Background: Type IV pili (T4P) are virulence factors composed of major and minor pilins.
Results: Minor pilins prime pilus assembly and traffic anti-retraction protein PilY1 to the surface; FimU and GspH are structurally similar.
Conclusion: Minor pilins are essential for pilus assembly and function.
Significance: This work expands the structural and functional similarities between the T4P and T2S systems.
Keywords: Adhesin, Bacterial Adhesion, Crystal Structure, Pseudomonas aeruginosa (P. aeruginosa), Type II Secretion System (T2SS), Type IV Pili, Pilin, Pseudopilin, Twitching Motility
Abstract
Type IV pili (T4P) contain hundreds of major subunits, but minor subunits are also required for assembly and function. Here we show that Pseudomonas aeruginosa minor pilins prime pilus assembly and traffic the pilus-associated adhesin and anti-retraction protein, PilY1, to the cell surface. PilV, PilW, and PilX require PilY1 for inclusion in surface pili and vice versa, suggestive of complex formation. PilE requires PilVWXY1 for inclusion, suggesting that it binds a novel interface created by two or more components. FimU is incorporated independently of the others and is proposed to couple the putative minor pilin-PilY1 complex to the major subunit. The production of small amounts of T4P by a mutant lacking the minor pilin operon was traced to expression of minor pseudopilins from the P. aeruginosa type II secretion (T2S) system, showing that under retraction-deficient conditions, T2S minor subunits can prime T4P assembly. Deletion of all minor subunits abrogated pilus assembly. In a strain lacking the minor pseudopilins, PilVWXY1 and either FimU or PilE comprised the minimal set of components required for pilus assembly. Supporting functional conservation of T2S and T4P minor components, our 1.4 Å crystal structure of FimU revealed striking architectural similarity to its T2S ortholog GspH, despite minimal sequence identity. We propose that PilVWXY1 form a priming complex for assembly and that PilE and FimU together stably couple the complex to the major subunit. Trafficking of the anti-retraction factor PilY1 to the cell surface allows for production of pili of sufficient length to support adherence and motility.
Introduction
Type IV pili (T4P)3 are long, thin hair-like protein polymers that mediate functions ranging from attachment to host cells and surfaces, flagella-independent twitching motility, biofilm formation, and DNA uptake to electron transfer in bacteria and archaea (1–3). There are two major classes of T4P: T4aP and T4bP. Twitching motility, which involves repeated rounds of pilus assembly, adherence, and disassembly that pull the cells toward the point of pilus attachment, is associated mainly with the T4aP class (1, 2, 4).
The T4P assembly system is evolutionarily related to the type II secretion (T2S) system, and both are proposed to function by the dynamic assembly and disassembly of pilin subunits. In the T2S system, the resulting short pseudopilus is thought to span only the width of the periplasm of a Gram-negative bacterium. The pilins (called pseudopilins in the T2S system) have a long hydrophobic N-terminal α-helix preceded by a characteristic type III leader sequence and a globular C-terminal domain (5, 6). The subunits are inserted into the membrane by the Sec system as pre-pilins, with the C termini outside the cytoplasmic membrane (7, 8). The pilins are processed by a dedicated pre-pilin peptidase (PilD in Pseudomonas aeruginosa) that cleaves the leader peptide at the cytoplasmic face of the membrane and methylates the mature pilin (9–11). Mature pilins are retained in the membrane prior to assembly and, in the case of T4aP, are thought to return to this compartment upon disassembly for reuse in subsequent assembly cycles (4, 12, 13).
In P. aeruginosa, T4aP are composed mainly of the highly abundant major pilin subunit, PilA, but minor pilins FimU and PilVWXE are also present at low abundance (14, 15). Similar to the major pilin, minor pilins are processed by the pre-prepilin peptidase, PilD (15, 16). The proposed functions of the minor pilins include priming of assembly, counteracting retraction, contributing to adhesion, and/or modulating the balance between pilus extension and retraction (15, 17–20). The P. aeruginosa minor pilins FimU and PilVWXE are encoded in a polycistronic operon with PilY1, a large (∼125 kDa) non-pilin protein implicated in anti-retraction, attachment, and other T4P-related functions (17, 21–24) (Fig. 1). In Neisseria meningitidis and P. aeruginosa, minor pilins are required for surface piliation (15, 22, 25). In Neisseria gonorrhoeae, the minor pilins are dispensable, although mutants have a 10-fold or greater reduction in surface piliation (16). In both P. aeruginosa and N. meningitidis, a few pili are assembled in retraction-deficient double mutants, suggesting that the role of the minor pilins is to optimize assembly (15, 16, 22, 25). Minor pilins are also involved in surface expression of PilY1 and its PilC1/PilC2 counterparts in Neisseria, although the specific mechanisms and components required have not been defined (16, 22).
FIGURE 1.
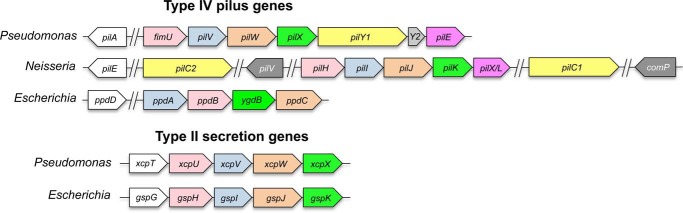
Gene organization of T4P and T2S minor components. The organization of pilin, minor pilin, pseudopilin, and minor pseudopilin genes in type IV pili and type II secretion systems of model organisms P. aeruginosa, E. coli, and Neisseria spp. Equivalent genes are similarly colored. Major subunit genes are shown in white. P. aeruginosa adhesin and anti-retraction factor PilY1 (yellow) is encoded in the minor pilin operon, whereas its Neisseria equivalents, PilC1 and PilC2, are not. Neisseria-specific minor pilin genes pilV and comP (gray) are not clustered with other minor pilin genes. P. aeruginosa gene pilY2, encoded between pilY1 and pilE, may be a pseudogene. Line breaks (//) denote noncontiguous genomic organization.
Minor pilins FimU-PilVWX are homologs of the T2S minor pseudopilins GspHIJK (called XcpUVWX in P. aeruginosa, EpsHIJK in Vibrio spp. and PulHIJK in Klebsiella spp.), suggesting that these four proteins are core components of the assembly machinery (26). There are no equivalents of PilE and PilY1 in the T2S system (27, 28) (Fig. 1). In N. meningitidis, the PilE homolog PilXNm was proposed to antagonize pilus retraction, promoting bacterial cell-cell aggregation (29, 30), and to allow conformational changes in the pilus fiber that are important for T4P-mediated signaling (31).
The minor pseudopilins interact via their globular and transmembrane domains (32, 33). The soluble C-terminal domains of Escherichia coli GspIJK were crystallized as a heterotrimer (34) whose large size and architecture suggested that it forms the tip of the pseudopilus, with GspK at the distal position to avoid steric clashes. Studies of P. aeruginosa minor pseudopilins revealed similar interactions between the C-terminal domains of XcpVWX and XcpUW (35). However, no interactions between truncated versions of the major pseudopilin XcpT and any of the minor pseudopilins were detected (35), although cross-linking studies showed interactions between full-length major pseudopilins and minor pseudopilin XcpU in P. aeruginosa or its Xanthomonas campestris equivalent, XpsH (36, 37). These data imply that the hydrophobic domains normally truncated for biochemical and structural studies are important for interactions between major and minor pilins.
In a recombinant E. coli strain overexpressing the Klebsiella oxytoca major pseudopilin PulG, the PulHIJK minor pseudopilin complex initiated pseudopilus assembly (38–41). In the absence of the minor pseudopilins, only rare pseudopili were observed (41). The PulHIJK complex was proposed to initiate assembly by priming the addition of major subunits or by stimulating the assembly motor through formation of a membrane-distorting pseudohelix (33). The first scenario, which implies that the priming complex becomes part of the resulting filament, was not supported experimentally in that study. Interestingly, Cisneros et al. (41) later showed that the T4P minor pilins of E. coli were interchangeable with the Klebsiella T2S minor pseudopilins for priming of either pilus or pseudopilus assembly. However, the T4aP system of E. coli is atypical, with only four minor pilins instead of five (no PilE equivalent) and no PilY1 homolog, making it more similar to the T2S system. These features may have provided unusually permissive conditions for cross-complementation.
Here, we used genetic, biochemical, and structural studies to investigate the specific roles of the T4aP minor pilins in P. aeruginosa. PilV, PilW, PilX, and the adhesin PilY1 were mutually dependent on each other for inclusion in pili, and all four were required to recruit PilE. In contrast, FimU was incorporated independently and interacted directly in vivo with the major pilin PilA, as well as with minor pilins PilV and PilE. A 1.4 Å high resolution x-ray crystal structure of FimU revealed its strong resemblance to minor pseudopilin GspH, despite minimal sequence identity. P. aeruginosa minor pilins or minor pseudopilins expressed from their native loci primed T4aP assembly and were incorporated into surface-exposed pili, although the full set of T4aP components including PilY1 was required to produce pili in retraction-proficient backgrounds. Together, the data suggest that the T4aP minor pilins form a complex that primes pilus assembly and traffics the T4aP-specific anti-retraction component PilY1 to the cell surface.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Bacterial strains were stored at −80 °C in Luria-Bertani broth with 15% glycerol. P. aeruginosa strains were grown at 37 °C on LB agar plates supplemented with gentamicin (30 μg/ml) or carbenicillin (200 μg/ml). E. coli strains were grown at 37 °C on LB agar plates supplemented with gentamicin (15 μg/ml), ampicillin (100 μg/ml), or kanamycin (50 μg/ml), as appropriate.
Generation of Mutants and Complementation Constructs
Minor pilin and minor pseudopilin operon deletion mutants of P. aeruginosa were generated by biparental mating with E. coli SM10. 1 kb upstream and downstream of the minor pilin or minor pseudopilin operon were amplified from mPAO1 and cloned into pEX18Gm or pEX18Ap with a GmFRT cassette inserted between the fragments for selection. The pEX18Gm-ΔMP and pEX18Ap-ΔMPP::GmFRT were transformed into E. coli SM10 for biparental mating with mPAO1 wild-type and pilT::Tn strains. The gentamicin resistance cassette was excised using Flp recombinase. Mutants were verified through PCR.
Minor pilin complementation constructs were made previously (15). The pilY1 complementation construct and minor pseudopilin xcpUVWX complementation constructs were created by PCR-amplifying genes from mPAO1 using Taq polymerase. pilY1 was ligated into pCR 2.1 (Invitrogen) and subcloned into pBADGr using the EcoRI restriction sites. The xcpUVWX PCR product was digested with EcoRI and HindIII (Thermo Scientific) and ligated with T4 DNA Ligase (Invitrogen) into pUCp20 linearized with the same enzymes. Ligated constructs were transformed into E. coli DH5α and verified by DNA sequencing.
Isolation of Sheared Surface Proteins, SDS-PAGE Analysis
Surface proteins were isolated as previously described (42) with the following modifications. Bacterial strains were streaked in a cross-hatched fashion on a large (150 × 15 mm) 1.5% LB-agar plate containing gentamicin (30 mg/L) or no antibiotic and grown overnight at 37 °C. Centrifugation was performed at 16,100 × g. The pellets for each sample were pooled and resuspended in 120 μl of 1× SDS loading buffer.
Western Blot Analysis of Sheared Surface Fractions
Sheared surface protein samples were separated on 15% SDS-PAGE gels for α-FimU, PilV, PilX, and PilE blots; 12.5% gels for α-PilW and α-XcpW blots; and 7.5% gels for α-PilY1 blots. Proteins were transferred to nitrocellulose membrane and blocked with 5% (w/v) skim milk at room temperature for 3 h. Blots were incubated with rabbit polyclonal α-FimU (15), PilV, PilW (15), PilY1 (gift from Matthew Wolfgang) (24), PilE (15), and XcpW (gift from Romé Voulhoux) at a 1:1000 dilution or α-PilX antibody at a 1:500 dilution overnight at 4 °C. Blots were washed with 1× PBS three times for 10 min each and incubated with secondary goat anti-rabbit IgG-alkaline phosphatase conjugated antibody (Bio-Rad) for 1 h at room temperature. Blots were washed as above and developed using NBT/BCIP.
Interaction of FimU with PilA and Minor Pilins
Direct protein-protein interactions were tested using the bacterial adenylate cyclase two-hybrid assay as described in Ref. 43 with modifications. Briefly, full-length mature FimU was N-terminally tagged with T18 by cloning it in frame into pUT18C, whereas full-length mature PilA and PilV/W/X and PilE were N-terminally tagged with T25 using pKT25. pUT18C-fimU was co-transformed with pKT25-pilA/V/W/X/E into the cya mutant strain BTH101 (F-, cya-99, araD139, galE15, galK16, rpsL1 (Str r), hsdR2, mcrA1, and mcrB1). Three colonies of similar size were selected and grown overnight at 30 °C followed by subculturing with 1 mm isopropyl β-d-thiogalactopyranoside for induction. Cells were grown to A600 = 0.6. These were spot plated on LB Agar + X-gal and McConkey + maltose and incubated at 30 °C for 24 h. 1 ml of cells was harvested to test β-galactosidase activity using a modified β-galactosidase enzyme assay system with reporter lysis buffer (Promega) where cells were resuspended in 1× PBS and lysed with FastPrep for 20 s prior to following the manufacturer's protocol.
Expression and Purification of FimU, PilV, and PilX
Truncated form of PAO1 fimU encoding 1–28 amino acid N-terminally truncated mature protein was PCR-amplified and ligated into the pET151/d-TOPO vector (Invitrogen) to generate an N-terminally His6 and V5 epitope-tagged protein with TEV protease cleavage site for subsequent removal of the tags. Expression and purification of FimU was performed previously (15) with a few changes. Briefly, E. coli Origami B (DE3) cells expressing HisV5-FimUΔ1–28 were grown overnight at 16 °C following induction of protein expression with isopropyl β-d-thiogalactopyranoside (final concentration, 1.0 mm). Cells were lysed by French Press, and proteins were purified by nickel affinity chromatography, followed by TEV cleavage and a second nickel affinity purification step. Fractions from the second nickel purification containing purified protein were pooled, buffer-exchanged using a HiPrep 26/10 desalting column (GE Healthcare Canada) into 20 mm Tris, pH 8, 50 mm NaCl, and concentrated to 25 mg/ml using Vivaspin 15R (Sartorius Stedim Biotech) concentrators. For SAD phasing, selenomethionine (SeMet)-labeled proteins were expressed in Origami B (DE3) using SeMet high yield M9 minimal medium (Shanghai Medicilon) according to the manufacturer's instructions and purified as above.
For polyclonal antibody production, truncated forms of pilV and pilX, encoding, respectively, 1–28-amino acid and 1–25-amino acid N-terminally truncated proteins were PCR-amplified and ligated into pET151/d-TOPO as above. PilV was expressed and purified as above and concentrated to 1 mg/ml. PilX was expressed and lysed as above. To purify the protein from inclusion bodies under denaturing conditions, the pellet was resuspended in 100 mm NaH2PO4, 10 mm Tris, pH 8, 6 m guanidine HCl, and centrifuged for 20 min at 11,952 × g. The supernatant was loaded onto a nickel affinity chromatography column and purified protein eluted with 250 mm imidazole following a wash with 25 mm imidazole. Both purified PilV and PilX were dialyzed into 1× PBS and sent to Cedarlane Laboratories (Burlington, Canada) for immunization of rabbits.
Structure Determination
FimU crystals were grown using the hanging drop/vapor diffusion method. Native protein crystals grew in a 1:1 ratio of protein (25 mg/ml FimU in 20 mm Tris, pH 8, 50 mm NaCl) and precipitant solution (0.2 m sodium chloride, 0.1 m Tris-HCl, pH 8.5, and 25% (w/v) PEG 3350) at 4 °C. FimU SeMet crystals were grown at 20 °C using a 2:1 ratio of protein: precipitant (0.2 m ammonium formate, pH 6.6, and 20% (w/v) PEG 3350). All crystals were flash frozen directly in a nitrogen cold stream (100 K) with no further cryo-protection. Diffraction data sets for native and SeMet crystals were collected at wavelengths of 1.1 and 0.979 Å, respectively at the Beamlines X29 and X25 of the National Synchrotron Light Source (Brookhaven, NY).
Data sets were processed using the HKL2000 program suite (44). Using HYSS (45), the expected SeMet sites were located. Phasing, density modification, auto model building, and refinement were carried out using the PHENIX suite of programs (45, 46). Using Coot (47), iterative rounds of manual model building and refinement were performed until R and Rfree values converged and could no longer be improved. The structure of native FimUΔ1–28 was determined by molecular replacement using the FimUΔ1–28-SeMet structure as an initial search model. Molecular replacement and model refinement were performed using the PHENIX suite of programs (45, 46). Stereochemical quality of the models were verified using PROCHECK (48). Data collection and model refinement statistics for both are listed in greater detail in Table 1. Structural illustrations were generated with PyMOL (49).
TABLE 1.
Data collection and refinement statistics
| SeMet-FimU | Native FimU | |
|---|---|---|
| Data collection | ||
| Beamline | NSLS X25 | NSLS X29 |
| Wavelength | 0.979 | 1.100 |
| Space group | P21 | P65 |
| Unit cell parameters | ||
| a, b, c (Å) | 61.98, 37.25, 103.1 | 40.69, 40.69, 272.92 |
| α, β, γ (°) | 90.0, 105.3, 90.0 | 90.0, 90.0, 120.0 |
| No. of mol in ASU | 4 | 2 |
| Resolution range (Å)a | 50.0–1.35 (1.37–1.35) | 50.0–1.40 (1.42–1.40) |
| Unique reflections | 99, 928 | 49, 829 |
| Data redundancya | 6.3 (4.7) | 15.4 (8.7) |
| Completeness (%)a | 99.8 (96.9) | 99.4 (93.9) |
| I/σ(I)a | 20.5 (3.2) | 53.8 (5.9) |
| Rmerge (%)a | 6.5 (42.4) | 4.4 (35.4) |
| Wilson B | 14.1 | 12.94 |
| Model and refinement | ||
| Resolution range (Å) | 49.76–1.35 | 35.25–1.40 |
| Rwork (%) | 17.63 | 16.42 |
| Rfree (%) | 20.27 | 19.59 |
| No. of reflections | 99, 216 | 48, 996 |
| No. of amino acid residues/atoms | 4,733 | 2,315 |
| No. of waters | 821 | 528 |
| Root mean square deviation bond lengths (Å) | 0.006 | 0.005 |
| Root mean square deviation bond angles (°) | 1.052 | 1.022 |
| Average B (Å2) | 26.19 | 24.35 |
| Ramachandran statistics (%) | ||
| Favored | 99.45 | 100.0 |
| Allowed | 0.37 | 0.0 |
| PDB code | 4IPU | 4IPV |
a Values in parentheses represent the highest-resolution shell.
RESULTS
PilVWX and PilY1 Are Dependent on One Another for Incorporation into Pili
We and others showed previously that minor pilins and PilY1 are present in sheared pilus fractions and that association of PilY1 with T4P depends on one or more of the minor pilins (15, 22, 24). Those data suggested that the minor pilins might form a complex with PilY1 that is subsequently incorporated into T4P. To test this idea, sheared surface proteins that were recovered from retraction-deficient mutants lacking individual minor pilins or PilY1 were probed with specific antibodies to determine how the absence of each component affected incorporation of the others into pili.
All the minor pilins (less FimU) and PilY1 were present in sheared pili from a fimU pilT mutant, and FimU was incorporated into pili when any of the other minor pilins or PilY1 was absent (Fig. 2). We showed previously that pilus assembly was severely impaired in retraction-deficient pilV, pilW, and pilX mutants (15) and saw the same trend in the absence of pilY1 (Fig. 2). PilV, PilW, and PilX were each dependent on the others for incorporation into pili, and when PilY1 was missing, none of PilV, PilW, or PilX was present. Similarly, PilY1 was detected in sheared surface fractions only when PilV, PilW, and PilX were present. Although full-length PilY1 has a predicted mass of ∼125 kDa, we and others (21) routinely detected a smaller fragment (∼80 kDa) associated with sheared T4P. This fragment is recognized by an antibody raised against a C-terminal 60-kDa fragment of PilY1 (24), suggesting that it contains this region of the protein. It is not clear whether the truncated form of PilY1 is functionally relevant or simply an artifact of the pilus purification procedure. Finally, although PilE was not required for inclusion of PilVWX or PilY1, the absence of any of PilVWX or PilY1 prevented incorporation of PilE into pili (Fig. 2). In summary, the data suggest that PilVWX and PilY1 form a subcomplex that recruits PilE and that FimU is not essential for formation of the putative PilVWXY1E complex or its incorporation into pili.
FIGURE 2.
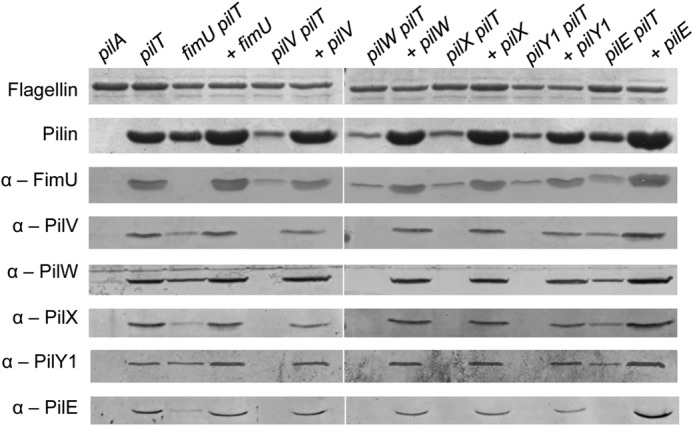
Incorporation of minor pilins into pili. Pili were sheared from the surface of minor pilin-pilT mutants and fractions probed by Western blot for incorporation of the minor pilins using anti-FimU (1/1000), anti-PilV (1/1000), anti-PilW (1/1000), anti-PilX (1/500), anti-PilY1 (1/1000), and anti-PilE (1/1000) antibodies. Proteins were detected at their expected masses except for PilY1, which had an apparent mass of ∼80 kDa.
FimU Interacts with the Major Pilin and Specific Minor Pilins
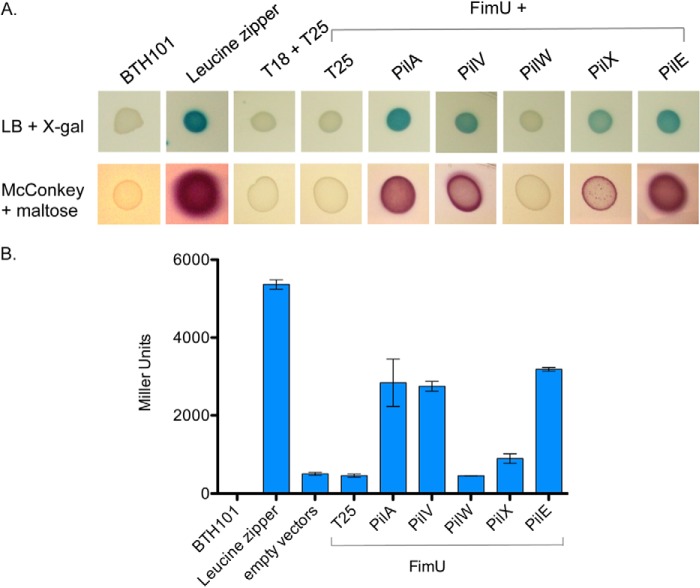
Because FimU was incorporated into pili in the absence of other minor components, we reasoned that it must interact directly with PilA, the major pilin subunit. The interaction of full-length FimU with PilA and the other minor pilins was tested using a bacterial adenylate cyclase two-hybrid assay (43), in which the T18 and T25 fragments were fused to the N termini of mature pilins. FimU interacted with PilA, but also with the minor pilins, particularly PilV and PilE (Fig. 3). According to the indicator plate assay, FimU interacts with PilX, but the interaction resulted in β-galactosidase activity values only slightly higher than the negative control (Fig. 3). These data, plus the pilus incorporation data, suggest that FimU might couple the putative minor pilin-PilY1 complex to the major pilin through direct interactions with PilE, PilV, and PilA.
FIGURE 3.
Interaction of FimU with major pilin and minor pilins. Direct protein-protein interactions of FimU with the major pilin and other minor pilins were probed using a bacterial adenylate cyclase two-hybrid assay. FimU was N-terminally tagged with the T18 fragment and the major pilin, and other minor pilins were N-terminally tagged with the T25 fragment. A, E. coli BTH101 recombinants were grown on LB agar + X-gal and McConkey + maltose, producing blue or red colonies, respectively, if there is an interaction. B, β-galactosidase activity of the same strains was measured as described under “Experimental Procedures.” The leucine zipper constructs T18-zip and T25-zip were used as positive controls.
High Resolution Crystal Structure of FimU
To gain further insight into FimU function, we solved the x-ray crystal structures of native and SeMet-substituted versions of its soluble C-terminal domain lacking the N-terminal 28 hydrophobic residues (numbering based on the mature protein; FimUΔ1–28). The protein crystallized in space group P21 for the SeMet form and P65 for the native form, containing four or two monomers, respectively, in the asymmetric unit. In both crystal forms, FimU was arranged as a tilted antiparallel dimer, with each monomer rotated ∼135° relative to the other. The final structures were refined to 1.35 Å (SeMet) and 1.4 Å (native) resolution. In the SeMet model, all residues of FimUΔ1–28 could be built into the electron density except for the first two in monomer A, and the first four in monomer B. The details of data collection and model refinement are summarized in Table 1.
FimU has a modified type IV pilin fold (Fig. 4), with an N-terminal α-helix leading into a total of eight β-strands, forming two four-stranded antiparallel β-sheets with noncontinuous strand connectivity. Strands β1, β2, β3, and β5 comprise the first β-sheet, and strands β4, β6, β7, and β8 comprise the second β-sheet. The strands are joined by short loops lacking regular secondary structure. Residues Cys127 and Cys158 are connected via a disulfide bond, generating a loop encompassing β7 and β8 that connects the C terminus to β6.
FIGURE 4.
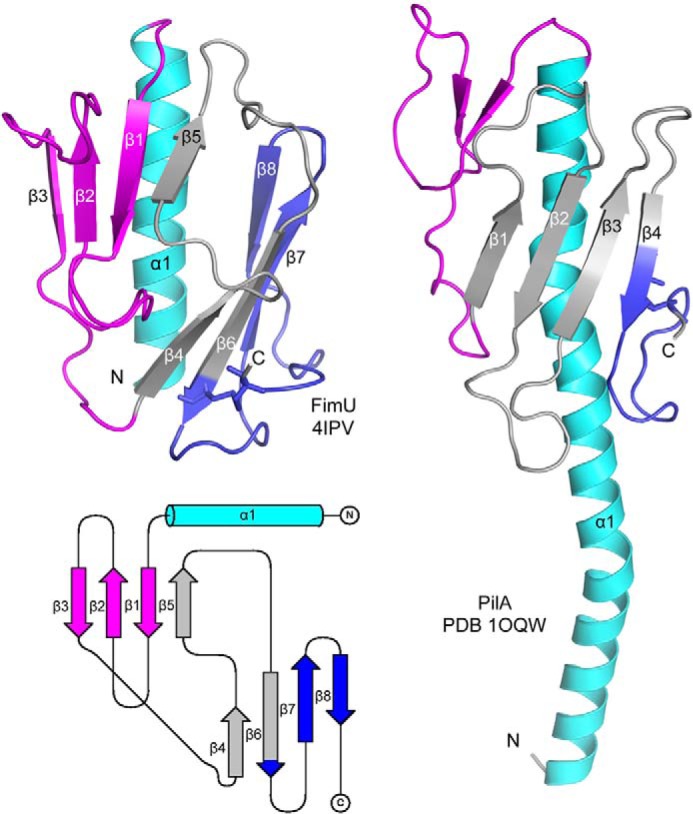
Crystal structure of FimUΔ1–28. The x-ray crystal structure of native FimUΔ1–28 was solved to 1.4 Å (top left panel). For comparison, the full-length structure of P. aeruginosa PilA (PDB code 1OQW) is shown in the right panel. The N-terminal α-helix is colored cyan, the αβ-loop is magenta, and the disulfide-bonded loop is blue. The intervening β-strands are colored gray. A topology diagram of FimU (produced using the TopDraw algorithm) depicting the organization of the structural elements is shown in the bottom left panel.
FimU Is Structurally Similar to T2S Minor Pseudopilin GspH
The equivalent of FimU in the P. aeruginosa T2S system is XcpU (GspH; Fig. 1), which was previously proposed to connect the minor pseudopilin complex to the major pseudopilin (35). Three structures of GspH homologs have been solved: EpsH from Vibrio cholerae, by x-ray crystallography (PDB codes 2QV8 and 4DQ9) (50, 51), and GspH from E. coli (PDB code 2KNQ) by NMR spectroscopy. Although there is less than 20% sequence identity between FimU and its GspH homologs, the proteins have similar architectures, characterized by two four- or five-stranded β-sheets with a central β-strand swap (Fig. 5).
FIGURE 5.
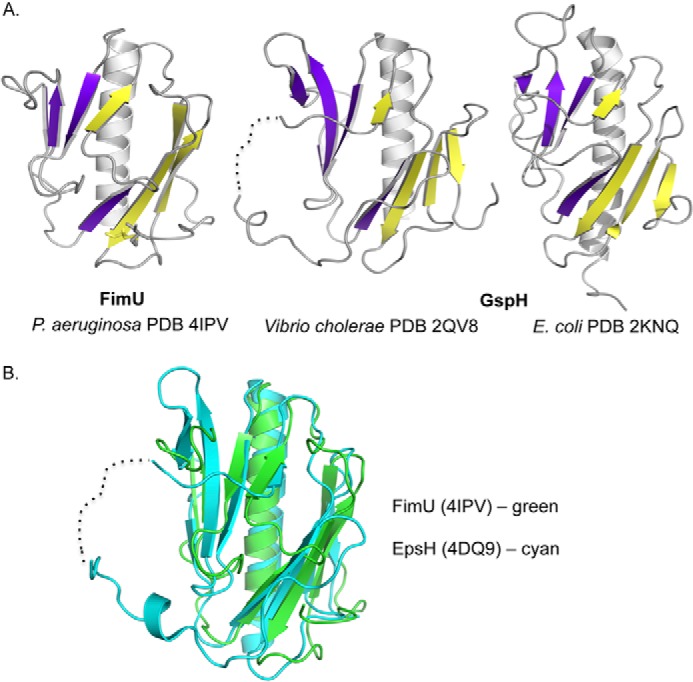
Structural comparison of FimU and T2S pseudopilin GspH. A, comparison of FimUΔ1–28 (left panel) with the x-ray crystal structure (middle panel) and NMR solution structure (right panel) of T2S pseudopilin GspH (EpsH) reveals similar architecture. The first four β-strands (five for V. cholerae EpsH) are colored in purple. The remaining four are colored in yellow. B, chain A of FimU (green) and EpsH (cyan) align with a root mean square deviation of 2.2 Å.
FimU (PDB code 4IPV, chain A) aligns with EpsH (PDB code 4DQ9, chain A) with a root mean square deviation of 2.2 Å over 106 residues, although EpsH has five β-strands in the first β-sheet, whereas FimU has four (Fig. 5B). In addition, EpsH has a long, partly disordered loop between swapped β-strands 5 and 6, which extends from the core of the protein. Those residues that could be resolved in this disordered region have different conformations in the two structures, as well as in separate monomers in the asymmetric unit (51). Based on its charged and apparently dynamic properties, this region of EpsH was suggested to be involved in recognition of T2S system secretion substrates (51). In FimU, the corresponding segment (connecting β-strands 4 and 5) is much shorter (five amino acids) and lacks charged residues, resulting in a more compact protein. Despite this difference, the overall striking similarity of the FimU and EpsH/GspH structures and the ability of FimU to interact directly with major and minor pilin subunits support the proposed role for FimU in connecting the major pilin to a putative minor pilin-PilY1 complex.
Type IVa Pilus Assembly Can Be Primed by Minor Pseudopilins
We showed previously that a retraction-deficient mutant lacking the positive regulator of minor pilin operon expression, AlgR, had a small amount of T4aP on the surface, implying that some assembly proceeds in the absence of minor subunits (42, 52). Similarly, Heiniger et al. (22) recovered a small amount of pili in a retraction-deficient strain when all the minor pilin genes were deleted. However, a recent study (41) showed that heterologous expression of T2S minor pseudopilins from K. oxytoca supported assembly of the E. coli major pilin PpdD in the absence of its cognate minor pilins. P. aeruginosa PAO1 encodes a T4aP, a T4bP (Tad) and two T2S systems, Xcp and Hxc (homologous to Xcp), although the Tad and Hxc systems are not expressed under laboratory growth conditions (53, 54). To learn whether the few pili recovered from retraction-deficient mutants lacking the minor pilins could be the result of cross-priming by T2S components, we made mutants lacking the minor pilins and/or the Xcp minor pseudopilins.
Consistent with previous studies (16, 22, 42), when the entire minor pilin operon was deleted (denoted ΔMP) in a retraction-deficient strain, the ΔMP pilT double mutant retained a small amount of surface pili (Fig. 6A). There was no effect on piliation when the minor pseudopilin genes were deleted (denoted ΔMPP) in the pilT background. However, when both sets of minor subunit genes were deleted, no surface pili were recovered (Fig. 6A). Therefore, pili assembled in the absence of the minor pilin operon were primed by the minor pseudopilins, and at least one set of minor components is necessary for pilus assembly. Levels of surface pili commensurate with the pilT control were restored upon complementation with either the minor pilin or the minor pseudopilin genes in trans. In contrast, piliation of the ΔMP pilT double mutant—in which the minor pseudopilins are expressed at native (low) levels from their chromosomal locus—was reduced compared with the pilT control (Fig. 6A). Expression of the minor pseudopilins from a multicopy vector increases their levels in the cell and thus the amount of pili assembled (Fig. 6, A and B, compare the third and last lanes).
FIGURE 6.
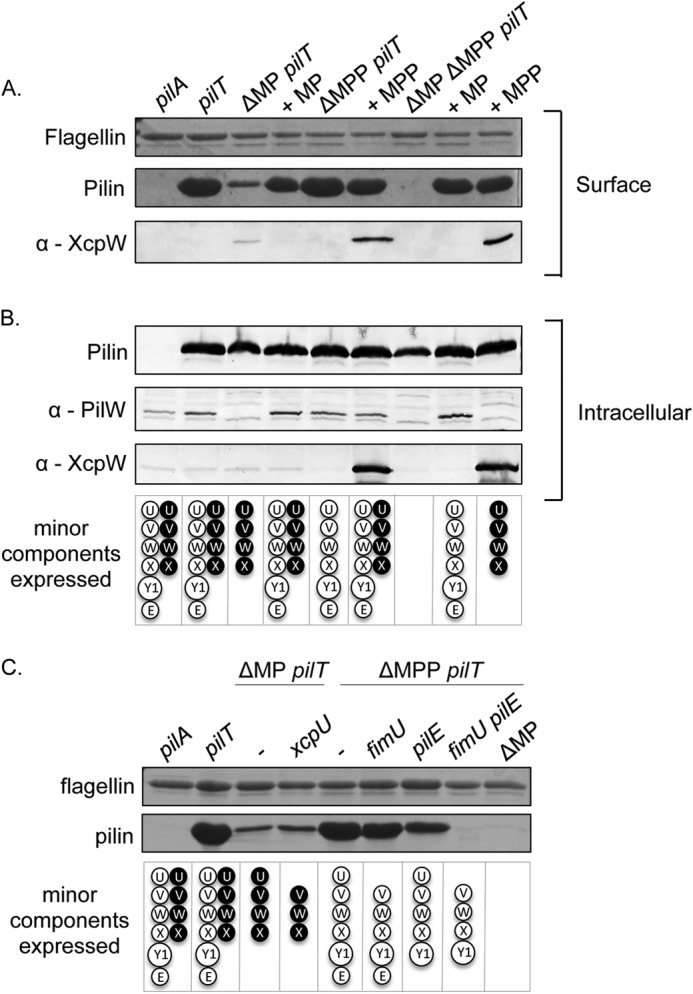
Pilus assembly in the absence of minor components. A, pilus assembly in the absence of minor components was probed by shearing surface proteins from a pilT mutant lacking the minor pilin fimUpilVpilWpilXpilY1pilE and minor pseudopilin xcpUVWX operons compared with single operon mutants and complementation with either operon and analyzing by SDS-PAGE. The flagellin band was used a loading control. The sheared surface protein samples were probed for the incorporation of minor pseudopilin XcpW into the pilus by Western blot analysis with an XcpW specific antibody (1/1000). B, cells recovered following shearing of surface proteins were lysed, separated by SDS-PAGE, and probed for levels of PilA, PilW, and XcpW using PilA (1/5000), PilW (1/1000), and XcpW (1/1000) specific antibodies. C, mutants made on minor pilin or minor pseudopilin retraction-deficient backgrounds were used to identify the minimum number of minor components required for pilus assembly by shearing the surface proteins from the cell and analyzing the surface pilus fractions by SDS-PAGE. The flagellin band was used a loading control.
In a previous study (41), E. coli T4aP assembly could be primed by Klebsiella minor pseudopilins, but it was unclear whether the minor subunits were incorporated into the pili or simply activated the assembly machinery (41, 55). To resolve this uncertainty, we probed sheared pilus fractions from retraction-deficient mutants with an antibody to the minor pseudopilin, XcpW. In the absence of the T4aP minor pilins, XcpW was found in sheared surface protein fractions, suggesting it was incorporated into pili (Fig. 6A). However, when both sets of minor components were expressed at native levels from their chromosomal loci, in the pilT single mutant, XcpW was not detectable in sheared pili. Taken together, the data suggest that the T2S minor pseudopilins can prime T4aP assembly and as a result become part of the filament. However, the T4aP minor pilins are used preferentially for assembly when both sets of minor subunits are expressed at physiological levels.
A Subset of Minor Components Can Prime Assembly
To determine how many and which minor components are required for pilus assembly, additional mutations were introduced into ΔMP pilT or ΔMPP pilT mutant backgrounds. In the absence of both the putative connectors FimU and XcpU, assembly of surface pili could still be initiated by the minor pilins PilVWXE plus PilY1 or by the minor pseudopilins, XcpVWX (Fig. 6C). In the absence of both FimU and PilE, where only PilVWXY1 are expressed, surface piliation was abolished (Fig. 6C). These data suggest that despite being the nominal equivalent of the XcpVWX subcomplex, PilVWXY1 alone are insufficient for priming of surface piliation and that FimU and PilE may cooperate to stably couple the priming complex to the major subunit (Fig. 7).
FIGURE 7.
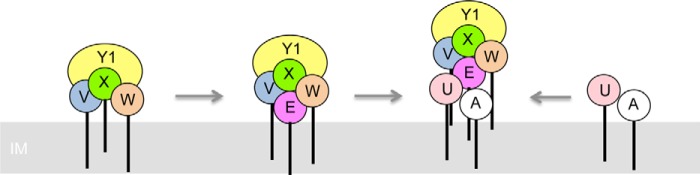
Model of pilus assembly. PilVWX and PilY1 interact, creating an interface that is bound by PilE. FimU interacts directly with PilA and connects it with the PilVWXY1E subcomplex through interactions with PilV and PilE.
DISCUSSION
Identification of a Putative Minor Pilin-PilY1 Complex
It has been challenging to assign specific functions to minor pilins because individual mutants have a similar “no pili” phenotype (15, 17–20, 25). We found that PilVWX and the large ∼125-kDa non-pilin protein PilY1 depend on one another for pilus incorporation, implying that they form a putative complex that becomes incorporated into pili. The late recruit PilE is encoded by the last gene in the minor pilin operon (Fig. 1), positioning it as a potential quality control checkpoint; it appears to recognize an interface formed by the putative PilVWXY1 subcomplex and does not appear in sheared pili if any of those components is missing. PilE interacts with the other minor pilins in the BTH assay, supporting this idea.4 Finally, under retraction-deficient conditions, FimU is not essential for formation or incorporation of PilVWXY1E into pili, and vice versa, consistent with its proposed role as a stabilizing “connector” subunit (below).
PilY1 is implicated in a number of pilus-related functions including adhesion, calcium-dependent antagonism of pilus retraction, and integrin binding (22, 24, 56). Our results suggest that PilVWX and PilY1 form a subcomplex that associates with PilE, although the exact nature of their interactions is not yet clear. P. aeruginosa PAO1 PilY1 was previously suggested to have a type IV pilin-like N terminus with a potential PilD cleavage site (57), but the N-terminal sequence is not conserved among P. aeruginosa strains (58). Instead, its hydrophobic N-terminal signal is more likely cleaved by signal peptidase 1, as predicted by SignalP (59).
Based on its size and adhesin function, we propose that PilY1 could be located at the distal position of a putative PilVWXY1E complex, placing it ultimately at the tip of a pilus. This hypothesis is based in part on the structure of the E. coli minor pseudopilin complex, where GspK, a subunit with a large α-domain insertion between β2 and β3 that would prohibit the addition of subunits above it because of potential steric clashes, occupies the distal position (34, 60). The equivalent of GspK in the T4aP system, PilX, is significantly smaller (185 residues versus 316), which may allow for an additional component at the distal position. The exact nature and duration of PilY1 association with the minor pilins remains under investigation, but positioning PilY1 at the tip of the pilus could facilitate its anti-retraction function, which requires a conserved calcium-binding motif in the C-terminal portion of the protein (24). We propose that calcium binding by PilY1 causes a conformational change, similar to opening of a drywall anchor, that enhances adhesiveness and/or prevents re-entry into the cell upon retraction. As a result, a minor pilin-PilY1 complex may be retracted only as far as the outside of the secretin but not re-enter the cell, explaining why PilY1 has been reported to be both T4aP- and cell surface-associated (21, 22). This scenario could allow for efficient pilus re-extension, because major subunits need only be added at the base of an existing pilus stub.
A link between minor pilins and PilY1-like proteins has also been reported in Neisseria. In N. gonorrhoeae, the PilY1 homologs PilC1 and PilC2 were undetectable in purified pili fractions in the absence of minor subunits PilHIKL (equivalent to FimU-PilVXE) and when pilC1 or pilC2 was deleted, the minor pilins PilHIKL were missing from purified pili (16). In addition, PilHIJKL are required for adherence (16), which we suggest is potentially due to association with PilC1/2. In N. meningitidis, pilIJK (equivalent to pilVWX) retraction-deficient mutants are also defective for adherence, although whether PilC1/2 was present in those mutant pili was not determined (25).
Unfortunately, we currently lack biochemical evidence for a minor pilin-PilY1 complex. Extensive efforts to purify individual components for protein interaction studies, as was done for the T2S minor subunits (35), have not been successful in all cases. PilX in particular has been insoluble in all forms tested so far, and purification of full-length PilY1 by our lab and others has also proven difficult (24). We are now working toward co-purifying all six components expressed from a single construct to preserve their native stoichiometry and interactions.
The Putative Connector Subunit, FimU
Our FimU structure (Fig. 4) is the first for a T4P minor pilin from P. aeruginosa, and for the FimU/PilH family. The structures of N. meningitidis minor pilins PilXNm (equivalent to PilE) and ComP (no equivalent in P. aeruginosa) have been solved (30, 61); however, deletion of these proteins does not affect piliation, suggesting that they modulate pilus properties, rather than controlling pilus assembly (29, 62, 63). FimU strongly resembles minor pseudopilin GspH (Fig. 5), proposed to link the GspIJK tip subcomplex to the major pseudopilin (35).
One notable difference between FimU and GspH is the length of the loop between the swapped β-strands (Fig. 8). In the pseudopilin, this region was proposed to participate in selection of secretion substrates (51). There is no evidence that the T4aP system of P. aeruginosa is involved in secretion, and the short uncharged loop in the corresponding position of FimU may reflect this divergence in function. A Phyre2 (64) model of the FimU equivalent from N. meningitidis, PilHNm, suggests that it too has a short loop between the central swapped β strands, but a predicted loop insertion between β-strands 1 and 2 accounts for its larger size (213 residues for mature PilHNm compared with 159 for FimU; Fig. 8).
FIGURE 8.
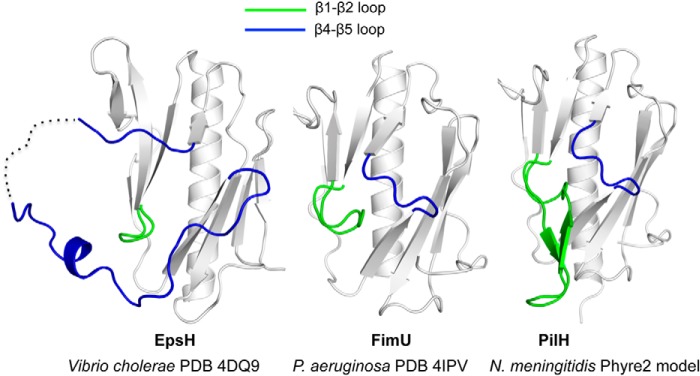
Comparison of loops in FimU, PilHNm, and EpsH. Comparison of truncated V. cholerae T2S pseudopilin EpsH (left panel) with P. aeruginosa FimU (center panel) and a Phyre2-generated model of N. meningitidis PilH (right panel) showing differences in the β1-β2 (colored green) and β4-β5 loops (colored blue).
Although incorporation of FimU into surface-exposed pili was independent of the other minor pilins and PilY1 (Fig. 2), this pattern is not conserved among species. In N. gonorrhoeae, the PilJNg protein (equivalent to P. aeruginosa PilW) was the only component found in surface pili in the absence of the others. Incorporation of PilHNg (equivalent to FimU) into surface pili depends on PilIJK (PilVWX) (16). The reasons for these differences are unclear but may reflect the need for the Neisseria minor pilin complex to accommodate more than one PilC variant and/or the minor pilins PilV and ComP (65–67), not present in P. aeruginosa. The predicted insertion between β1-β2 in PilHNm may also be related to these differences between species.
Although our data support a role for FimU in connecting a minor pilin-PilY1 complex to the major pilin, it was dispensable for pilus assembly in a retraction-deficient background. In fimU pilT mutants, PilA likely interacts with the minor pilin-PilY1 subcomplex via PilE and PilV, its closest homologs among the minor pilins, to allow for assembly. When both of FimU and PilE were absent, no pili were assembled (Fig. 6C). Interestingly, FimU and PilE are the only minor pilins with a Pro22 residue, similar to the major pilin. Pro22 creates a kink in the N-terminal helix, controlling intersubunit packing. The conformation conferred by this residue may facilitate interactions of FimU and PilE with one another and with major pilin subunits. These similarities may also lead to the occasional incorporation of individual minor subunits into the pilus that we reported previously (15).
Minor Subunits Prime Pilus Assembly
Because a few pili were still produced in minor pilin-deficient strains when retraction was blocked, the roles of minor pilins have been unclear (16, 22, 42). Here we found that pilus assembly is abolished when both the minor pilins (plus PilY1) and minor pseudopilins are missing (Fig. 6A) and that the minor pilins plus PilY1 or the minor pseudopilins can restore piliation (Fig. 6A), suggesting a conserved role in initiation of pilus assembly. Cisneros et al. (41) showed that K. oxytoca minor pseudopilins primed assembly of E. coli PpdD pili and E. coli minor pilins primed assembly of K. oxytoca pseudopili. As a caveat, E. coli has only four minor pilins (equivalent to FimU-PilVWX) and lacks PilY1 and PilE homologs (Fig. 1), making its T4aP system more similar to the T2S system and potentially facilitating the reported cross-complementation. Our data show that the Xcp minor pseudopilins, expressed at native levels from their chromosomal locus in the ΔMP pilT mutant, can prime pilus assembly (Fig. 6A), confirming the findings in E. coli and showing that a subcomplex containing the T4aP-specific components PilY1 and PilE is not essential for initiation of P. aeruginosa T4aP assembly.
Minor Subunits Are Part of the Pilus Filament
Cisneros et al. (33, 41) suggested that priming of pilus assembly by minor subunits could occur by creation of a priming complex to which major subunits were subsequently added or by stimulation of motor activity, without minor subunits becoming part of the resulting filament (55). Our data and that of Winther-Larsen et al. (16) support a model (Fig. 7) in which the minor subunits and PilY1 form a complex that becomes part of the pilus, because all components are found in sheared fractions (Fig. 2). In the case of priming by the minor pseudopilins, we had antisera for only XcpW (Fig. 6). However, XcpUVWX have been shown to form a complex (35); thus XcpUVX are likely also present. Despite this evidence for incorporation of minor subunits into pili, we note recent reports suggesting that minor pilins can exert specific functions from within the periplasm. When fused to a bulky fluorescent protein that prevented them from being displayed on the cell surface, N. meningitidis minor pilins PilXNm (equivalent of PilE) and PilVNm (no P. aeruginosa equivalent) modulated pilus assembly independent of pilus incorporation (63). Although these proteins were not detected in surface pilus fractions, processing by the prepilin peptidase PilD was required for their function, suggesting that at least partial extraction from the inner membrane was necessary. In P. aeruginosa, PilW, PilX, and PilY1 repressed swarming motility independent of PilD processing and pilus assembly, by modulating levels of secondary messenger c-di-GMP (23, 68). The signaling pathways involved in these novel phenotypes require further investigation.
Minor Pseudopilins Cannot Replace Minor Pilins when Retraction Is Active
When retraction is active, P. aeruginosa cells become non-piliated if they are missing even a single minor pilin or PilY1 (15, 17–20), even though the Xcp minor pseudopilins are concurrently expressed (based on detection of XcpW in whole cell lysates; data not shown). Increased expression of XcpUVWX from a plasmid in trans does not restore surface piliation or twitching motility in retraction-proficient minor pilin mutants (data not shown). There are at least two reasons why the minor pseudopilins may be unable to substitute for minor pilin function under conditions where retraction is active.
First, the minor pilins are used preferentially for assembly when both sets of minor components are expressed at physiological levels (Fig. 6A), suggesting a higher affinity for the major pilin. Lower affinity of the minor pseudopilins for the major pilin may lead to inefficient pilus extension that is unable to compete with retraction. We showed previously (42) that compatibility between major and minor subunits was important for efficient piliation. Each P. aeruginosa strain encodes one of two different sets of minor pilins, each associated with specific major pilin alleles. When the major subunit from one strain is expressed in another that has heterologous minor pilins, both assembly and twitching motility are impaired (42). Thus, optimal interactions among the major and minor subunits have an important role in regulating extension-retraction dynamics.
Alternatively, incorporation of a stable subcomplex containing the anti-retraction factor PilY1 may be mandatory for pili to remain on the cell surface in retraction-proficient strains. Although the minor pseudopilins can prime assembly in retraction-deficient backgrounds, the resulting pili lack PilY1 and PilE (data not shown), suggesting that those components are unable to interact with the Xcp subunits. Thus, pili primed by the T2S minor subunits lack factors that oppose retraction. In P. aeruginosa fimU or pilE single mutants that express pili containing PilVWX-PilY1 complexes (Fig. 2), we speculate that stable interaction of the PilVWX-PilY1 subcomplex with the major subunit, as well as the resulting capacity to oppose retraction, is compromised. The evolution of a pilus-associated subcomplex containing the T4aP-specific components PilY1 and PilE, coupled with the high expression level of major subunits compared with the T2S system, likely represent adaptations of the ancestral secretion system to promote formation of long surface-exposed filaments that perform the myriad of functions associated with T4aP.
Acknowledgments
We thank Rami El-Sebai and Colin Rous for generating pilY1 mutants and complementation constructs and Drs. Romé Voulhoux and Matthew Wolfgang for providing antisera.
This work was funded by Canadian Institutes of Health Research Grants MOP 93585 (to L. L. B.) and MOP 89903 (to M. S. J.).
Y. Nguyen, S. D. Bell, S. Sugiman-Marangos, M. S. Junop, and L. L. Burrows, manuscript in preparation.
- T4P
- type IV pili
- T2S
- type II secretion
- PDB
- Protein Data Bank
- SeMet
- selenomethionine.
REFERENCES
- 1. Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 [DOI] [PubMed] [Google Scholar]
- 2. Burrows L. L. (2012) Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 [DOI] [PubMed] [Google Scholar]
- 3. Craig L., Pique M. E., Tainer J. A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 4. Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 [DOI] [PubMed] [Google Scholar]
- 5. Szabó Z., Stahl A. O., Albers S. V., Kissinger J. C., Driessen A. J., Pohlschröder M. (2007) Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189, 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sastry P. A., Finlay B. B., Pasloske B. L., Paranchych W., Pearlstone J. R., Smillie L. B. (1985) Comparative studies of the amino acid and nucleotide sequences of pilin derived from Pseudomonas aeruginosa PAK and PAO. J. Bacteriol. 164, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Francetic O., Buddelmeijer N., Lewenza S., Kumamoto C. A., Pugsley A. P. (2007) Signal recognition particle-dependent inner membrane targeting of the PulG Pseudopilin component of a type II secretion system. J. Bacteriol. 189, 1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arts J., van Boxtel R., Filloux A., Tommassen J., Koster M. (2007) Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189, 2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nunn D. N., Lory S. (1991) Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. U.S.A. 88, 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strom M. S., Lory S. (1991) Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. J. Biol. Chem. 266, 1656–1664 [PubMed] [Google Scholar]
- 11. Strom M. S., Nunn D. N., Lory S. (1993) A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. U.S.A. 90, 2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craig L., Li J. (2008) Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skerker J. M., Berg H. C. (2001) Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 98, 6901–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mattick J. S., Whitchurch C. B., Alm R. A. (1996) The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa: a review. Gene 179, 147–155 [DOI] [PubMed] [Google Scholar]
- 15. Giltner C. L., Habash M., Burrows L. L. (2010) Pseudomonas aeruginosa minor pilins are incorporated into the type IV pilus. J. Mol. Biol. 398, 444–461 [DOI] [PubMed] [Google Scholar]
- 16. Winther-Larsen H. C., Wolfgang M., Dunham S., van Putten J. P., Dorward D., Løvold C., Aas F. E., Koomey M. (2005) A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol. Microbiol. 56, 903–917 [DOI] [PubMed] [Google Scholar]
- 17. Alm R. A., Hallinan J. P., Watson A. A., Mattick J. S. (1996) Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 22, 161–173 [DOI] [PubMed] [Google Scholar]
- 18. Alm R. A., Mattick J. S. (1996) Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178, 3809–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alm R. A., Mattick J. S. (1995) Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16, 485–496 [DOI] [PubMed] [Google Scholar]
- 20. Russell M. A., Darzins A. (1994) The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol. Microbiol. 13, 973–985 [DOI] [PubMed] [Google Scholar]
- 21. Bohn Y. S., Brandes G., Rakhimova E., Horatzek S., Salunkhe P., Munder A., van Barneveld A., Jordan D., Bredenbruch F., Häussler S., Riedel K., Eberl L., Jensen P. O., Bjarnsholt T., Moser C., Hoiby N., Tümmler B., Wiehlmann L. (2009) Multiple roles of Pseudomonas aeruginosa TBCF10839 PilY1 in motility, transport and infection. Mol. Microbiol. 71, 730–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heiniger R. W., Winther-Larsen H. C., Pickles R. J., Koomey M., Wolfgang M. C. (2010) Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell Microbiol. 12, 1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuchma S. L., Ballok A. E., Merritt J. H., Hammond J. H., Lu W., Rabinowitz J. D., O'Toole G. A. (2010) Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J. Bacteriol. 192, 2950–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orans J., Johnson M. D., Coggan K. A., Sperlazza J. R., Heiniger R. W., Wolfgang M. C., Redinbo M. R. (2010) Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility. Proc. Natl. Acad. Sci. U.S.A. 107, 1065–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carbonnelle E., Helaine S., Nassif X., Pelicic V. (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61, 1510–1522 [DOI] [PubMed] [Google Scholar]
- 26. Giltner C. L., Nguyen Y., Burrows L. L. (2012) Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76, 740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bleves S., Voulhoux R., Michel G., Lazdunski A., Tommassen J., Filloux A. (1998) The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol. Microbiol. 27, 31–40 [DOI] [PubMed] [Google Scholar]
- 28. Nunn D. N., Lory S. (1993) Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 175, 4375–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hélaine S., Carbonnelle E., Prouvensier L., Beretti J. L., Nassif X., Pelicic V. (2005) PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol. Microbiol. 55, 65–77 [DOI] [PubMed] [Google Scholar]
- 30. Helaine S., Dyer D. H., Nassif X., Pelicic V., Forest K. T. (2007) 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 104, 15888–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brissac T., Mikaty G., Duménil G., Coureuil M., Nassif X. (2012) The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect. Immun. 80, 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yanez M. E., Korotkov K. V., Abendroth J., Hol W. G. (2008) The crystal structure of a binary complex of two pseudopilins: EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus. J. Mol. Biol. 375, 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cisneros D. A., Bond P. J., Pugsley A. P., Campos M., Francetic O. (2012) Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 31, 1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korotkov K. V., Hol W. G. (2008) Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat. Struct. Mol. Biol. 15, 462–468 [DOI] [PubMed] [Google Scholar]
- 35. Douzi B., Durand E., Bernard C., Alphonse S., Cambillau C., Filloux A., Tegoni M., Voulhoux R. (2009) The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J. Biol. Chem. 284, 34580–34589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuo W. W., Kuo H. W., Cheng C. C., Lai H. L., Chen L. Y. (2005) Roles of the minor pseudopilins, XpsH, XpsI and XpsJ, in the formation of XpsG-containing pseudopilus in Xanthomonas campestris pv. campestris. J. Biomed. Sci. 12, 587–599 [DOI] [PubMed] [Google Scholar]
- 37. Lu H. M., Motley S. T., Lory S. (1997) Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol. Microbiol. 25, 247–259 [DOI] [PubMed] [Google Scholar]
- 38. Durand E., Bernadac A., Ball G., Lazdunski A., Sturgis J. N., Filloux A. (2003) Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185, 2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sauvonnet N., Vignon G., Pugsley A. P., Gounon P. (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 19, 2221–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vignon G., Köhler R., Larquet E., Giroux S., Prévost M. C., Roux P., Pugsley A. P. (2003) Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185, 3416–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cisneros D. A., Pehau-Arnaudet G., Francetic O. (2012) Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol. Microbiol. 86, 805–818 [DOI] [PubMed] [Google Scholar]
- 42. Giltner C. L., Rana N., Lunardo M. N., Hussain A. Q., Burrows L. L. (2011) Evolutionary and functional diversity of the Pseudomonas type IVa pilin island. Environ. Microbiol. 13, 250–264 [DOI] [PubMed] [Google Scholar]
- 43. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 45. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 46. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 48. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 49. DeLano W. L. (2002) The PyMOL User's Manual, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 50. Yanez M. E., Korotkov K. V., Abendroth J., Hol W. G. (2008) Structure of the minor pseudopilin EpsH from the type 2 secretion system of Vibrio cholerae. J. Mol. Biol. 377, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raghunathan K., Vago F. S., Grindem D., Ball T., Wedemeyer W. J., Bagdasarian M., Arvidson D. N. (2014) The 1.59A resolution structure of the minor pseudopilin EpsH of Vibrio cholerae reveals a long flexible loop. Biochim. Biophys. Acta 1844, 406–415 [DOI] [PubMed] [Google Scholar]
- 52. Belete B., Lu H., Wozniak D. J. (2008) Pseudomonas aeruginosa AlgR regulates type IV pilus biosynthesis by activating transcription of the fimU-pilVWXY1Y2E operon. J. Bacteriol. 190, 2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Bentzmann S., Aurouze M., Ball G., Filloux A. (2006) FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188, 4851–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ball G., Durand E., Lazdunski A., Filloux A. (2002) A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43, 475–485 [DOI] [PubMed] [Google Scholar]
- 55. Burrows L. L. (2012) Prime time for minor subunits of the type II secretion and type IV pilus systems. Mol. Microbiol. 86, 765–769 [DOI] [PubMed] [Google Scholar]
- 56. Johnson M. D., Garrett C. K., Bond J. E., Coggan K. A., Wolfgang M. C., Redinbo M. R. (2011) Pseudomonas aeruginosa PilY1 binds integrin in an RGD- and calcium-dependent manner. PLoS One 6, e29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lewenza S., Gardy J. L., Brinkman F. S., Hancock R. E. (2005) Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15, 321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., Yu N. Y., Hancock R. E., Brinkman F. S. (2011) Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 60. Forest K. T. (2008) The type II secretion arrowhead: the structure of GspI-GspJ-GspK. Nat. Struct. Mol. Biol. 15, 428–430 [DOI] [PubMed] [Google Scholar]
- 61. Cehovin A., Simpson P. J., McDowell M. A., Brown D. R., Noschese R., Pallett M., Brady J., Baldwin G. S., Lea S. M., Matthews S. J., Pelicic V. (2013) Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. U.S.A. 110, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brown D. R., Helaine S., Carbonnelle E., Pelicic V. (2010) Systematic functional analysis reveals that a set of seven genes is involved in fine-tuning of the multiple functions mediated by type IV pili in Neisseria meningitidis. Infect. Immun. 78, 3053–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Imhaus A. F., Duménil G. (2014) The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J. 33, 1767–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 65. Winther-Larsen H. C., Hegge F. T., Wolfgang M., Hayes S. F., van Putten J. P., Koomey M. (2001) Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc. Natl. Acad. Sci. U.S.A. 98, 15276–15281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rudel T., Facius D., Barten R., Scheuerpflug I., Nonnenmacher E., Meyer T. F. (1995) Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 92, 7986–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aas F. E., Wolfgang M., Frye S., Dunham S., Løvold C., Koomey M. (2002) Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46, 749–760 [DOI] [PubMed] [Google Scholar]
- 68. Kuchma S. L., Griffin E. F., O'Toole G. A. (2012) Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J. Bacteriol. 194, 5388–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]