Abstract
Background
Control of methicillin-resistant Staphylococcus aureus (MRSA) is difficult in select populations. We used molecular typing to study the effect of universal surveillance and decolonization of carriers, without isolation, on MRSA transmission in a specialized unit.
Methods
Patients admitted to the unit were screened for nasal MRSA at admission and discharge. Those who acquired MRSA during their stay were identified and linked to carriers with shared time in unit. Molecular typing of isolates was performed to identify transmission.
Results
Of 3285 admissions, 82% were tested for MRSA nasal carriage; the discharge screening compliance was 64.7%. Admission prevalence was 2.3% among patients screened, and 7 (0.42%) acquired nasal MRSA during their stay. All patients who acquired MRSA shared time in the unit with a colonized patient. There were 3.9 MRSA acquisitions per 1000 at-risk days. Isolates from 5 patients that acquired MRSA during their stay as well as their potential donors (11 donor: recipient patient pairs) were available for typing. Pulsed-field gel electrophoresis matched 1 acquisition isolate to a colonized patient isolate. There were no MRSA infections during the study period.
Conclusions
Despite less than perfect nasal screening compliance and exemption from traditional isolation precautions, acquisition of MRSA was 0.42% in this patient population over a course of 4.75 years, including a single case of acquisition, genetically similar to a known potential donor source. Screening for MRSA colonization and decolonizing of carriers was sufficient in reducing transmission in this vulnerable population.
Keywords: molecular typing, nosocomial MRSA, psychiatry units, transmission
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of healthcare-associated infection that is often transmitted by asymptomatic carriers, with the risk of invasive disease being greatest shortly after acquisition of a new strain [1–3]. Control of acquisition and spread of nosocomial MRSA is a public health challenge that requires sustained effort and constant monitoring with considerable associated costs [4]. Strategies for MRSA control are targeted toward the detection and decolonization or isolation of reservoirs, reduction of transmission by appropriate hand hygiene, and lowering of antimicrobial pressure [5]. Several controlled and quasi-experimental studies have been conducted to evaluate the effectiveness of infection prevention measures with seemingly controversial results [6–8]. In particular, the use of universal screening and contact isolation, one of the most laborious and expensive practices, has been strongly debated [1, 9–11]. NorthShore University HealthSystem (NorthShore) implemented universal surveillance in August, 2005 to detect nasal carriage of MRSA on all admissions, followed by contact isolation and decolonization of colonized patients.
A mental health unit located within the acute care Evanston Hospital, a part of this program, was exempt from contact isolation; decolonization alone was performed on MRSA carriers in this unit. Discharge screening tracked MRSA conversions. This study evaluates the prevalence of colonization and extent of MRSA transmission in that unit and further evaluates whether specific acquisition of MRSA could be linked to nasally colonized patients on the unit.
METHODS
Patient Selection
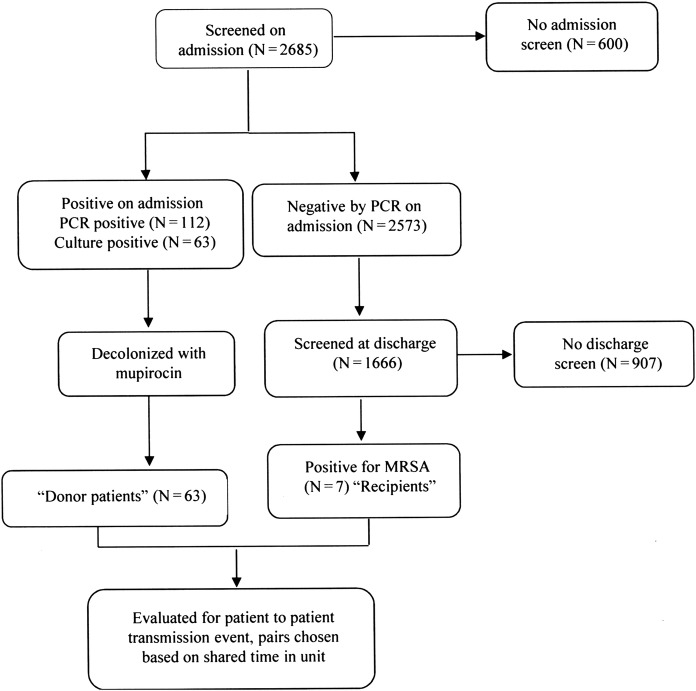
The NorthShore enterprise data warehouse was accessed to retrospectively identify patients that acquired MRSA during their hospital stay (defined as negative for MRSA nasal carriage upon admission, but subsequently became culture positive at discharge after a length of stay of >2 days). To investigate the possibility of patient-patient transmission of MRSA, the following terms were used to define epidemiologic relationships between patients. The algorithm of testing for MRSA and allocation of epidemiologic relations is detailed in Figure 1.
Patients positive by culture for nasal MRSA on admission screen were considered as potential “donors”.
Patients that tested negative for nasal MRSA on admission screen but were culture positive at discharge (at least 2 calendar days after admission) were identified as having become “colonized during their stay” and were considered to be potential “recipients”.
All nasal MRSA conversions were assumed to be due to in-unit transmissions. A recipient could potentially become colonized from any donor present on the same unit simultaneously. Matched donor and recipient isolates were identified by shared time on unit and selected for molecular typing.
Figure 1.
Algorithm of nasal methicillin-resistant Staphylococcus aureus screen for patients admitted to the mental health unit, and allocation of patients to different epidemiologic groups. The investigation did not monitor potential transmission from colonized healthcare workers or the environment.
Methicillin-Resistant Staphylococcus aureus Screening and Molecular Tests on Isolates
Nasal screening for MRSA at admission and discharge was accomplished by molecular testing using the BD GeneOhm MRSA Assay (Becton Dickinson, Sparks, MD) [12]. All polymerase chain reaction (PCR)-positive nasal swabs were subcultured on CHROMagar (BBL, Becton Dickinson, Sparks, MD) to confirm as positive. One swab head was also incubated in Trypticase soy broth (enrichment culture) which was incubated for 18–24 hours at 35 to 37°C, the broth was then streaked onto another CHROMagar MRSA plate, and the plate was incubated for 24–48 hours at 35 to 37°C as described above. Positive cultures were confirmed by performing a Staphaurex agglutination test (Remel, Lenexa, KS) on pure colonies. Only patients with culture-positive nasal swabs were considered true positive MRSA carriers for this analysis. All MRSA isolates were identified by conventional microbiologic techniques or by in-house PCR assay for mecA gene [12, 13]. Isolates were also tested for genes encoding high-level mupirocin resistance mupA and the Panton-Valentine leukocidin (PVL) toxin as previously described [13, 14].
Decolonization Protocol
As recommended by the NorthShore Department of Infection Control, all colonized patients were treated with a 5-day regimen comprising mupirocin calcium, 2% twice daily to the nares, and a chlorhexidine 4% wash or shower on days 1, 3, and 5 of decolonization.
Molecular Typing
Frozen MRSA isolates were inoculated onto Columbia Colistin Nalidixic Acid agar (BD) and incubated for 20–24 hours at 37°C to recover archived strains. Pulsed-field gel electrophoresis (PFGE) was performed using SmaI as described previously [15, 16], using S aureus NCTC 8325 as the control. Gel images were captured and digitalized as TIFF images using Gel Doc XR+ with Image Lab Software version 3.0 (Bio-Rad, Hercules, CA), and band comparisons were performed using BioNumerics software version 6.5 (Applied Maths Inc., Austin, TX). Results of comparisons were visually inspected to confirm potential strain associations. Dendograms were generated for each recipient isolate and their corresponding donor source isolates using unweighted-pair group method based on arithmetic averages using Dice coefficients. Band tolerance and optimization were set at 1.25% and 0.8%, respectively. Pulsed-field gel electrophoresis band patterns were considered to be within the same cluster if they had a similarity coefficient ≥80% and identical if the similarity coefficient was ≥95% [16, 17].
This study was conducted as a part of ongoing Quality Maintenance and Assurance and Infection Control Program approved by the NorthShore University HealthSystem Institutional Review Board.
RESULTS
The study included all patients admitted to the mental health unit over a period of 57 months (April 2007 through December 2011).
Prevalence of Methicillin-Resistant Staphylococcus aureus Nasal Colonization
During the study period, there were 3285 admissions in the study unit and 2685 patients were screened for nasal MRSA carriage, with an admission screening compliance rate of 81.7%. The prevalence of MRSA colonization as determined by positive PCR on nasal swabs was 112 of 2685 (4.2%); all PCR-positive patients were decolonized as recommended by infection control. For the transmission investigation, only culture-positive patients were considered as true potential donors; 56% of admission-positive PCR tests were culture positive, for a culture-positive MRSA admission prevalence of 2.3%.
Incidence of Methicillin-Resistant Staphylococcus aureus Acquisition During Hospitalization
There were 2573 patients that tested negative for nasal MRSA on admission. Discharge screening was performed on 1666 of these patients (discharge screening compliance of 64.7%). We identified 26 (1.6%) new nasal MRSA acquisitions by PCR; 26.9% of these samples tested grew MRSA for a true conversion rate among at-risk patients of 0.42%. Assessment of potential transmission and at-risk days were based on the number of colonized patients admitted and the number of at-risk days. There were 6589 potential at-risk days (ie, days when patients negative on admission were in the unit) and 259 potential transmission days (ie, days when patients positive on admission were in the unit); assuming all conversions were due to in-unit transmission, there were 3.9 conversions per 1000 at-risk days and 0.1 transmission/potential transmission day. Because clinical characteristics of patients were not collected, we were unable to ascertain risk factors associated with MRSA acquisition. No nosocomial MRSA infections were identified on this unit during the observation period. Repeat culture results were available in 79% of the patients' positive for nasal MRSA by culture. A total of 38 patients were confirmed to have cleared MRSA after decolonization regimen, whereas 12 (25%) were found to be continuously colonized despite therapy. The results support an earlier study conducted in our center where mupirocin-based therapy reduced colonization [18]. In addition, patients were only tested nasally and as has been documented in literature, MRSA could have persisted in other body sites.
Methicillin-Resistant Staphylococcus aureus Transmission and Molecular Typing of Isolates
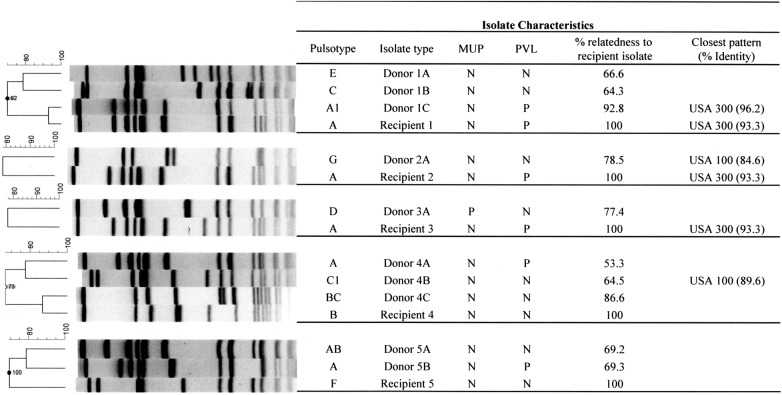
All recipients that had shared any time with potential donors were identified and matched; the algorithm assumed that all colonized patients with shared time on the unit could be potential donors. Figure 2 shows the details of each recipient patient and the corresponding potential donors. Five of the 6 available acquisition isolates had at least 1 potential donor source, available for typing (range 1–4 and median 2) with a total of 10 donor isolates. The PFGE image for each donor, ie, recipient(s) patient combinations are shown in Figure 2. One predominant PFGE pattern was common among the recipient isolates (3 of 5) and was also found in 2 of the donor isolates. This pattern had >90% similarity to the USA300 PFGE (community-associated) pulsotype. These 3 MRSA acquisitions with near-identical PFGE pattern were separated in time by years and so were not attributed to an epidemiologically linked transmission. Only 1 recipient isolate was found to have a potential donor source with a matching isolate, showing >90% identity, with both recipient and donor isolates resembling the USA300 PFGE pattern.
Figure 2.
Molecular characterization of “donor” and corresponding “recipient(s)” isolates. The pulsed-field gel electrophoresis (PFGE) dendogram compares fingerprint patterns of each recipient isolate (indicated by a letter) and their matching putative donor isolate(s) (indicated by the corresponding letter followed by numbers). Columns marked mupA and PVL (Panton-Valentine leukocidin) indicate the results for genetic tests performed to detect the mupA and PVL gene, respectively. Percentage relatedness to the recipient isolate and PFGE-type strains are indicated.
Prevalence of Community-Associated Methicillin-Resistant Staphylococcus aureus and Mupirocin Resistance
Of the 63 patients colonized on admission, 28 (44.4%) had isolates that were identical or closely related to the USA300 PFGE type, these were also positive for the PVL gene. High-level mupirocin resistance (mupA gene) was detected in 4 (5.6%) isolates, among those colonized on admission.
Among the 7 patients who acquired MRSA during their hospital stay, 3 (37.5%) had isolates closely related or identical to the USA300 PFGE pattern and were positive for PVL gene, and none were positive for the mupA gene.
DISCUSSION
Our study was conducted to evaluate the effectiveness of decolonization without isolation as a strategy for prevention of MRSA transmission in this specialized unit. This approach and resultant data suggest that transmission of MRSA on units where contact isolation might hinder care can be curtailed by simple decolonization (eg, “medical isolation”) of nasal carriers at admission. Our admission prevalence of 2.3% was lower than in another recent survey that found a prevalence of 5.2% when nares and axilla was screened on patients in a psychiatry unit [19]; however, this study included both asymptomatic colonized patients as well as those with MRSA infection that contributed to their higher admission prevalence—we had no MRSA infections among patients admitted to the nursing unit. Thus, both studies suggest that MRSA colonization may be at a modest level in uninfected patients admitted to mental health units.
The unique requirements of patients in mental health units make choice of potential strategies to prevent MRSA acquisition especially important. Contact isolation can be considered a deterrent to patient care that has been associated with depression [20], and stigmatization of such a protocol could negatively impact therapy in some patients. No intervention was given to colonized patients in the Farley et al study [19], and they reported an acquisition rate of 2.5% (7 of 282) that was 5-fold higher than what we found (7 of 1666) for our investigation (χ2 test, P = .001). The result of our study is especially important in this regard, because it demonstrates transmission in a mental health unit can be curtailed when decolonization alone (no isolation) is used to prevent spread of MRSA.
We did not detect any new MRSA clinical disease during the study period in this unit. Resistance to mupirocin was detected in 4 isolates (all being isolated from patients positive on admission); there was no increase in mupirocin resistance detected over the study period, which could negatively impact the control of MRSA in these units [21].
Jones et al [3] recently reported the detailed findings investigating the relationship between MRSA admission rate, transmission, and clinical disease at 112 Veterans Affairs Medical Centers across the United States. A significant decrease in the clinical culture rate (MRSA disease) occurred only in patients who were negative for MRSA at the time of admission (P = .002). Limiting transmission led to lower admission prevalence, and both were associated with reduced nosocomial clinical disease; in other words, reduction of disease relies on lowering transmission in the hospital. Although the populations are not comparable, this report supports our finding of low transmission in that during the final year of the Veterans Affairs study [3], data showed transmission at 4 acquisitions/1000 days at risk—virtually identical to the 3.9 transmissions/1000 days at risk we observed. Our study is novel in that we studied the transmission of MRSA in a unique environment that requires special infection prevention practices due to the vulnerability of patients. In addition, we included molecular typing along with epidemiologic patient contact data in ascertaining the likelihood of person-to-person transmission of nasal MRSA.
The study is limited in that we did not include information regarding length of stay, underlying disease, or demographic data in the patient group, all of which can impact MRSA prevalence [22]. However, obtaining this information from a psychiatric unit is very challenging due to unique patient confidentiality protections. Because our healthcare system is similar to hospitals throughout the United States, we have no reason to suspect that the demographics on this unit are unique. Although screening compliance was less than ideal at 81.7% on admission and 64.7% at discharge, and a proportion of isolates could not be recovered for typing analysis (which could have led to missed transmission(s)); the compliance levels are very similar to the report of Farley et al [19], who captured 60.2% of their enrolled subjects at discharge. Another limitation is that transmission might also have occurred from colonized healthcare workers who may have entered the unit from anywhere in the hospital or from the environment that would have been missed in our study; such transmission, although not definitively proven, has been described [23, 24]. In addition, colonization in body sites other than the nares, although debated, could have served as a potential source of transmission [25].
Finally, we used PFGE for typing instead of whole-genome sequencing (WGS) as recently reported for MRSA epidemiology in acute care hospital [26]. Although WGS provides extensive information regarding the strain type, in the study by Price et al [26] rates of transmission were the same using WGS compared with conventional epidemiologic methods, although the patients identified were different. The results of the study by Price et al [26] is very similar to our study in that they also found that majority of the acquisitions could not be linked to colonized inpatients on the same nursing unit.
In conclusion, our findings, taken with other published information, suggest that there is evidence of ongoing transmission of MRSA in psychiatry units and that intervention is possible. Our study supports the concept that tailored infection prevention practices can be effective in reducing person-to-person transmission of MRSA even in these specialized nursing units. In particular, we observed decolonization of patients found to be MRSA nasally colonized at the time of admission was successful and that this approach was associated with a very low rate of transmission on the unit.
Acknowledgments
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Nelson RE, Samore MH, Smith KJ, et al. Cost-effectiveness of adding decolonization to a surveillance strategy of screening and isolation for methicillin-resistant Staphylococcus aureus carriers. Clin Microbiol Infect. 2010;16:1740–6. doi: 10.1111/j.1469-0691.2010.03324.x. [DOI] [PubMed] [Google Scholar]
- 2.Wertheim HF, Vos MC, Ott A, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet. 2004;364:703–5. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 3.Jones M, Ying J, Huttner B, et al. Relationships between the importation, transmission, and nosocomial infections of methicillin-resistant Staphylococcus aureus: an observational study of 112 Veterans Affairs Medical Centers. Clin Infect Dis. 2014;58:32–9. doi: 10.1093/cid/cit668. [DOI] [PubMed] [Google Scholar]
- 4.Tansarli GS, Karageorgopoulos DE, Kapaskelis A, et al. Impact of antimicrobial multidrug resistance on inpatient care cost: an evaluation of the evidence. Expert Rev Anti Infect Ther. 2013;11:321–31. doi: 10.1586/eri.13.4. [DOI] [PubMed] [Google Scholar]
- 5.Lee AS, Huttner B, Harbarth S. Control of methicillin-resistant Staphylococcus aureus. Infect Dis Clin North Am. 2011;25:155–79. doi: 10.1016/j.idc.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Bode LG, Kluytmans JA, Wertheim HF, et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med. 2010;362:9–17. doi: 10.1056/NEJMoa0808939. [DOI] [PubMed] [Google Scholar]
- 7.Huskins WC, Huckabee CM, O'Grady NP, et al. Intervention to reduce transmission of resistant bacteria in intensive care. N Engl J Med. 2011;364:1407–18. doi: 10.1056/NEJMoa1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurup A, Chlebicka N, Tan KY, et al. Active surveillance testing and decontamination strategies in intensive care units to reduce methicillin-resistant Staphylococcus aureus infections. Am J Infect Control. 2010;38:361–7. doi: 10.1016/j.ajic.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Huang SS, Septimus E, Kleinman K, et al. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med. 2013;368:2255–65. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhar S, Marchaim D, Tansek R, et al. Contact precautions: more is not necessarily better. Infect Control Hosp Epidemiol. 2014;35:213–21. doi: 10.1086/675294. [DOI] [PubMed] [Google Scholar]
- 11.Harris AD, Pineles L, Belton B, et al. Universal glove and gown use and acquisition of antibiotic-resistant bacteria in the ICU: a randomized trial. JAMA. 2013;310:1571–80. doi: 10.1001/jama.2013.277815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paule SM, Hacek DM, Kufner B, et al. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus test before and during high-volume clinical use. J Clin Microbiol. 2007;45:2993–8. doi: 10.1128/JCM.00670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paule SM, Pasquariello AC, Hacek DM, et al. Direct detection of Staphylococcus aureus from adult and neonate nasal swab specimens using real-time polymerase chain reaction. J Mol Diagn. 2004;6:191–6. doi: 10.1016/S1525-1578(10)60509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacek DM, Robb WJ, Paule SM, et al. Staphylococcus aureus nasal decolonization in joint replacement surgery reduces infection. Clin Orthop Relat Res. 2008;466:1349–55. doi: 10.1007/s11999-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaggi P, Paule SM, Peterson LR, et al. Characteristics of Staphylococcus aureus infections, Chicago Pediatric Hospital. Emerg Infect Dis. 2007;13:311–4. doi: 10.3201/eid1302.060295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDougal LK, Steward CD, Killgore GE, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limbago B, Fosheim GE, Schoonover V, et al. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J Clin Microbiol. 2009;47:1344–51. doi: 10.1128/JCM.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robicsek A, Beaumont JL, Thomson RB, Jr, et al. Topical therapy for methicillin-resistant Staphylococcus aureus colonization: impact on infection risk. Infect Control Hosp Epidemiol. 2009;30:623–32. doi: 10.1086/597550. [DOI] [PubMed] [Google Scholar]
- 19.Farley JE, Ross T, Krall J, et al. Prevalence, risk factors, and molecular epidemiology of methicillin-resistant Staphylococcus aureus nasal and axillary colonization among psychiatric patients on admission to an academic medical center. Am J Infect Control. 2013;41:199–203. doi: 10.1016/j.ajic.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 20.Day HR, Perencevich EN, Harris AD, et al. Depression, anxiety, and moods of hospitalized patients under contact precautions. Infect Control Hosp Epidemiol. 2013;34:251–8. doi: 10.1086/669526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter SS, Diekema DJ, Heilmann KP, et al. Activities of vancomycin, ceftaroline, and mupirocin against Staphylococcus aureus isolates collected in a 2011 National Surveillance Study in the United States. Antimicrob Agents Chemother. 2014;58:740–5. doi: 10.1128/AAC.01915-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suetens C, Niclaes L, Jans B, et al. Determinants of methicillin-resistant Staphylococcus aureus carriage in nursing homes. Age Ageing. 2007;36:327–30. doi: 10.1093/ageing/afm013. [DOI] [PubMed] [Google Scholar]
- 23.Hawkins G, Stewart S, Blatchford O, et al. Should healthcare workers be screened routinely for methicillin-resistant Staphylococcus aureus? A review of the evidence. J Hosp Infect. 2011;77:285–9. doi: 10.1016/j.jhin.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 24.Stein M, Navon-Venezia S, Chmelnitsky I, et al. An outbreak of new, nonmultidrug-resistant, methicillin-resistant Staphylococcus aureus strain (sccmec type iiia variant-1) in the neonatal intensive care unit transmitted by a staff member. Pediatr Infect Dis J. 2006;25:557–9. doi: 10.1097/01.inf.0000219407.31195.44. [DOI] [PubMed] [Google Scholar]
- 25.Matheson A, Christie P, Stari T, et al. Nasal swab screening for methicillin-resistant Staphylococcus aureus--how well does it perform? A cross-sectional study. Infect Control Hosp Epidemiol. 2012;33:803–8. doi: 10.1086/666639. [DOI] [PubMed] [Google Scholar]
- 26.Price JR, Golubchik T, Cole K, et al. Whole-genome sequencing shows that patient-to-patient transmission rarely accounts for acquisition of Staphylococcus aureus in an intensive care unit. Clin Infect Dis. 2014;58:609–18. doi: 10.1093/cid/cit807. [DOI] [PMC free article] [PubMed] [Google Scholar]