Background: In contrast to their yeast orthologues, the mechanism by which mammalian ORMDLs regulate serine palmitoyltransferase is not understood.
Results: Overexpression of serine palmitoyltransferase in HEK293 cells results in increased long-chain base synthesis and an activity-dependent increase in ORMDL expression.
Conclusion: A product of ceramide synthase mediates regulation of ORMDL expression and inhibition of serine palmitoyltransferase.
Significance: Serine palmitoyltransferase activity indirectly regulates ORMDL expression.
Keywords: Enzyme, Lipid Metabolism, Metabolic Regulation, Serine Palmitoyltransferase, Sphingolipid
Abstract
The relationship between serine palmitoyltransferase (SPT) activity and ORMDL regulation of sphingolipid biosynthesis was investigated in mammalian HEK293 cells. Each of the three human ORMDLs reduced the increase in long-chain base synthesis seen after overexpression of wild-type SPT or SPT containing the C133W mutation in hLCB1, which produces the non-catabolizable sphingoid base, 1-deoxySa. ORMDL-dependent repression of sphingoid base synthesis occurred whether SPT was expressed as individual subunits or as a heterotrimeric single-chain SPT fusion protein. Overexpression of the single-chain SPT fusion protein under the control of a tetracycline-inducible promoter in stably transfected cells resulted in increased endogenous ORMDL expression. This increase was not transcriptional; there was no significant increase in any of the ORMDL mRNAs. Increased ORMDL protein expression required SPT activity since overexpression of a catalytically inactive SPT with a mutation in hLCB2a had little effect. Significantly, increased ORMDL expression was also blocked by myriocin inhibition of SPT as well as fumonisin inhibition of the ceramide synthases, suggesting that increased expression is a response to a metabolic signal. Moreover, blocking ORMDL induction with fumonisin treatment resulted in significantly greater increases in in vivo SPT activity than was seen when ORMDLs were allowed to increase, demonstrating the physiological significance of this response.
Introduction
Regulation of serine palmitoyltransferase (SPT)2 activity in yeast by the Orms has recently been the subject of extensive study, revealing multiple levels of regulation. Based on the response of yeast cells to ER stress, Gururaj et al. proposed that there is complex transcriptional regulation (1). In addition, post-translational regulation is also important. By identifying phosphopeptides derived from the N termini of the yeast Orm proteins and showing that mutation of the phosphorylated residues resulted in increased myriocin sensitivity, Breslow et al. proposed that phosphorylation of the N-terminal domain of the yeast Orms resulted in derepression of SPT (2). Subsequently, Roelants et al. demonstrated that the yeast Ypk1 and Ypk2 kinases phosphorylate this domain and that their deletion results in loss of viability, presumably as a result of constitutive repression of long-chain base (LCB) synthesis. This hypothesis is supported by the fact that deletion of the Orms rescues the ypk1Δypk2Δ kinase knock-out mutant (3). Dephosphorylation of the Orms by Cdc55p protein phosphatase 2A (PP2A) has also been shown to be necessary for the reversal of Ypk1 phosphorylation (4). In addition, Shimobayashi et al. have shown that yeast Npr1 kinase, whose activity is regulated by TorC1, also phosphorylates the Orms, but in this case apparently to stimulate complex sphingolipid synthesis (5). Thus, the yeast Orms integrate the input from multiple signaling pathways to regulate sphingolipid biosynthesis.
In mammalian cells, the three ORMDL proteins have also been implicated in regulation of sphingolipid biosynthesis (2, 6). However, while highly homologous to their yeast counterparts, they lack the major portion of the N-terminal domain of the yeast Orm proteins that is phosphorylated by Ypk1, Ypk2, and Npr1 and far less is known about the mechanisms by which they regulate SPT activity. Transcriptional regulation of ORMDL3, polymorphisms in which are associated with an increased risk of childhood asthma, has been reported (7, 8). In addition, Breslow et al. reported that silencing of all three ORMDL mRNAs resulted in increased ceramide levels (2). Siow and Wattenberg further showed that silencing of all three ORMDLs increased the rate of ceramide synthesis and relieved the ORMDL-mediated repression of SPT in response to treatment with C6-ceramide (6). Based on these results, they hypothesized that a complex sphingolipid regulates SPT activity by acting on the ORMDLs. Nevertheless, the identity of the signaling molecule and the mechanism by which it regulates ORMDL function is unknown. Regulation of mammalian SPT has also been reported to result from phosphorylation of Tyr-164 of hLCB1 (9). This phosphorylation reduces SPT activity and can be blocked by inhibiting (with imatinib mesylate) or silencing the c-abl kinase in CML cells. Thus, like SPT regulation in yeast, SPT regulation in mammalian cells is complex. However, the specific mechanisms are clearly different.
In the present study, we have employed a different approach to study ORMDL regulation of sphingolipid synthesis. Using a stable HEK293 cell line in which a previously described hLCB2a-ssSPTa-hLCB1 SPT fusion (10) is expressed under the control of a tetracycline inducible promoter, ORMDL expression and in vivo SPT activity were studied. Our results show that upon induction of SPT, ORMDL mRNA levels were relatively unchanged, but a striking increase in the level of ORMDL protein was observed. This increase was dependent upon ceramide or a downstream product.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
Dulbecco's modified Eagle's medium (DMEM), F12 Ham's Medium, Opti-MEM® Reduced Serum Medium, penicillin, streptomycin, trypsin-EDTA (0.05%), Lipofectamine, Flp-In T-Rex HEK293 cells, pOG44 recombinase, and 4 to 12% Bis-Tris SDS gels were purchased from Invitrogen (Grand Island, NY). Protease inhibitors and PhosSTOP-phosphatase inhibitor mixture were purchased from Roche Diagnostics GmbH (Indianapolis, IN). Sphingolipid standards were obtained from Avanti Polar Lipids, Inc. (Alabaster, Alabama). [3H]serine was from Perkin Elmer (Boston, MA). The Expand Long Range PCR amplification kit and the QuickChange Mutagenesis kit were from Roche Applied Science and Agilent Technologies, Inc. (Santa Clara, CA), respectively. Plasmids containing the human LCB1 and LCB2a cDNAs under the control of the CMV promoter were purchased from OriGene Technologies, Inc. (Rockville, MD). The plasmids expressing NubG-HA tagged ssSPTa (11) and the hLCB2a-ssSPTa-hLCB1 fusion SPT (10) were previously described. N-terminally NubG-HA-tagged human ORMDL1, ORMDL2, and ORMDL3 were PCR amplified from a yeast split-ubiquitin 2-hybrid pR3N library containing human cDNAs (11) and the fragments were inserted into pcDNA. The C-terminally tagged ORMDL1-GFP in the pCMV6-AC-GFP vector was purchased from OriGene Technologies, Inc. pcDNA5/FRT/TO-Lcb2a-ssSPTa-Lcb1 was constructed by ligating a BglII/SalI-ended PCR fragment containing the Lcb2a-ssSPTa-Lcb1 fusion between the BamHI and XhoI sites of pcDNA5/FRT/TO (Invitrogen). pcDNA5/FRT/TO-Lcb2a-ssSPTa-Lcb1C133W was generated by replacing the BamHI-XhoI fragment of pcDNA5/FRT/TO-Lcb2a-ssSPTa-Lcb1 with the same fragment from pDualCMV Lcb2a-ssSPTa-Lcb1C133W (10).
Cell Culture and Transfections
HEK293 cells (American Type Culture Collection, Manassas, VA) were cultured in DMEM with high glucose, pyruvate, and glutamax, supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. Flp-In™ 293 T-REx cells cultured in the same medium supplemented with zeocin (100 μg/ml) and blasticidin (2.5 μg/ml), were used for generating the stable cell lines expressing the hLCB2a-ssSPTa-LCB1 fusion SPTs. The cells were co-transfected with the pOG44 vector encoding Flp recombinase, and pcDNA5/FRT/TO-, pcDNA5/FRT/TO-Lcb2a-ssSPTa-Lcb1 or pcDNA5/FRT/TO-Lcb2a-ssSPTa-Lcb1C133W using Lipofectamine 2000. Two days after transfection the cells were replated in selective growth medium containing 100 μg/ml hygromycin B and 2.5 μg/ml blasticidin. The selective medium was changed every 3 to 4 days until colonies appeared on the plates. The colonies were subcultured and maintained in medium containing 50 μg/ml hygromycin B and 2.5 μg/ml blasticidin, induced with 2 μg/ml tetracycline, and cell lysates were analyzed by immunoblotting for expression of the fusion SPTs. CHO-LY-B cells were grown in Ham's F12 medium supplemented with 10% FBS, penicillin, and streptomycin as described previously (11).
Cell Lysate Preparation and Immunoblot Analysis
Cells were washed twice with ice-cold phosphate-buffered saline containing 1 mm EGTA, 1 mm EDTA (PBSE), and lysed in 50 mm Tris-HCl, pH 8.0, containing 10% glycerol, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1% deoxycholate, 0.1% SDS, 1 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 mm PMSF, 2 mm NaF, 1 mm orthovanadate, 25 mm β-glycerophosphate, and PhosSTOP-phosphatase inhibitor mixture (1 tablet/50 ml). The lysates were sonicated and centrifuged at 3000 × g for 4 min to remove debris. 40 μg of protein were resolved on 4–12% Bis-Tris SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were incubated with mouse anti-mLCB1 (BD Biosciences, San Jose, CA), rabbit anti-CHO LCB2a (12), goat anti-human ORMDL E15, or mouse anti-human GAPDH (both from Santa Cruz Biotechnology, Santa Cruz, CA). The blots were washed and incubated with HRP-conjugated secondary antibodies obtained from Bio-Rad, and proteins were detected by enhanced chemiluminescence (Western Lightning Plus-ECL, PerkinElmer, Shelton, CT) and visualized on x-ray film. Films were scanned with a Microtek ScanMaker 9800XL and images analyzed using ImageJ. Expression of the ORMDL proteins was normalized to GAPDH or calnexin.
Long Chain Base Analysis
HEK293 or CHO-LY-B cells were harvested from 100 mm tissue culture dishes in ice-cold PBSE containing 1 mm PMSF, pelleted and resuspended in 8 ml PBS. 6 ml were transferred to a glass tube, pelleted, and dried under nitrogen. Following acid hydrolysis, total LCBs were extracted after adding C17 sphingosine as an internal standard and derivatized using AccQ-Fluor (Waters, Milford, MA) as previously described (13). The remainder of the cells was centrifuged and cell lysates were prepared for protein determination and immunoblotting as described above. Samples equivalent to 400 μg of protein were analyzed by reverse phase HPLC on a 4 μm C18 column (GraceVydac, Hesperia, CA) using an HP Series II 1090 liquid chromatography system with HP Chemstation coupled to an Agilent 1100 series fluorescence detector as previously described (14). The areas under the peaks, measured in luminescence units (LU), were normalized to the internal C17 sphingosine standard and the amounts of sphingosine (So), sphinganine (Sa), and 1-deoxysphinganine (1-deoxySa) were determined using standard curves for each of the LCBs and are reported as nmol/mg protein.
Microsomal SPT Assays
HEK293 or CHO-LY-B cells grown in 100 mm dishes were harvested by scraping into 4 ml of PBSE buffer. Cells were pelleted, washed once with the same buffer, and resuspended in TEGM buffer (50 mm Tris-HCl, pH 7.5 containing 1 mm EGTA, 1 mm β-mercaptoethanol, 1 mm PMSF, and a protease inhibitor mixture). Microsomes were prepared by sonication and differential centrifugation essentially as described previously (12). Microsomal SPT activity was assayed by measuring acyl-CoA-dependent incorporation of 3[H]serine into long chain bases as previously described (11).
RT-PCR and Quantitative Real-time PCR
RNA was isolated from HEK293 cells using the RNAqueous-4PCR kit according to the manufacturer's instructions (Invitrogen). cDNA was generated using the Superscript III First-Strand Synthesis SuperMix two step RT-PCR kit (Invitrogen) according to the manufacturer's instructions. PCR was conducted using Econo Taq Plus Green 2× master Mix (Lucigen Corporation, Middleton, WI). qPCR was conducted with the Power SYBR Green 2× PCR Master mix (Applied Biosystems, Foster City, CA) using the ABI 7900 HT Fast Real-Time PCR System. Relative gene expression was compared using real-time qPCR analysis, and the changes in mRNA expression levels were calculated following normalization to GAPDH using RQ Manager Version 1.2 as described by the manufacturer (Applied Biosystems).
RESULTS
The ORMDLs Inhibit in Vivo SPT Activity
We have previously shown that overexpression of hLCB1 and hLCB2a results in a modest increase in catalytic activity of the heterodimer in vitro, which can be increased nearly 100-fold by the coexpression of an ssSPT isoform (11). However, despite clear evidence that SPT activity is tightly regulated, there is no evidence that either of the catalytic subunits or the ssSPTs are themselves regulated in response to changes in cellular sphingolipid levels at the level of transcription or translation (15), leading to the conclusion that there must be other regulatory components of the SPT complex responsible for sphingolipid homeostasis. Indeed, in yeast, SPT copurifies with Orm1 and Orm2, regulators of sphingolipid synthesis, which themselves are subject to regulation by phosphorylation of their N termini (1–4, 16, 17). In mammalian cells, there are three ORMDL proteins, which though they lack the N-terminal extension of the yeast Orms, copurify with mammalian SPT and have been shown to negatively regulate sphingolipid synthesis (2, 6).
To study regulation of mammalian SPT by the ORMDLs, we have taken advantage of the fact that SPT heterotrimers containing the hLCB1C133W mutant subunit condense alanine with palmitoyl-CoA more efficiently than heterotrimers containing wild type hLCB1 (10, 18). This condensation results in the synthesis of 1-deoxySa, which is not present in the growth medium and which, although it can be acylated on the amino group of C2 (19), cannot be phosphorylated and catabolized. Thus, following hydrolysis of all sphingolipid species, measurement of 1-deoxySa provides a sensitive measure of de novo LCB synthesis.
The value of this approach is clearly seen in Fig. 1, where after overexpressing the mutant enzyme, the alanine-containing LCB (1-deoxySa) increased to a far greater extent than the serine-containing LCBs (sphingosine, So and dihydrosphinganine, Sa) (Fig. 1D). When the ORMDL2 and ORMDL3 proteins were coexpressed with the subunits of the SPT heterotrimer, LCB accumulation was significantly attenuated (Fig. 1, B and D), confirming previous reports that like the yeast Orm proteins, the mammalian ORMDLs are negative regulators of SPT (2, 6). For reasons that remain unclear, despite the fact that ORMDL1 is highly homologous to ORMDL2 and ORMDL3, this protein could only be efficiently expressed when C-terminally epitope-tagged (Fig. 1, A and C). However, when expressed with the epitope tag, it too repressed SPT. Comparable results were obtained when single-chain hLCB2a-ssSPTa-LCB1 fusion SPTs were used instead of the individual subunits (Fig. 2). Thus, the ORMDLs regulate the fusion SPT in the same manner as the native heterotrimer. Taken together, these results suggest that regulation of ORMDL expression rather than regulation of SPT expression is responsible for maintaining sphingolipid homeostasis.
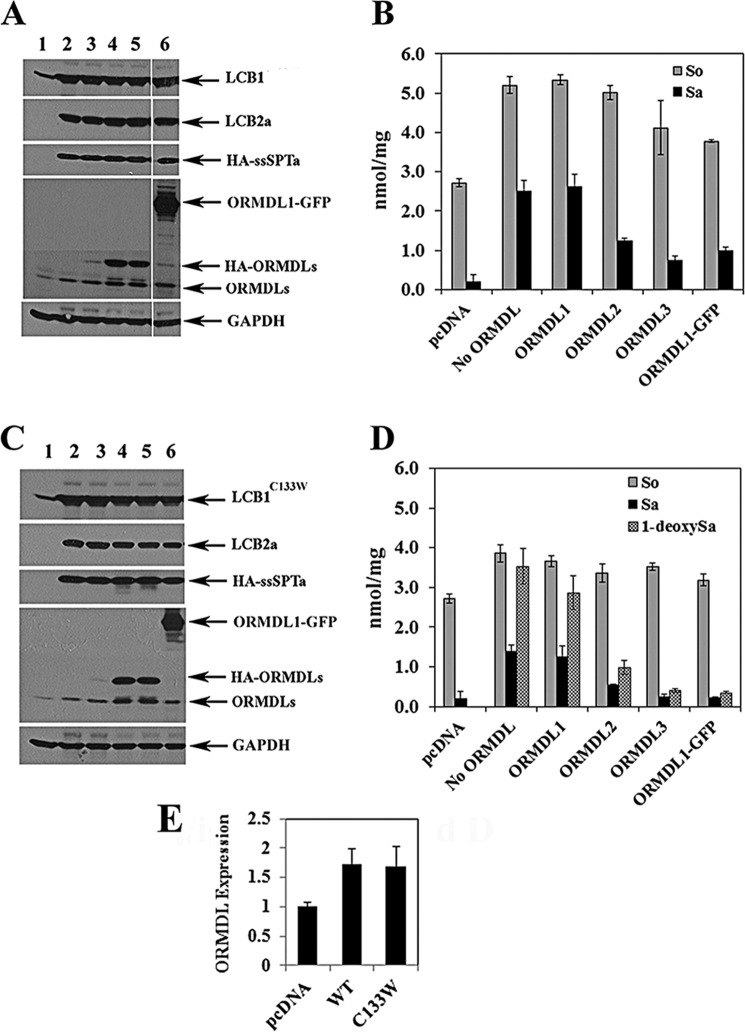
FIGURE 1.
The ORMDLs suppress in vivo activity of wild-type and mutant SPT. A, HEK293 cells were transfected with empty vector (lane 1) or hLCB1, hLCB2a, and HA-ssSPTa in the absence (lane 2) or presence of HA-ORMDL1 (lane 3), HA-ORMDL2 (lane 4), HA-ORMDL3 (lane 5), or ORMDL1-GFP (lane 6). Protein expression was verified by immunoblotting. B, total levels of sphingosine (So), dihydrosphinganine (Sa), and 1-deoxySa were determined in cells transfected with pcDNA, or hLCB1, hLCB2a, ssSPTa, and the indicated ORMDLs as described under “Experimental Procedures.” Results reported are the average of three independent experiments ± S.D. C and D, same as A and B with the exception that the hLCB1C133W mutant subunit, which results in the efficient synthesis of 1-deoxySa, was used. E, ORMDL expression was quantified as described under “Experimental Procedures.” Results are presented as the mean ± S.D. from three independent experiments.
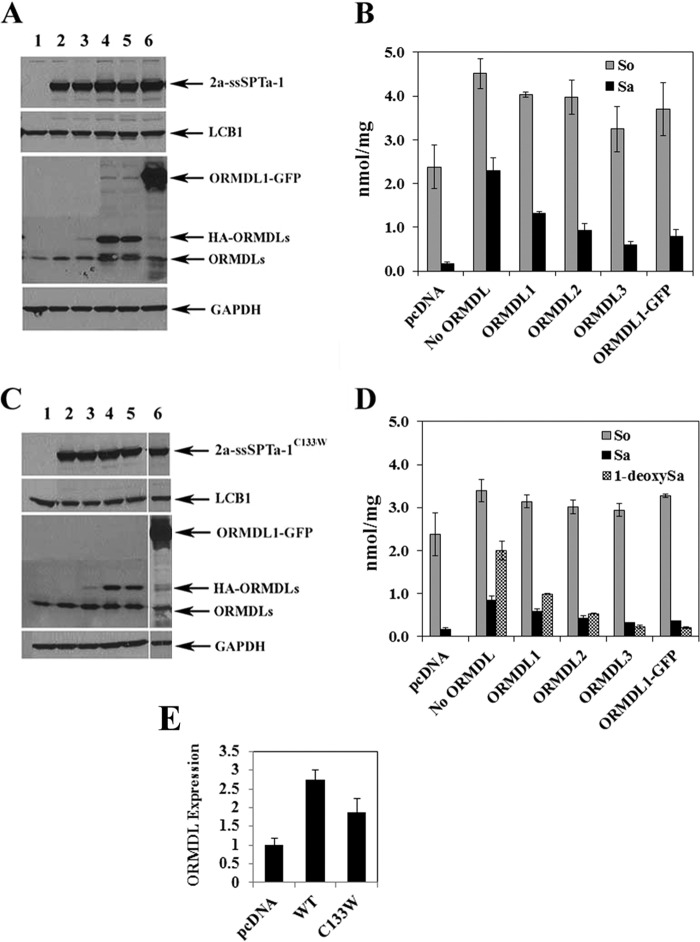
FIGURE 2.
The ORMDLs suppress in vivo activity of the wild-type and mutant hLCB2a-ssSPTa-LCB1 fusion SPTs. A, HEK293 cells were transfected with empty vector (lane 1) or wild-type fusion SPT in the absence (lane 2) or presence of HA-ORMDL1 (lane 3), HA-ORMDL2 (lane 4), HA-ORMDL3 (lane 5), or ORMDL1-GFP (lane 6). Protein expression was verified by immunoblotting. B, total levels of sphingosine (So), dihydrosphinganine (Sa), and 1-deoxySa were determined in cells transfected with pcDNA, or hLCB1, hLCB2a, ssSPTa, and the indicated ORMDLs as described under “Experimental Procedures.” Results are reported are the average of three independent experiments ± S.D. C and D, same as A and B with the exception that the hLCB1C133W mutant fusion SPT was used. E, ORMDL expression was quantified as described under “Experimental Procedures.” Results are presented as the mean ± S.D. from three independent experiments.
Elevated SPT Activity Results in Increased ORMDL Expression
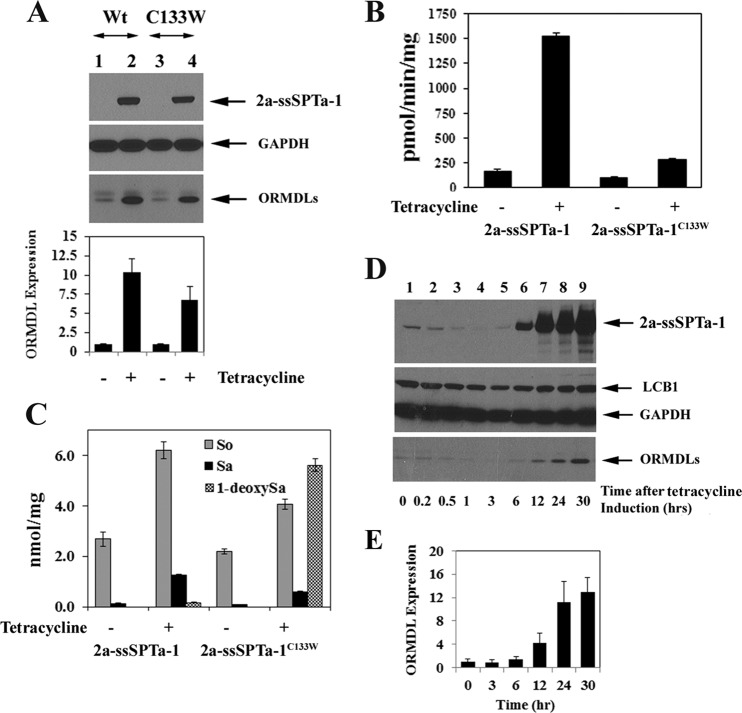
If this were the case, then increasing SPT activity might be expected to alter ORMDL expression in an attempt to maintain physiological concentrations of LCBs. Indeed, there is a small increase in endogenous ORMDL expression after transfection of the individual subunits of SPT (Fig. 1E) or the heterotrimeric fusion SPT (Fig. 2E). This occurs in vivo as well as in vitro. Injection of adenovirus expressing the fusion SPT into the tail vein of mice resulted in hepatic overexpression of both SPT and ORMDLs (data not shown). To more carefully study the response of ORMDL expression to changes in SPT activity, stable cell lines in which either a wild-type or mutant (hLCB1C133W) single-chain hLCB2a-ssSPTa-LCB1 fusion was integrated downstream of a common tetracycline-inducible promoter were generated. As expected, tetracycline treatment of these cell lines for 48 h resulted in a substantial increase in expression of the fusion SPT proteins (Fig. 3A). The increase in either wild-type or mutant SPT was accompanied by an increase in SPT activity both in vitro (Fig. 3B) and in vivo (Fig. 3C). More importantly, ORMDL expression, assessed by immunoblotting with a pan ORMDL antibody, increased ∼10-fold after induction of the wild-type fusion SPT (Fig. 3A). This increase was slightly more than after tetracycline induction of the mutant protein, a result consistent with the lower enzymatic activity of the mutant enzyme (Fig. 3B). Interestingly, the increase in ORMDL levels lagged slightly behind the induction of SPT (Fig. 3D), a result consistent with the hypothesis that increased ORMDL expression is a response to an increase in long-chain bases or a downstream product. It is also noteworthy that expression of endogenous hLCB1, an obligatory subunit of SPT, did not decrease in response to increased SPT activity (Fig. 3D), indicating that autoregulation of SPT expression is not a major mechanism for regulation of sphingolipid biosynthesis.
FIGURE 3.
Increased SPT activity induces ORMDL expression. Stably transfected HEK293 cell lines expressing wild-type or mutant heterotrimeric SPT fusions under the control of a tetracycline-inducible promoter were grown in the absence or presence of tetracycline for various periods of time. A, SPT and ORMDL expression after growth for 42 h was determined by immunoblotting. B, in vitro SPT activity was measured in microsomes prepared from cells grown in the absence or presence of tetracycline for 42 h. C, total long chain bases were determined as in Fig. 1 after growth in tetracycline for 42 h. D, fusion SPT and endogenous hLCB1 and ORMDL immunoreactivity were determined after growth in tetracycline for the indicated periods of time. E, ORMDL expression (panel D) was quantified as described under “Experimental Procedures.” Results are presented as the mean ± standard deviation from three independent experiments.
To determine whether the increase in ORMDL protein was the result of increased transcription of one or more of the ORMDL genes, ORMDL mRNA was quantified by RT-PCR. The results showed that although ORMDL protein increased ∼10-fold after tetracycline induction of SPT (Fig. 3, A, D, and E), there was less than a 2-fold increase in ORMDL1, ORMDL2, or ORMDL3 mRNA (Fig. 4). Thus, transcriptional regulation cannot account for the increase in ORMDL protein.
FIGURE 4.
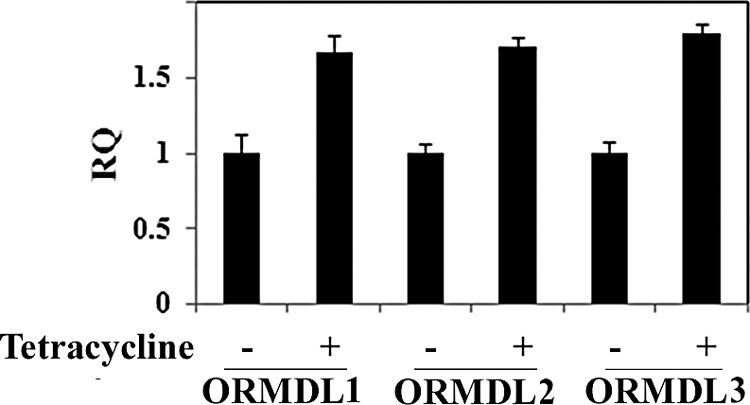
ORMDL mRNA levels are not significantly affected by increased SPT activity. mRNA was isolated from cells grown in the absence or presence of tetracycline for 30 h and each ORMDL mRNA analyzed by qPCR and relative quantification (RQ) was calculated as described under “Experimental Procedures.” RQ represents the ratio of induced to uninduced ORMDL mRNA after normalization to GAPDH. Results represent the mean ± S.D. from three independent experiments.
SPT Activity Is Required for ORMDL Induction
To determine whether it is simply the increase in the amount of SPT protein or the increase in enzymatic activity that is responsible for ORMDL induction, a catalytically dead fusion SPT in which the PLP-binding lysine of hLCB2a has been replaced with threonine was expressed in CHO-LY-B cells, which lack endogenous SPT activity (20). Measurement of SPT activity in these cells, and in cells transfected with the wild-type SPT fusion protein, confirmed the lack of catalytic activity of the mutant protein (data not shown). More importantly, whereas the wild-type SPT induced a significant increase in ORMDL expression, no such increase in ORMDL expression was elicited by the mutant SPT protein (Fig. 5A). The dependence upon an increase in catalytic activity was confirmed by the observation that addition of myriocin, an inhibitor of SPT, to stably transfected HEK293 cells during tetracycline induction completely abolished ORMDL induction without affecting the increase in SPT fusion protein (Fig. 5B). Treatment with myriocin also appeared to result in a small decrease in the amount of immunoreactive ORMDL in uninduced cells (Fig. 5B), suggesting that not only does increased SPT activity elevate ORMDL expression, but decreases in endogenous activity have an opposing effect. Thus, it is clear that a product of SPT regulates ORMDL expression.
FIGURE 5.
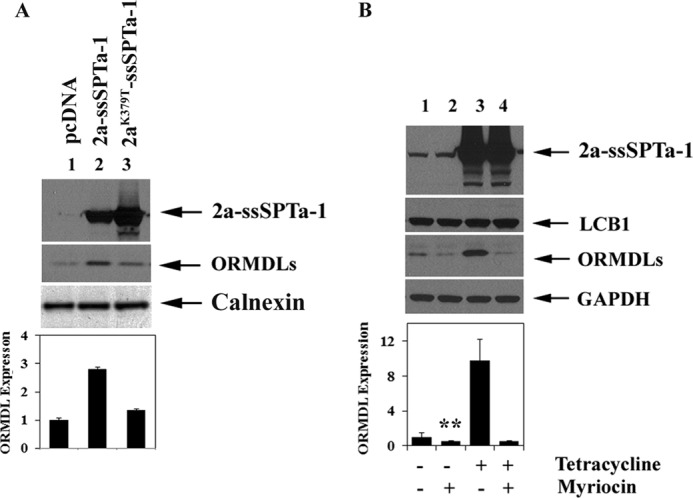
SPT activity is required for ORMDL induction. A, ORMDL expression was determined by immunoblotting in CHO-LY-B cells transfected with a wild-type heterotrimeric SPT fusion protein or a catalytically inactive fusion protein in which the pyridoxal-phosphate binding lysine (K379 of hLCB2a) was replaced with threonine as described under “Experimental Procedures.” B, stably transfected HEK293 cells expressing the wild-type heterotrimeric SPT fusion were treated with tetracycline and/or myriocin as indicated and ORMDL immunoreactivity determined as in panel A. ORMDL immunoreactivity was quantified as described under “Experimental Procedures.” Results represent the mean ± S.D. from three independent experiments. Asterisks indicate a statistically significant difference (p < 0.01).
Inhibition of Ceramide Synthase Also Blocks ORMDL Induction
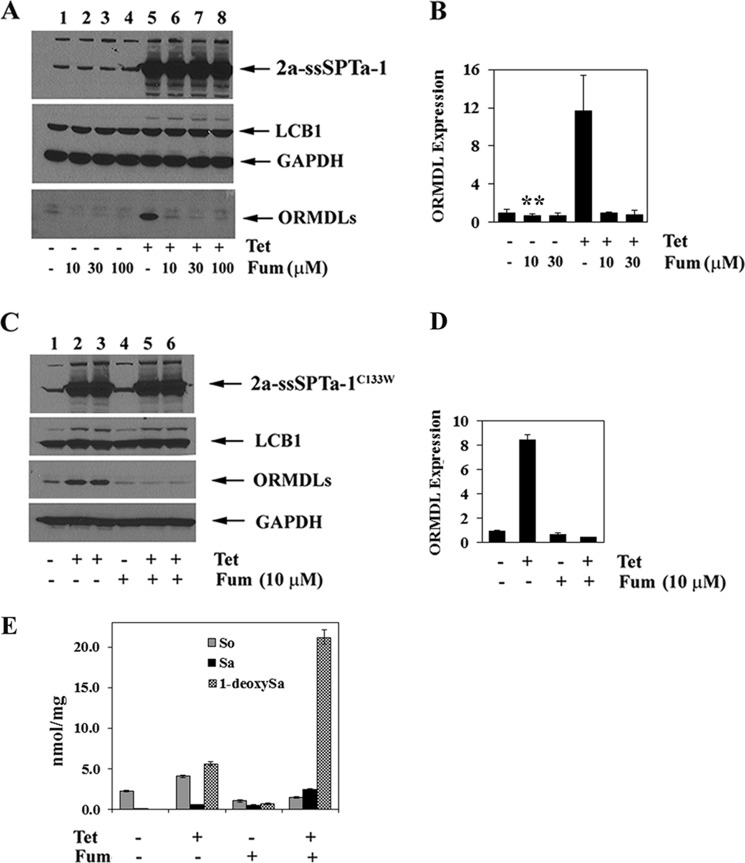
Fumonisin, an inhibitor of the ceramide synthases (21), also blocked the increase in ORMDL expression after tetracycline induction (Fig. 6, A, B, C, and D). It therefore appears that it is not a de novo long-chain base, but rather ceramide, a downstream product, or a metabolite that is required to elicit ORMDL induction. Since the ORMDLs are negative regulators of SPT, the results described above predict that in vivo SPT activity should be higher in fumonisin-treated cells than in untreated cells because the former have less ORMDL protein than the latter. To test this prediction, cells were induced with tetracycline in the absence or presence of fumonisin and in vivo SPT activity was measured. For these experiments, the cell line containing the hLCB2a-ssSPTa-LCB1C133W mutant SPT was used because, for the reasons described above, differences in in vivo SPT activity are most accurately reflected by comparing 1-deoxySa levels. The results showed that total 1-deoxySa levels were ∼4-fold higher in the fumonisin-treated cells than in the untreated cells (Fig. 6E). Since induction of the fusion SPT was unaffected by fumonisin (Fig. 6, A and B), the increase in in vivo SPT activity must be the result of the lack of ORMDL induction. In addition, the fact that fumonisin treatment also resulted in increased 1-deoxySa in tetracycline-uninduced cells suggests that even at the basal level of SPT activity the ORMDLs are responding to ceramide or a downstream metabolite (Fig. 6E). These results provide a mechanistic explanation for the fumonisin-induced increase in 1-deoxySa reported by Zitomer et al. (19). Moreover, our results clearly show that there is a physiologically significant consequence of the SPT/sphingolipid-induced increase in ORMDL expression and further support the conclusion that the level of ORMDL expression is an important regulator of in vivo SPT activity.
FIGURE 6.
Complex sphingolipids regulate ORMDL expression. Stably transfected HEK293 cells expressing wild-type (A and B) or mutant (C and D) heterotrimeric SPT fusion protein were grown in the absence or presence of tetracycline with the indicated concentrations of fumonisin. The amount of immunoreactive ORMDL (B and D) was determined as described under “Experimental Procedures.” Results represent the mean ± S.D. from three independent experiments. E, HEK293 cells expressing the mutant heterotrimeric fusion SPT were grown in the absence and presence of tetracycline with or without 10 μm fumonisin. Total So, Sa, and 1-deoxySa were determined as described under “Experimental Procedures,” and results are the average of three experiments ± S.D. Asterisks indicate a statistically significant difference (p < 0.01).
DISCUSSION
Recent studies in yeast have shown that phosphorylation of the Orm proteins by Ypk1 and Ypk2 is an important component of SPT regulation (2–4, 16). In addition, Npr1 and Npr2 phosphorylate the Orm proteins at different sites and have been implicated in regulation of complex sphingolipid biosynthesis (5). While the mammalian ORMDL proteins have also been shown to regulate sphingolipid biosynthesis (2, 6), little work has been done to identify the mechanisms by which they do so. To address this question, we created a stable cell line in which SPT could be induced 10-fold by the addition of tetracycline. This cell line provides four important advantages. First, it allows direct regulation of SPT expression rather than relying upon indirect perturbations, such as addition of exogenous long-chain bases to the medium or inhibition of SPT activity with compounds such as myriocin, which might themselves alter ORMDL expression or its interaction with the SPT heterotrimer. Second, by integrating an SPT fusion protein, stoichiometric expression of hLCB1, hLCB2a, and ssSPTa is assured. This is essential for accurate analysis because nonstoichiometric expression of either hLCB1 or hLCB2 results in mislocalization of the subunits,3 raising the possibility that regulatory components might also mislocalize. Third, unlike transient transfection, which underestimates ORMDL induction because only a fraction of the cells overexpress SPT, integration of an inducible fusion SPT insures that all cells respond equivalently. Thus, the magnitude of increase in ORMDL expression seen in Fig. 3 is a more accurate reflection of the biological response to increased sphingolipids than are the smaller increases seen in Figs. 1 and 2. Last, by incorporating the C133W mutation into hLCB1 it was possible to use the synthesis of 1-deoxySa as a readout of in vivo SPT activity.
The importance of the last point is most clearly illustrated by comparing the effect of ORMDL expression on alanine- and serine-containing long-chain bases in Figs. 1D, and 2D, where the magnitude of the ORMDL-mediated decrease in 1-deoxySa synthesis exceeds the decrease in either Sa or So. We attribute this to the fact that unlike Sa or So, 1-deoxySa is not subject to phosphorylation and subsequent degradation. Thus, its intracellular levels are directly proportional to its rate of synthesis. In addition, by measuring 1-deoxySa, the contribution of the two wild-type hLCB1 genes in HEK293 cells does not need to be considered, thereby maximizing the dynamic range of the in vivo SPT assay.
Breslow et al. showed that yeast ORMs associate with yeast SPT to form a SPOTS complex and that mammalian ORMDLs are similarly associated with mammalian SPT (2). The observation that induction of a catalytically dead SPT mutant, or myriocin inhibition of wild-type SPT during induction, both failed to elicit an increase in ORMDL expression provide strong evidence that the mechanism of ORMDL regulation is not direct stabilization of the ORMDLs upon binding to SPT. In addition, the observation that fumonisin blocks ORMDL induction indicates that the mechanism regulating increased expression is responsive to ceramide, a more complex sphingolipid or a ceramide metabolite, which could indirectly result in ORMDL stabilization. However, levels of transiently transfected ORMDLs did not increase upon induction of SPT (data not shown). Thus, the increase in ORMDL expression does not appear to involve post-translational modification and indirect stabilization.
Neither does the increase in ORMDL expression appear to be the result of transcriptional regulation. None of the ORMDL mRNA levels changed significantly after tetracycline-induction of SPT and transcriptional regulation has only been reported for ORMDL3 (7, 8, 22, 23), making it unlikely that a common transcriptional mechanism can account for the regulation we have observed. One possible explanation for these results is that regulation involves alternative splicing or modulation of translational efficiency. Indeed, Hjelmqvist et al. identified alternatively spliced transcripts for ORMDL1 (24). However, the fact that all three ORMDLs appear to be regulated suggests that whatever the mechanism is, it must coordinately regulate all of their mRNAs or proteins. This is consistent with the conclusions of Siow and Wattenberg who examined ORMDL regulation by complex sphingolipids (6).
In these experiments we have assessed the effects of the ORMDLs by in vivo assay of SPT because measurement of in vitro SPT activity in microsomes prepared from cells in which in vivo SPT is either high or low shows little difference. This lack of concordance is not the result of ORMDL dissociation during microsomal preparation; the ORMDLs coimmunoprecipitate with the subunits of SPT under both conditions (data not shown). One explanation for this observation is that, as suggested by Breslow et al. (2), the ORMs alter the suborganellar localization of SPT. If this is the case, then the implication is that SPT activity is different in different subdomains of the ER. Clearly, this adds another layer of complexity whose resolution will require additional experimentation.
A single nucleotide polymorphism in the non-coding region of ORMDL3 that alters the levels of mRNA expression has been associated with an increased incidence of childhood asthma (25–28). It has been suggested that this is a result of decreased SPT activity since treatment with myriocin results in increased airway reactivity in mice that is also seen when one of the hLCB2a alleles is deleted (29). Thus, it appears that asthmatic patients carrying this allele lack homeostatic mechanisms to restore normal sphingolipid levels. This is entirely consistent with numerous observations that there is little if any regulation of expression of any of the SPT subunits and suggests that cells may simply lack a mechanism for adapting to dysregulation of ORMDL expression. In contrast, we studied ORMDL regulation by altering SPT expression. Our results clearly show that in response to increased SPT activity there is a concomitant increase in ORMDL expression while in response to decreased activity there is a decrease in ORMDL protein. Taken together, our results and those of Siow and Wattenberg and Worgall et al. (6, 29) suggest that regulation of SPT activity is mediated by the ORMDLs and not regulation of the expression of the catalytic components of the enzyme. However, it is unclear whether the three ORMDLs transduce the same or different signals. If the latter is the case, then simple binding of the ORMDLs to SPT is insufficient to account for its complex regulation.
Acknowledgments
We thank the laboratory of Dr. Roopa Biswas for advice on quantifying mRNA levels and Dr. Jennifer Martindale in the laboratory of Dr. Myriam Gorospe, National Institute of Aging, for helpful discussions about regulation of protein expression.
This work was supported by Uniformed Services University Grants R071KD and CO75PI, by National Institutes of Health Grants R01NS072446 and R21HD080181, and by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
P. Moitra, J. M. Harmon, and T. M. Dunn, unpublished data.
- SPT
- serine palmitoyltransferase
- 1-deoxySa
- 1-deoxysphinganine
- HSAN1
- hereditary sensory autonomic neuropathy, type I
- LCB
- long-chain base
- LU
- luminescence units
- Sa
- dihydrosphinganine
- So
- sphingosine
- SPT
- serine palmitoyltransferase
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Gururaj C., Federman R., Chang A. (2013) Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 288, 20453–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roelants F. M., Breslow D. K., Muir A., Weissman J. S., Thorner J. (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sun Y., Miao Y., Yamane Y., Zhang C., Shokat K. M., Takematsu H., Kozutsumi Y., Drubin D. G. (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 23, 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shimobayashi M., Oppliger W., Moes S., Jenö P., Hall M. N. (2013) TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell 24, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siow D. L., Wattenberg B. W. (2012) Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 287, 40198–40204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller M., Tam A. B., Cho J. Y., Doherty T. A., Pham A., Khorram N., Rosenthal P., Mueller J. L., Hoffman H. M., Suzukawa M., Niwa M., Broide D. H. (2012) ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc. Natl. Acad. Sci. U.S.A. 109, 16648–16653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhuang L. L., Jin R., Zhu L. H., Xu H. G., Li Y., Gao S., Liu J. Y., Zhou G. P. (2013) Promoter characterization and role of cAMP/PKA/CREB in the basal transcription of the mouse ORMDL3 gene. PLoS One 8, e60630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taouji S., Higa A., Delom F., Palcy S., Mahon F. X., Pasquet J. M., Bosse R., Ségui B., Chevet E. (2013) Phosphorylation of serine palmitoyltransferase long chain-1 (SPTLC1) on tyrosine 164 inhibits its activity and promotes cell survival. J. Biol. Chem. 288, 17190–17201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gable K., Gupta S. D., Han G., Niranjanakumari S., Harmon J. M., Dunn T. M. (2010) A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J. Biol. Chem. 285, 22846–22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Jr., Harmon J. M., Dunn T. M. (2009) Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. U.S.A. 106, 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. (2004) The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J. Biol. Chem. 279, 53707–53716 [DOI] [PubMed] [Google Scholar]
- 13. Harmon J. M., Bacikova D., Gable K., Gupta S. D., Han G., Sengupta N., Somashekarappa N., Dunn T. M. (2013) Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 288, 10144–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsegaye Y., Richardson C. G., Bravo J. E., Mulcahy B. J., Lynch D. V., Markham J. E., Jaworski J. G., Chen M., Cahoon E. B., Dunn T. M. (2007) Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J. Biol. Chem. 282, 28195–28206 [DOI] [PubMed] [Google Scholar]
- 15. Hojjati M. R., Li Z., Jiang X. C. (2005) Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim. Biophys. Acta 1737, 44–51 [DOI] [PubMed] [Google Scholar]
- 16. Han S., Lone M. A., Schneiter R., Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu M., Huang C., Polu S. R., Schneiter R., Chang A. (2012) Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J. Cell Sci. 125, 2428–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Penno A., Reilly M. M., Houlden H., Laurá M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H., Jr., von Eckardstein A., Hornemann T. (2010) Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285, 11178–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., Merrill A. H., Jr., Riley R. T. (2009) Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284, 4786–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanada K., Hara T., Fukasawa M., Yamaji A., Umeda M., Nishijima M. (1998) Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J. Biol. Chem. 273, 33787–33794 [DOI] [PubMed] [Google Scholar]
- 21. Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H., Jr. (1991) Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266, 14486–14490 [PubMed] [Google Scholar]
- 22. Qiu R., Yang Y., Zhao H., Li J., Xin Q., Shan S., Liu Y., Dang J., Yu X., Gong Y., Liu Q. (2013) Signal transducer and activator of transcription 6 directly regulates human ORMDL3 expression. Febs J. 280, 2014–2026 [DOI] [PubMed] [Google Scholar]
- 23. Jin R., Xu H. G., Yuan W. X., Zhuang L. L., Liu L. F., Jiang L., Zhu L. H., Liu J. Y., Zhou G. P. (2012) Mechanisms elevating ORMDL3 expression in recurrent wheeze patients: role of Ets-1, p300 and CREB. Int. J. Biochem. Cell Biol. 44, 1174–1183 [DOI] [PubMed] [Google Scholar]
- 24. Hjelmqvist L., Tuson M., Marfany G., Herrero E., Balcells S., Gonzàlez-Duarte R. (2002) ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 3, RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akhabir L., Sandford A. J. (2011) Genome-wide association studies for discovery of genes involved in asthma. Respirology 16, 396–406 [DOI] [PubMed] [Google Scholar]
- 26. Cantero-Recasens G., Fandos C., Rubio-Moscardo F., Valverde M. A., Vicente R. (2010) The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum. Mol. Genet. 19, 111–121 [DOI] [PubMed] [Google Scholar]
- 27. Galanter J., Choudhry S., Eng C., Nazario S., Rodríguez-Santana J. R., Casal J., Torres-Palacios A., Salas J., Chapela R., Watson H. G., Meade K., LeNoir M., Rodriguez-Cintrón W., Avila P. C., Burchard E. G. (2008) ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am. J. Respir. Crit. Care Med. 177, 1194–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halapi E., Gudbjartsson D. F., Jonsdottir G. M., Bjornsdottir U. S., Thorleifsson G., Helgadottir H., Williams C., Koppelman G. H., Heinzmann A., Boezen H. M., Jonasdottir A., Blondal T., Gudjonsson S. A., Jonasdottir A., Thorlacius T., Henry A. P., Altmueller J., Krueger M., Shin H. D., Uh S. T., Cheong H. S., Jonsdottir B., Ludviksson B. R., Ludviksdottir D., Gislason D., Park C. S., Deichmann K., Thompson P. J., Wjst M., Hall I. P., Postma D. S., Gislason T., Kong A., Jonsdottir I., Thorsteinsdottir U., Stefansson K. (2010) A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur. J. Hum. Genet. 18, 902–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Worgall T. S., Veerappan A., Sung B., Kim B. I., Weiner E., Bholah R., Silver R. B., Jiang X. C., Worgall S. (2013) Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci. Transl. Med. 5, 186ra167. [DOI] [PubMed] [Google Scholar]