EHEC O157:H7 clade 6 strains harboring stx2a and/or stx2c and clade 8 strains harboring stx2a or stx2a/stx2c were frequently associated with childhood HUS cases in Japan. Rapid and specific detection of such lineages are required for infection control measures.
Keywords: clade, EHEC, HUS, O157, stx
Abstract
Background
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 infection causes severe diseases such as bloody diarrhea and hemolytic uremic syndrome (HUS). Although EHEC O157:H7 strains have exhibited high genetic variability, their abilities to cause human diseases have not been fully examined.
Methods
Clade typing and stx subtyping of EHEC O157:H7 strains, which were isolated in Japan during 1999–2011 from 269 HUS patients and 387 asymptomatic carriers (ACs) and showed distinct pulsed-field gel electrophoresis patterns, were performed to determine relationships between specific lineages and clinical presentation.
Results
Clades 6 and 8 strains were more frequently found among the isolates from HUS cases than those from ACs (P = .00062 for clade 6, P < .0001 for clade 8). All clade 6 strains isolated from HUS patients harbored stx2a and/or stx2c, whereas all clade 8 strains harbored either stx2a or stx2a/stx2c. However, clade 7 strains were predominantly found among the AC isolates but less frequently found among the HUS isolates, suggesting a significant association between clade 7 and AC (P < .0001). Logistic regression analysis revealed that 0–9 year old age is a significant predictor of the association between clade 8 and HUS. We also found an intact norV gene, which encodes for a nitric oxide reductase that inhibits Shiga toxin activity under anaerobic condition, in all clades 1–3 isolates but not in clades 4–8 isolates.
Conclusions
Early detection of EHEC O157:H7 strains that belonged to clades 6/8 and harbored specific stx subtypes may be important for defining the risk of disease progression in EHEC-infected 0- to 9-year-old children.
Over the last 3 decades, enterohemorrhagic Escherichia coli (EHEC), one of the major categories of diarrheagenic E coli, has been frequently reported to cause severe diseases such as hemorrhagic colitis, hemolytic uremic syndrome (HUS), and encephalopathy [1, 2]. The serogroup O157 (serotype O157:H7/H−) was identified as the major EHEC responsible for sporadic cases and outbreaks in several countries [3–7]. In Japan, more than 3000 annual cases of EHEC infections (including asymptomatic carriers [ACs]) were reported during 2004–2012 [8]. Among the isolates collected during 2008–2012, serogroup O157 predominated (62.2%) followed by O26 (22.5%), O111 (3.25%), and O103 (3%) [8–12]. During this time period, serogroup O157 was found in more than 85% of total HUS cases [8–12].
Shiga toxin (Stx) is the most important and critical virulence factor of EHEC that is known to cause these severe diseases. There are 2 antigenically distinct types of Stx, Stx1 and Stx2. Several studies have suggested that, compared with strains harboring stx1 gene, strains with stx2 are more often associated with HUS [13–15]. These 2 genes have been further divided into several subtypes: subtypes stx1a, stx1c, and stx1d for the stx1 gene, and subtypes stx2a–g for the stx2 gene [16]. The subtypes stx2a and/or stx2c were more often found to be associated with HUS than the other stx types [17, 18].
Genomic diversity of serotype O157:H7 has been extensively studied by using several molecular subtyping methods including pulsed-field gel electrophoresis (PFGE) [19–21], multilocus variable tandem repeat analysis (MLVA) [22–24], octamer-based genome sequencing [25], lineage-specific polymorphism assay-6 (LSPA-6) [26], and single-nucleotide polymorphism (SNP) typing [27]. Although PFGE and MLVA have revealed considerable genetic diversity of O157:H7 strains and have been used for epidemiological analysis, they are not applicable to phylogenetic or population genetic analyses. Octamer-based genome sequencing analysis initially identified 2 distinct lineages of EHEC O157:H7 strains, designated as lineage I and lineage II [25]. Lineage-specific polymorphism assay analysis, which can subtype strains by the amplicon sizes of 6 polymerase chain reaction (PCR) products, showed that most of the strains belonging to lineage I were LSPA type 111111, whereas the strains belonging to lineage II were consisted of LSPA types 222222, 222211, 222212, and 222221 [26]. A microarray-based comparative genome hybridization analysis further identified another lineage, termed lineage I/II, which contains LSPA type 211111 [28]. An SNP-based subtyping method was also used to classify O157:H7 strains into 9 genetic clades (clades 1–9) [27]. The lineage I described above corresponded to clades 1, 2, 3, and 4 [29, 30]; lineage I/II corresponded to clades 6, 7 [30], and 8 [29–31]; and lineage II (LSPA type 222222 and 212111) corresponded to clade 7 and 9 [30].
A detailed epidemiological analysis demonstrated that patients with HUS were more likely to be infected with strains belonging to clade 8 than with strains belonging to other clades [27]. However, only 11 isolates from HUS patients were available for characterization in this study, and thus the association between clade membership and HUS requires confirmation, particularly for strains isolated from different geographic locations. Follow-up studies demonstrated that some of the clade 8 isolates showed elevated expression levels of stx2 and locus of enterocyte effacement genes (LEE, which is responsible for the intimate adhesion of EHEC to the epithelial cells), as well as increased adhesion of these isolates to cultured epithelial cells relative to strains of clades 1–3. However, not all clade 8 isolates screened exhibited the same phenotype, suggesting that genetic variability exists within these clade 8 strains [32–34].
In the present study, we have determined the clade distribution and analyzed the stx subtypes of 656 EHEC O157:H7 isolates, which were collected in Japan during the years 1999–2011 for evaluating their public health significance.
METHODS
Escherichia coli O157:H7 Isolates Used in This Study
We collected more than 320 O157:H7 isolates from HUS patients during 1999–2011 in cooperation with local public health laboratories in Japan. Among these isolates, we chose a total of 296 isolates, consisting of all isolates from sporadic cases and 1 representative isolate from each outbreak. We also chose a total of 392 O157:H7 isolates from ACs including food handlers or workers in daycare centers, who were required by law to undergo periodic stool examination. Table 1 lists the ages of all HUS patients and ACs included in this study.
Table 1.
Age-Wise Distribution in HUS Patients and AC
| Age (years) | Strains Derived From: |
|
|---|---|---|
| HUS | AC | |
| 0–9 | 192 | 29 (137*) |
| 10–19 | 20 | 24 |
| 20–64 | 27 | 312 |
| >65 | 30 | 22 |
| Total | 269 | 387 (495*) |
Abbreviations: AC, asymptomatic carrier; HUS, hemolytic uremic syndrome.
* Total number of test strains after addition of 108 strains (see text).
Because the above-described set of strains contained only 29 isolates from ACs 0 to 9 years old, we included another 108 O157:H7 strains, each one of which was isolated from a 0- to 9-year-old AC and exhibited a distinct PFGE pattern, for our analysis.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis analysis was carried out as described previously [21]. Pulsed-field gel electrophoresis patterns were analyzed by BioNumerics software, version 6.6 (Applied Maths, Belgium).
Mismatch Amplification Mutation Assay-Polymerase Chain Reaction for Detecting Clade 8 Strains
Based on the available information that the clade 8-specific SNP found in the ECs2357 gene (open reading frame number of E coli O157:H7 Sakai strain [accession no. BA000007.2]) was C539A [38], we developed a PCR assay, called the mismatch amplification mutation assay (MAMA)-PCR here, to specifically detect clade 8 isolates. For the PCR amplification, a common primer, 2357-F3 (5′-GAAGTGTGCGATCTGTCAGAA-3′), and 2 specific primers, 539A-R (5′-AAGAGCGTTTTCCAGTGGCTCTT-3′) and 539C-R (5′-CAGAGCGTTTTCCAGTGGCTCTG-3′), were designed to detect clade 8 and other clades, respectively. The 3′-end nucleotides of 539A-R and 539C-R primers (T and G, respectively) were complementary to A (clade 8) and C (other clades), respectively, at position 539, but a mismatched nucleotide was introduced at the penultimate base at the 3′ ends of both primers (designed T for C) to enhance the specificity. The underlined nucleotides in 539A-R and 539C-R primers, as shown above, indicate the hairpin regions, which could also increase the specificity. The condition to amplify the 222 base pair (bp)-long PCR product was as follows: 95°C for 2 minutes, followed by 25 cycles of 95°C for 30 seconds, 65°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 5 minutes. ExTaq polymerase (Takara, Japan) was used for the PCR amplification, and PCR was performed using a thermal cycler (T1 Thermocycler, Biometra, Germany).
stx Subtyping and Detection of Internal Deletion in norV
Subtyping of stx1 and stx2 was performed as described previously [16]. To detect an internal deletion of 204 bp-long DNA in the norV gene, PCR amplification was performed using the following 2 primers: NORV-F1 (5′-ATGTCGAATAACACCCGCAT-3′) and NORV-R1 (5′-GCCTTTTGCCGGATCGTAAA-3′). The condition used for the PCR amplification was as follows: 94°C for 2 minutes, followed by 25 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 1 minute. Using this condition, we expected to obtain either a 540 bp DNA fragment (intact norV) or a 336 bp DNA fragment (norV containing an internal deletion of 204 bp).
Clade Typing
Clades 1–3 and 8 strains were subtyped by DNA sequencing based on their SNP profiles as described previously [27]. The remaining clades were identified on the basis of SNPs or combinations of SNPs specific for individual clade [27]. Minimal requirements for determining clades 4 or 5, 6, 7, and 9 were as follows: clade 9 was determined by using the SNP T281C of ECs0654; clade 7 was determined by using the SNP A247G of ECs0517 for strains not belonging either to clade 8 or to clade 9; clade 6 was determined by using the SNP A348C of ECs3942 for strains not belonging to clade 3; and clades 4 or 5 were determined by using the SNP G776A of ECs4380 for strains not belonging to clades 6–9.
Statistical Analysis
Statistical analysis was performed using χ2 and Fisher's exact tests (EpiInfo version 7.1.1.14 or on-line calculator available at the online Website for statistical computation [http://vassarstats.net/odds2x2.html]). Logistic regression analysis was performed on the online calculator at http://statpages.org/logistic.html. Two-tailed P values < .01 were considered significant.
RESULTS
Pulsed-Field Gel Electrophoresis Analysis to Determine Genetic Relatedness of Tested Strains
We examined the phylogenetic relationship of EHEC O157:H7 isolates obtained from patients with HUS and ACs to determine whether specific lineages could be correlated with the strain source. For this purpose, we examined epidemiologically unlinked EHEC O157:H7 isolates, collected in Japan during 1999–2011, from 296 HUS patients and 392 ACs. We initially performed a cluster analysis of all strains using PFGE to determine how many of them shared the same PFGE patterns. Among all the isolates, 253 strains from HUS patients and 386 strains from ACs showed distinct PFGE patterns (results not shown). There were also multiple strains sharing the same PFGE pattern; thus, 46 strains from HUS patients showed 16 distinct PFGE patterns and 6 strains from ACs showed 1 PFGE pattern (results not shown). Therefore, from each group of strains with identical PFGE pattern, only 1 representative strain was chosen for clade typing.
Clade Typing of Isolates With Distinct Pulsed-Field Gel Electrophoresis Patterns
A total of 656 isolates (269 isolates from HUS patients and 387 isolates from ACs), all having distinct PFGE patterns as described above, were used for the clade typing (clades 1–9). Results shown in Table 2 suggest that clade 3 (n = 84, 31.2%) strains predominated among the HUS isolates, which was followed by clade 2 (n = 75, 27.9%) and clade 8 (n = 50, 18.6%) strains. In contrast, clade 7 (n = 216, 55.8%) strains predominated among the AC isolates, which was followed by clade 2 (n = 75, 19.4%) and clade 3 (n = 66, 17.1%) strains. We did not detect clade 9 strains in either group. The number of clade 8 strains found among the AC isolates was 13 (3.4%). Therefore, the frequency of clade 8 strains found among the HUS isolates was more than 5 times higher than those found among the AC isolates. This result suggests a statistically significant association between the clade 8 strains and HUS, and that this association was better than that with other clades (clades 1–7) combined (odds ratio [OR], 6.57; confidence interval [CI], 3.49–12.4; P < .0001). In addition to clade 8, statistically significant association was also observed between clade 3 (OR, 2.21; CI, 1.53–3.2; P < .0001) and clade 6 (OR, 3.69; CI, 1.74–7.86; P = .00062) strains and HUS (Table 2). Although the clade 8 strains were more likely to be isolated from HUS patients than the clade 3 (OR, 3.02; CI, 1.52–6.03; P = .0022) strains, no significant difference was observed between the clades 6 and 8 (OR, 1.6; CI, 0.62–4.17; P = .47) strains. Therefore, clades 6 and 8 strains were not very different with respect to their association with HUS. In contrast to these results, clade 7 strains were predominantly found among the AC isolates but were less frequently found among the HUS isolates (n = 23, 8.6%). In fact, the frequency of finding clade 7 strains among the AC isolates was 6 times higher than those among the HUS isolates. Thus, this result suggests a statistically significant association between the clade 7 strains and AC, and that this association was better than that with other clades (clades 1–6, 8) combined (OR, 0.074; CI, 0.046–0.12; P < .0001).
Table 2.
Distribution of Clade Type of O157:H7 Strains Isolated From HUS Patients and ACs
| Clade | Strains Derived From: |
OR (95% CI), P Value for HUS | |
|---|---|---|---|
| HUS (n = 269) | AC (n = 387) | ||
| 1 | 9 | 5 | 2.64 (0.88–7.98), .13 |
| 2 | 75 | 75 | 1.61 (1.11–2.32), .014 |
| 3 | 84 | 66 | 2.21 (1.53–3.2), <.0001 |
| 4/5 | 4 | 2 | 2.91 (0.53–16), .23* |
| 6 | 24 | 10 | 3.69 (1.74–7.86), .00062 |
| 7 | 23 | 216 | .074 (0.046–0.12), <.0001 |
| 8 | 50 | 13 | 6.57 (3.49–12.4), <.0001 |
Abbreviations: AC, asymptomatic carrier; CI, confidence interval; HUS, hemolytic uremic syndrome; OR, odds ratio.
* Fisher's exact test value (2-tailed).
stx Subtypes and Their Association With Clade and Hemolytic Uremic Syndrome Cases
We next analyzed the stx subtypes of strains used for the clade analysis. As shown in Table 3, stx1a/stx2a was the most frequent stx subtype found among the isolates from both HUS patients (152, 56.5%) and ACs (139, 35.9%). The stx subtypes stx2c and stx1a/stx2c were found more frequently among the AC isolates than among the HUS isolates (Table 3). This result is mainly due to the high prevalence of clade 7 among these stx subtypes (Table 4). The frequencies of finding stx subtypes stx1a/stx2a and stx2a/stx2c among the HUS isolates were higher than those from the AC isolates (Table 3). Because most of the strains harboring stx1a/stx2a belonged to clade 2 or clade 3 (275 of 291; 94.5%), higher association of these stx subtypes with HUS, rather than that with AC, was due to the high prevalence of their respective clades. The frequency of finding the stx2a/stx2c subtype was higher among the HUS isolates than among the AC isolates because most isolates harboring this stx subtype belonged to either clade 6 or to clade 8 (57 of 72; 79.2%). In contrast, the frequency of finding the stx2a subtype among the HUS and AC isolates were not significantly different (Table 3). However, 23 of 30 (76.7%) clade 8 strains were isolated from HUS patients, suggesting a statistically significant association between the clade 8 strains harboring only stx2a and HUS (OR, 4.37; CI, 1.14–16.69; P = .0012). Likewise, 40 of 51 (78.5%) clade 7 strains were isolated from ACs, suggesting a statistically significant association between the clade 7 strains harboring only stx2a and AC (OR, 11.75; CI, 4.72–29.3; P < .0001) (Table 4). Taken together, these results suggest that the clade typing rather than the stx2a genotype is a determinant for the association of strains with HUS or AC.
Table 3.
Association of stx Genotypes With Clinical Outcome
| stx Genotype | Strains From: |
OR (95% CI) P Value for HUS | |
|---|---|---|---|
| HUS (n = 269) | AC (n = 387) | ||
| stx1a | 1 | 3 | .48 (0.049–4.62) .65* |
| stx2a | 53 | 53 | 1.55 (1.02–2.35) .04 |
| stx2c | 10 | 131 | .075 (0.04–0.15) <.0001 |
| stx1a/stx2a | 152 | 139 | 2.32 (1.69–3.19) <.0001 |
| stx1a/stx2c | 2 | 30 | .06 (0.02–0.27) <.0001* |
| stx1a/stx2a/stx2c | 1 | 9 | .16 (0.02–1.24) .05* |
| stx2a/stx2c | 50 | 22 | 3.79 (2.23–6.43) <.0001 |
Abbreviations: AC, asymptomatic carrier; CI, confidence interval; HUS, hemolytic uremic syndrome; OR, odds ratio.
* Fisher's exact test value (2-tailed).
Table 4.
stx Genotype of Strains Belonging to Each Clade Group
| Clade |
stx Genotype of Isolates From HUS/AC |
||||||
|---|---|---|---|---|---|---|---|
| stx1a | stx2a | stx2c | stx1a/stx2a | stx1a/stx2c | stx1a/stx2a/stx2c | stx2a/stx2c | |
| 1 | 0/0 | 1/0 | 0/0 | 8/5 | 0/0 | 0/0 | 0/0 |
| 2 | 1/1 | 11/3 | 0/0 | 62/69 | 0/0 | 1/2 | 0/0 |
| 3 | 0/2 | 2/2 | 0/0 | 82/62 | 0/0 | 0/0 | 0/0 |
| 4/5 | 0/0 | 3/0 | 0/2 | 0/0 | 0/0 | 0/0 | 1/0 |
| 6 | 0/0 | 2/1 | 3/1 | 0/0 | 0/3 | 0/0 | 19/5 |
| 7 | 0/0 | 11/40 | 7/128 | 0/3 | 2/27 | 0/7 | 3/11 |
| 8 | 0/0 | 23/7 | 0/0 | 0/0 | 0/0 | 0/0 | 27/6 |
| Total | 1/3 | 53/53 | 10/131 | 152/139 | 2/30 | 1/9 | 50/22 |
Abbreviations: AC, asymptomatic carrier; HUS, hemolytic uremic syndrome.
Development of Mismatch Amplification Mutation Assay-Polymerase Chain Reaction to Detect Further Clade 8 Strains
A previous study has developed a real-time PCR assay for detecting clade 8 strains [35]. In the present study, we have developed an alternative PCR-based assay, called MAMA-PCR, which allowed inexpensive detection of clade 8 strains without performing any DNA sequencing analysis. As shown in Figure 1, clade 8 was specifically detected by a specific primer set but not by the other primer set that can detect all other clades (clades 1–7). Using this assay, we were able to confirm all 63 clade 8 strains (listed in Table 2), which were subtyped by DNA sequencing (results not shown).
Fig. 1.
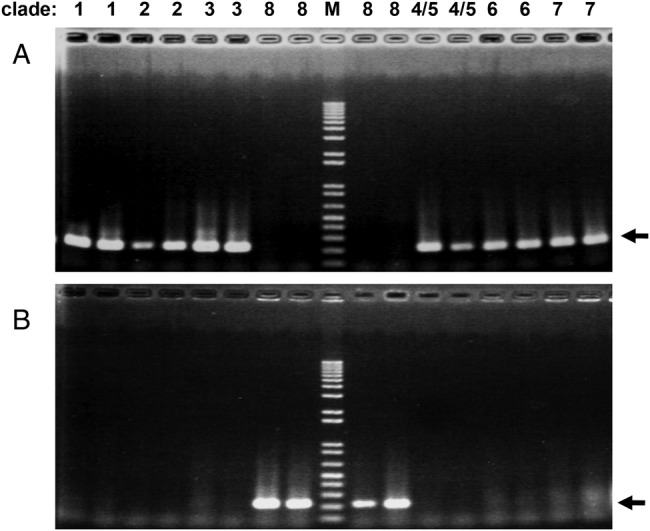
Development of a clade 8-specific mismatch amplification mutation-polymerase chain reaction (PCR) assay. Lane M shows the DNA size markers (Tracklt 1 kb Plus DNA Ladder, Life Technologies) including DNA fragments sized 100, 200, 300, 400, 500, 650, 850, 1000 bp, and so on. The black arrows on the right side of both panels indicate positions of the PCR products. Two different strains from each clade were used. (A) Polymerase chain reaction performed using the 539C-R primer detected strains belonging to clades 1–7, but not strains belonging to clade 8. (B) Polymerase chain reaction performed using the 539A-R primer detected strains belonging to clade 8, but not strains belonging to clades 1–7.
The 0- to 9-Year-Old Age Group Is a Significant Predictor for the Association Between Clade 8 and Hemolytic Uremic Syndrome
In Japan, incidence of EHEC infection was highest among the 0- to 4-year-old age group followed by those in the 5- to 9-year-old age group [8]. As shown in Table 1, 192 of 269 (71.4%) strains isolated from HUS patients were from children of ages between 0 and 9 years; in contrast, only 29 isolates were from ACs, which did not contain any clade 8 strain. Therefore, for further analyzing the frequency of occurrence of clade 8 strains among the 0- to 9-year-old age group, we included another 108 test strains, which were obtained from 0- to 9-year-old children without any symptoms, and each one of these strains exhibited different PFGE patterns (data not shown). Mismatch amplification mutation assay-PCR analysis (Table 5) demonstrated that 6 of 108 strains belonged to clade 8, suggesting that the association between clade 8 and HUS for the 0- to 9-year-old age group was still statistically significant (OR, 6.11; CI, 2.52–14.84; P < .0001). On the other hand, this association was not significant when strains were isolated from HUS patients or ACs who were over 10 years old (OR, 3.08; CI, 1.23–7.7; P = .019 [Fisher's exact test value]). Logistic regression analysis indicated that the 0- to 9-year-old age group was a significant predictor for the association between clade 8 and HUS; thus, crude and adjusted OR values for this age group were 5.72 and 4.44, and difference between them was more than 10%. However, no association was observed between gender and HUS status (OR, 1.02; CI, 0.74–1.41; P = 1). Gender was not a predictor for the association between clade 8 and HUS; in this case, the crude OR was equal to that of the adjusted OR (both were 6.3).
Table 5.
Clade 8 Strains Isolated From Patients With HUS or ACs (Age-Wise Distribution)
| Age (years) | Number of Clade 8/Other Clades |
OR (95% CI), P Value of Clade 8 for HUS | |
|---|---|---|---|
| HUS | AC | ||
| 0–9 | 42/150 | 6/131 | 6.11 (2.52–14.84), <.0001 |
| >10 | 8/69 | 13/345 | 3.08 (1.23–7.7), .019* |
| Total | 50/219 | 19/476 | 5.72 (3.29–9.93), <.0001 |
Abbreviations: AC, asymptomatic carrier; CI, confidence interval; HUS, hemolytic uremic syndrome; OR, odds ratio.
* Fisher's exact probability test value (2-tailed).
Clade Specific Deletion of norV
The norV gene, which encodes an anaerobic nitric oxide (NO) reductase, has been shown to be a putative virulence determinant in certain O157:H7 strains because NO inhibited Stx2 expression under anaerobic condition [36, 37]. In the reference strains EDL933 (belonging to clade 3) and Sakai (belonging to clade 1), the NO reductase activity was abolished because of an internal 204-bp deletion in the norV gene; however, in the spinach outbreak-derived clade 8 strain TW14359, the norV gene was found to be intact [38].
We designed specific PCR primers (see Methods) to examine the distribution of this 204-bp deletion in norV in each clade group. We found that 84 of 145 strains (57.9%) isolated from HUS patients and 177 of 223 strains (79.4%) isolated from AC harbored the intact norV gene. Therefore, the presence of norV gene by itself does not cause HUS. We found that all tested isolates belonging to clade 1 (n = 12), clade 2 (n = 47), and clade 3 (n = 48) had the same norV deletion as that found in the Sakai and EDL933 strains [39]. On the other hand, all tested isolates belonging to clade 4 or 5 (n = 4), clade 6 (n = 33), clade 7 (n = 173), and clade 8 (n = 51) had intact norV gene. Therefore, presence of an intact norV is rather nonrandomly distributed across O157:H7 lineages (clades 1–3 vs clades 4–8).
DISCUSSION
A previous study, for which only 11 isolates from HUS patients were available, suggested that HUS patients were 7 times more likely to be infected with a clade 8 strain than with a strain belonging to other clades (clades 1–7) [27]. In the present study, clade typing of 656 epidemiologically independent O157:H7 isolates, collected from 269 HUS patients and 387 ACs in Japan during the years 1999–2011, revealed that the degree of association of clade 8 strains with HUS was statistically more significant than that with AC. Using the same strain set, we also found significant association between the strains belonging to clade 6 and HUS. To the best of our knowledge, this is the first report that uses a large set of O157 strains and demonstrates a significant association not only between clade 8 strains and HUS cases but also between clade 6 strains and HUS cases. In addition to these results, we also found statistically significant association between clade 7 strains and AC, which confirmed the earlier observation that the clade 7 strains caused less severe disease [27, 39].
To assess the potential contributions of high virulent lineages of EHEC O157 to HUS, it is highly desirable to have suitable methods available for their early detection. Therefore, detection of clade 6 and clade 8 strains by using a rapid and specific method, such as reverse transcription (RT)-PCR, is not only important to have but is also useful for clinical laboratories. Availability of such methods would also help in understanding the environmental reservoirs and sources of such high virulent strains. In the present study, we have developed MAMA-PCR, an inexpensive PCR-based method for detecting clade 8 strains, as an alternative to the RT-PCR-based detection method that was developed previously [35].
Several previous studies showed that some of the clade 8 strains expressed elevated levels of several virulence-related genes and demonstrated enhanced ability to attach to epithelial cells compared with several other strains belonging to clades 1–3 [33–35]. However, further studies using a large set of strains, including the clade 6 strains, would be required to understand the molecular mechanism of lineage-specific variability in virulence of EHEC O157 strains.
Subtyping of stx gene in each clade revealed that all clade 6 strains isolated from HUS patients harbored stx2a and/or stx2c, whereas all clade 8 strains harbored either stx2a or stx2a/stx2c. In most clade 2 and clade 3 strains, the observed stx subtype was stx1a/stx2a (86.2% and 96%, respectively). Thus, there seems to be close association between these clades and stx subtypes of strains, which contribute to the observed high degree of association between the stx subtypes and HUS cases.
Clade 7 strains harbored several different stx subtypes, including stx2a, stx2c, stx1a/stx2a, stx1a/stx2c, stx1a/stx2a/stx2c, and stx2a/stx2c. Among these stx subtypes, the major stx subtypes found in clade 7 strains were stx2c and stx1a/stx2c. Thus, these subtypes showed higher degree of association with AC than with HUS.
Hemolytic uremic syndrome was more often found to be associated with the stx subtype stx2a than with any other stx subtypes [17, 18]. However, results shown in this study indicated that the frequency of finding the stx2a subtype among the HUS and AC isolates was comparable. On the other hand, our findings that the clade 7 strains harboring only the stx subtype stx2a showed higher association with ACs rather than with HUS cases (P < .0001) and the clade 6 or clade 8 strains harboring the stx subtype stx2a showed higher association with the HUS cases rather than with ACs (P = .00079), suggested that the clade typing is essential for determining the virulence potential of EHEC O157 strains harboring only the stx subtype stx2a.
In the present study, we also observed that the 0- to 9-year-old age group was a significant predictor for the association between clade 8 strains and HUS cases. In addition to the clade 8 strains, close association between the clade 6 strains and HUS was also observed for this age group. Thus, 12% (23 of 192) and 1.3% (1 of 77) of clade 6 isolates from HUS patients were actually obtained from the 0- to 9-year-old and >10-year-old age groups, respectively; in contrast, numbers of clade 6 strains isolated from ACs were comparable (4.4% vs 2.8%) for both age groups. Contrary to these observations, clade 7 strains isolated from patients with HUS were comparable in number (8.3% vs 9.1%) for both 0- to 9-year-old and >10-year-old age groups, whereas those isolated from ACs were 23.4% (32 of 137) and 58.1% (208 of 358) for the 0- to 9-year-old and >10-year-old age groups, respectively. Therefore, we found significant association between the clade 7 strains isolated from ACs and >10-year-old age group (P < .0001).
All of the clades 1–3 strains used in this study carried an internal 204 bp deletion in the norV gene, which encodes a putative virulence factor; however, all the tested strains belonging to clades 4–8 carried the intact norV gene. Consistent with these results, a previous study examining O157 strains isolated from geographically different places showed that all strains isolated in Argentina (n = 57) and Australia (n = 58) and harboring the intact norV gene belonged to clade 4, 6, 7, or 8 [40]. These results suggest that the presence of intact norV does not have any association with HUS.
In conclusion, the present study using a large set of EHEC O157 strains isolated from HUS patients and ACs showed a significant association of clade 6 or 8 and clade 7 strains with HUS and ACs, respectively. All Clade 6 strains isolated from HUS patients harbored stx2a and/or stx2c, whereas clade 8 strains harbored either stx2a or stx2a/stx2c but not stx2c. Taken together, we believe that our observed results would prompt further investigations that would help us find the reservoirs and sources of these high virulent strains among geographically different samples and also help us develop molecular methods to rapidly and specifically identify clades with or without stx subtype.
Acknowledgments
We thank Hitomi Satou, Nobuko Takai, Yasunori Saitoh, Yukie Nakajima, and Ai Yoshida for technical assistances. We also thank the following public health institutes in Japan for O157 strains: Yamagata Prefectural Institute of Public Health; Fukui Prefectural Institute of Public Health and Environmental Science; Health Center of Kanazawa City; Amagasaki City Institute of Public Health; Chiba Prefectural Institute of Public Health; Hiroshima City Institute of Public Health; Public Health Institute of Kochi Prefecture; Nagasaki City Public Health Center; Wakayama City Institute of Public Health; Nara Prefectural Institute of Health; Kagawa Prefectural Research Institute for Environmental Sciences and Public Health; Shimane Prefectural Institute of Public Health and Environmental Science; Gunma Prefectural Institute of Public Health and Environmental Sciences; Sakai City Institute of Public Health; Kumamoto City Environmental Research Institute; Yamanashi Institute for Public Health and Environment; Aomori Prefectural Institute of Public Health; Tokushima Prefectural Institute of Public Health; Ibaraki Prefectural Institute of Public Health; Hiroshima Prefectural Technology Research Institute; Toyohashi City Public Health Center; Toyota City Public Health Center; Kyoto City Institute of Health and Environmental Sciences; Shizuoka City Institute of Environmental Sciences and Public Health; Sendai City Institute of Public Health; Hokkaido Institute of Public Health; Sagamihara City Laboratory of Public Health; Higashiosaka City Public Health; Ishikawa Prefectural Institute of Public Health and Environmental Science.
Financial support. This work was supported by grants-in-aids for scientific research from the Ministry of Education, Culture, Sport, Science and Technology of Japan (grant number 33800600; to S. I.) and from the Ministry of Health, Labour and Welfare (grant numbers H24-Shinkou-Ippan-005 and H24-Shinkou-Ippan-012) of Japan.
Potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
APPENDIX
EHEC working group in Japan: Reiko Arai, Masao Kawase (Niigata Prefectural Institute of Public Health and Environmental Sciences); Yukiko Asano (Ehime Prefectural Institute of Public Health and Environmental Science); Nanami Asoshima (Fukuoka City Institute for Hygiene and the Environment); Kazuki Chiba (Fukushima Prefectural Institute of Public Health); Ichiro Furukawa, Toshiro Kuroki (Kanagawa Prefectural Institute of Public Health); Madoka Hamada (Kagoshima Prefectural Institute for Environmental Research and Public Health); Seiya Harada (Kumamoto Prefectural Institute of Public-Health and Environmental Science); Takashi Hatakeyama (Miyagi Prefectural Institute of Public Health and Environment); Takashi Hirochi, Yumiko Sakamoto (Sapporo City Institute of Public Health, Hokkaido); Midori Hiroi, Takashi Kanda (Shizuoka Institute of Environment and Hygiene); Kazumi Horikawa (Fukuoka Institute of Health and Environmental Sciences); Kaori Iwabuchi (Research Institute for Environmental Sciences and Public Health of Iwate Prefecture); Mitsuhiro Kameyama (Yamaguchi Prefectural Institute of Public Health and Environment); Hitomi Kasahara (Nagano Environmental Conservation Research Institute); Shinya Kawanishi (Himeji City Institute of Environment and Health, Hyogo); Koji Kikuchi, Hiroyuki Ueno (Saitama-City Institute of Health Science and Research, Saitama); Tomoko Kitahashi (Chiba City Institute of Health and Environment, Chiba); Yuka Kojima (Kawasaki City Institute for Public Health, Kanagawa); Noriko Konishi, Hiromi Obata, Akemi Kai (Tokyo Metropolitan Institute of Public Health); Tomomi Kono (Shiga Prefectural Institute of Public Health); Takayuki Kurazono (Saitama Institute of Public Health); Masakado Matsumoto (Aichi Prefectural Institute of Public Health); Yuko Matsumoto (Yokohama City Institute of Health, Kanagawa); Yuhki Nagai (Mie Prefecture Health and Environment Research Institute, Japan); Hideki Naitoh (Tochigi Prefectural Institute of Public Health and Environmental Science); Hiroshi Nakajima (Okayama Prefectural Institute for Environmental Science and Public Health); Hiromi Nakamura (Department of Microbiology, Osaka City Institute of Public Health and Environmental Sciences); Kunihiko Nakane (Okazaki City Public Health Center, Aichi); Keiko Nishi (Saga Prefectural Institute of Public Health and Pharmaceutical Research); Etsuko Saitoh (Hyogo Prefectural institute of Public Health and Consumer Sciences, Public Health Science Research Center); Hiroaki Satoh, Mitsuteru Takamura (Morioka City Health Center, Iwate); Yutaka Shiraki (Gifu Prefectural Research Institute for Health and Environmental Sciences); Junichi Tanabe (Niigata City Institute of Public Health and Environment); Keiko Tanaka (Wakayama Prefectural Research Center of Environment and Public Health); Yuki Tokoi (Utsunomiya City Institute of Public Health and Environmental Sciences, Tochigi); Jun Yatsuyanagi (Akita Research Center for Public Health and Environment).
References
- 1.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–79. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivas M, Miliwebsky E, Chinen I, et al. Characterization and epidemiologic subtyping of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina. Foodborne Pathog Dis. 2006;3:88–96. doi: 10.1089/fpd.2006.3.88. [DOI] [PubMed] [Google Scholar]
- 4.Jenke C, Harmsen D, Weniger T, et al. Phylogenetic analysis of enterohemorrhagic Escherichia coli O157, Germany, 1987–2008. Emerg Infect Dis. 2010;16:610–6. doi: 10.3201/eid1604.091361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) Incidence and trends of infection with pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. sites, 1996–2012. MMWR Morb Mortal Wkly Rep. 2013;62:283–7. [PMC free article] [PubMed] [Google Scholar]
- 6.Eurosurveillance Editorial Team. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2012. Available at: http://www.efsa.europa.eu/en/efsajournal/pub/3129.htm . Accessed 17 June 2014. [PubMed] [Google Scholar]
- 7.Vally H, Hall G, Dyda A, et al. Epidemiology of Shiga toxin producing Escherichia coli in Australia, 2000–2010. BMC Public Health. 2012;12:63. doi: 10.1186/1471-2458-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Infectious Agents Surveillance Report (IASR) Enterohemorrhagic Escherichia coli infection in Japan as of April 2013. Available at: http://www.nih.go.jp/niid/en/iasr-vol34-e/865-iasr/3570-tpc399.html . Accessed 17 June 2014. [Google Scholar]
- 9.Infectious Agents Surveillance Report (IASR) Enterohemorrhagic Escherichia coli infection in Japan as of April 2009. Available at: http://idsc.nih.go.jp/iasr/30/351/tpc351.html . Accessed 17 June 2014.
- 10.Infectious Agents Surveillance Report (IASR) Enterohemorrhagic Escherichia coli infection in Japan as of May 2010. Available at: http://idsc.nih.go.jp/iasr/31/364/tpc364.html . Accessed 17 June 2014. [Google Scholar]
- 11.Infectious Agents Surveillance Report (IASR) Enterohemorrhagic Escherichia coli infection in Japan as of April 2011. Available at: http://idsc.nih.go.jp/iasr/32/375/tpc375.html . Accessed 17 June 2014. [Google Scholar]
- 12.Infectious Agents Surveillance Report (IASR) Enterohemorrhagic Escherichia coli infection in Japan as of April 2012. Available at: http://www.nih.go.jp/niid/en/iasr-vol33-e/865-iasr/2134-tpc387.html . Accessed 17 June 2014. [Google Scholar]
- 13.Boerlin P, McEwen SA, Boerlin-Petzold F, et al. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol. 1999;37:497–503. doi: 10.1128/jcm.37.3.497-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich AW, Bielaszewska M, Zhang WL, et al. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. 2002;185:74–84. doi: 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- 15.Ostroff SM, Tarr PI, Neill MA, et al. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–8. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 16.Scheutz F, Teel LD, Beutin L, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol. 2012;50:2951–63. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orth D, Grif K, Khan AB, et al. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn Microbiol Infect Dis. 2007;59:235–42. doi: 10.1016/j.diagmicrobio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Persson S, Olsen KE, Ethelberg S, et al. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol. 2007;45:2020–4. doi: 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett TJ, Lior H, Green JH, et al. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–7. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumiya H, Terajima J, Wada A, et al. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:1675–80. doi: 10.1128/jcm.35.7.1675-1680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terajima J, Izumiya H, Iyoda S, et al. Effectiveness of pulsed-field gel electrophoresis for the early detection of diffuse outbreaks due to Shiga toxin-producing Escherichia coli in Japan. Foodborne Pathog Dis. 2006;3:68–73. doi: 10.1089/fpd.2006.3.68. [DOI] [PubMed] [Google Scholar]
- 22.Noller AC, McEllistrem MC, Stine OC, et al. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:675–9. doi: 10.1128/JCM.41.2.675-679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyytiä-Trees E, Smole SC, Fields PA, et al. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157) Foodborne Pathog Dis. 2006;3:118–31. doi: 10.1089/fpd.2006.3.118. [DOI] [PubMed] [Google Scholar]
- 24.Izumiya H, Pei Y, Terajima J, et al. New system for multilocus variable-number tandem-repeat analysis of the enterohemorrhagic Escherichia coli strains belonging to three major serogroups: O157, O26, and O111. Microbiol Immunol. 2010;54:569–77. doi: 10.1111/j.1348-0421.2010.00252.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Nietfeldt J, Benson AK. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA. 1999;96:13288–93. doi: 10.1073/pnas.96.23.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Kovar J, Kim J, et al. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl Environ Microbiol. 2004;70:6846–54. doi: 10.1128/AEM.70.11.6846-6854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning SD, Motiwala AS, Springman AC, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci USA. 2008;105:4868–73. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Laing C, Steele M, et al. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics. 2007;8:121. doi: 10.1186/1471-2164-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laing CR, Buchanan C, Taboada EN, et al. In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics. 2009;10:287. doi: 10.1186/1471-2164-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K, Knabel SJ, Dudley EG. rhs genes are potential markers for multilocus sequence typing of Escherichia coli O157:H7 strains. Appl Environ Microbiol. 2009;75:5853–62. doi: 10.1128/AEM.00859-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartzell A, Chen C, Lewis C, et al. Escherichia coli O157:H7 of genotype lineage-specific polymorphism assay 211111 and clade 8 are common clinical isolates within Pennsylvania. Foodborne Pathog Dis. 2011;8:763–8. doi: 10.1089/fpd.2010.0762. [DOI] [PubMed] [Google Scholar]
- 32.Abu-Ali GS, Ouellette LM, Henderson ST, et al. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One. 2010;5:e10167. doi: 10.1371/journal.pone.0010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abu-Ali GS, Ouellette LM, Henderson ST, et al. Differences in adherence and virulence gene expression between two outbreak strains of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 2010;156:408–19. doi: 10.1099/mic.0.033126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neupane M, Abu-Ali GS, Mitra A, et al. Shiga toxin 2 overexpression in Escherichia coli O157:H7 strains associated with severe human disease. Microb Pathog. 2011;51:466–70. doi: 10.1016/j.micpath.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riordan JT, Viswanath SB, Manning SD, et al. Genetic differentiation of Escherichia coli O157:H7 clades associated with human disease by real-time PCR. J Clin Microbiol. 2008;46:2070–3. doi: 10.1128/JCM.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vareille M, de Sablet T, Hindré T, et al. Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli. Proc Natl Acad Sci USA. 2007;104:10199–204. doi: 10.1073/pnas.0702589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu T, Tsutsuki H, Matsumoto A, et al. The nitric oxide reductase of enterohaemorrhagic Escherichia coli plays an important role for the survival within macrophages. Mol Microbiol. 2012;85:492–512. doi: 10.1111/j.1365-2958.2012.08122.x. [DOI] [PubMed] [Google Scholar]
- 38.Kulasekara BR, Jacobs M, Zhou Y, et al. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect Immun. 2009;77:3713–21. doi: 10.1128/IAI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanaja SK, Springman AC, Besser TE, et al. Differential expression of virulence and stress fitness genes between Escherichia coli O157:H7 strains with clinical or bovine-biased genotypes. Appl Environ Microbiol. 2010;76:60–8. doi: 10.1128/AEM.01666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellor GE, Sim EM, Barlow RS, et al. Phylogenetically related Argentinean and Australian Escherichia coli O157 isolates are distinguished by virulence clades and alternative Shiga toxin 1 and 2 prophages. Appl Environ Microbiol. 2012;78:4724–31. doi: 10.1128/AEM.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


