Background: PHGDH encodes a key metabolic enzyme in the serine biosynthesis pathway and is frequently amplified in melanomas.
Results: p53 transcriptionally represses PHGDH expression. Serine starvation promotes p53-mediated apoptosis in melanomas through PHGDH suppression.
Conclusion: p53-mediated repression of PHGDH enhances the apoptotic response upon serine starvation in melanoma cells.
Significance: The combination of the drugs activating p53 and serine deprivation could be a better treatment for melanomas.
Keywords: Apoptosis, Cancer Biology, Cell Death, Melanoma, Metabolism, p53, Tumor Suppressor Gene
Abstract
Although p53 is frequently mutated in human cancers, about 80% of human melanomas retain wild-type p53. Here we report that PHGDH, the key metabolic enzyme that catalyzes the rate-limiting step of the serine biosynthesis pathway, is a target of p53 in human melanoma cells. p53 suppresses PHGDH expression and inhibits de novo serine biosynthesis. Notably, upon serine starvation, p53-mediated cell death is enhanced dramatically in response to Nutlin-3 treatment. Moreover, PHGDH has been found recently to be amplified frequently in human melanomas. We found that PHGDH overexpression significantly suppresses the apoptotic response, whereas RNAi-mediated knockdown of endogenous PHGDH promotes apoptosis under the same treatment. These results demonstrate an important role of p53 in regulating the serine biosynthesis pathway through suppressing PHGDH expression and reveal serine deprivation as a novel approach to sensitize p53-mediated apoptotic responses in human melanoma cells.
Introduction
Cancer cells are known to have an altered metabolism. As described by Otto Warburg, the majority of tumor cells use aerobic glycolysis instead of mitochondrial respiration to produce energy and support cell growth and proliferation (1). In addition, the metabolic reprogramming toward macromolecule biosynthesis, which is thought to sustain rapid cell growth, is also being recognized as a hallmark of cancer (2). There is accumulating evidence indicating that p53 plays a critical role in modulating metabolism in human cancers (3). However, the precise mechanisms by which the metabolic activities contribute to overall p53 responses are not well understood (4–7).
Recently, the serine synthesis pathway has been shown to be essential in certain human cancers, and it contributes to oncogenesis through diverting glycolytic flux and further promoting biosynthesis (8, 9). l-serine is an important precursor for macromolecule biosynthesis. It is an essential amino acid for protein synthesis and synthesis of sphingolipids and phospholipids, which are important components of cellular membranes (10, 11). The serine synthesis pathway converts the glycolytic intermediate 3-phosphoglycerate into serine via a three-step reaction involving three metabolic enzymes: phosphoglycerate dehydrogenase (PHGDH),3 phosphoserine aminotransferase 1 (PSAT1), and phosphate ester hydrolysis (12). l-serine can then be converted into glycine by serine hydroxymethyltransferase, which results in the formation of one-carbon units that contribute to de novo synthesis of purines and pyridines (13). Specifically, PHGDH catalyzes the first and rate-limiting step of the serine synthesis pathway from 3-phosphoglycerate to phospho-hydroxypyruvate and is potentially important during tumor development. The human PHGDH gene is located in a highly amplified region of chromosome 1p (8). It is frequently amplified and overexpressed in 70% of the estrogen receptor-negative breast cancers, 39% of melanoma, and cervical cancers (8, 9, 14, 15). Depletion of PHGDH in PHGDH-amplified cells leads to an impairment of cell proliferation and serine synthesis in both breast cancer and melanoma cell lines (8, 9, 16), suggesting that the repression of the serine synthesis pathway may have therapeutic roles toward suppressing certain types of cancers.
Cancer cells can acquire l-serine from both de novo synthesis and the extracellular environment. Upon serine starvation, PHGDH and PSAT1 are significantly up-regulated, and, thereby, increase conversion of 3-phosphoglycerate to serine (17). Furthermore, serine starvation also leads to p53-dependent metabolic remodeling in cancer cells (17). In this study, we investigated the contribution of p53 to the regulation of serine metabolism. p53 is widely accepted as a tumor suppressor and responds to varieties of cellular stresses (18). It mainly functions as a transcription factor to monitor signaling pathways through the promoter-specific regulation of target genes involved in cell growth arrest, apoptosis, and senescence (19). Recent studies have suggested that p53 can also regulate metabolic reprogramming through numerous target genes, including TIGAR, GLUT1/3/4, GLS2, PARK3, and so forth (3, 5, 20–24). Therefore, further understanding of the regulation of the serine metabolism pathway by p53 might be crucial for revealing its mechanisms in tumor suppression.
Although p53 is frequently mutated in human cancers, 80% of human melanomas retain wild-type p53 (7, 25). Furthermore, PHGDH is frequently amplified and highly expressed in melanoma (9). As such, we focused our study on the interaction between p53 and PHGDH in melanoma cell lines. We found that PHGDH is a novel p53 repressing gene in the serine biosynthesis pathway. PHGDH expression is significantly down-regulated upon treatment of Nutlin-3, a p53 agonist that has been developed to suppress tumor growth through interrupting the interaction of p53 and its negative regulator, Mdm2 (26). Although Nutlin-3 only induces p53-mediated growth arrest, but not apoptosis (27–29), in melanoma cell lines expressing wild-type p53, we observed that serine starvation further sensitizes Nutlin-3 to induce apoptosis in melanoma cells through repression of PHGDH by p53.
EXPERIMENTAL PROCEDURES
Plasmids
pGIPZ shRNA against TP53 and pTRIPZ shRNA against PHGDH were purchased from Thermo Scientific. A control hairpin in the pGIPZ vector that targeted GFP (shGFP) was used. pBabe-puro-FLAG-PHGDH was generated by PCR-based subcloning.
Cell Culture and Stable Lines
All cells were cultured in a 37 °C incubator with 5% CO2. All media used were supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (all Invitrogen). For serine starvation experiments, cells were washed with PBS twice and fed with serine- and glycine-free medium consisting of minimum Eagle's medium (catalog no. 11095, Invitrogen) supplemented with additional 1× minimum Eagle's medium vitamins (catalog no. 11120, Invitrogen), 10% dialyzed FBS (catalog no. 26400, Invitrogen) and additional d-glucose to 25 mm (catalog no. 25-037-Cl, Cellgro). For control medium, serine and glycine (Sigma) were added back to a final concentration of 0.4 mm. According to Maddocks et al. (17), serine is the major factor in serine and glycine starvation. Therefore, all starvation experiments were described as serine starvation. To generate the A375 cell line with stable knockdown of p53, HEK293T cells were transfected with pGIPZ shRNA vectors against TP53 and lentiviral packaging vectors, and lentiviruses produced from 293T were used to infect A375 cells. Selection under 1 μg/ml puromycin was carried out 2 days after infection. The A375-PHGDH stable cell line and A375 inducible knockdown of the PHGDH cell line (A375-shPHGDH) were generated by a similar procedure with the pBabe-puro-FLAG-PHGDH and pTRIPZ-shPHGDH vectors. To induce knockdown of PHGDH, 5 μg/ml of doxycycline (Sigma) was added to culture medium.
siRNA-mediated Ablation of ATF4
Knockdown of ATF4 was performed by transfection of A375-shPHGDH cells with siRNA duplex oligoset (On-Target-Plus SMARTpool, catalog no. L-00512500, Dharmacon) using Lipofectamine RNAiMAX (catalog no. 13778030, Invitrogen) according to the protocol of the manufacturer. Control siRNA (On-Target-Plus siControl nontargeting pool, catalog no. D00181010, Dharmacon) was also used for transfection.
Western Blotting and Antibodies
Cell lysates were prepared in FLAG lysis buffer with fresh protease inhibitor mixture. Protein extracts were analyzed by Western blotting according to standard protocols using primary antibodies specific for PHGDH (catalog no. HPA021241, Sigma), p53 (human, catalog no. DO-1, Santa Cruz Biotechnology), Mdm2 (catalog no. Ab5, Millipore), Tigar (catalog no. E-2, Santa Cruz Biotechnology), p21 (catalog no. SX118, Santa Cruz Biotechnology), Puma (catalog no. H-136, Santa Cruz Biotechnology), cleaved caspase 3 (Asp-175, Cell Signaling Technologies), ATF4 (catalog no. sc-200, Santa Cruz Biotechnology), and β-Actin (catalog no. A3853, Sigma-Aldrich). HRP-conjugated anti-mouse and anti-rabbit secondary antibodies (GE Healthcare) were used, and signals were detected on autoradiographic films with an ECL Western blotting detection system (GE Healthcare) or SuperSignal West Dura reagents (Thermo Scientific).
RNA Extraction and qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the protocol of the manufacturer. cDNA was synthesized from total RNA using an M-MulV reverse transcriptase kit (New England Biolabs). PCR analysis was performed using an Applied Biosystems 7500 fast system. For the qRT-PCR analysis of human transcripts, the following primers were used: PHGDH forward, 5′-ATCTCTCACGGGGGTTGTG-3′; PHGDH reverse, 5′-AGGCTCGCATCAGTGTCC-3′; ATF4 forward, 5′-GGTCAGTCCCTCCAACAACA-3′; ATF4 reverse, 5′-CTATACCCAACAGGGCATCC-3′; PUMA forward, 5′-GGTCCTCAGCCCTCGCTCTC-3′; PUMA reverse, 5′-CTTGTCTCCGCCGCTCGTAC-3′; NOXA forward, 5′-CGTGTGTAGTTGGCATCTCCG-3′; NOXA reverse, 5′-GACGCGAGCTGAACACGAAC-3′; BAX forward, 5′-TTCTGACGGCAACTTCAACTGG-3′; BAX reverse, 5′-CCCGGAGGAAGTCCAATGTC-3′; GAPDH forward, 5′-ATCAATGGAAATCCCATCACCA-3′; and GAPDH reverse, 5′-GACTCCACGACGTACTCAGCG-3′.
ChIP
ChIP assays were performed in transfected H1299 cells or A375 cells as described previously (30). The primers used for p53 the ChIP assay were as follows: PHGDH forward, 5′-TGAGAATATGCGGTGTTTGG-3′; PHGDH reverse, 5′-GGGTAAATGTGCAAGGCACT-3′; TIGAR forward, 5′-CGGCAGGTCTTAGATAGCTT-3′; and TIGAR reverse, 5′-GGCAGCCGGCATCAAAAACA-3′.
EMSA
Purified FLAG-p53 protein was obtained from transfected H1299 cells. The 234-bp DNA probe containing p53-responsive elements was PCR-amplified from the PHGDH promoter using PHGDH ChIP primers labeled by T4 kinase (catalog no. M0201S, New England Biolabs) and purified using Bio-Spin 30 columns (Bio-Rad). The protein-DNA binding reactions were performed as described previously (30). In supershift assays, 100 ng of pAb421 (Millipore) antibody was added to the reactions.
Cell Death Count, Drugs, and Inhibitors
Cells were trypsinized, collected, and stained with trypan blue, followed by counting with a hemocytometer using a standard protocol. Cells stained blue under the microscope were considered dead cells. Nutlin-3 (Sigma) was used in the experiments at a concentration of 10 μm unless indicated otherwise. The DNA-damaging agent doxorubicin (Dox, Sigma) was used at 0.2 μg/ml. Specific cell death inhibitors were used in the experiments with indicated concentrations as follows: Z-VAD-fmk (10 μg/ml, a caspase 3 inhibitor, Sigma), 3-methyladenine (2 mm, an autophagy inhibitor, Sigma), Necrostatin 1 (10 μg/ml, a necroptosis inhibitor, Sigma), and Ferrostatin 1 (2 μm, a ferroptosis inhibitor, Xcess Biosciences).
Metabolite Measurements
To measure steady-state metabolite levels, prechilled (on dry ice) 80% methanol was added to cells in 10-cm culture dishes, followed by incubation on dry ice for 15 min. Cells were then collected by scraping, and cellular metabolite was extracted by collecting the supernatant following centrifugation at 4000 rpm for 10 min (4 °C). The supernatant was then dried down using lyophilizer (Labconco) and stored at −80 °C before being subjected to LC-MS/MS-based metabolite analysis at the Beth Israel Deaconess Medical Center Mass Spectrometry Core Facility as described previously (31). Briefly, samples were suspended in HPLC-grade water and analyzed using a 5500 QTRAP hybrid triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence Ultra Fast liquid chromatography HPLC system (Shimadzu) via selected reaction monitoring of a total of 254 endogenous water-soluble metabolites using positive and negative polarity switching. Peak areas from the total ion current for each metabolite were integrated using MultiQuant v2.0 software (AB/SCIEX). Three biological triplicates were analyzed for each treatment. In addition, two parallel dishes of cells were collected and measured for protein concentration using a Bradford assay (Bio-Rad). Subsequent metabolite measurements were normalized to the total protein amount.
RESULTS
Nutlin-3 Induces p53-mediated Repression of PHGDH
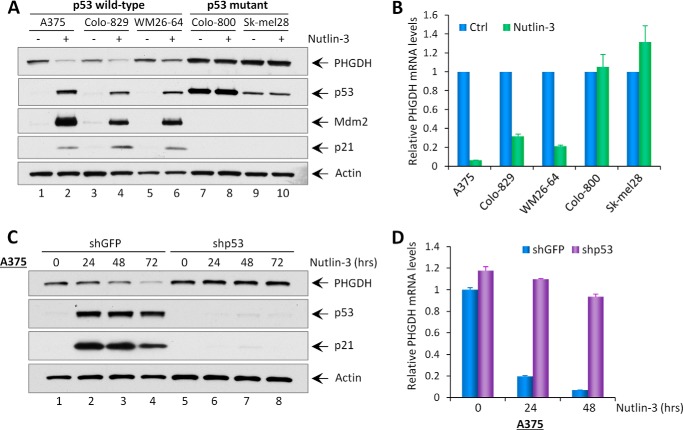
To evaluate the regulation of PHGDH by endogenous p53, we analyzed human melanoma cell lines containing wild-type p53 or mutant p53. Wild-type p53 cell lines (A375, Colo-829, and WM26–64) and mutant p53 cell lines (Colo-800 and Sk-mel28) were treated with Nutlin-3, a non-genotoxic drug that activates p53 by interrupting the Mdm2-p53 interaction. Total cell lysates were then harvested and subjected to Western blot analysis, and mRNA was extracted for quantitative real-time PCR (qRT-PCR). As expected, in p53 wild-type cell lines, the protein levels of p53 and its transcriptional targets Mdm2 and p21 were increased in response to Nutlin-3 treatment (Fig. 1A). Notably, a significant decrease in both PHGDH protein and mRNA levels was also observed (Fig. 1, A and B). However, we did not observe a change in PHGDH expression levels in p53-mutant melanoma cell lines after the same treatment (Fig. 1, A and B). To further determine p53-dependent regulation on PHGDH expression, we compared A375 cells with p53 depletion to cells with mock depletion. Cells that underwent mock depletion and were treated with Nutlin-3 showed a time-dependent reduction of both PHGDH protein and mRNA levels (Fig. 1, C and D). In contrast, the expression of PHGDH in p53-depleted cells remained unchanged after Nutlin-3 treatment (Fig. 1, C and D). Together, these results indicate that PHGDH is transcriptionally down-regulated upon Nutlin-3 treatment in a p53-dependent manner.
FIGURE 1.
Nutlin-3 induces p53-mediated repression of PHGDH. A, Western blot analysis of the protein levels of PHGDH, p53, Mdm2, p21, and Actin in indicated melanoma cell lines (A375, Colo-829, and WM26–64, p53 wild-type; Colo-800 and Sk-mel28, p53 mutant) without (−) or with (+) Nutlin-3 treatment (10 μm) for 2 days. B, the mRNA expression levels of PHGDH in the indicated melanoma cell lines treated with Nutlin-3 (10 μm) were measured using qRT-PCR. Ctrl, control. C, A375-shGFP and A375-shp53 stable knockdown cell lines were treated with Nutlin-3 (10 μm) for the indicated times, and total protein lysates were subjected to Western blot analysis for the expression of PHGDH, p53, p21, and Actin. D, qRT-PCR analysis of PHGDH mRNA levels in A375-shGFP and A375-shp53 cell lines treated with Nutlin-3 (10 μm). All mRNA expression levels were normalized with GAPDH. Error bars represent mean ± S.D. from three experiments.
DNA Damage Down-regulates PHGDH Protein and mRNA Levels in a p53-dependent Manner
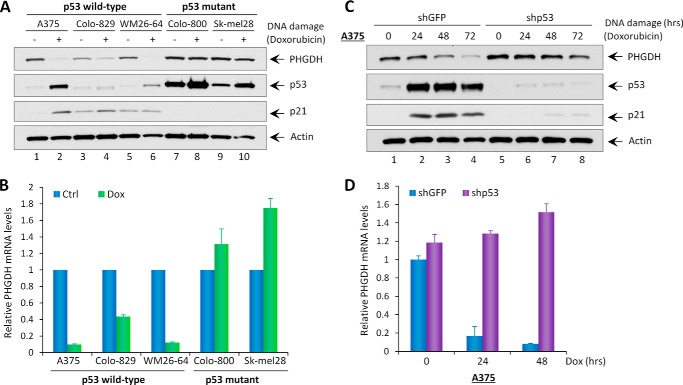
To further confirm that PHGDH is a p53-repressing target, we examined PHGDH expression in response to DNA damage. As expected, treatment with the DNA-damaging agent Dox induced activation of p53 (demonstrated by up-regulation of its downstream target, p21) in p53 wild-type cell lines but not in p53 mutant cell lines (Fig. 2A). Notably, in all three p53 wild-type melanoma cell lines, the levels of PHGDH protein and mRNA were decreased upon doxorubicin treatment (Fig. 2, A and B). In contrast, PHGDH expression remained unchanged in p53-mutant melanoma cell lines despite undergoing the same treatment (Fig. 2, A and B). Similarly, A375 cells that underwent mock depletion and were treated with doxorubicin showed a time-dependent reduction of both PHGDH protein and mRNA levels. However, the expression of PHGDH was not affected upon the same treatment in p53-depleted cells (Fig. 2, C and D). These data collectively suggest that the expression of PHGDH is transcriptionally repressed by p53 upon p53 stabilization after treatment with DNA-damaging agents.
FIGURE 2.
DNA damage down-regulates PHGDH protein and mRNA levels in a p53-dependent manner. A, expression of endogenous PHGDH, p53, p21, and Actin in the indicated melanoma cell lines without (−) or with (+) treatment of the DNA-damaging agent doxorubicin (0.2 μg/ml). B, qRT-PCR analysis of PHGDH mRNA levels in the indicated melanoma cell lines treated with Dox (0.2 μg/ml). Ctrl, control. C, A375-shGFP and A375-shp53 stable knockdown cell lines were treated with doxorubicin (0.2 μg/ml) for the indicated times, and total protein lysates were analyzed by Western blotting for the expression of PHGDH, p53, p21, and Actin. D, qRT-PCR analysis of PHGDH mRNA levels in A375-shGFP and A375-shp53 cell lines treated with doxorubicin (0.2 μg/ml) for the indicated times. All mRNA expression levels were normalized with GAPDH. Error bars represent mean ± S.D. from three experiments.
The PHGDH Promoter Contains One p53 Binding Site
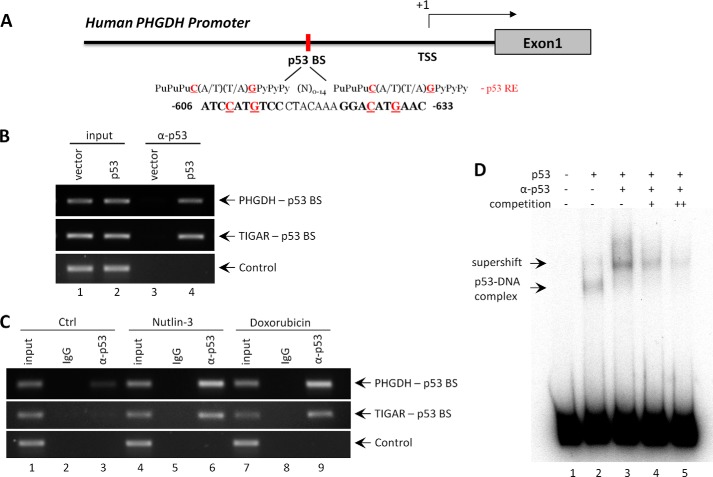
As a transcription factor, p53 binds to its responsive elements in its target genes and regulates gene transcription in response to various stresses. As shown in Fig. 3A, we found a consensus binding sequence in the promoter region of the PHGDH gene that matches the p53 binding site sequence (32). To verify that p53 binds to its responsive element in the promoter region of the PHGDH gene, ChIP assays were performed. H1299 p53-null cells were transfected with the p53 expression vector, cross-linked with formaldehyde, and p53-bound DNA was immunoprecipitated via anti-p53 antibody (α-p53). The resulting DNA fragments were amplified using primers flanking the predicted p53 binding site in the PHGDH promoter. In this assay, we observed enrichment of PHGDH and TIGAR promoter sequences in the presence of p53 (Fig. 3B). Furthermore, binding of p53 at the PHGDH promoter was significantly enriched in A375 cells treated with either Nutlin-3 or doxorubicin (Fig. 3C). These results demonstrate that p53 interacts with the p53-responsive element in the PHGDH promoter in vivo.
FIGURE 3.
The PHGDH promoter contains one p53 binding site. A, schematic of the promoter region in human PHGDH gene. The p53 binding site upstream of the first exon is indicated and compared with the consensus p53 responsive element (p53 RE). Pu, purine; Py, pyramidine; N, any nucleotide; BS, binding site; TSS, transcription start site. B, ChIP semiquantitative analysis of p53 enrichment at the promoter regions of PHGDH and TIGAR in H1299 cells transfected with empty or p53 vector for 24 h. The cross-linked protein-DNA complex was immunoprecipitated using full-length anti-p53 antibody (α-p53). GAPDH was used as a negative control. C, A375 cells were treated with vehicle, Nutlin-3 (10 μm), or doxorubicin (0.2 μg/ml) for 24 h, and ChIP assays for determining the enrichment of p53 on PHGDH and TIGAR promoters were carried out using anti-p53 antibody or control rabbit IgG. GAPDH was used as a negative control (Ctrl). D, EMSA showing the binding of p53 on 32P-labeled oligonucleotides containing the p53-binding site in the human PHGDH promoter region. The DNA binding activity of purified p53 protein was enhanced with the C-terminal p53 antibody pAb421 (α-p53). The binding specificity was verified using an excess of non-radiolabeled wild-type probes (competition).
We next performed an EMSA to determine whether p53 directly binds to the PHGDH promoter in vitro. Notably, a p53-DNA complex was identified upon incubation of radiolabeled oligonucleotide probes containing the responsive element with purified full-length human p53 protein (Fig. 3D). Moreover, the p53-DNA complex was enhanced and supershifted in the presence of anti-p53 antibody (ab421) (Fig. 3D). Furthermore, binding of p53 to the radiolabeled fragments was diminished and outcompeted by the addition of unlabeled probes (Fig. 3D). Together, these results indicate that the PHGDH gene is a transcriptional target of p53 and that the consensus p53 binding site on the PHGDH promoter is responsible for p53-dependent transcriptional repression.
Serine Starvation Promotes Nutlin-3-induced Cell Death through Repression of PHGDH
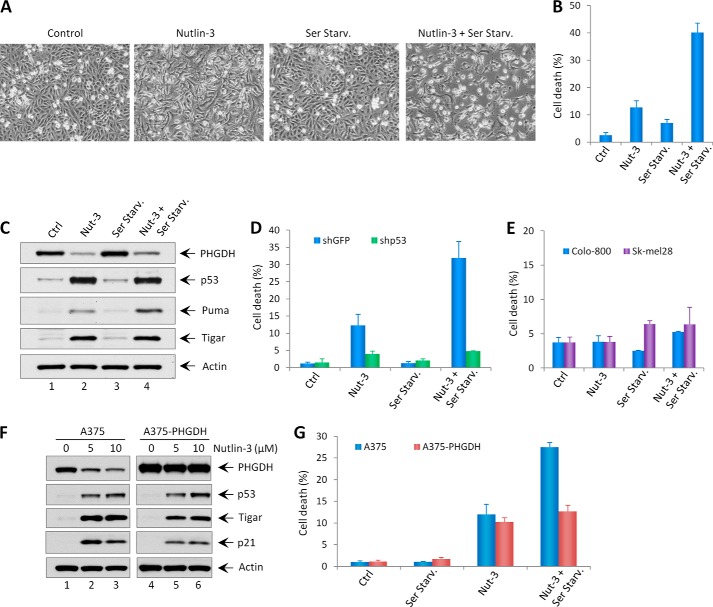
As a non-essential amino acid, serine can be acquired through de novo serine synthesis and from an exogenous nutrient source. Because serine is an important building block for biosynthesis and contributes to cancer cell growth, our findings that PHGDH is a p53 target suggest that PHGDH may mediate p53 function in controlling cell fate upon serine starvation. Previous studies have shown that treatment of Nutlin-3 on melanoma cell lines expressing wild-type p53 only induces p53-mediated cell cycle arrest but not apoptosis (27–29). As expected, A375 cells (p53 wild-type) treated with Nutlin-3 only showed significant growth arrest with no obvious cell death (Fig. 4, A and B). Surprisingly, although serine starvation alone did not induce obvious growth arrest nor cell death, a combination of serine starvation and Nutlin-3 treatment induced significant cell death in A375 cells (Fig. 4, A and B). Notably, Puma, a major p53 up-regulated apoptosis modulator, was further activated upon combination treatments compared with Nutlin-3 treatment alone, whereas p53 levels were the same under these two conditions, indicating that serine deprivation may lead to p53-independent Puma activation (Fig. 4C). To ascertain whether the cell death is dependent on p53, p53-depleted A375 cells and p53-mutant melanoma cell lines (Colo-800 and Sk-mel28) were treated with Nutlin-3 and/or serine starvation. Notably, Nutlin-3 did not induce growth arrest nor cell death upon serine starvation in these cell lines (Fig. 4, D and E), indicating that Nutlin-induced cell death upon serine starvation is p53-dependent.
FIGURE 4.
Serine starvation promotes Nutlin-3-induced cell death through repression of PHGDH. A, representative phase-contrast images of A375 cells treated with Nutlin-3 (10 μm) in the presence (Control) or absence (Ser Starv.) of serine in culture medium. B, the percentages of cell death for all treatments shown in A were quantified by trypan blue exclusion assay. Ctrl, control; Nut-3, Nutlin-3. C, Western blot analysis of expression levels of PHGDH, p53, Puma, Tigar, and Actin in A375 cells treated with Nutlin-3 (10 μm) in the presence or absence of serine. D, the percentages of cell death in A375-shGFP and A375-shp53 stable knockdown cells treated with Nutlin-3 (10 μm) in the presence or absence of serine. E, the percentages of cell death in p53 mutant cell lines (Colo-800 and Sk-mel28) treated with Nutlin-3 (10 μm) in the presence or absence of serine. F, the A375 and A375-PHGDH cell lines were treated with the indicated concentrations of Nutlin-3 for 48 h, and the expression levels of PHGDH, p53, Tigar, p21, and Actin were measured by Western blotting. G, the percentages of cell death in A375 and A375-PHGDH cell lines treated with Nutlin-3 (10 μm) under normal or serine starvation condition. Error bars represent mean ± S.D. from three different experiments.
To further explore the effect on cell fate by p53-mediated repression of PHGDH, we generated a PHGDH overexpression stable cell line in A375 (A375-PHGDH). As shown in Fig. 4F, although the expression of PHGDH was decreased upon Nutlin-3 treatment in A375 cells, it remained unchanged and overexpressed in A375-PHGDH cells after the same treatment, indicating that the high expression level of exogenous PHGDH negates the repressive effect of p53 on endogenous PHGDH expression. Upon Nutlin-3 treatment on A375-PHGDH cells, serine starvation did not sensitize Nutlin-3 to induce more cell death (Fig. 4G), indicating that the cell death induced by the combination of Nutlin-3 and serine starvation is mediated through repression of PHGDH by p53. Together, our results indicate that the function of p53 in repressing PHGDH mediates its role in regulating cell fate upon serine starvation.
Mechanistic Insights into the Apoptotic Response Regulated by PHGDH upon Serine Starvation
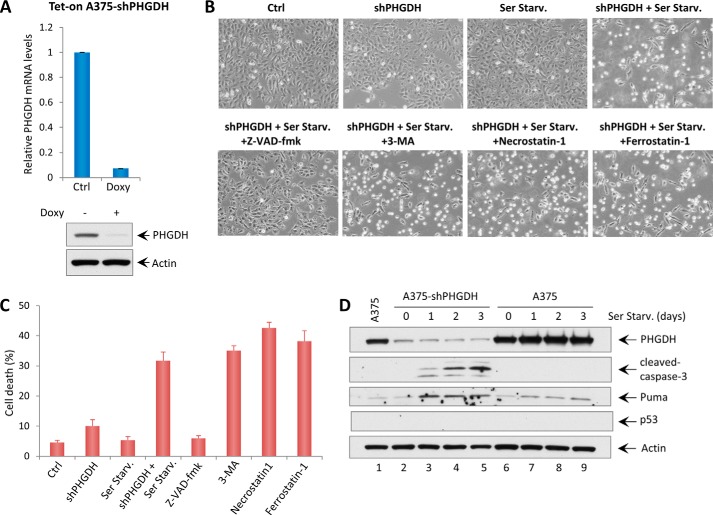
To understand the mechanisms of how serine starvation sensitizes cells to cell death upon Nutlin-3 treatment in p53 wild-type melanoma cell lines, we investigated whether PHGDH depletion is sufficient to induce cell death upon serine removal. Therefore, a doxycycline-inducible shRNA cell line in A375 targeting PHGDH (Tet-On A375-shPHGDH) was generated. As shown in Fig. 5A, PHGDH expression was efficiently depleted upon addition of doxycycline. Cells that underwent mock depletion or PHGDH depletion were then incubated in control medium or serine starvation medium for an additional 48 h. Notably, PHGDH depletion in A375 cells induced significant cell death upon serine starvation, whereas PHGDH depletion or serine starvation alone did not affect cell survival (Fig. 5, B and C). Interestingly, the cell death induced by PHGDH depletion and serine starvation was completely rescued by the caspase 3 inhibitor Z-VAD-fmk but not by inhibitors of other forms of cell death, including autophagy (3-methyladenine), necroptosis (Necrostatin 1), and ferroptosis (Ferrostatin 1) (Fig. 5, B and C). Western blot analysis also showed elevated levels of cleaved caspase 3 and Puma in cells that were undergoing cell death (Fig. 5D). These results indicate that the synergistic effect of PHGDH depletion and serine starvation leads to apoptosis in A375 cells.
FIGURE 5.
PHGDH knockdown induces apoptosis upon serine starvation. A, qRT-PCR analysis of PHGDH mRNA levels in the Tet-On A375-shPHGDH stable cell line after adding doxycycline (Doxy, 5 μg/ml) for 3 days. The Western blot analysis also shows the expression levels of PHGDH and Actin in Tet-On A375-shPHGDH cells after adding doxycycline. Ctrl, control. B, representative phase-contrast images of A375 cells expressing doxycycline-inducible shRNA against PHGDH with or without serine starvation (Ser Starv.) for 48 h. The images also show A375-shPHGDH cells under serine starvation conditions with the addition of cell death inhibitors (Z-VAD-fmk, a caspase 3 inhibitor, 10 μg/ml; 3-methyladenine (3-MA), an autophagy inhibitor, 2 mm; Necrostatin 1, a necroptosis inhibitor, 10 μg/ml; Ferrostatin 1, a ferroptosis inhibitor, 2 μm). C, the percentages of cell death for all experiments shown in B were measured by trypan blue exclusion assay. D, A375 shPHGDH cells and A375 cells were incubated in serine starvation medium for the indicated times, and total protein lysates were subjected to Western blot analysis for the expression of PHGDH, cleaved caspase 3, Puma, p53, and Actin.
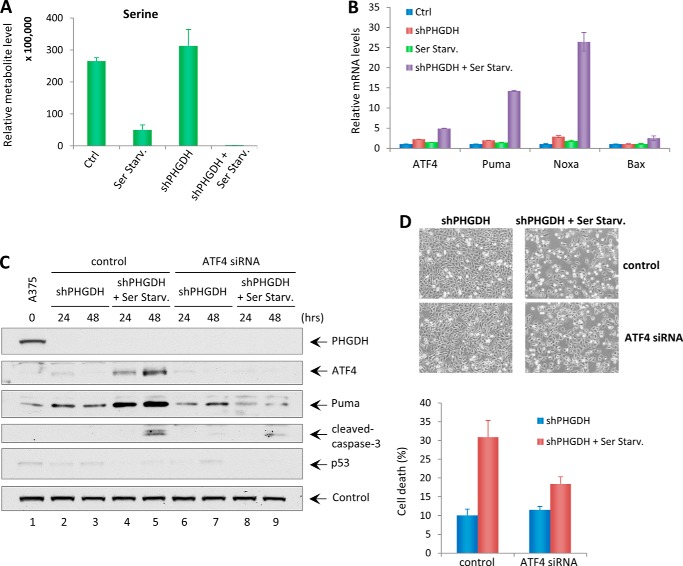
Although the link between apoptotic pathways and amino acid starvation is not fully understood, recent studies suggested that activation transcription factor 4 (ATF4) plays an important role in regulating amino acid metabolism and tumor cell death under stress conditions (33). Qing et al. (33) found that glutamine starvation induces apoptosis through ATF4-dependent, but p53-indepdent, Puma and Noxa induction in MYC-transformed cells. In our study, metabolite analysis showed that steady-state serine levels were decreased markedly in PHGDH-depleted A375 cells upon serine starvation (Fig. 6A). Meanwhile, a significant up-regulation of ATF4, Puma, and Noxa levels and a slight induction of the apoptosis regulator Bax were observed in A375 cells undergoing apoptosis (Fig. 6B). However, no p53 activation was present under this condition (Fig. 5D). To further prove that PHGDH knockdown-induced apoptosis upon serine starvation is indeed dependent on ATF4, PHGDH-depleted A375 cells were transfected with control siRNA or ATF4 siRNA and then incubated in control medium or serine-free medium. Although ATF4 expression was significantly up-regulated upon serine starvation in PHGDH knockdown A375 cells, its expression was completely ablated upon ATF4 knockdown (Fig. 6C). Notably, activation of Puma was abolished, and lower levels of cleaved caspase 3 were observed upon ATF4 knockdown, whereas no p53 activation was present under any conditions (Fig. 6C). Furthermore, ATF4 siRNA significantly reduced serine starvation-induced cell death in PHGDH-depleted A375 cells (Fig. 6D). These results collectively indicate that the apoptosis induced by PHGDH depletion upon serine starvation is dependent on ATF4.
FIGURE 6.
ATF4 mediates PHGDH knockdown-induced apoptosis upon serine starvation. A, metabolite analysis showing the intracellular steady levels of serine in A375-shPHGDH cells cultured in complete medium or serine starvation medium (Ser Starv.). Ctrl, control. B, qRT-PCR analysis of mRNA levels of ATF4, Puma, Noxa, and Bax in A375-shPHGDH cells cultured in control medium or serine starvation medium. C, A375 shPHGDH cells were first transfected with control siRNA or ATF4 siRNA for 24 h and then incubated in complete medium or serine starvation medium for up to 48 h. Total cell lysates were harvested and subjected to Western blot analysis for the expression of PHGDH, ATF4, Puma, cleaved caspase 3, and p53. Vinculin was used as a loading control. D, representative phase-contrast images of A375-shPHGDH cells transfected with control or ATF4 siRNA in the presence or absence of serine at 48 h. The percentages of cell death were measured by trypan blue exclusion assay. Error bars represent mean ± S.D. from three experiments.
DISCUSSION
p53 is a bona fide tumor suppressor and serves as a guardian of the genome to prevent malignant transformation. It mainly functions as a transcription factor to monitor signaling pathways through transcriptional regulation of target genes involved in cell cycle arrest, cell death, and metabolism (19). Although p53 is well known for its transcriptional activation function, there are more and more studies suggesting that p53 is also able to repress gene transcription and that such regulation is important for its ability to regulate cell death and metabolism (34, 35). In this study, we identified that PHGDH, the key serine metabolism gene, is a repression target of p53 and mediates its apoptotic response upon serine starvation. PHGDH has recently been shown to play an important role in tumor development and is frequently amplified or overexpressed in melanoma, a type of skin cancer that rarely has p53 mutations. Here, in melanoma cell lines, we demonstrated that PHGDH mRNA and protein levels were significantly down-regulated in response to Nutlin-3 and DNA-damaging agents in a p53-dependent manner. Furthermore, we found a p53 consensus sequence in the PHGDH promoter region, and, therefore, p53 binds directly to this response element to repress gene transcription. Although the precise mechanisms of transcription repression remain largely unknown, a number of studies suggest that p53 represses gene transcription through interfering with the functions of DNA-binding transcriptional activators or recruiting transcriptional repressors, such as histone deacetylases (36). Therefore, it would be interesting to explore whether histone deacetylase inhibitors such as trichostatin A can block p53-mediated transcriptional repression of PHGDH.
PHGDH catalyzes the first and rate-limiting step in the serine synthesis pathway (14). In light of the recent findings that some melanoma and breast cancer cell lines with an amplified PHGDH locus are highly dependent on PHGDH expression for their proliferation (8, 9), PHGDH inhibitors are being developed (in preclinical studies) to treat these types of cancers (37). In our study, we revealed that a combination of PHGDH depletion and serine starvation not only inhibits proliferation but also induces significant apoptosis in melanoma cells. We also demonstrated that the mechanisms underlying apoptosis triggered by inhibiting PHGDH in the presence of serine starvation are mediated through ATF4-dependent but p53-independent Puma and Noxa activation. These results imply that inhibitors of serine metabolism, when utilized in the setting of appropriate nutrient (specifically serine) restriction, may have therapeutic potential against cancer.
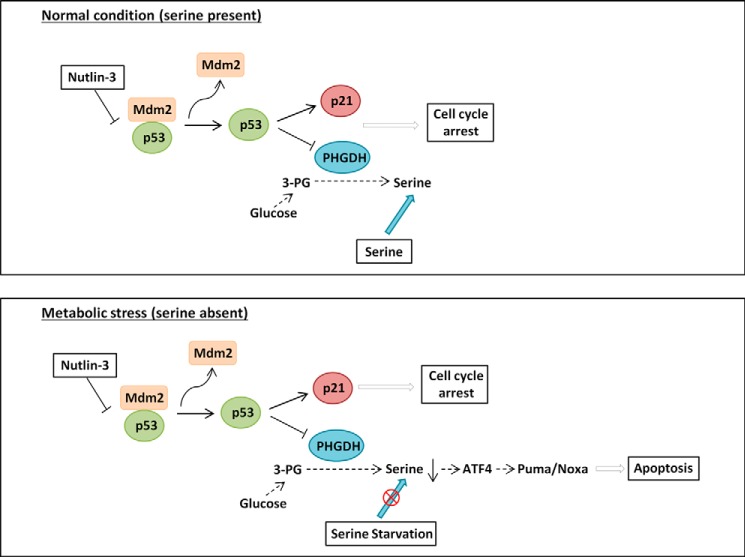
Given our results showing that PHGDH is a novel repression target of p53, activators of p53 can be used to inhibit PHGDH expression. Nutlin-3 is a non-genotoxic drug that specifically activates p53 transcription through disrupting its interaction with Mdm2. However, Nutlin-3 is not an ideal drug for eradicating melanoma cells that retain wild-type p53 because it only induces p53-mediated cell cycle arrest without triggering apoptosis (27). Therefore, the possibility of reactivating the apoptotic function of p53 in melanoma cell lines could have profound implications in future therapeutic directions (6, 7). Here we identified a novel p53 downstream pathway that modulates serine metabolism, which can sensitize melanoma cells to apoptosis. As shown in the proposed model in Fig. 7, activation of p53 in melanoma cell lines by Nutlin-3 can suppress PHGDH expression through transcriptional repression. Under normal conditions in which serine is present, activation of p53 by Nutlin-3 only induces cell cycle arrest through p21 activation and PHGDH down-regulation. However, under metabolic stress conditions in which serine is absent, suppression of PHGDH by Nutlin-3 can further promote apoptosis through ATF4-dependent Puma and Noxa activation. Our findings implied that small molecules that interrupt serine uptake may be combined with Nutlin-3 to enhance the killing of human melanoma cells. In conclusion, our study provides novel mechanistic insights into how p53 promotes apoptosis in cancer cells through the regulation of the oncogenic serine metabolism pathway.
FIGURE 7.
Proposed models demonstrating the differential effects of p53 on its apoptotic response through regulating PHGDH under normal condition or serine starvation conditions. See text for details.
This work was supported, in whole or in part, by NCI, National Institutes of Health Awards 5RO1CA085533, 5RO1CA172023, 5RO1 CA166294, 5RO1CA085533, and 2P01CA080058 (to W. G.) and R01 CA166717 (to B. Z.).
- PHGDH
- phosphoglycerate dehydrogenase
- qRT-PCR
- quantitative real-time PCR
- Dox
- doxorubicin
- TIGAR
- TP53-induced glycolysis and apoptosis regulator
- Z-VAD-fmk
- benzyloxycarbonyl-VAD-fluoromethyl ketone.
REFERENCES
- 1. Hsu P. P., Sabatini D. M. (2008) Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 [DOI] [PubMed] [Google Scholar]
- 2. Ward P. S., Thompson C. B. (2012) Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liang Y., Liu J., Feng Z. (2013) The regulation of cellular metabolism by tumor suppressor p53. Cell Biosci. 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Y., He Y., Jin A., Tikunov A. P., Zhou L., Tollini L. A., Leslie P., Kim T. H., Li L. O., Coleman R. A., Gu Z., Chen Y. Q., Macdonald J. M., Graves L. M., Zhang Y. (2014) Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U.S.A. 111, E2414–E2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang C., Liu J., Liang Y., Wu R., Zhao Y., Hong X., Lin M., Yu H., Liu L., Levine A. J., Hu W., Feng Z. (2013) Tumour-associated mutant p53 drives the Warburg effect. Nat. Commun. 4, 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mandinova A., Lee S. W. (2011) The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci. Transl. Med. 3, 64rv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jochemsen A. G. (2014) Reactivation of p53 as therapeutic intervention for malignant melanoma. Curr. Opin. Oncol. 26, 114–119 [DOI] [PubMed] [Google Scholar]
- 8. Possemato R., Marks K. M., Shaul Y. D., Pacold M. E., Kim D., Birsoy K., Sethumadhavan S., Woo H. K., Jang H. G., Jha A. K., Chen W. W., Barrett F. G., Stransky N., Tsun Z. Y., Cowley G. S., Barretina J., Kalaany N. Y., Hsu P. P., Ottina K., Chan A. M., Yuan B., Garraway L. A., Root D. E., Mino-Kenudson M., Brachtel E. F., Driggers E. M., Sabatini D. M. (2011) Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Locasale J. W., Grassian A. R., Melman T., Lyssiotis C. A., Mattaini K. R., Bass A. J., Heffron G., Metallo C. M., Muranen T., Sharfi H., Sasaki A. T., Anastasiou D., Mullarky E., Vokes N. I., Sasaki M., Beroukhim R., Stephanopoulos G., Ligon A. H., Meyerson M., Richardson A. L., Chin L., Wagner G., Asara J. M., Brugge J. S., Cantley L. C., Vander Heiden M. G. (2011) Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Futerman A. H., Riezman H. (2005) The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15, 312–318 [DOI] [PubMed] [Google Scholar]
- 11. Kuge O., Nishijima M. (2003) Biosynthetic regulation and intracellular transport of phosphatidylserine in mammalian cells. J. Biochem. 133, 397–403 [DOI] [PubMed] [Google Scholar]
- 12. Snell K. (1984) Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv. Enzyme Regul. 22, 325–400 [DOI] [PubMed] [Google Scholar]
- 13. de Koning T. J., Snell K., Duran M., Berger R., Poll-The B. T., Surtees R. (2003) l-serine in disease and development. Biochem. J. 371, 653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mullarky E., Mattaini K. R., Vander Heiden M. G., Cantley L. C., Locasale J. W. (2011) PHGDH amplification and altered glucose metabolism in human melanoma. Pigment Cell Melanoma Res. 24, 1112–1115 [DOI] [PubMed] [Google Scholar]
- 15. Jing Z., Heng W., Aiping D., Yafei Q., Shulan Z. (2013) Expression and clinical significance of phosphoglycerate dehydrogenase and squamous cell carcinoma antigen in cervical cancer. Int. J. Gynecol. Cancer 23, 1465–1469 [DOI] [PubMed] [Google Scholar]
- 16. Chen J., Chung F., Yang G., Pu M., Gao H., Jiang W., Yin H., Capka V., Kasibhatla S., Laffitte B., Jaeger S., Pagliarini R., Chen Y., Zhou W. (2013) Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget 4, 2502–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maddocks O. D., Berkers C. R., Mason S. M., Zheng L., Blyth K., Gottlieb E., Vousden K. H. (2013) Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levine A. J., Oren M. (2009) The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 20. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H. (2006) TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 [DOI] [PubMed] [Google Scholar]
- 21. Schwartzenberg-Bar-Yoseph F., Armoni M., Karnieli E. (2004) The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 64, 2627–2633 [DOI] [PubMed] [Google Scholar]
- 22. Hu W., Zhang C., Wu R., Sun Y., Levine A., Feng Z. (2010) Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. U.S.A. 107, 7455–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki S., Tanaka T., Poyurovsky M. V., Nagano H., Mayama T., Ohkubo S., Lokshin M., Hosokawa H., Nakayama T., Suzuki Y., Sugano S., Sato E., Nagao T., Yokote K., Tatsuno I., Prives C. (2010) Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 107, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang C., Lin M., Wu R., Wang X., Yang B., Levine A. J., Hu W., Feng Z. (2011) Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. U.S.A. 108, 16259–16264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Albino A. P., Vidal M. J., McNutt N. S., Shea C. R., Prieto V. G., Nanus D. M., Palmer J. M., Hayward N. K. (1994) Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 4, 35–45 [DOI] [PubMed] [Google Scholar]
- 26. Vu B. T., Vassilev L. (2011) Small-molecule inhibitors of the p53-MDM2 interaction. Curr. Top. Microbiol. Immunol. 348, 151–172 [DOI] [PubMed] [Google Scholar]
- 27. de Lange J., Ly L. V., Lodder K., Verlaan-de Vries M., Teunisse A. F., Jager M. J., Jochemsen A. G. (2012) Synergistic growth inhibition based on small-molecule p53 activation as treatment for intraocular melanoma. Oncogene 31, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 28. Ji Z., Njauw C. N., Taylor M., Neel V., Flaherty K. T., Tsao H. (2012) p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J. Invest. Dermatol. 132, 356–364 [DOI] [PubMed] [Google Scholar]
- 29. Tseng H. Y., Jiang C. C., Croft A., Tay K. H., Thorne R. F., Yang F., Liu H., Hersey P., Zhang X. D. (2010) Contrasting effects of nutlin-3 on TRAIL- and docetaxel-induced apoptosis due to upregulation of TRAIL-R2 and Mcl-1 in human melanoma cells. Mol. Cancer Ther. 9, 3363–3374 [DOI] [PubMed] [Google Scholar]
- 30. Wang S. J., Yu G., Jiang L., Li T., Lin Q., Tang Y., Gu W. (2013) p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle 12, 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan M., Breitkopf S. B., Yang X., Asara J. M. (2012) A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 33. Qing G., Li B., Vu A., Skuli N., Walton Z. E., Liu X., Mayes P. A., Wise D. R., Thompson C. B., Maris J. M., Hogarty M. D., Simon M. C. (2012) ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer cell 22, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McKenzie L., King S., Marcar L., Nicol S., Dias S. S., Schumm K., Robertson P., Bourdon J. C., Perkins N., Fuller-Pace F., Meek D. W. (2010) p53-dependent repression of polo-like kinase-1 (PLK1). Cell Cycle 9, 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang P., Du W., Mancuso A., Wellen K. E., Yang X. (2013) Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ho J., Benchimol S. (2003) Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 10, 404–408 [DOI] [PubMed] [Google Scholar]
- 37. Vander Heiden M. G. (2011) Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 10, 671–684 [DOI] [PubMed] [Google Scholar]