. Values represent luciferase activity normalized to β-gal expression. C, NHMs were electroporated using the AMAXA NucleofectorTM with either CRD-BP-overexpressing plasmid or empty vector. 72 h after transfection, cells were collected and assayed for levels of MITF mRNA by qRT-PCR, normalized to GAPDH expression, and presented in percentage to control (pBABE). D, NHMs were electroporated using the AMAXA NucleofectorTM with the indicated plasmids. 48 h after electroporation, cells were stained for senescence-associated β-gal, and the percentage of β-gal-positive cells was calculated. E, 451Lu cells were co-transfected with Tet-Off plasmid, p-BI-G-MITF, and either pcDNA control or CRD-BP-overexpressing plasmid. The turnover of MITF transcripts was analyzed by qRT-PCR after stopping transcription by doxycycline treatment for the indicated times. Normalization was done with respect to GAPDH expression. All results are representative of three separate experiments and expressed as the mean ± S.D. (error bars). The average half-lives of mRNAs are presented in Table 3. F, FLAG immunoprecipitation of UV-cross-linked RNP complexes. Protein extracts from 293T cells transfected with FLAG-CRD-BP were incubated with internally 32P-labeled RNA from three fragments of the MITF mRNA coding region, the full-length MITF mRNA, and the short 3′-UTR. Fragment 1 contains nucleotides 1–421, fragment 2 consists of nucleotides 422–841, and fragment 3 consists of nucleotides 842–1260 of the MITF coding region. RNP complexes were precipitated with anti-FLAG antibodies and analyzed on PAGE and autoradiographed.
CRD-BP Interferes with miR-340 Function, Resulting in the Stabilization of MITF mRNA
Based on the finding that CRD-BP directly interacts with the short 3′-UTR of MITF mRNA, we investigated whether CRD-BP prevents miR-340-dependent regulation of MITF mRNA in a similar fashion to what was reported earlier for different mRNAs (21). Munro et al. (43) characterized the sequence motif required for IMP binding in Drosophila as UUUAY, and we found this motif either within or juxtaposed to both miR-340 binding sites. Subsequently, when CRD-BP was overexpressed, the half-life of MITF mRNA increased, similar to the effect on MITF mRNA when miR-340 function was inhibited (Fig. 5A and Table 3). However, CRD-BP overexpression could not further increase the half-life of short 3′-UTR MITF mRNA with concurrent inhibition of miR-340 function because miR-340 inhibition already stabilized the mRNA (Fig. 5A). Similarly, when miR-340 function was inhibited, there was an increase in the endogenous MITF mRNA level, which was not further altered due to CRD-BP overexpression (Fig. 5B). Further, overexpression of CRD-BP increased the levels of MITF mRNA in a dose-dependent manner but failed to do so when miR-340 was also ectopically overexpressed (Fig. 5C). Last, knockdown of CRD-BP decreased MITF-dependent transcription by more than 2-fold but had no effect when miR-340 function was already inhibited (Fig. 5D). These data suggest that CRD-BP exerts its effect on MITF mRNA by preventing the access of miR-340 to its sites. When miR-340 action is blocked, the mRNA is stable by itself and becomes unresponsive to the availability of CRD-BP. In order to further confirm the mechanism of CRD-BP interference with the function of miR-340, we measured the half-life of MITF mRNAs where miR-340 sites were deleted. We found that the half-life of mutated MITF mRNA increased significantly compared with wild type MITF mRNA, and overexpression of CRD-BP had no effect on the stability of MITF mRNAs lacking miR-340 binding sites (Fig. 5E and Table 3). The finding that CRD-BP could stabilize MITF mRNA only when miR-340 is functional demonstrates that CRD-BP acts by blocking miR-340-dependent regulation of MITF mRNA. Additionally, ectopic expression of CRD-BP does not affect the levels of miR-340 and overexpression of miR-340 failed to change CRD-BP levels (Fig. 5, F and G), ruling out the possibility that the observed changes to MITF expression are due to the effects of CRD-BP and miR-340 on each other. To analyze the effect of miR-340 inhibition on cell proliferation, we performed colony formation assays and found that inhibition of miR-340 function using specific sponge constructs resulted in a significant increase in the number of colonies formed by both 928 mel and 451Lu melanoma cells (Fig. 5H). A similar increase was detected when CRD-BP was overexpressed, but no additive effect on colony formation was observed when CRD-BP and sponge-340 were co-expressed. As expected, knockdown of CRD-BP resulted in a drastic inhibition in the number of colonies; however, inhibition of miR-340 rescued the effect of CRD-BP knockdown on the growth of 928 mel and 451Lu melanoma cells (Fig. 5H).
FIGURE 5.
CRD-BP counteracts miR-340 action and stabilizes MITF mRNA. A, wild type MITF expressing plasmid p-BI-G-MITFwt was co-transfected with Tet-Off plasmid and the indicated constructs in 451Lu cells. The turnover of MITF transcripts was analyzed as in Fig. 4E. B, 451Lu cells were transfected with the indicated constructs. 48 h after transfection, cells were collected and assayed for levels of endogenous MITF mRNA by qRT-PCR, normalized to GAPDH expression, and presented as a percentage of control. C, 451Lu cells were transfected with either control pcDNA 3.1, CRD-BP-overexpressing, control pCMV-MIR, or miR-340 overexpressing (pCMV-MIR340) plasmids, as indicated. 48 h after transfection, cells were collected and assayed for levels of MITF mRNA by qRT-PCR, normalized to GAPDH expression, and presented as a percentage of control. D, 451Lu cells were co-transfected with MITF-dependent luciferase expressing vector pHTRPL4, indicated shRNA, miR-sponge-expressing plasmids, and β-gal-expressing plasmid. Values correspond to luciferase activity normalized to β-gal expression. E, construct expressing MITF with both of the miR-340 sites deleted (p-BI-G-MITFδmiR-340) was co-transfected with Tet-Off plasmid and the indicated constructs in 451Lu cells. The turnover of MITF transcript was analyzed as in Fig. 4E. All results are representative of three separate experiments and expressed as the mean ± S.D. (error bars). The average half-lives of mRNAs are presented in Table 3. F, 451Lu cells were transfected as indicated. 48 h after transfection, cells were collected and assayed for levels of mature miR-340 transcript by ddPCR. Results are presented as the number of copies/1 μg of total RNA. G, 451Lu cells were transfected as indicated. 48 h after transfection, cells were collected and assayed for levels of CRD-BP mRNA by qRT-PCR, normalized to GAPDH expression, and presented as -fold change compared with control. H, 451Lu and 928 mel cells were co-transfected with the indicated constructs and pTk-Puro. The colonies were selected for puromycin resistance, stained with crystal violet, and counted.
DISCUSSION
MITF is widely regarded as the master regulator of melanocyte biology because of its involvement in the regulation of important melanogenic proteins as well as its contribution toward melanoblast survival, melanocyte lineage commitment, and melanoma pathogenesis (reviewed in Ref. 44). Therefore, it is the importance of MITF that potentiates the necessity to study its regulatory mechanisms. Post-transcriptional regulation of gene expression involving RBPs is pivotal in regulating the expression of several genes and the discovery of miRNAs added a new dimension to this regulatory scheme. MITF is also post-transcriptionally regulated and two recent studies show evidence that miR-137 and miR-182 target MITF mRNA (19, 20). In the context of this study, we set out to investigate the post-transcriptional regulation of MITF mRNA expression in further detail.
One key mechanism underlying the process of malignant transformation is the activation of proto-oncogenes. Loss of miRNA binding sites from the 3′-UTRs of oncogenic mRNAs is considered an important mode of oncogene activation (45, 46). This was supported by recent observations demonstrating that cancer cells and other proliferating cells favor the expression of mRNA isoforms containing shorter 3′-UTRs in order to escape miRNA-mediated regulation (22, 23). MITF mRNA also has several isoforms with different length 3′-UTRs and, most interestingly, the isoform with a short 3′-UTR does not have binding sites for the previously reported miRNAs (Fig. 1A). Here, we show that melanoma cells preferentially express MITF mRNA isoforms with short 3′-UTRs (Fig. 1B) and comply with other fast proliferating cells in terms of the regulation of oncogenic mRNAs. This finding, thus, designates the MITF mRNA isoform with a short 3′-UTR as the most prevalent isoform among melanoma cell lines. Moreover, our results show that although the MITF mRNA isoform with a short 3′-UTR escapes regulation by previously reported miRNAs, it is still being regulated by a different miRNA, miR-340. Interaction of miR-340 with its two target sites present on the short 3′-UTR of MITF mRNA results in the destabilization of MITF mRNA and a decrease in MITF expression and transcriptional activity (Figs. 2, 3, and 4B). Interestingly, miR-340 was found to be expressed in primary melanoma cell lines, suggesting its importance in the regulation of melanoma-specific target mRNAs (47). Because the fragment containing the miR-340 binding sites is part of all three known MITF 3′-UTRs, the regulation of MITF by miR-340 appears to be independent of APA and therefore more universal (shared by both melanocytes and melanoma cells).
Results of this study establish CRD-BP as an important positive regulator of MITF expression and function. CRD-BP stabilizes MITF mRNA and increases MITF expression as well as its transcriptional activity (Fig. 4B). This effect of CRD-BP is mediated by counteracting the miR-340-mediated degradation of MITF mRNA (Fig. 5). This is not surprising because we demonstrated previously that CRD-BP also interferes with miR-183 function, resulting in the stabilization of βTrCP1 mRNA (21). Our findings here are in line with recent reports describing an interplay between RBPs and miRNAs, where RBPs interfere with miRNA function, thereby highlighting a novel mode of post-transcriptional regulation of gene expression (Fig. 6) (48, 49).
FIGURE 6.
Model for interference of miR-340 function by CRD-BP. A, in the absence of CRD-BP, the miR-340-guided miRNA-induced silencing complex (miRISC) interacts with the 3′-UTR of MITF mRNA and accelerates its degradation. B, when CRD-BP is present, it binds to the 3′-UTR of MITF mRNA in the vicinity of miR-340 binding sites, shielding the MITF mRNA from miR-340-mediated down-regulation. The resulting elevated levels of MITF contribute to melanoma cell proliferation.
CRD-BP is important for the growth and survival of many types of cancer cells (28, 42, 50). It is reported to be involved in mammalian development and linked to effects on cellular adhesion and invasion taking place during development and malignancy (39, 51). These effects of CRD-BP are attributed to its regulation of different target mRNAs (c-myc, βTrCP1, Gli1, etc.). We reported previously that CRD-BP is overexpressed in a majority of malignant melanomas (24). However, knockdown of CRD-BP in melanoma cells had a much more robust effect than in other tumor cells and thus it was hypothesized that an additional melanoma-specific factor must be involved. Data provided in this work suggest that the regulation of MITF by CRD-BP may contribute to the observed effects of CRD-BP on melanoma survival and progression and also indicate that CRD-BP is a potential target for the prevention and treatment of this deadly disease.
Acknowledgments
We thank Drs. K. Kinzler, P. Robbins, J. Ross, P. Sharp, T. Tuschl, and B. Vogelstein for the generous gifts of reagents and Dr. N. Sanek for help with data collection.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grants AR063361, CA191550, and CA121851 (to V. S. S.) and CA12509 (to V. S.) and National Institutes of Health Grant GM076244 (to Y. G.).
- MITF
- microphthalmia-associated transcription factor
- APA
- alternative cleavage and polyadenylation
- CRD-BP
- coding region determinant-binding protein
- miRNA or miR
- microRNA
- NHM
- normal human melanocyte
- PSM
- antibiotic-antimycotic solution, containing penicillin, streptomycin, and amphotericin B
- nt
- nucleotide(s)
- qRT-PCR
- quantitative RT-PCR
- ddPCR
- droplet digital PCR
- RBP
- RNA-binding protein.
REFERENCES
- 1. Hallsson J. H., Haflidadóttir B. S., Stivers C., Odenwald W., Arnheiter H., Pignoni F., Steingrímsson E. (2004) The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bentley N. J., Eisen T., Goding C. R. (1994) Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol. Cell. Biol. 14, 7996–8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hemesath T. J., Steingrímsson E., McGill G., Hansen M. J., Vaught J., Hodgkinson C. A., Arnheiter H., Copeland N. G., Jenkins N. A., Fisher D. E. (1994) Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770–2780 [DOI] [PubMed] [Google Scholar]
- 4. Yasumoto K., Yokoyama K., Shibata K., Tomita Y., Shibahara S. (1994) Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol. Cell. Biol. 14, 8058–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carreira S., Liu B., Goding C. R. (2000) The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 275, 21920–21927 [DOI] [PubMed] [Google Scholar]
- 6. Vance K. W., Carreira S., Brosch G., Goding C. R. (2005) Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 65, 2260–2268 [DOI] [PubMed] [Google Scholar]
- 7. Du J., Widlund H. R., Horstmann M. A., Ramaswamy S., Ross K., Huber W. E., Nishimura E. K., Golub T. R., Fisher D. E. (2004) Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6, 565–576 [DOI] [PubMed] [Google Scholar]
- 8. McGill G. G., Horstmann M., Widlund H. R., Du J., Motyckova G., Nishimura E. K., Lin Y. L., Ramaswamy S., Avery W., Ding H. F., Jordan S. A., Jackson I. J., Korsmeyer S. J., Golub T. R., Fisher D. E. (2002) Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109, 707–718 [DOI] [PubMed] [Google Scholar]
- 9. McGill G. G., Haq R., Nishimura E. K., Fisher D. E. (2006) c-Met expression is regulated by Mitf in the melanocyte lineage. J. Biol. Chem. 281, 10365–10373 [DOI] [PubMed] [Google Scholar]
- 10. Saito H., Yasumoto K., Takeda K., Takahashi K., Yamamoto H., Shibahara S. (2003) Microphthalmia-associated transcription factor in the Wnt signaling pathway. Pigment Cell Res. 16, 261–265 [DOI] [PubMed] [Google Scholar]
- 11. Yasumoto K., Takeda K., Saito H., Watanabe K., Takahashi K., Shibahara S. (2002) Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J. 21, 2703–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takeda K., Yasumoto K., Takada R., Takada S., Watanabe K., Udono T., Saito H., Takahashi K., Shibahara S. (2000) Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J. Biol. Chem. 275, 14013–14016 [DOI] [PubMed] [Google Scholar]
- 13. Dorsky R. I., Raible D. W., Moon R. T. (2000) Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 14, 158–162 [PMC free article] [PubMed] [Google Scholar]
- 14. Price E. R., Ding H. F., Badalian T., Bhattacharya S., Takemoto C., Yao T. P., Hemesath T. J., Fisher D. E. (1998) Lineage-specific signaling in melanocytes: c-Kit stimulation recruits p300/CBP to microphthalmia. J. Biol. Chem. 273, 17983–17986 [DOI] [PubMed] [Google Scholar]
- 15. Watanabe A., Takeda K., Ploplis B., Tachibana M. (1998) Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat. Genet. 18, 283–286 [DOI] [PubMed] [Google Scholar]
- 16. Hemesath T. J., Price E. R., Takemoto C., Badalian T., Fisher D. E. (1998) MAP kinase links the transcription factor microphthalmia to c-Kit signalling in melanocytes. Nature 391, 298–301 [DOI] [PubMed] [Google Scholar]
- 17. Wu M., Hemesath T. J., Takemoto C. M., Horstmann M. A., Wells A. G., Price E. R., Fisher D. Z., Fisher D. E. (2000) c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14, 301–312 [PMC free article] [PubMed] [Google Scholar]
- 18. Khaled M., Larribere L., Bille K., Aberdam E., Ortonne J. P., Ballotti R., Bertolotto C. (2002) Glycogen synthase kinase 3β is activated by cAMP and plays an active role in the regulation of melanogenesis. J. Biol. Chem. 277, 33690–33697 [DOI] [PubMed] [Google Scholar]
- 19. Segura M. F., Hanniford D., Menendez S., Reavie L., Zou X., Alvarez-Diaz S., Zakrzewski J., Blochin E., Rose A., Bogunovic D., Polsky D., Wei J., Lee P., Belitskaya-Levy I., Bhardwaj N., Osman I., Hernando E. (2009) Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. U.S.A. 106, 1814–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bemis L. T., Chen R., Amato C. M., Classen E. H., Robinson S. E., Coffey D. G., Erickson P. F., Shellman Y. G., Robinson W. A. (2008) MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 68, 1362–1368 [DOI] [PubMed] [Google Scholar]
- 21. Elcheva I., Goswami S., Noubissi F. K., Spiegelman V. S. (2009) CRD-BP protects the coding region of βTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell 35, 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayr C., Bartel D. P. (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elcheva I., Tarapore R. S., Bhatia N., Spiegelman V. S. (2008) Overexpression of mRNA-binding protein CRD-BP in malignant melanomas. Oncogene 27, 5069–5074 [DOI] [PubMed] [Google Scholar]
- 25. Craig E. A., Weber J. D., Spiegelman V. S. (2012) Involvement of the mRNA binding protein CRD-BP in the regulation of metastatic melanoma cell proliferation and invasion by hypoxia. J. Cell Sci. 125, 5950–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Craig E. A., Spiegelman V. S. (2011) Inhibition of CRD-BP sensitizes melanoma cells to chemotherapeutic agents. Pigment Cell Melanoma Res. 25, 83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cummins J. M., He Y., Leary R. J., Pagliarini R., Diaz L. A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A. E., Labourier E., Raymond C. K., Roberts B. S., Juhl H., Kinzler K. W., Vogelstein B., Velculescu V. E. (2006) The colorectal microRNAome. Proc. Natl. Acad. Sci. U.S.A. 103, 3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noubissi F. K., Elcheva I., Bhatia N., Shakoori A., Ougolkov A., Liu J., Minamoto T., Ross J., Fuchs S. Y., Spiegelman V. S. (2006) CRD-BP mediates stabilization of βTrCP1 and c-myc mRNA in response to β-catenin signalling. Nature 441, 898–901 [DOI] [PubMed] [Google Scholar]
- 29. Ebert M. S., Neilson J. R., Sharp P. A. (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poenitzsch Strong A. M., Setaluri V., Spiegelman V. S. (2014) microRNA-340 as a modulator of RAS-RAF-MAPK signaling in melanoma. Arch. Biochem. Biophys. 563, 118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spiegelman V. S., Slaga T. J., Pagano M., Minamoto T., Ronai Z., Fuchs S. Y. (2000) Wnt/β-catenin signaling induces the expression and activity of βTrCP ubiquitin ligase receptor. Mol. Cell 5, 877–882 [DOI] [PubMed] [Google Scholar]
- 33. Fang D., Tsuji Y., Setaluri V. (2002) Selective down-regulation of tyrosinase family gene TYRP1 by inhibition of the activity of melanocyte transcription factor, MITF. Nucleic Acids Res. 30, 3096–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yasumoto K., Yokoyama K., Takahashi K., Tomita Y., Shibahara S. (1997) Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem. 272, 503–509 [DOI] [PubMed] [Google Scholar]
- 35. Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A. H., Wewer U. M., Nielsen F. C. (1999) A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nielsen F. C., Nielsen J., Christiansen J. (2001) A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand. J. Clin. Lab. Invest. Suppl. 234, 93–99 [PubMed] [Google Scholar]
- 37. Runge S., Nielsen F. C., Nielsen J., Lykke-Andersen J., Wewer U. M., Christiansen J. (2000) H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem. 275, 29562–29569 [DOI] [PubMed] [Google Scholar]
- 38. Atlas R., Behar L., Elliott E., Ginzburg I. (2004) The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 89, 613–626 [DOI] [PubMed] [Google Scholar]
- 39. Hansen T. V., Hammer N. A., Nielsen J., Madsen M., Dalbaeck C., Wewer U. M., Christiansen J., Nielsen F. C. (2004) Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol. Cell. Biol. 24, 4448–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prokipcak R. D., Herrick D. J., Ross J. (1994) Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J. Biol. Chem. 269, 9261–9269 [PubMed] [Google Scholar]
- 41. Tessier C. R., Doyle G. A., Clark B. A., Pitot H. C., Ross J. (2004) Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 64, 209–214 [DOI] [PubMed] [Google Scholar]
- 42. Noubissi F. K., Goswami S., Sanek N. A., Kawakami K., Minamoto T., Moser A., Grinblat Y., Spiegelman V. S. (2009) Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res. 69, 8572–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Munro T. P., Kwon S., Schnapp B. J., St Johnston D. (2006) A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J. Cell Biol. 172, 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Levy C., Khaled M., Fisher D. E. (2006) MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12, 406–414 [DOI] [PubMed] [Google Scholar]
- 45. Lee Y. S., Dutta A. (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mayr C., Hemann M. T., Bartel D. P. (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mueller D. W., Rehli M., Bosserhoff A. K. (2009) miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Invest. Dermatol. 129, 1740–1751 [DOI] [PubMed] [Google Scholar]
- 48. Kedde M., Agami R. (2008) Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 7, 899–903 [DOI] [PubMed] [Google Scholar]
- 49. Kim H. H., Kuwano Y., Srikantan S., Lee E. K., Martindale J. L., Gorospe M. (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kawakami Y., Kubota N., Ekuni N., Suzuki-Yamamoto T., Kimoto M., Yamashita H., Tsuji H., Yoshimoto T., Jisaka M., Tanaka J., Fujimura H. F., Miwa Y., Takahashi Y. (2009) Tumor-suppressive lipoxygenases inhibit the expression of c-myc mRNA coding region determinant-binding protein/insulin-like growth factor II mRNA-binding protein 1 in human prostate carcinoma PC-3 cells. Biosci. Biotechnol. Biochem. 73, 1811–1817 [DOI] [PubMed] [Google Scholar]
- 51. Vikesaa J., Hansen T. V., Jønson L., Borup R., Wewer U. M., Christiansen J., Nielsen F. C. (2006) RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 25, 1456–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]