Background: External Na+ inhibits ENaC.
Results: Mutations centered about a key aspartate in an acidic cleft weakened Na+ inhibition and altered inhibitor selectivity.
Conclusion: The acidic cleft hosts an inhibitory Na+ binding site.
Significance: The acidic cleft harbors a key Na+ binding site for ENaC and perhaps sites for ligands that regulate other members of the ENaC/degenerin family.
Keywords: Acid-sensing Ion Channel (ASIC), Allosteric Regulation, Epithelial Sodium Channel (ENaC), Hypertension, Ion Channel, Membrane Transport, Protein Cross-linking, Effector Specificity, pH-dependent Activation
Abstract
The epithelial Na+ channel (ENaC) has a key role in the regulation of extracellular fluid volume and blood pressure. ENaC belongs to a family of ion channels that sense the external environment. These channels have large extracellular regions that are thought to interact with environmental cues, such as Na+, Cl−, protons, proteases, and shear stress, which modulate gating behavior. We sought to determine the molecular mechanism by which ENaC senses high external Na+ concentrations, resulting in an inhibition of channel activity. Both our structural model of an ENaC α subunit and the resolved structure of an acid-sensing ion channel (ASIC1) have conserved acidic pockets in the periphery of the extracellular region of the channel. We hypothesized that these acidic pockets host inhibitory allosteric Na+ binding sites. Through site-directed mutagenesis targeting the acidic pocket, we modified the inhibitory response to external Na+. Mutations at selected sites altered the cation inhibitory preference to favor Li+ or K+ rather than Na+. Channel activity was reduced in response to restraining movement within this region by cross-linking structures across the acidic pocket. Our results suggest that residues within the acidic pocket form an allosteric effector binding site for Na+. Our study supports the hypothesis that an acidic cleft is a key ligand binding locus for ENaC and perhaps other members of the ENaC/degenerin family.
Introduction
The epithelial Na+ channel (ENaC)2 is a highly Na+-selective heterotrimeric channel whose activity is modified by a variety of extracellular factors (1, 2). ENaC-mediated Na+ conductance through the apical membrane is rate-limiting for transepithelial Na+ absorption in many Na+-transporting epithelia (2). In the distal nephron, this transporter has a crucial role in extracellular fluid volume regulation and is under the control of the volume-regulatory hormones aldosterone and vasopressin (2, 3). The role of ENaC in transepithelial Na+ transport is broad because the channel is expressed in many transporting epithelia, including the distal nephron, lung airway and alveoli, distal colon, sweat glands, lingual epithelia, and inner ear (4–6). ENaC subunits are also expressed in vascular smooth muscle and endothelial cells, where they appear to modulate vascular tone (7).
Sensitivity to extracellular factors is a characteristic of members of the ENaC/degenerin family. For ENaC, a high extracellular [Na+] inhibits the channel through at least two mechanisms. First, external Na+ rapidly inhibits ENaC by directly binding to extracellular allosteric effector sites within the channel. This allosteric inhibition, referred to as Na+ self-inhibition, reflects a low-affinity (tens of micromolar), pH-sensitive, non-voltage-sensitive reduction in ENaC open probability (8–13). This inhibitory effect is also cation-selective because both Na+ and Li+ inhibit the channel, whereas K+ has a modest inhibitory effect at best, and protons activate the channel (9, 10). Second, an elevated intracellular Na+ concentration inhibits the channel over a slow time course (14, 15).
The extracellular domains of members of the ENaC gene family likely host allosteric effector sites that sense factors in the extracellular environment. For example, acid-sensing ion channels (ASICs) are proton-activated. The resolved structure of ASIC1 features an acidic pocket in each subunit that was proposed to contain proton binding sites competent to trigger gating (16). A major challenge in elucidating ENaC gene family function has been the ability to distinguish ligand binding from downstream transduction steps. The crystal structure of ASIC1 suggests several putative proton binding sites, and mutations at select sites affect acid activation of ASIC1 (17–19). However, one cannot readily distinguish a proton binding site from sites involved in transmitting conformational changes to the channel gate. In this work, we used inhibitor specificity to tackle this challenge.
ENaCs are activated by proteases, which cleave the α and γ subunits at specific sites, releasing imbedded inhibitory tracts (1, 20–23). On the basis of ASIC1 homology and our work that identified the binding site of a peptide derived from the α subunit inhibitory tract, we developed a structural model of the ENaC α subunit (24, 25). We found that the α subunit inhibitory tract lies adjacent to an acidic region that is homologous to the ASIC1 acidic cleft. We hypothesized that specific residues within the acidic cleft of the α subunit form a Na+ binding site required for the Na+ self-inhibition response.
Na+ self-inhibition is an allosteric response. Channel modifications that affect the apparent strength of Na+ self-inhibition could alter channel behavior at different steps in this regulatory process. Experimental manipulations that affect Na+ self-inhibition could alter the Na+ binding site, transduction steps subsequent to Na+ binding, or the stability of the open state relative to the closed state of the channel independent of Na+ binding. The fact that ENaC is both regulated by external Na+ and conducts Na+ complicates our ability to differentiate the Na+ binding step from other steps in the Na+ self-inhibition mechanism. To define sites within the channel periphery that participate in Na+ binding, we studied properties that directly reflect this event. Accordingly, we measured the effect of mutations on the effector or cation specificity of the Na+ self-inhibition response. Select mutations within the acidic cleft weakened Na+ self-inhibition and resulted in channels that were more effectively inhibited by Li+ or K+ than by Na+, in contrast to the wild-type channel phenotype, where Na+ is the most potent inhibitor. We identified a single titratable residue whose mutation abrogated Na+ self-inhibition as well as channel activation by acidic pH. Through cross-linking, we demonstrate that constraining movements of adjacent structures within the acidic cleft reduced channel activity. Our data show that the acidic cleft of the α subunit of ENaC hosts an allosteric effector binding site for Na+ and provide strong support for the concept that interactions of external factors with the extracellular region of ENaC have an important role in modulating channel activity.
EXPERIMENTAL PROCEDURES
ENaC Expression and Mutagenesis
Mouse ENaC subunits were mutated and expressed in Xenopus oocytes as described previously (26). All mutations were confirmed by direct sequencing.
Current Measurement
Electrophysiological measurements were performed using a GeneClamp 500B voltage clamp amplifier (Axon Instruments, Foster City, CA) and pClamp 10.2 software (Axon Instruments). Oocytes were placed in a 20-μl recording chamber (AutoMate Scientific, Berkeley, CA) with perfusion (3–5 ml/min) controlled by an eight-channel pinch valve system (AutoMate Scientific). For experiments that did not examine the cation specificity of ENaC inhibition, we used a high Na+ bath solution (110 mm NaCl, 2 mm KCl, 2 mm CaCl2, and 10 mm HEPES (pH 7.4)). We replaced HEPES with MES for lower pH versions of the high-Na+ bath solution. To make a low-Na+ bath solution, we replaced NaCl with 109 mm NMDG and 1 mm NaCl. For experiments that examined the cation specificity of ENaC inhibition, we used buffers that included K+ channel blockers as described previously (9) (100 mm NMDG-Cl, 0.82 mm MgCl2, 0.41 mm CaCl2, 10 mm NMDG-HEPES, 5 mm BaCl2, and 10 mm TEA-Cl for the NMDG+ solution and with 100 mm NaCl, 100 mm LiCl, and 100 mm KCl replacing NMDG-Cl for the Na+, Li+, and K+ solutions, respectively). Amiloride was dissolved in dimethyl sulfoxide at 100 mm and diluted in bath solution to 10 μm for fixed-voltage experiments or 100 μm for voltage ramp experiments.
Current-Voltage Curve Analysis
The comparison of ENaC inhibition by extracellular Na+, Li+, and K+ is complicated by the fact that ENaC is permeable to these cations by a 1.6:1:<0.01 (Li+:Na+:K+) ratio (27). To measure ENaC inhibition by these ions, we determined the relative Na+ permeability (PNa) in the presence of each ion from the amiloride-sensitive steady-state current-voltage (I-V) curves by fitting each curve to the Goldman-Hodgkin-Katz model as described previously (9) with some modifications. When Na+ was the only permeant ion present, we fit I-V curves to equation 1,
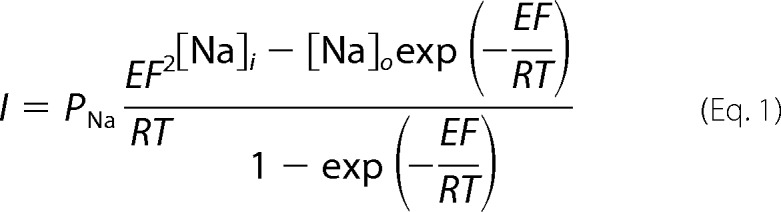 |
where E, F, R, T, [Na]i, and [Na]o are the membrane potential, Faraday constant, gas constant, absolute temperature, intracellular [Na+], and extracellular [Na+], respectively. When the bath solution was switched to Li+, we fit I-V curves to equation 2,
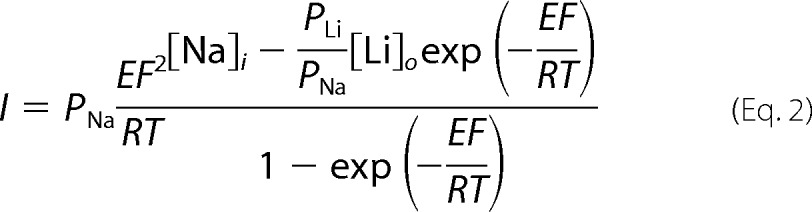 |
where [Li]o is extracellular [Li+] and PLi/PNa was assumed to equal 1.6 on the basis of the ion selectivity of the pore (9, 27). We assumed that the mutations examined here, far from the pore, did not affect the value of PLi/PNa. We also assumed that [Na]i remained constant throughout the experiment and that intracellular [Li+] was zero. To minimize changes in [Na]i during the experiment, we clamped oocytes at −20 mV between voltage ramps. We collected five amiloride-sensitive I-V curves for each oocyte, each using bath solutions with different cations (NMDG+, Na+, Li+, K+, and NMDG+ again) for 1 min, followed by the same solution supplemented with 100 μm amiloride for 20 s. We fit the I-V curves from each oocyte simultaneously using a global fitting routine and Igor Pro 6 (Wavemetrics, Oswego, OR). This allowed us to use data under all conditions to determine a global value for [Na]i while independently determining local values for PNa under each condition. We assigned values for the global parameters F, R, and T and the local parameters [Na]o or [Li]o·PLi/PNa. E was the independent variable. Each fit produced five PNa values, one for each condition. We normalized each PNa value to a corrected PNa determined with bath NMDG+ at the beginning and end of the experiment to account for channel rundown, which we assumed was linear over the course of the experiment.
Statistical Analyses
Values presented are mean ± S.E. We performed multiple group comparisons using analysis of variance followed by Tukey post hoc test. We adjusted the significance threshold from p = 0.05 for multiple comparisons using the Bonferroni correction. We performed pairwise comparisons using Student's t test. We considered p values of less than 0.05 significant.
RESULTS
Protons Activate Mouse ENaC
Collier and Snyder (10) and Collier et al. (28) reported previously that human ENaC is activated by extracellular acidification with an apparent pH50 of activation of 6.5. They found that rat ENaC was also activated by acid but at a lower pH (28). Because acidification was associated with reduced Na+ self-inhibition, they suggested that extracellular acidification activated ENaC by relieving Na+ self-inhibition. We examined the effect of extracellular acidification on mouse ENaC expressed in Xenopus oocytes by two-electrode voltage clamp (Fig. 1A). Although modest acidification (to pH 6.5) did not alter channel activity (see below), lowering the extracellular pH from 7.4 to 4.7 stimulated currents by 44 ± 3% (n = 6, p < 0.05 using paired Student's t test). This is similar to the increase observed for human ENaC by lowering the pH from 7.4 to 6.5 (10). When currents are measured under voltage clamp conditions, currents decay over time. We found that the rate of decay of ENaC currents increased from 3.4 ± 0.9%/min to 10.3 ± 0.5%/min with acidification to pH 4.7 (p < 0.05 using paired Student's t test). Channel activation by reducing extracellular pH to the range that should titrate acidic residues suggested that residues in the acidic cleft could be involved in Na+ sensing.
FIGURE 1.
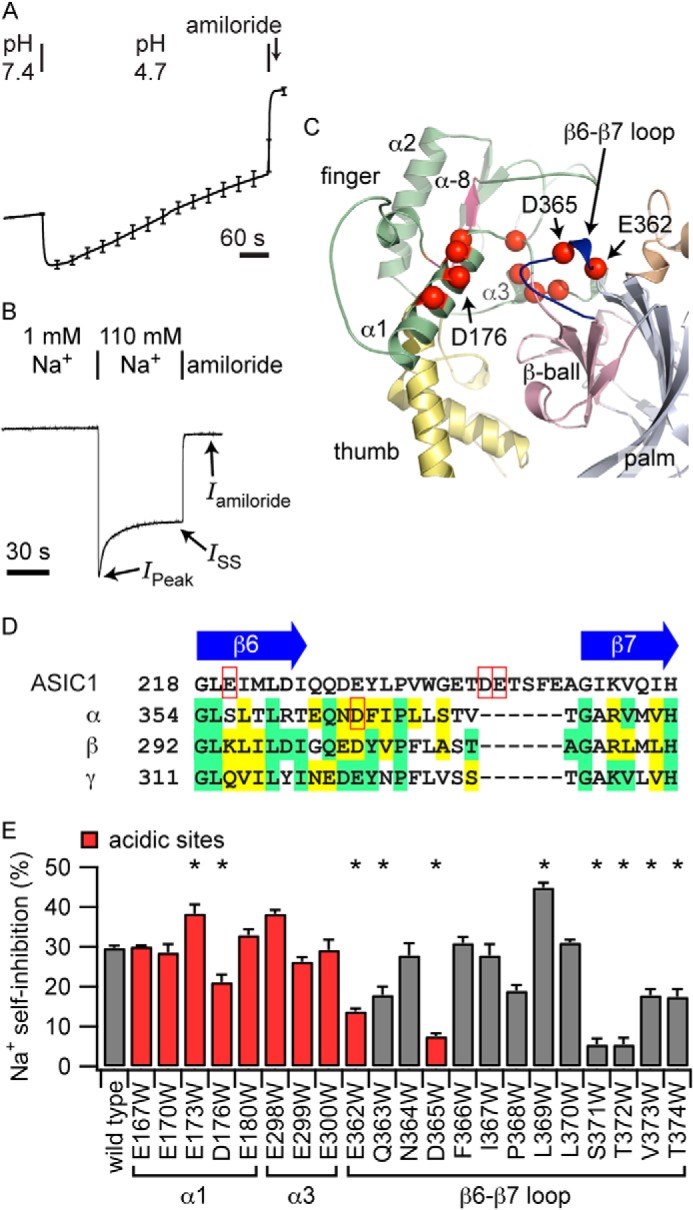
Mutations of acidic cleft residues affect Na+ self-inhibition. A, acidification activates mouse ENaC. Recordings were normalized to the average current prior to acidification. The average of six normalized current tracings is shown, with periodic error bars indicating the mean ± S.E. The average starting current was 1.8 ± 0.4 μA. B, representative recording of the Na+ self-inhibition response of wild-type mouse ENaC (−100 mV holding potential). Bathing solutions and the addition of amiloride (10 μm) are noted. IPeak and ISS currents following the transition to a high [Na+] solution are also noted. C, model of the acidic cleft. Acidic residues (red spheres) and the β6-β7 loop (blue) are highlighted. An imbedded inhibitory tract from which an inhibitory peptide can be derived (α-8, pink) is located near the acidic cleft. D, sequence alignment of the β6-β7 loop region. αAsp-365 and ASIC1 residues implicated in the response to H+ are highlighted (red boxes). E, Na+ self-inhibition of the Trp mutants is reported as 100 × [1 − (ISS − Iamiloride)/(IPeak − Iamiloride)]. Trp mutation at sites flanking αLeu-369 strongly reduced Na+ self-inhibition, including at acidic residue αAsp-365. *, p < 0.002 versus the wild type (n = 6–15 for mutants and 115 for the wild type). Model positions of mutations are noted.
Mutation of Sites in the Acidic Cleft Affect Na+ Self-inhibition
Our model of the α subunit features an acidic region at the juncture of the finger, β-ball, and thumb domains (Fig. 1C), which has the appearance of an acidic cleft (24). A homologous region in ASIC1 has been proposed to play a role in proton sensing (16). We hypothesized that residues in the vicinity of the acidic cleft of ENaC subunits coordinate Na+, the first step in the Na+ self-inhibition response.
To examine whether mutations of residues in the acidic cleft weakened Na+ self-inhibition, we performed a Trp mutagenesis screen of acidic cleft Asp and Glu residues (Fig. 1C, red spheres) and measured the effect of mutation on Na+ self-inhibition. Wild-type or mutant ENaCs were expressed in Xenopus laevis oocytes, and ENaC currents were measured by two-electrode voltage clamp. Because ENaC Na+ self-inhibition manifests slowly relative to bath solution exchange, acutely raising the bath [Na+] from a low concentration (where the channel open probability (Po) is high) results in a peak inward current that subsequently declines (8, 11), reflecting Na+ binding events at extracellular allosteric site(s) that are transduced to reduce channel Po (i.e. Na+ self-inhibition) (Fig. 1B) (12, 13, 29). Of the acidic sites tested, we found that the Trp mutation at αAsp-176, αGlu-362, and αAsp-365 reduced Na+ self-inhibition (Fig. 1E, red columns). The latter two lie at the beginning of a loop connecting the sixth and seventh β strands (Fig. 1C; β6-β7 loop), whereas αAsp-176 on the α1 helix interacts with the β6-β7 loop (Fig. 2A).
FIGURE 2.
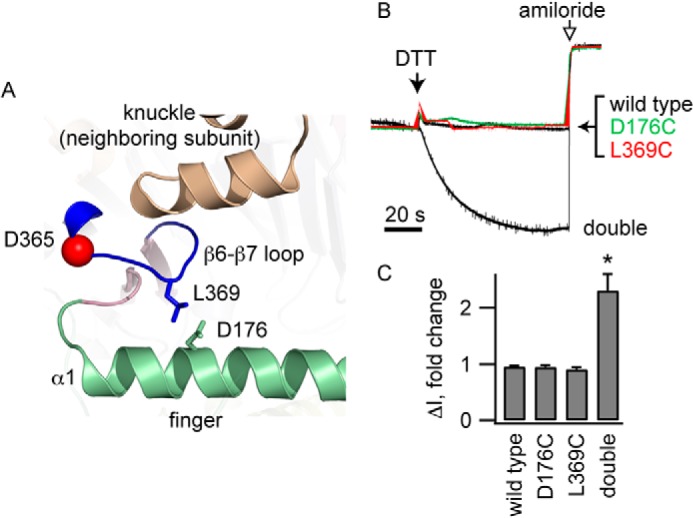
Cross-linking the β6-β7 loop to the α1 helix reduces channel activity. A, Leu-369 on the β6-β7 loop and Asp-176 on helix α1 are adjacent in our model of the α subunit. B, oocytes expressing wild type ENaC, either of the α subunit single mutants (αD176C or αL369C) or the αD176C/L369C double mutant, were exposed to 10 mm DTT and then amiloride (10 μm) while currents were measured at a holding potential of −100 mV. Representative recordings are shown. Currents were normalized to the current immediately prior to DTT addition. Average currents prior to DTT were −1.7 ± 0.4 μA (wild type), −1.5 ± 0.3 μA (αD176C), −1.9 ± 0.4 μA (αL369C), and −1.1 ± 0.2 μA (αD176C/L369C). C, amiloride-sensitive currents following DTT addition normalized to currents prior to DTT (n = 7–10). *, p < 0.01.
We hypothesized that β6-β7 loop residues participate in Na+ binding. We performed Trp scanning mutagenesis of the residues lining the β6-β7 loop and determined the effects of individual mutations on Na+ self-inhibition (Fig. 1E). In our model, the β6-β7 loop extends from the β-ball toward helix α1 of the finger domain, with αLeu-369 at the apex of the loop interacting with αAsp-176 from helix α1 of the finger domain (Figs. 1C and 2A). We found that a Trp mutation at multiple residues flanking αLeu-369 reduced Na+ self-inhibition, whereas αL369W enhanced Na+ self-inhibition.
If Na+ binding to residues in the β6-β7 loop results in a decrease in channel activity, then the conformation of the β6-β7 loop must be linked to the conformation of the channel gate. It follows that constraining movement of the loop should alter channel activity. We used a cross-linking approach to test this hypothesis. Our model suggested that αLeu-369 at the apex of the β6-β7 loop interacts with αAsp-176 from helix α1 of the finger domain (Fig. 2A). We introduced Cys at αLeu-369 in the β6-β7 loop and, simultaneously, to αAsp-176 in helix α1 of the finger domain. Because αL369C and αD176C are predicted to be in close proximity, we examined whether these introduced Cys residues formed a natural disulfide bond, linking the β6-β7 loop with the α1 helix. DTT addition activated ENaC-bearing Cys residues at both sites (Fig. 2, B and C). This activation required the presence of both introduced Cys residues because DTT did not affect the wild type or channels with a single Cys substitution. The DTT-treated double mutant had modestly reduced Na+ self-inhibition compared with DTT-treated wild-type channels (22.8 ± 0.5% versus 38 ± 2%, n = 4, p < 0.05), suggesting that the introduced cysteines residues, per se, modestly altered channel function. These data suggest that conformational changes that involve the β6-β7 loop affect channel activity.
Residues in the Acidic Cleft Determine the Cation Specificity of Inhibition
To identify residues involved in the Na+ binding step, we measured the effect of specific mutations on the ligand specificity of Na+ self-inhibition. As noted above, mutations that alter the Na+ self-inhibition response could reflect changes in the Na+ binding step, transduction steps subsequent to Na+ binding, or the relative stability of the open and/or closed states. We reasoned that the ability to discriminate Na+ and Li+ from other cations reflects ligand binding.
To compare the inhibition of ENaC by Na+, Li+, and K+, we examined steady-state Na+ permeability in the presence of different extracellular cations using a method adapted from Bize and Horisberger (9). Amiloride sensitive I-V curves were obtained at steady state in oocytes sequentially perfused with buffers containing NMDG+, Na+, Li+, K+, and then NMDG+ again as the predominant cation (Figs. 3, A and B). We then fit the I-V curves to equations derived from the Goldman-Hodgkin-Katz model of ion flux to determine the relative permeability of ENaC to Na+ (PNa) under each condition (Fig. 3B and equations 1 and 2, see “Experimental Procedures”). Reductions in PNa in Na+, Li+, or K+ relative to PNa in NMDG+ reflect ENaC inhibition by these effectors. We assumed that the Li+/Na+ permeability ratio (PLi/PNa, i.e. the pore selectivity) is 1.6 on the basis of published reports (27, 30) and is not affected by these mutations far from the pore. We assumed that the K+ permeability is 0. We also assumed that intracellular [Na+] ([Na]i) remains steady through the course of the experiment. With these assumptions, fitting each curve to the Goldman-Hodgkin-Katz model depends on two variables, PNa and [Na]i, which have little interdependency for curve fitting purposes (Fig. 3B). We corrected for channel rundown using PNa measurements in NMDG+ at the beginning and end of the experiment and assuming a linear rundown.
FIGURE 3.
Mutations in the acidic cleft alter the cation selectivity of ENaC inhibition. A, ENaC expressing oocytes were perfused for 1 min with each solution, after which currents were determined at varying holding potentials (−140 to 60 mV in 20-mV steps, 0.5 s/step). Amiloride (amil)-supplemented solutions were then perfused for 20 s, followed by measurement of currents at the different holding potentials. B, resultant I-V curves were fit with equation 1 or 2 to derive PNa values in each solution (red, initial NMDG+; green, Na+; blue, Li+; black, K+; orange, final NMDG+). Measurements in NMDG+ were taken at the beginning and end of each experiment to estimate channel rundown (∼2–7%/min), which was used to adjust PNa values for channel rundown. C, correlation between Na+ self-inhibition measured under a constant voltage clamp (Fig. 1B) and the ratio of Na+ permeabilities in Na+ and NMDG+. D, ratio of Na+ permeabilities measured in different bath solutions: Li+ versus Na+ (top panel) and K+ versus Na+ (bottom panel). *, p < 0.003 versus wild type (n = 40 for the wild type and 5–12 for mutants).
The extent of Na+ inhibition measured by this method correlated with Na+ self-inhibition measured by rapidly switching solutions from NMDG+ to Na+ (Fig. 1B). Na+ self-inhibition is given by the ratio of PNa with extracellular Na+ to PNa with extracellular NMDG+ (PNa(Na)/PNa(NMDG)). We plotted the results for the wild type, αD176W, and the β6-β7 loop Trp mutants (Fig. 3C). The fitted line gave a Pearson's correlation coefficient of −0.75 (p < 0.005). Notably, the fitted line is shifted from its theoretical position (the diagonal from (0, 100) to (1, 0)) toward the origin. This may reflect, in part, an underestimation of the peak current (e.g. Fig. 1B), which depends upon the efficiency of solution exchange and the kinetics of channel inhibition.
Wild-type mouse ENaC showed a slightly different effector specificity for channel inhibition (Na+ > Li+ ≫ K+, Fig. 3D) than what has been reported for human ENaC (Na+ ≈ Li+ > K+) (9). Trp substitutions at sites toward the beginning or end of the β6-β7 loop changed this order. For αQ363W, αD365W, and αV373W, Li+ was a more potent inhibitor than Na+, whereas K+ and Na+ had similar inhibitory effects. The mutants αI367W, αS371W, and αT374W exhibited weakened Na+ inhibition relative to K+, whereas αL370W exhibited weakened Li+ inhibition relative to Na+. Trp mutation at other sites neighboring αGln-363 and αVal-373 showed trends toward changing inhibitor specificity but were not significantly different from the wild type.
To corroborate these results, we measured effector specificity using a second method that adapted a fixed-voltage protocol for measuring Na+ self-inhibition (Fig. 1B). First we examined Na+ inhibition by rapidly switching from an NMDG+ solution to a Na+ solution while voltage-clamping the oocyte at −100 mV (Fig. 4). As described above, this leads to a rapid inward peak current (IPeak) that declines to a steady-state level (ISS). The fall from IPeak to ISS reflects Na+ self-inhibition. Because ENaC also conducts Li+, we measured Li+ inhibition in a similar manner by rapidly switching from an NMDG+ solution to a Li+ solution. Because ENaC does not appreciably conduct K+, we measured K+ inhibition indirectly by examining the effect of K+ pretreatment on Na+ inhibition. Instead of initially bathing the oocyte in an NMDG+ solution, where ENaC Po is high (12), we used a K+ solution. If channel inhibition by K+ is much weaker than by Na+, switching from NMDG+ or K+ to Na+ should produce similar Na+ self-inhibition responses. This is what we observed for wild-type channels (Fig. 4). If channel inhibition by K+ and Na+ are similar, then pretreatment with K+ should attenuate the inhibitory response to Na+, as we observed with the αV373W and αD365W mutants (Fig. 4). If K+ inhibits an ENaC mutant more effectively than Na+, then switching from a K+ to an Na+ bath should induce a rise in current that represents Na+ relief of K+ inhibition. This was seen with the αQ363W mutant (Fig. 4). Using this fixed-voltage protocol, we also found that Na+ was a more effective inhibitor than Li+ of wild-type mouse ENaC. αN364W exhibited a pattern similar to the wild type. For αV373W and αD365W, this pattern was reversed. For αQ363W, Li+ and Na+ were similarly effective inhibitors. For the wild type and αN364W, K+ pretreatment did not alter Na+ self-inhibition when compared with NMDG+ pretreatment.
FIGURE 4.
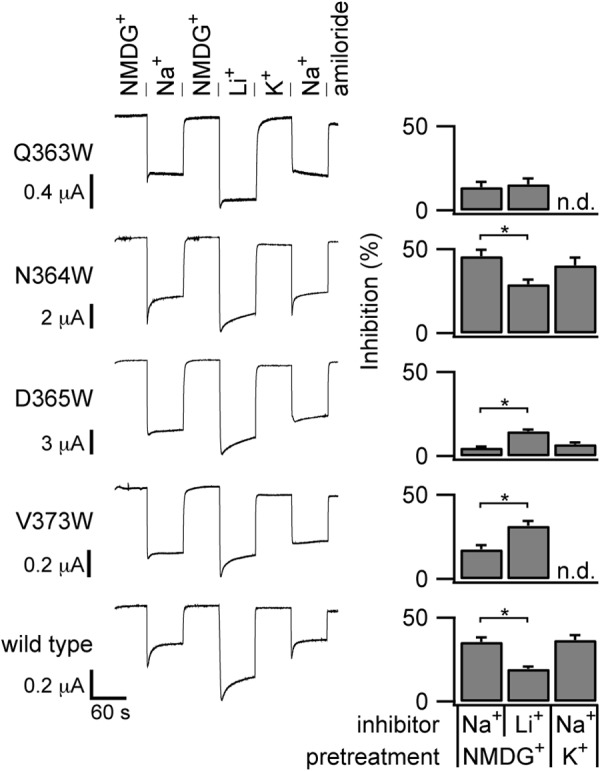
Cation specificity of the inhibitory response measured with a constant voltage clamp. ENaC-expressing oocytes were voltage-clamped at −100 mV during current measurements. Perfusates with 100 mm of the indicated cation were switched every 60 s. Amiloride was added to the Na+ solution at the end of each experiment. Inhibition upon Na+ or Li+ addition was calculated as in Fig. 1E. Quantified results are from 18 wild-type experiments and six to nine experiments for each mutant. *, p < 0.05 by paired Student's t test. n.d., not determined.
Sites Adjacent to the β6-β7 Loop Affect Inhibitor Specificity
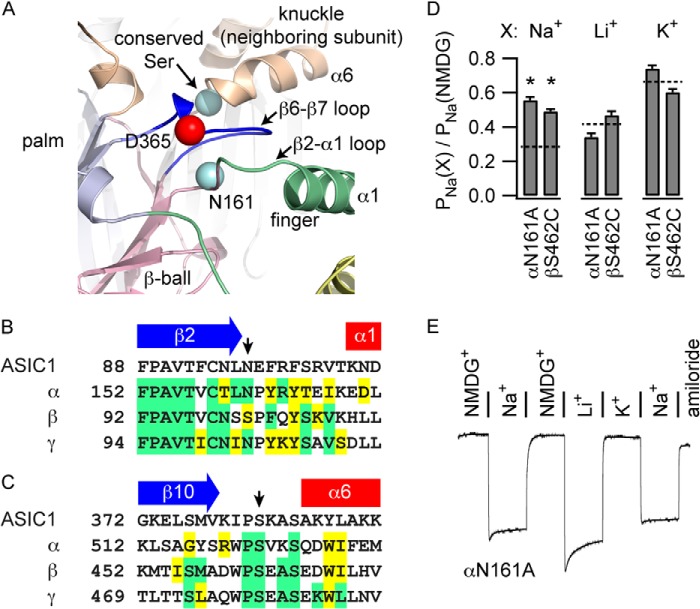
Mutations in the β6-β7 loop affect the cation selectivity of channel inhibition. Our model of a hypothetical ENaC α trimer suggested that adjacent sites could coordinate a bound ion in concert with sites in the β6-β7 loop (Fig. 5A). αAsn-161 is at the end of a highly conserved sequence in the β2-α1 loop connecting the β ball and the finger domain in the α subunit and lies in proximity to the β6-β7 loop (Fig. 5B). The first Ser within a WPSXXS motif in the knuckle domain of a neighboring subunit lies across the plane of the α subunit β6-β7 loop (Fig. 5C). According to recent reports, the neighboring knuckle domain is part of the β subunit (31, 32). Notably, a polymorphism of the Trp residue in the WPSXXS motif of the human α subunit (αW493R) was associated with a loss of Na+ self-inhibition and an increased channel Po (33). We mutated αAsn-161 to Ala and βSer-462 to Cys and assessed the resultant inhibitor specificity. We compared PNa values derived from steady state I-V curves using different perfusion buffers as described above. Both αN161A and βS462C weakened Na+ inhibition but did not affect Li+ or K+ inhibition (Fig. 5D). We corroborated these results for αN161A using the fixed-voltage protocol for measuring self-inhibition induced by Na+ or Li+ (Fig. 5E), which showed a pattern of inhibition similar to αD365W and αV373W and different from wild-type ENaC (Fig. 4).
FIGURE 5.
Specific conserved sites on structures near αD365 weaken Na+ inhibition. A, structures near the β6-β7 loop in an ENaC α trimer model, including the β2-α1 loop of the α subunit and the beginning of helix α6 of the neighboring β subunit. B and C, sequence alignments of the β2-α1 loop (B) and the β10-α6 loop (C), with sites of interest indicated by arrows. D, PNa in various bath solutions (X) was determined as described for Fig. 3. PNa values where Na+, Li+, or K+ were the predominant cation were normalized to PNa determined in NMDG+. *, p < 0.003 versus wild (n = 40 for the wild type and 8 for mutants). Values for the wild type are indicated by dashed lines. E, representative experiment examining the effector specificity of inhibition of αN161A under a constant voltage clamp. Quantification of eight experiments gave values of 10.9 ± 0.9% for Na+ inhibition following NMDG+ pretreatment, 25 ± 1% for Li+ inhibition, and 7 ± 1% for Na+ inhibition following K+ pretreatment. Comparable wild type values were 36 ± 3%, 19 ± 2%, and 34 ± 3%, respectively (n = 6). Na+ inhibition measurements following NMDG+ or K+ perfusion was weaker in both cases for αN161A compared with the wild type (p < 0.05, Student's t test). K+ pretreatment did not affect Na+ inhibition of either the wild type or αN161A compared with NMDG+ pretreatment (p = not significant, paired Student's t test). In contrast to the wild type, Li+ inhibition was greater than Na+ inhibition for αN161A channels (p < 0.05, paired Student's t test).
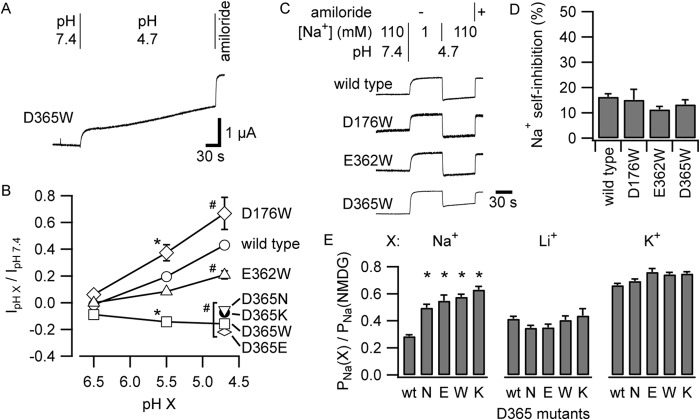
Protonation of αAsp-365 Activates ENaC
Acidification of the extracellular solution increased mouse ENaC currents (Fig. 1A). We hypothesized that protonation of a Na+-coordinating residue weakens Na+ binding, diminishing Na+ self-inhibition and activating the channel. To identify titratable residues responsible for this activation, we performed titrations of wild-type channels and three mutants: αD176W, αE362W, and αD365W (Fig. 6B). We performed each experiment as described for Fig. 1A and measured the acute amiloride-sensitive change in current upon acidification from pH 7.4. Although each mutant blunts Na+ self-inhibition (Fig. 1), we observed that only αD365W reversed the effect of acidification on ENaC currents, causing currents to modestly fall instead of rise (Figs. 1A and 6, A and B). Channels bearing αD176W or αE362W responded to acidification similarly to wild-type. For all groups tested, effects at pH 5.5 were intermediate to effects at pH 4.7. In contrast to human ENaC (10), effects at pH 6.5 were minor for all groups tested.
FIGURE 6.
αAsp-365 mutants lack acid-dependent channel activation. A, acidification inhibits αD365W currents. B, effects of pH titration on the wild type and select mutants of acidic residues in the acidic cleft. Each experiment was performed as in Figs. 1A and 6A while varying the final pH. The acute amiloride-sensitive change in current upon acidification to pH 6.5, 5.5, or 4.7 was quantified. For experiments in which currents increased as a result of acidification, we used the maximal increase in current, which occurred at 10 ± 1 s after initiating solution exchange (Fig. 1A). For experiments in which the current decreased as a result of acidification, there was no transient nadir because both the fast and slow phases of the change affected the current in the same direction (A). For quantification of the acute effect of changing pH on current, we used the current value at 10 s after initiating solution exchange. *, p < 0.01 versus the wild type at pH 5.5; #, p < 0.005 versus the wild type at pH 4.7. For all groups, n = 5–8. C, acidification weakens Na+ self-inhibition. Shown are representative current recordings of oocytes expressing the wild type or α subunit mutants and voltage-clamped at −100 mV. Solutions were exchanged rapidly as indicated. D, Na+ self-inhibition was quantified at pH 4.7 as for Fig. 1E. No significant differences were detected (n = 5–8). E, experiments were performed as described for Fig. 3. PNa values where Na+, Li+, or K+ were the principal cation were normalized to PNa determined in NMDG+. *, p < 0.003 versus the wild type (n = 40 for the wild type and 5–8 for mutants).
We also examined the effect of acidification on the Na+ self-inhibition response. At pH 4.7, the wild type and the three Trp mutants exhibited low Na+ self-inhibition responses (Fig. 6, C and D). These data, taken together, suggest that reducing the pH to 4.7 or altering the Asp group at αAsp-365 removes an important coordinating group from one of the putative Na+ binding sites of ENaC. To further address this hypothesis, we examined the response of the channel to acidification after introducing conservative Glu or Asn substitutions at αAsp-365 or after reversing the charge by Lys substitution. All three αAsp-365 mutants (Glu, Asn, and Lys) prevented channel activation by acidification, echoing our result with αD365W (Fig. 6B). We also examined the effect of these mutations on Na+, Li+, or K+ inhibition at steady state using the voltage ramp method (Fig. 3). We found that each mutant specifically weakened Na+ inhibition (Fig. 6E). Our data suggest that the specific moiety at position α365, rather than charge per se, is needed to confer “full” sensitivity to external Na+.
DISCUSSION
ENaCs are members of a family of ion channels that respond to factors in the extracellular environment. One of the external factors that regulate ENaC is Na+. There are multiple lines of evidence supporting the notion that Na+ binding to one or more effector sites within the extracellular domains results in the inhibition of channel activity in an allosteric manner (1, 12, 13, 29, 34). Given that allosteric transitions are required for this process, it is not surprising that many substitutions throughout the extracellular regions of ENaC subunits altered the Na+ self-inhibition response. For example, we and others have reported mutations that weaken Na+ self-inhibition in the palm domain, thumb domain, β ball-thumb domain interface, knuckle domain, finger domain, and subunit interfaces and among the Cys involved in disulfide bridges (13, 25, 28, 31, 33, 35–39). Furthermore, it is not surprising that mutations in transmembrane helical residues that interfere with pore closure and increase channel Po (e.g. βS518K) significantly weaken Na+ self-inhibition (38, 40). Likewise, other maneuvers that increase channel Po, e.g. Zn2+ addition (41) or Cl− removal (42), weaken apparent Na+ self-inhibition. In summary, on the basis of previous studies, it has been difficult to discern whether specific mutations that change Na+ self-inhibition do so by altering Na+ binding, downstream transduction steps, or the baseline stability of the open or closed states of the channel.
We postulated that Na+ binds to a site defined, in part, by acidic residues, consistent with Na+-bound protein structures (43, 44) and the predominance of oxygen atoms coordinating Na+ in model small molecules (45). We also reasoned that the preference of the channel for Na+ over other cations as an inhibitory effector originates from the binding site(s) rather than from subsequent transduction steps. On these premises, we tested the ability of α subunit acidic cleft mutants to both affect Na+ self-inhibition and alter the relative inhibitory efficacy of Na+, Li+, and K+. Mutation of αAsp-365 or neighboring residues in our α subunit model weakened inhibition by external Na+ and altered the cation selectivity of channel inhibition. Even conservative αAsp-365 mutations (Glu or Asn) had this effect. αAsp-365 substitutions also prevented channel activation by acidification regardless of whether a conservative or non-conservative substitution was used. These results support the notion that αAsp-365 has a role in Na+ binding and the subsequent reduction in channel Po, and that acidification activates ENaC by protonating αAsp-365 and disrupting Na+ binding at an effector site that includes αAsp-365.
Our results also suggest that there are multiple effector sites for Na+ self-inhibition. All mutants tested at or near αAsp-365 retained at least a small Na+ self-inhibition response. Acidification had no effect on the Na+ self-inhibition response of αD365W, but reduced the response of the wild type and other mutants to similarly low but measurable values (compare Figs. 1E and 6D). We propose that there are additional Na+ effector sites. Because αD365W has a small but measurable Na+ self-inhibition response, at least one of the additional effector sites does not depend on residues titratable in the pH 7.4–4.7 range. Accordingly, conservative mutations of the β and γ sites that correspond to αAsp-365 (βD302N and γE322Q) exhibited Na+ self-inhibition similar to the wild type (βD302N, 33 ± 4%; γE322Q, 27 ± 3%; wild type, 32 ± 2%; n = 5–6; p = not significant).
Other members of the ENaC/Deg family have an Asp or Glu residue at the position equivalent to mouse αAsp-365 (e.g. Glu-229 in chicken ASIC1, Fig. 1D). This residue lies at the beginning of the β6-β7 loop, which connects the β-ball and palm domains (1, 16). Electrostatic calculations for the ASIC1 Glu (equivalent to αAsp-365) predicted a pKa of 7.4, a prerequisite for proton sensing in the physiological range (17). However, mutating this residue in ASIC1 to Gln had little effect on the pH sensitivity of channel activation (17, 18). The δ subunit of ENaC replaces the α subunit in the channel complex expressed in some tissues of several species, including humans (but not rodents). Interestingly, human δβγ channels are poorly inhibited by external Na+ (46). The δ subunit has a Pro residue at the equivalent site to αAsp-365. However, mutating δPro-314 to Asp did not sensitize human δβγ ENaC to external Na+ (data not shown). This observation suggests that there are residues in addition to αAsp-365 that are needed to form a Na+ binding site in the α subunit.
The β6-β7 loop of ASIC1 interacts with the finger and thumb domains and is six residues longer than its ENaC subunit counterparts (Fig. 1D). In our α subunit model, the shorter β6-β7 loop approaches the α1 helix in the finger domain but does not abut thumb domain residues (Fig. 1C) (24). Our results demonstrating that αD176C in the α1 helix and αL369C in the β6-β7 loop form a disulfide bond suggest that these structures are indeed in close proximity and that stabilizing interactions between the α1 helix and the β6-β7 loop is inhibitory. The ASIC1 β6-β7 loop includes Asp-238 and Glu-239 (cASIC1 numbering, Fig. 1D) that pair with thumb domain residues Asp-350 and Asp-346, respectively. These thumb domain residues have been proposed to sense protons (16). Krauson et al. (19) showed that mutating Asp-346 in mouse ASIC1a results in a biphasic pH activation curve, suggesting that this mutation specifically weakened of one of multiple proton sensing sites. ASIC1 residues Asp-238 and Glu-239 and ENaC residues α371–373 are located in a similar region of the β6-β7 loop (Fig. 1D), where α subunit Trp substitutions weaken Na+ self-inhibition and alter the cation selectivity of this process (Figs. 1, 3, and 4). Baconguis and Gouaux (47) provided additional evidence for the role of the β6-β7 loop in modulating channel gating in response to external factors. They found that psalmotoxin, an ASIC1 inhibitor, extends an Arg-rich hairpin into the acidic pocket and forms hydrogen bonds with the β6-β7 loop backbone (47). These previous observations and our current findings, taken together, suggest that the β6-β7 loop has evolved within members of the ENaC/degenerin family members to facilitate the sensing of specific factors.
This work was supported, in whole or in part by National Institutes of Health Grants R01 DK051391 (to T. R. K.), R01 DK098204 and K01 DK078734 (to O. B. K.), P30 DK079307, and T32 DK061296.
- ENaC
- epithelial Na+ channel
- ASIC
- acid-sensing ion channel 1
- PNa
- Na+ permeability
- I-V
- current-voltage
- NMDG
- N-methyl-d-glucamine
- Po
- open probability
- IPeak
- peak current
- ISS
- steady-state current.
REFERENCES
- 1. Kashlan O. B., Kleyman T. R. (2011) ENaC structure and function in the wake of a resolved structure of a family member. Am. J. Physiol. Renal Physiol. 301, F684–F696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhalla V., Hallows K. R. (2008) Mechanisms of ENaC regulation and clinical implications. J. Am. Soc. Nephrol. 19, 1845–1854 [DOI] [PubMed] [Google Scholar]
- 3. Pearce D., Soundararajan R., Trimpert C., Kashlan O. B., Deen P. M., Kohan D. E. (2014) Collecting duct principal cell transport processes and their regulation. Clin. J. Am. Soc. Nephrol. in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duc C., Farman N., Canessa C. M., Bonvalet J. P., Rossier B. C. (1994) Cell-specific expression of epithelial sodium channel α, β, and γ subunits in aldosterone-responsive epithelia from the rat: localization by in situ hybridization and immunocytochemistry. J. Cell Biol. 127, 1907–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kretz O., Barbry P., Bock R., Lindemann B. (1999) Differential expression of RNA and protein of the three pore-forming subunits of the amiloride-sensitive epithelial sodium channel in taste buds of the rat. J. Histochem. Cytochem. 47, 51–64 [DOI] [PubMed] [Google Scholar]
- 6. Gründer S., Müller A., Ruppersberg J. P. (2001) Developmental and cellular expression pattern of epithelial sodium channel α, β, and γ subunits in the inner ear of the rat. Eur. J. Neurosci. 13, 641–648 [DOI] [PubMed] [Google Scholar]
- 7. Drummond H. A., Gebremedhin D., Harder D. R. (2004) Degenerin/epithelial Na+ channel proteins: components of a vascular mechanosensor. Hypertension 44, 643–648 [DOI] [PubMed] [Google Scholar]
- 8. Fuchs W., Larsen E. H., Lindemann B. (1977) Current-voltage curve of sodium channels and concentration dependence of sodium permeability in frog skin. J. Physiol. 267, 137–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bize V., Horisberger J. D. (2007) Sodium self-inhibition of human epithelial sodium channel: selectivity and affinity of the extracellular sodium sensing site. Am. J. Physiol. Renal Physiol. 293, F1137–F1146 [DOI] [PubMed] [Google Scholar]
- 10. Collier D. M., Snyder P. M. (2009) Extracellular protons regulate human ENaC by modulating Na+ self-inhibition. J. Biol. Chem. 284, 792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheng S., Bruns J. B., Kleyman T. R. (2004) Extracellular histidine residues crucial for Na+ self-inhibition of epithelial Na+ channels. J. Biol. Chem. 279, 9743–9749 [DOI] [PubMed] [Google Scholar]
- 12. Sheng S., Carattino M. D., Bruns J. B., Hughey R. P., Kleyman T. R. (2006) Furin cleavage activates the epithelial Na+ channel by relieving Na+ self-inhibition. Am. J. Physiol. Renal Physiol. 290, F1488–F1496 [DOI] [PubMed] [Google Scholar]
- 13. Maarouf A. B., Sheng N., Chen J., Winarski K. L., Okumura S., Carattino M. D., Boyd C. R., Kleyman T. R., Sheng S. (2009) Novel determinants of epithelial sodium channel gating within extracellular thumb domains. J. Biol. Chem. 284, 7756–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anantharam A., Tian Y., Palmer L. G. (2006) Open probability of the epithelial sodium channel is regulated by intracellular sodium. J. Physiol. 574, 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kellenberger S., Gautschi I., Rossier B. C., Schild L. (1998) Mutations causing Liddle syndrome reduce sodium-dependent downregulation of the epithelial sodium channel in the Xenopus oocyte expression system. J. Clin. Invest. 101, 2741–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 17. Liechti L. A., Bernèche S., Bargeton B., Iwaszkiewicz J., Roy S., Michielin O., Kellenberger S. (2010) A combined computational and functional approach identifies new residues involved in pH-dependent gating of ASIC1a. J. Biol. Chem. 285, 16315–16329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paukert M., Chen X., Polleichtner G., Schindelin H., Gründer S. (2008) Candidate amino acids involved in H+ gating of acid-sensing ion channel 1a. J. Biol. Chem. 283, 572–581 [DOI] [PubMed] [Google Scholar]
- 19. Krauson A. J., Rued A. C., Carattino M. D. (2013) Independent contribution of extracellular proton binding sites to ASIC1a activation. J. Biol. Chem. 288, 34375–34383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 21. Hughey R. P., Bruns J. B., Kinlough C. L., Kleyman T. R. (2004) Distinct pools of epithelial sodium channels are expressed at the plasma membrane. J. Biol. Chem. 279, 48491–48494 [DOI] [PubMed] [Google Scholar]
- 22. Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the g-subunit. J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 23. Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., Kleyman T. R. (2006) The epithelial Na+ channel is inhibited by a peptide derived from proteolytic processing of its a subunit. J. Biol. Chem. 281, 18901–18907 [DOI] [PubMed] [Google Scholar]
- 24. Kashlan O. B., Adelman J. L., Okumura S., Blobner B. M., Zuzek Z., Hughey R. P., Kleyman T. R., Grabe M. (2011) Constraint-based, homology model of the extracellular domain of the epithelial Na+ channel a subunit reveals a mechanism of channel activation by proteases. J. Biol. Chem. 286, 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kashlan O. B., Boyd C. R., Argyropoulos C., Okumura S., Hughey R. P., Grabe M., Kleyman T. R. (2010) Allosteric inhibition of the epithelial Na+ channel (ENaC) through peptide binding at peripheral finger and thumb domains. J. Biol. Chem. 285, 35216–35223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kashlan O. B., Blobner B. M., Zuzek Z., Carattino M. D., Kleyman T. R. (2012) Inhibitory tract traps the epithelial Na+ channel in a low activity conformation. J. Biol. Chem. 287, 20720–20726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kellenberger S., Gautschi I., Schild L. (1999) A single point mutation in the pore region of the epithelial Na+ channel changes ion selectivity by modifying molecular sieving. Proc. Natl. Acad. Sci. U.S.A. 96, 4170–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Collier D. M., Peterson Z. J., Blokhin I. O., Benson C. J., Snyder P. M. (2012) Identification of extracellular domain residues required for epithelial Na+ channel activation by acidic pH. J. Biol. Chem. 287, 40907–40914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chraïbi A., Horisberger J. D. (2002) Na self inhibition of human epithelial Na channel: temperature dependence and effect of extracellular proteases. J. Gen. Physiol. 120, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ishikawa T., Marunaka Y., Rotin D. (1998) Electrophysiological characterization of the rat epithelial Na+ channel (rENaC) expressed in MDCK cells: effects of Na+ and Ca2+. J. Gen. Physiol. 111, 825–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collier D. M., Snyder P. M. (2011) Identification of epithelial Na+ channel (ENaC) intersubunit Cl− inhibitory residues suggests a trimeric agb channel architecture. J. Biol. Chem. 286, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen J., Myerburg M. M., Passero C. J., Winarski K. L., Sheng S. (2011) External Cu2+ inhibits human epithelial Na+ channels by binding at a subunit interface of extracellular domains. J. Biol. Chem. 286, 27436–27446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rauh R., Diakov A., Tzschoppe A., Korbmacher J., Azad A. K., Cuppens H., Cassiman J. J., Dötsch J., Sticht H., Korbmacher C. (2010) A mutation of the epithelial sodium channel associated with atypical cystic fibrosis increases channel open probability and reduces Na+ self inhibition. J. Physiol. 588, 1211–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horisberger J. D., Chraïbi A. (2004) Epithelial sodium channel: a ligand-gated channel? Nephron Physiol. 96, p37–p41 [DOI] [PubMed] [Google Scholar]
- 35. Chen J., Kleyman T. R., Sheng S. (2013) Gain-of-function variant of the human epithelial sodium channel. Am. J. Physiol. Renal Physiol. 304, F207–F213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheng S., Maarouf A. B., Bruns J. B., Hughey R. P., Kleyman T. R. (2007) Functional role of extracellular loop cysteine residues of the epithelial Na+ channel in Na+ self-inhibition. J. Biol. Chem. 282, 20180–20190 [DOI] [PubMed] [Google Scholar]
- 37. Edelheit O., Hanukoglu I., Dascal N., Hanukoglu A. (2011) Identification of the roles of conserved charged residues in the extracellular domain of an epithelial sodium channel (ENaC) subunit by alanine mutagenesis. Am. J. Physiol. Renal Physiol. 300, F887–F897 [DOI] [PubMed] [Google Scholar]
- 38. Winarski K. L., Sheng N., Chen J., Kleyman T. R., Sheng S. (2010) Extracellular allosteric regulatory subdomain within the g subunit of the epithelial Na+ channel. J. Biol. Chem. 285, 26088–26096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edelheit O., Ben-Shahar R., Dascal N., Hanukoglu A., Hanukoglu I. (2014) Conserved charged residues at the surface and interface of epithelial sodium channel subunits-roles in cell surface expression and the sodium self-inhibition response. FEBS J. 281, 2097–2111 [DOI] [PubMed] [Google Scholar]
- 40. Condliffe S. B., Zhang H., Frizzell R. A. (2004) Syntaxin 1A regulates ENaC channel activity. J. Biol. Chem. 279, 10085–10092 [DOI] [PubMed] [Google Scholar]
- 41. Sheng S., Perry C. J., Kleyman T. R. (2004) Extracellular Zn2+ activates epithelial Na+ channels by eliminating Na+ self-inhibition. J. Biol. Chem. 279, 31687–31696 [DOI] [PubMed] [Google Scholar]
- 42. Collier D. M., Snyder P. M. (2009) Extracellular chloride regulates the epithelial sodium channel. J. Biol. Chem. 284, 29320–29325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu W., Chun E., Thompson A. A., Chubukov P., Xu F., Katritch V., Han G. W., Roth C. B., Heitman L. H., Ijzerman A. P., Cherezov V., Stevens R. C. (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Whorton M. R., MacKinnon R. (2013) X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature 498, 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harding M. M. (2002) Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr. D Biol. Crystallogr. 58, 872–874 [DOI] [PubMed] [Google Scholar]
- 46. Ji H. L., Su X. F., Kedar S., Li J., Barbry P., Smith P. R., Matalon S., Benos D. J. (2006) δ-Subunit confers novel biophysical features to α β γ-human epithelial sodium channel (ENaC) via a physical interaction. J. Biol. Chem. 281, 8233–8241 [DOI] [PubMed] [Google Scholar]
- 47. Baconguis I., Gouaux E. (2012) Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]