Abstract
Background
We performed human immunodeficiency virus type 1 (HIV-1) transmitted/founder (T/F) virus analysis of the VAX003 vaccine efficacy trial participants to characterize the transmission bottleneck and test for vaccine-associated reduction or enhancement of infection in this injection drug user (IDU) cohort.
Methods
We performed single genome sequencing of plasma vRNA from 50 subjects sampled in early HIV infection. Sequences were analyzed phylogenetically, T/F viruses enumerated, and a sieve analysis performed.
Results
Eight of 19 (42%) placebo recipients were productively infected by more than 1 virus (range 1–5, median 1, mean 1.7). This frequency of multiple virus transmission was greater than reported for heterosexual cohorts (19%, P = .03) but not statistically different from vaccine recipients (22.6%, P > .05), where the range was 1–3, median 1, and mean 1.3 (P > .05 for all comparisons). An atypical sieve effect was detected in Env V2 but was not associated with reduction or enhancement of virus acquisition.
Conclusions
The number of T/F viruses in IDUs was surprising low, with 95% of individuals infected by only 1–3 viruses. This finding suggests that a successful vaccine or other prevention modality generally needs to protect against only one or a few viruses regardless of risk behavior. T/F analysis identified an atypical genetic sieve in the V2 region of Envelope and found no evidence for vaccine-mediated enhancement in VAX003.
Keywords: HIV transmission, VAX003, injection drug users, multiple virus transmission, genetic sieve
The development of a safe and effective human immunodeficiency virus (HIV) vaccine is of critical importance in the fight against HIV/AIDS, with injection drug users (IDUs) representing a high-risk population. IDUs are significant drivers of the HIV epidemic, representing approximately 10% of HIV-infected individuals worldwide and more than 40% of new infections in some high-incidence regions, including eastern Europe and central Asia [1]. Recent studies have demonstrated a stringent population bottleneck in heterosexual (mucosal) transmission of HIV-1, with approximately 80% of infections initiated by a single transmitted/founder (T/F) virus [2–4]. Biological and epidemiological considerations suggest that HIV-1 acquisition could differ substantially when occurring via injection rather than sexual activity. Though employing high inocula, nonhuman primate studies of simian immunodeficiency virus (SIV) infection demonstrate orders of magnitude differences in the number of T/F viruses between intravenous and mucosal inoculation [5–8], and epidemiologic studies ascribe a higher per event transmission risk to parenteral exposures [9, 10]. Direct examination of the HIV-1 transmission bottleneck in IDUs, however, has been limited. Just 2 small studies have quantified T/F viruses in IDUs, demonstrating a wide range in the number of T/F viruses (1–16) with conflicting results as to the mean (1.46 vs 3) and median (1 vs 3) number of T/F viruses in IDUs [11, 12]. A third study of a high-risk needlestick exposure found a minimum of 15 T/F viruses established infection despite post-exposure prophylaxis [13]. Thus, critical questions remain as to what extent the HIV-1 transmission barrier differs between injection and sexual cohorts and how the IDU transmission route might influence vaccine trial design and assessment and, ultimately, vaccine efficacy.
A single vaccine efficacy trial, VAX003, enrolled an IDU cohort [14]. VAX003 was a randomized, double-blinded, placebo-controlled trial of the AIDSVAX (recombinant gp120) B/E vaccine with an HIV-1 incidence of 3.4/100 person-years [14]. This trial demonstrated no vaccine efficacy (VE) in preventing HIV-1 acquisition [14]. AIDSVAX B/E was subsequently used in conjunction with a canarypox-vector prime (ALVAC) in the RV144 trial conducted in Thai heterosexuals [15], where it demonstrated a 31.2% VE [15]. Post hoc analyses further identified increased VE in the lowest risk RV144 subjects, highlighting the possibility that higher risk modes of acquisition, such as IDU, may affect vaccine trial outcomes [14–16].
The molecular identification of discrete T/F genomes responsible for productive clinical infection by single genome sequencing (SGS) allows for a precise quantitative and qualitative analysis of HIV-1 transmission, including factors that might inhibit or enhance virus infection. These include potential T-cell or antibody mediated pressures resulting in selective virus outgrowth (sieving)[17, 18] or enhancement [19, 20]. In the present study, we applied T/F analyses to the VAX003 trial to examine 3 key questions: (1) What is the multiplicity of HIV-1 infection in IDUs, ie, how high is the bar set for clinical vaccine protection? (2) Is there evidence of protection or enhancement of virus acquisition associated with an Env containing vaccine? (3) Is there evidence of vaccine-associated sieving of viruses leading to selective virus transmission?
METHODS
Study Subjects
Deidentified plasma samples were obtained from Global Solutions for Infectious Diseases. The estimated date of infection was determined as midpoint between HIV-specific antibody positive and negative testing or 10 days prior to HIV viral RNA (vRNA) positive/antibody negative sampling [21]. For 7 subjects, we had access to 2 samples obtained within the first 100 days of infection (subjects 3022, 3025, 3046, 3090, 3112, 3126, 3210). The VAX003 trial was conducted in accordance with the declaration of Helsinki and local institutional review boards [14].
Viral RNA Extraction, cDNA Synthesis, SGS
Viral RNA extraction, complementary DNA (cDNA) synthesis, and SGS were performed to amplify gp41 or gp160 env as previously described [2, 22]. Amplicons were directly sequenced and chromatograms were inspected for evidence of priming from multiple templates or introduction of polymerase chain reaction (PCR) error in early cycles [2, 22].
Phylogenetic Analyses
Sequences were aligned with ClustalW and hand-checked using MacClade 4.08. The composite maximum-likelihood (ML) phylogenetic tree with ML bootstrap support was estimated using RAxML-HPC-SSE3 version 7.6.3 with a Γ model of rate heterogeneity and ML estimate of the α parameter [23]. All other phylogenetic trees were generated by the neighbor-joining (NJ) method using ClustalW or ML method using PhyML v.3. Sequences were analyzed by a previously described model of neutral virus evolution [2, 24]. Under this model, low-diversity sequence lineages display a star-like phylogeny and a Poisson distribution of mutations and coalesce to an unambiguous T/F sequence [2, 24]. Sequences with evidence of APOBEC3G-mediated hypermutation were removed from model calculations and figures. All sequences were deposited in GENBANK (KJ952241–KJ953713).
Sieve Analysis
Sieve analysis of the envelope V1V2 region followed a prespecified approach based on the RV144 analysis protocol using the EPIMAP site set [25]. As per the RV144 analysis, we included only CRF01_AE sequences; a secondary analysis was performed excluding all but the first of phylogenetically linked infections. Two subjects with only gp41 sequences were excluded (subjects 3054 and 3192); subject 3110 had additional plasma available for V1V2 sequence amplification and was included. Multiplicity correction for site scanning methods used a Benjamini-Hochberg false discovery rate (q-value) threshold of 0.2 [26]. The prespecified analyses and associated multiplicity correction were conducted separately for each insert (A244 and MN) and statistical method [25].
Comparison Between Heterosexual and IDU Cohorts
A review of the literature revealed 5 manuscripts using single genome methods to enumerate T/F viruses in subjects with documented IDU and heterosexual transmission risks. These articles describe 174 heterosexuals[2–4] and 42 unvaccinated IDUs [11, 12].
RESULTS
Subject Characteristics
T/F analysis requires samples obtained early in infection so that viral sequence evolution conforms to a model of random or near-random evolution [2, 24]. Enumeration of T/F variants has been validated in samples obtained within approximately 100 days of infection [11]. We elected to study all VAX003 subjects with high-risk IDU practices and samples collected within 100 days of infection. Fifty (19 placebo- and 31 vaccine-recipients) out of a total of 211 HIV-infected subjects met these prespecified criteria. Subjects were largely male (88%), with ages ranging from 21 to 53 years, and estimated time since HIV-1 infection of between 10 and 100 days (mean 58 days; Table 1). Plasma samples were analyzed by HIV-1 enzyme-linked immunosorbent assay (ELISA) and Western blot, and the Fiebig clinicopathological stage was determined [2, 21]. In total there were 14 pre-antibody seroconversion samples and 36 post-seroconversion samples (Table 1) [21]. The placebo and vaccine groups had similar demographic and Fiebig stage composition, with 11% and 13% female representation, mean ages of 28.4 and 27.0 years, and 26% and 29% preseroconversion sampling, respectively; none of these characteristics differed statistically between groups.
Table 1.
Demographics and sequence analysis of VAX003 subjects
| Sample ID | Study arm | Sex | Age (years) | Estimated days since infectiona | Fiebig stage | HIV-1 subtype | Amplified gene | No. sequencesb | No. T/F virusesc | Max diversity | Max HDd | GOF P- valuee |
MRCA days poissonf | Star phylogenyg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3007 | Placebo | M | 20 | 83 | V | CRF01_AE | gp160 | 31 | 3 | 0.0522 | 9 | 0.2493 | 66 (52,80) | Near |
| 3017 | Placebo | M | 53 | 84 | V | CRF01_AE | gp160 | 32 | 2 | 0.00901 | 5 | 0.2222 | 36 (29,43) | Yes |
| 3022 | Placebo | M | 40 | 10, 40 | II, IV | CRF01_AE | gp160 | 56 (31/25) | 1 | 0.00272 | 5 | 0.932 | 24 (17,31) | Yes |
| 3025 | Placebo | M | 26 | 10, 47 | II, V | CRF01_AE | gp160 | 55 (20/35) | 1 | 0.00305 | 8 | 0.5484 | 58 (47,70) | Near |
| 3046 | Placebo | M | 26 | 10, 30 | II, V | CRF01_AE | gp160 | 52 (31/21) | 1 | 0.00592 | 5 | 0.8122 | 20 (12,27) | Yes |
| 3090 | Placebo | F | 44 | 10, 52 | II, V | CRF01_AE | gp160 | 55 (32/23) | 3 | 0.0355 | 4 | 0.7863 | 15 (9,21) | Yes |
| 3098 | Placebo | M | 23 | 71 | V | CRF01_AE | gp160 | 32 | 1 | 0.00509 | 13 | <0.05 | 117 (108,126) | No |
| 3112 | Placebo | F | 23 | 10, 47 | II, V | CRF01_AE | gp160 | 50 (20/30) | 3 | 0.0247 | 3 | 0.3381 | 17 (11,22) | Near |
| 3126 | Placebo | M | 21 | 10, 57 | II, V | CRF01_AE | gp160 | 63 (33/30) | 1 | 0.00308 | 7 | 0.7006 | 28 (20,37) | Yes |
| 3127 | Placebo | M | 27 | 86 | VI | CRF01_AE | gp160 | 31 | 5 | 0.027 | 4 | 0.753 | 28 (14,43) | Yes |
| 3139 | Placebo | M | 28 | 70 | V | CRF01_AE | gp160 | 26 | 1 | 0.0051 | 13 | <0.05 | 113 (99,127) | No |
| 3141 | Placebo | M | 44 | 85 | V | B | gp41 | 21 | 1 | 0.0071 | 6 | 0.5605 | 88 (64,1120) | Near |
| 3147 | Placebo | M | 23 | 62 | VI | CRF15_01B | gp160 | 31 | 2 | 0.0307 | 9 | <0.05 | 61 (46,76) | Near |
| 3155 | Placebo | M | 24 | 85 | VI | CRF01_AE | gp160 | 19 | 1 | 0.00582 | 15 | 0.2258 | 99 (78,121) | No |
| 3156 | Placebo | M | 38 | 89 | VI | CRF01_AE, B | gp160 | 31 | 2 | 0.226 | 10 | <0.05 | 70 (58,82) | No |
| 3181 | Placebo | M | 21 | 90 | VI | B | gp41 | 21 | 1 | 0.00868 | 8 | <0.05 | 165 (132,198) | Near |
| 3202 | Placebo | M | 24 | 22 | V | CRF01_AE | gp160 | 30 | 1 | 0.00275 | 7 | 0.9077 | 34 (25,43) | Yes |
| 3210 | Placebo | M | 24 | 10 | II, V | CRF01_AE | gp160 | 36 (22/14) | 2 | 0.0325 | 4 | 0.7774 | 48 (28,68) | Yes |
| 3220 | Placebo | M | 21 | 90 | V | CRF01_AE | gp160 | 30 | 1 | 0.00436 | 11 | 0.1754 | 94 (84,104) | Near |
| 3002 | Vaccine | F | 21 | 41 | IV | CRF01_AE | gp160 | 24 | 2 | 0.0143 | 11 | <0.05 | 90 (75,104) | No |
| 3003 | Vaccine | M | 31 | 10 | IV | B | gp41h | 17 | 1 | 0.00642 | 6 | 0.6465 | 122 (74,167) | No |
| 3019 | Vaccine | M | 24 | 10 | II | CRF01_AE | gp160 | 30 | 2 | 0.0318 | 5 | 0.8201 | 31 (22,40) | Yes |
| 3041 | Vaccine | M | 21 | 14 | V | CRF01_AE | gp41 | 18 | 1 | 0.00412 | 4 | 0.3018 | 56 (39,73) | No |
| 3043 | Vaccine | M | 29 | 14 | V | CRF01_AE | gp160 | 30 | 1 | 0.00195 | 5 | 0.9438 | 18 (12,24) | Yes |
| 3045 | Vaccine | M | 40 | 10 | II | B | gp160 | 27 | 2 | 0.0242 | 6 | 0.8627 | 31 (23,39) | Yes |
| 3054 | Vaccine | M | 22 | 80 | VI | CRF01_AE | gp41 | 41 | 1 | 0.00561 | 6 | 0.9008 | 53 (39,68) | Yes |
| 3059 | Vaccine | M | 21 | 50 | V | B | gp41 | 12 | 2 | 0.0496 | 3 | 0.7054 | 72 (40,103) | No |
| 3063 | Vaccine | M | 30 | 87 | V | CRF01_AE | gp160 | 22 | 1 | 0.00388 | 6 | 0.9514 | 38 (28,49) | No |
| 3067 | Vaccine | M | 32 | 98 | V | CRF01_AE | gp160 | 23 | 1 | 0.00510 | 13 | 0.4794 | 112 (95,129) | No |
| 3078 | Vaccine | M | 27 | 10 | II | CRF01_AE | gp160 | 20 | 1 | 0.00115 | 3 | 0.8695 | 11 (6,17) | Yes |
| 3084 | Vaccine | M | 26 | 91 | V | CRF01_AE | gp160 | 33 | 1 | 0.00386 | 10 | 0.5447 | 76 (66,86) | No |
| 3104 | Vaccine | M | 25 | 86 | V | CRF01_AE | gp160 | 23 | 3 | 0.0305 | >15 | na | na | No |
| 3110 | Vaccine | M | 31 | 98 | V | CRF01_AE | gp41h | 22 | 1 | 0.00523 | 3 | 0.7583 | 101 (71,130) | No |
| 3111 | Vaccine | F | 22 | 10 | II | CRF01_AE | gp160 | 21 | 1 | 0.00157 | 4 | 0.2906 | 30 (21,39) | Yes |
| 3118 | Vaccine | M | 20 | 99 | IV | CRF01_AE | gp160 | 23 | 1 | 0.00426 | 11 | 0.1104 | 84 (70,98) | No |
| 3131 | Vaccine | M | 26 | 94 | V | CRF01_AE | gp160 | 30 | 1 | 0.00573 | 11 | 0.4296 | 87 (76,99) | No |
| 3135 | Vaccine | M | 21 | 96 | V | CRF01_AE | gp160 | 30 | 1 | 0.00313 | 8 | 0.9266 | 49 (39,59) | Yes |
| 3151 | Vaccine | M | 21 | 92 | V | CRF01_AE | gp160 | 21 | 2 | 0.0210 | 13 | 0.3031 | 81 (50,113) | No |
| 3152 | Vaccine | M | 21 | 89 | V | CRF01_AE | gp160 | 35 | 1 | 0.00390 | 10 | 0.9472 | 53 (44,63) | Yes |
| 3153 | Vaccine | M | 23 | 87 | V | CRF01_AE | gp160 | 34 | 1 | 0.00154 | 4 | 0.3139 | 22 (16,28) | Yes |
| 3162 | Vaccine | M | 26 | 79 | V | CRF01_AE | gp160 | 21 | 1 | 0.00347 | 9 | 0.2576 | 57 (44,70) | No |
| 3184 | Vaccine | M | 44 | 94 | VI | CRF01_AE | gp160 | 30 | 1 | 0.00593 | 10 | 0.06063 | 91 (79, 104) | No |
| 3186 | Vaccine | M | 20 | 10 | II | CRF01_AE | gp160 | 21 | 1 | 0.000769 | 2 | 0.6605 | 5 (1,8) | Yes |
| 3189 | Vaccine | M | 24 | 80 | VI | CRF01_AE | gp160 | 22 | 3 | 0.0445 | >15 | na | na | No |
| 3192 | Vaccine | M | 26 | 90 | V | CRF01_AE | gp41 | 22 | 1 | 0.00374 | 4 | 0.5475 | 62 (44,81) | Yes |
| 3193 | Vaccine | M | 31 | 10 | V | CRF01_AE | gp160 | 23 | 1 | 0.00235 | 6 | 0.3764 | 41 (31,50) | Yes |
| 3203 | Vaccine | F | 21 | 95 | VI | CRF01_AE | gp160 | 20 | 1 | 0.00428 | 11 | <0.05 | 83 (68,98) | No |
| 3212 | Vaccine | M | 49 | 10 | IV | CRF01_AE | gp160 | 23 | 1 | 0.00312 | 8 | 0.5592 | 67 (58,76) | No |
| 3218 | Vaccine | M | 35 | 90 | V | CRF01_AE | gp160 | 30 | 1 | 0.00588 | 15 | <0.05 | 94 (77,110) | No |
| 3219 | Vaccine | F | 26 | 90 | VI | CRF01_AE | gp160 | 22 | 1 | 0.00232 | 6 | 0.143 | 59 (51,67) | No |
Abbreviations: HIV-1, human immunodeficiency virus type 1; T/F, transmitted/founder.
a The estimated date of infection was determined as midpoint between HIV-specific antibody positive and negative testing or as 10 days prior to a HIV viral RNA positive/antibody negative sample. For subjects with 2 samples, estimated dates are listed for both.
b Number of total sequences analyzed per subject. In subjects with 2 available samples obtained within the first 100 days of infection, the numbers of sequences analyzed from first and second timepoint are shown in parentheses.
c Estimate of minimum number of T/F viruses. In subjects with multiple T/F viruses, initial model testing indicated a GOF P value < .05 for the whole subject dataset, whereupon model analyses of maximum Hamming distance, Lambda, Goodness of Fit P value, MRCA days by Poisson, and Star phylogeny were conducted on the largest T/F lineage. In subjects with 2 samples, analyses were performed on the earlier timepoint.
d Maximum Hamming distance.
e Goodness of Fit P value. Low P values (<.05) indicate divergence from a Poisson distribution.
f Number of days since most recent common ancestor as determined by Poisson distribution, with 95% confidence intervals in parentheses.
g Star phylogeny defined as divergence of <10% between observed and estimated convolution values. Near Star phylogeny indicates the sequence set meets this criterion upon removal of up to 3 sites of shared polymorphisms.
h For subjects 3003 and 3110 additional sequences were generated spanning 1200 nucleotides of gp120, including the V1V2 region.
Phylogenetic Analyses
A total of 1473 env gp160 or gp41 sequences generated by SGS were obtained with a median of 26 per subject timepoint. Gp160 sequences were generated for 42 subjects. In 8 subjects (3/19 [16%] placebo and 5/31 [19%] vaccine arm), viral loads were lower or the plasma sample was of poorer quality such that we chose to amplify gp41 to increase sequencing efficiency. The gp41 env sequences from all 50 subjects were analyzed together in a ML phylogeny along with relevant reference sequences (Fig. 1). Sequences from 43 subjects (86%) clustered unambiguously with Thai CRF01_AE reference viruses, 5 subjects clustered with Thai Subtype B viruses, 1 subject (Subject 3156) clustered with both Subtype B and CRF01_AE viruses, and 1 subject (Subject 3147) clustered with CRF15_01B viruses (Fig. 1). The use of SGS enabled detection of minority variants, including individuals infected with multiple subtypes (subject 3156; see Fig. 1 and Supplementary material, Fig. S1). In 7 subjects, there was phylogenetic evidence of epidemiologically closely related infections, including 2 likely “acute-to-acute” transmissions (subjects 3017 and 3212 and subjects 3017 and 3213; Fig. 1, Supplementary material, Figs S2 and S3).
Fig. 1.
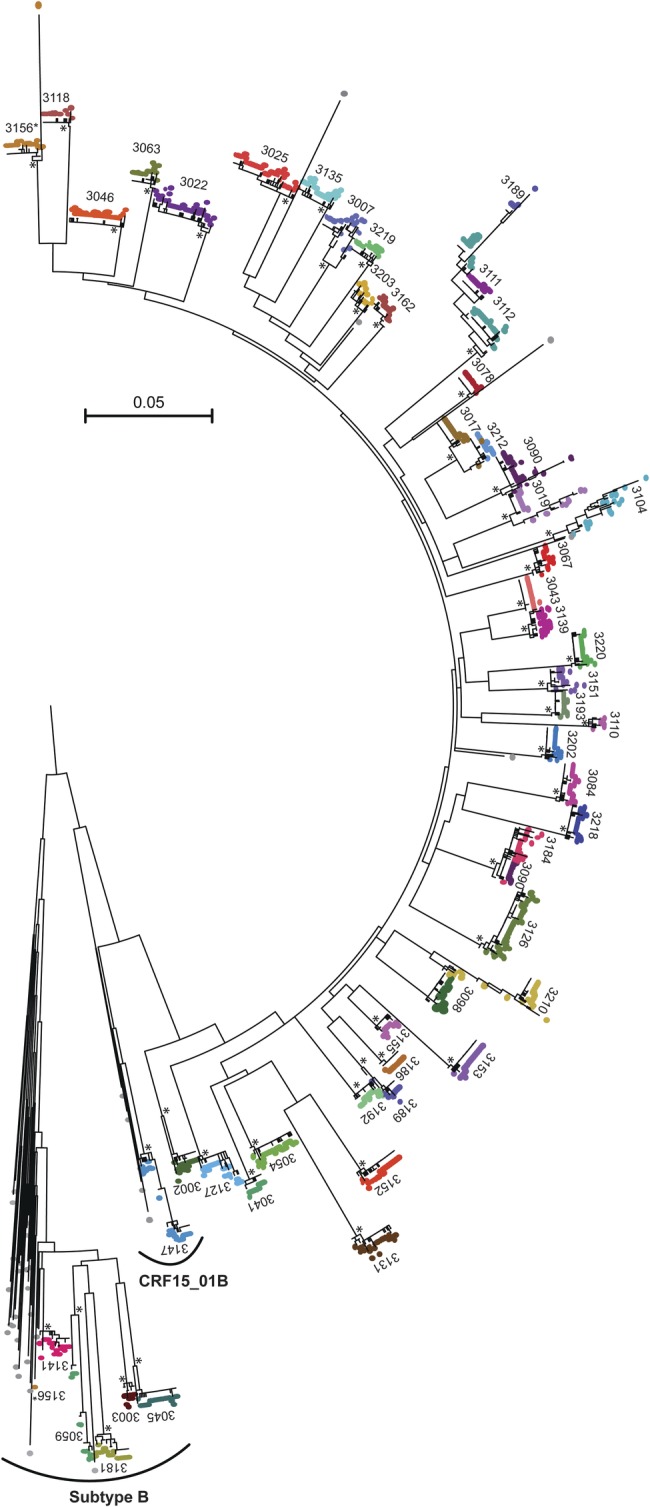
ML tree of HIV-1 gp41 env sequences from the 50 VAX003 subjects. Each subject's sequence set is shown in a different color and labeled with the subject identifier. Reference sequences are shown in gray. Forty-six subjects were infected by CRF01_AE viruses; CRF15_01B and subtype B clades are specifically indicated. Individual subject sequence sets with bootstrap values >95% are annotated with an asterisk. There are 8 individual subject sequence lineages with bootstrap values >75%: the 2 acute-to-acute transmissions (subjects 3212 and 3017 and subjects 3184 and 3090), 3 subjects with related but distinct lineages (subjects 3189, 3111 and 3112), and subject 3156. Subject 3156 (shown in mustard orange and annotated with a hashtag at the top and bottom of tree) has sequences clustering with CRF01_AE and subtype B. The single long branch in subject 3156's viral sequences at the top of the figure is a unique interlineage recombinant shown in greater detail in Supplementary material, Figure S1. Genetic distance is indicated by the scale bar. Abbreviations: HIV-1, human immunodeficiency virus type 1; ML, maximum-likelihood.
T/F Analysis of Placebo Recipients
To assess the stringency of the IDU transmission barrier, we analyzed the 19 placebo recipient's gp160 or gp41 env sequences from the first available timepoint to enumerate the viruses founding productive infection. For the placebo subjects, within-patient maximum env diversities ranged from 0.272% to 0.868%. Eleven of the placebo subjects' phylogenies demonstrated a single low-diversity lineage (maximum within-env diversity 0.27%–0.87%), as represented in Figure 2 and summarized in Table 1. For all but 2 of these 11 subjects, the diversity was within that attainable in env within the first 100 days of infection (<0.6%, 95% confidence interval [CI]: 0.54%–0.68%)[2]; 2 subjects sampled later in infection had marginally higher diversities in their single sequence lineages (0.71% and 0.868%; Table 1). This phylogenetic pattern of a single low-diversity lineage is characteristic of acute infection founded by 1 T/F virus, and the majority of these subjects' sequences conformed to a model of random virus evolution from a single template (Table 1). The sequences that did not conform to model predictions were sampled later (Fiebig stages V–VI) and deviated from model predictions due to few sites of shared mutations (Table 1) [2, 27].
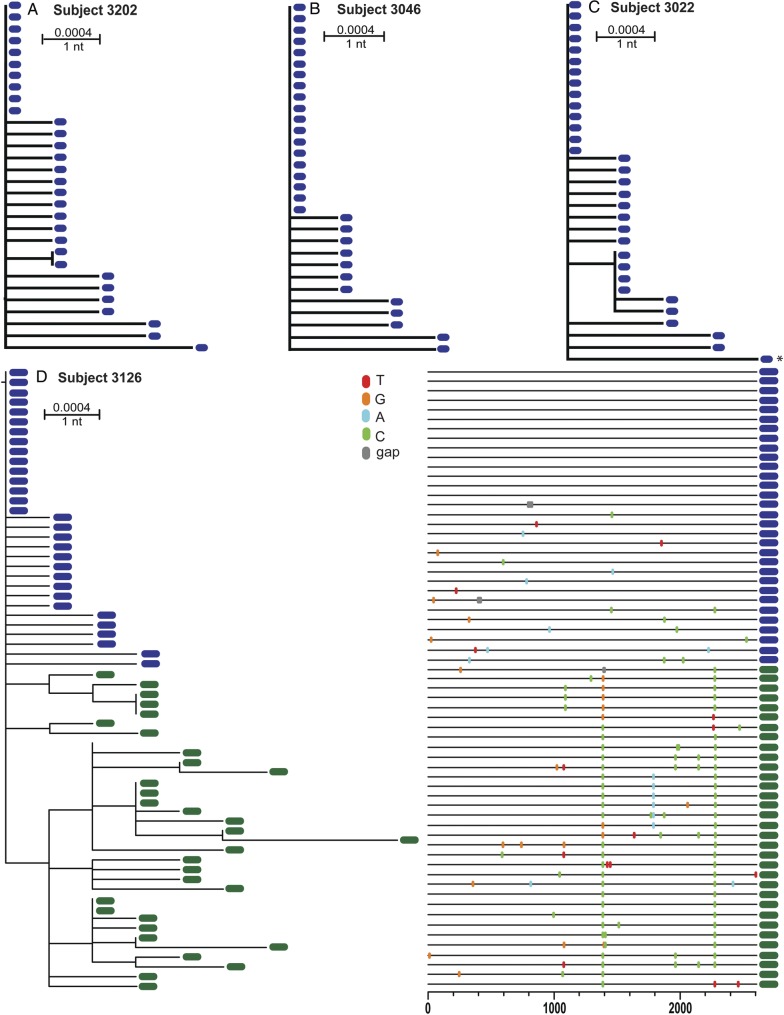
Fig. 2.
Single T/F virus transmission in VAX003 placebo recipients. ML trees of HIV-1 gp160 env sequences for placebo subjects 3202 (A), 3046 (B), and 3022 (C). ML tree and Highlighter plot for gp160 env sequences from placebo subject 3126 from 2 timepoints (Fiebig stage II sample sequences shown in blue, Fiebieg stage IV sample sequences, estimated 67 days post-infection, shown in green) (D). All 4 subjects' phylogenies reveal initial single, low-diversity lineages consistent with productive clinical infection by a single virus. Later sequences from 3126 reveal evidence of selection at several positions. Genetic distances are indicated by the scale bars. Abbreviations: HIV-1, human immunodeficiency virus type 1; ML, maximum likelihood; T/F, transmitted/founder.
In 4 of the placebo subjects with a single T/F virus (subjects 3022, 3025, 3046, 3126), we had access to additional plasma samples obtained 20–60 days after the first sample. As illustrated by subject 3126 (Fig. 2D), who was sampled preseroconversion and 57 days later, the longitudinal sequences clustered together and demonstrated the expected accrual of genetic diversity over time [2, 24, 27–30]. Importantly, in all 4 subjects, lineages remained discernible as single virus transmissions, although identification of exact T/F sequences was confounded by selection.
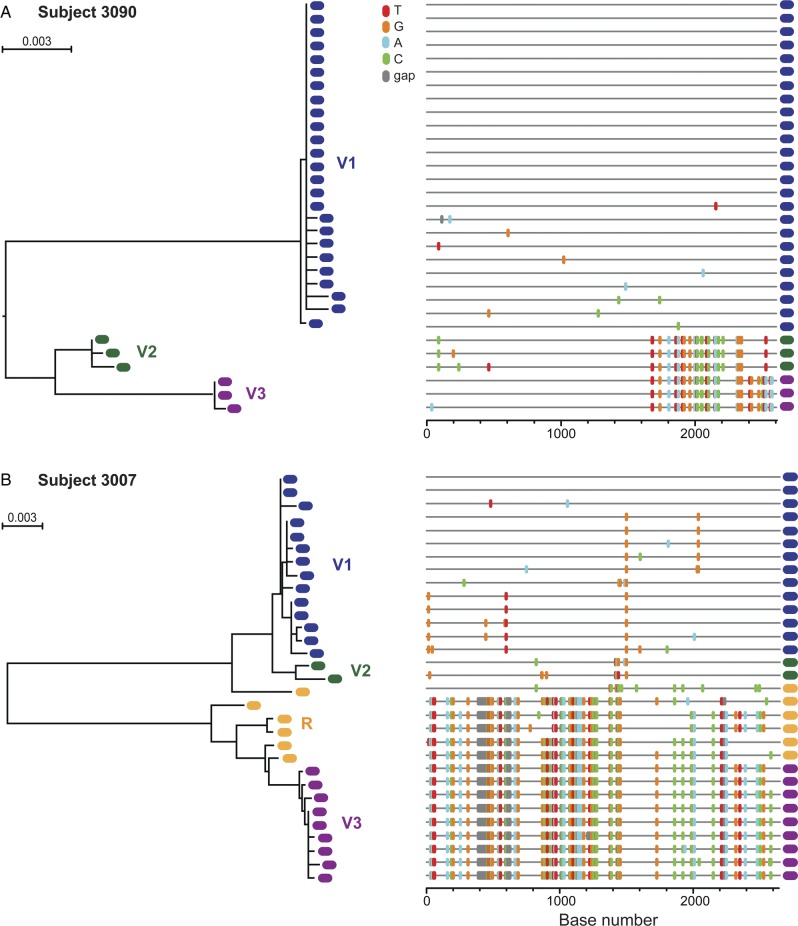
Eight of the placebo subjects (8/19, 42.1%) demonstrated phylogenetic evidence of multiple virus transmission (MVT). In these subjects, as represented by subjects 3090 and 3007 in Figure 3, 2 or more discrete low-diversity lineages were present, often with interlineage recombinant sequences. Maximum env diversities ranged from 0.901% to 22.6%. Seven subjects had maximum env diversities typical of the HIV quasispecies of an HIV-infected individual (0.901%–5.22%), whereas subject 3156 had >20% env diversity due to the presence of sequences from both subtype B and CRF01_AE (Supplementary material, Fig. S1). Despite the overall increased sequence diversity reflecting MVT, individual sequence lineages showed limited diversity and conformed to a model of random virus diversification from discrete T/F genomes, thus allowing for estimation the number of T/F viruses (Table 1). Importantly, these are minimum estimates of numbers of T/F viruses, with the sensitivity and precision of the estimates influenced by sequencing depth (see[2] for model predictions and power calculations).
Fig. 3.
Multiple T/F virus transmission in VAX003 placebo recipient subjects. ML trees and Highlighter plots of HIV-1 gp160 env sequences from subject 3090 (A) and subject 3007 (B), each phylogeny revealing productive infection by at least 3 viruses. T/F virus lineages are color-coded and labeled variant (V)1–3; recombinant (R) sequences shown in orange. Abbreviations: HIV-1, human immunodeficiency virus type 1; ML, maximum likelihood; T/F, transmitted/founder.
The range in the number of viruses initiating productive infection in the placebo recipients was 1–5. In total, 42% of the placebo recipients had HIV-1 infection founded by multiple viruses, with a mean of 1.7 T/F viruses per subject. When compared with 174 subjects from cohorts of sexual transmission analyzed by the same SGS methodology [2–4], this frequency of MVT is significantly higher than the 19% seen in heterosexuals (P = .03, Fisher exact test).
T/F Analysis of the Vaccine Recipients
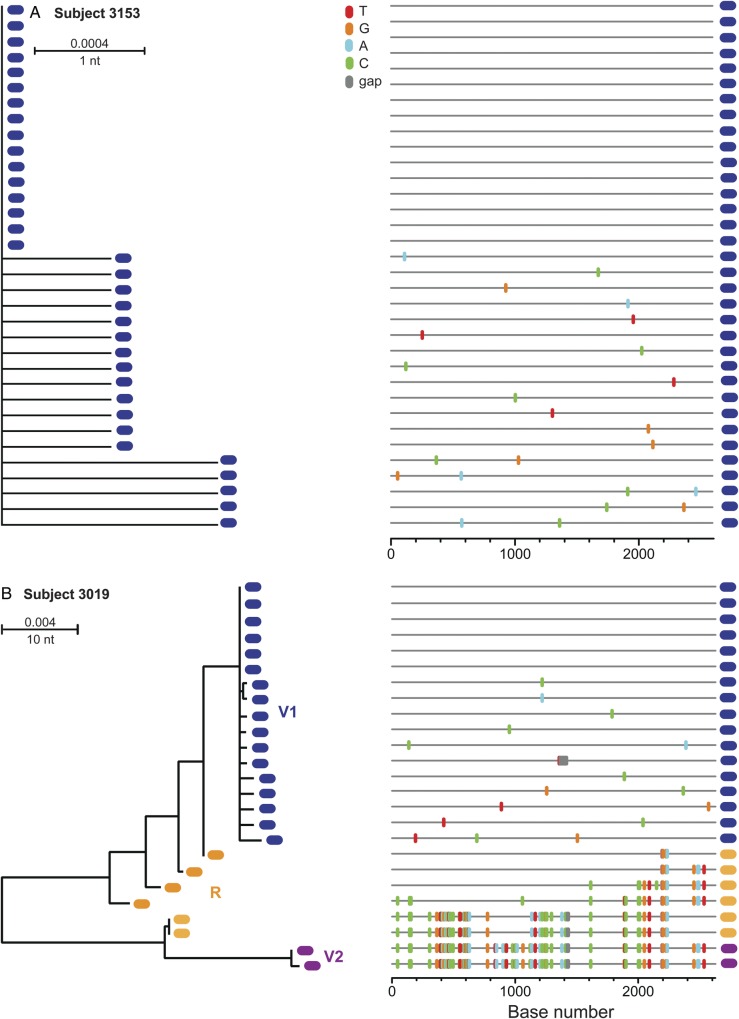
We next analyzed each of the 31 vaccine recipients' sequences for overall diversity and evidence of random virus evolution [2, 24, 27]. Twenty-four of the 31 subjects exhibited a phylogenetic pattern of single low-diversity lineages (maximum within-env diversity of 0.769% to 0%), as shown in Figure 4A. Phylogenies from 22 of these sequence sets met model criteria for a Poisson distribution of mutations, and 19 had star-like or near star-like phylogenies (Table 1). Seven of the vaccine recipients' sequence sets had greater env diversity than predicted for single virus transmission (1.43%–4.96%) and phylogenies consistent with MVT, as exemplified by subject 3019 in Figure 4B. In total, 22.6% (7/31) of the vaccine recipients had infection founded by multiple viruses, with a mean of 1.3 T/F viruses per subject and a range of 1–3. Comparing the 2 study arms, there was no evidence of vaccine-associated enhancement leading to increased numbers of T/F viruses. Instead, there was a statistically nonsignificant trend toward a reduction in the number of viruses establishing clinical infection (mean 1.3 vs 1.7, P = .133, Wilcoxin test) and the overall frequency of MVT (22.6% vs 42.1%, P = 0.21, Fisher exact test) in vaccinees compared with placebo-recipients.
Fig. 4.
ML tree and Highlighter plots of gp160 env sequences from VAX003 vaccine recipient subjects. Subject 3153 (A) demonstrates single virus transmission, while subject 3019 reveals multiple virus transmission by at least 2 viruses. T/F lineages are labeled and color-coded and recombinant sequences shown in orange. Abbreviations: ML, maximum likelihood; T/F, transmitted/founder.
Sieve Analysis
We next tested for genetic signatures of a vaccine effect through sieve analysis. Genetic sieve analyses look for sequence evidence that vaccine-induced immune responses selectively block the transmission of certain viruses [17, 18, 31]. This is accomplished by comparing the sequences of breakthrough viruses in the vaccine and placebo recipients to the vaccine insert(s) [31]. Sieve analysis of the RV144 trial demonstrated 2 sites in V2 with statistically different rates of amino acid mismatch (Env positions 169 and 181) [18]. Further, both a typical sieve effect (ie, greater mismatch in the vaccine-recipient sequences) at position 169 and an atypical effect (ie, greater mismatch in the placebo-recipient sequences) at position 181 were associated with increased VE [18, 32]. Because RV144 and VAX003 tested identical Env immunogens (with or without ALVAC), we used the RV144 sieve analysis as a template. To maximize power, we employed a prespecified statistical plan to test only sites determined to be of likely importance based on structural and binding models, termed EPIMAP sites [12]. Our sieve analysis differed from RV144 in that we compared the T/F virus sequences and full SGS-derived sequence sets obtained in the first 100 days of infection, which enabled us to mitigate some post-acquisition selection effects possible in studies evaluating all breakthrough infections [17, 18]. We performed 2 predetermined analyses of V1V2 mismatching; the first tested the signals seen in the RV144 analysis, and the second examined EPIMAP sites [18]. Each sample was compared against the CRF01_AE and Subtype B Env inserts, thus restricting comparisons to between 5 and 7 sites. We used the same 3 statistical methods to compare insert mismatches as did the RV144 analyses, (1) GWJ (a non-parametric T-test comparing insert mismatch in the major T/F sequence per subject), (2) MBS (a Bayesian model-based test comparing the major T/F sequence per subject), and (3) SMMB (a simplified bootstrap pairwise AA test using all the SGA-derived sequences) [18].
Analysis of the positions identified in the RV144 sieve analysis (Env 169 and 181) showed no evidence of selection in the VAX003 sequences. The EPIMAP sites, however, revealed a sieve effect at Env position 170 in the V2 crown. At this position, there was a greater frequency of insert mismatch with the placebo-recipient sequences than vaccinee sequences (37.5% [6/16] vs 5% [1/20]) (Fig. 5). This atypical sieve effect was statistically significant (P = .0368 for GWJ method, .035 for MBS method, .015 for SMMB method) and met the multiplicity correction of q < 0.2 for 4 of the 6 tests, with differences due to number of sites compared per insert (q = 0.258 and 0.184 for GWJ method, 0.245 and 0.1743 for MBS method, and 0.135 and 0.105 for SMMB method for comparison with the A244 and MN inserts, respectively). A secondary analysis excluding linked transmissions revealed similar trends (P = .053 for GWJ method, .051 for MBS, and .020 for SMMB) and (q = 0.355 and 0.253 for MBS, and 0.374 and 0.267 for GWJ, and 0.182 and 0.1414 for the SMMB for A244 and MN inserts, respectively).
Fig. 5.
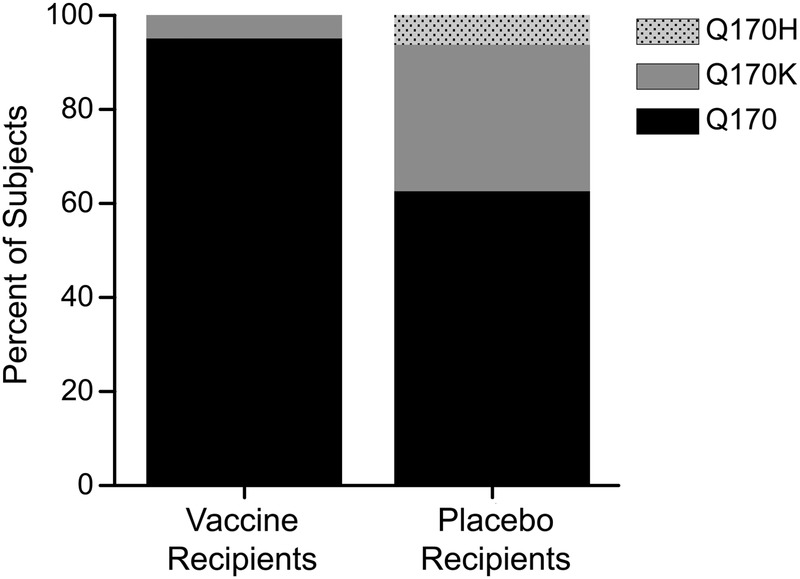
Sieve analysis at Env position 170. 95% (19/20) of T/F virus sequences from vaccine recipients matched the A244 and MN vaccine inserts at position 170 compared with 62.5% (10/16) of placebo recipient T/F virus sequences (P < .05 for all tests performed). Abbreviation: T/F, transmitted/founder.
DISCUSSION
HIV-1 transmission in IDUs is an important but understudied subject with relevance to multiple prevention efforts. Here we identified and quantified the T/F viruses from 50 VAX003 subjects, enabling a robust characterization of the transmission bottleneck in IDUs. Surprisingly, we found only a modest increase in the multiplicity of infection in these IDUs compared with heterosexuals.
HIV-1 transmission in heterosexuals is characterized by a strict genetic bottleneck, with 81% of subjects infected with a single T/F virus and the vast majority of the remainder infected with just 2 or 3 viruses (Fig. 6) [2–4]. The proportion of MVT in IDUs was higher in both the VAX003 placebo-recipients (n = 19, 42% MVT, P = .03, Fisher exact test) and in an aggregate of published unvaccinated IDUs with enumerated T/F viruses (n = 42, 43% MVT, P = .002), as shown in Figure 6. Importantly, 43% MVT is also greater than the 21% MVT reported for the 52 HIV-infected heterosexual participants in the placebo arm of RV144 (P = .027) [25].
Fig. 6.
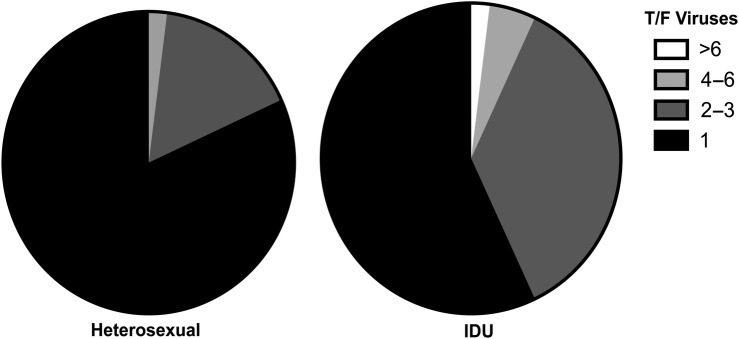
Increased multiplicity of HIV-1 infection in IDUs compared with heterosexuals. HIV-1 infections founded by a minimum of 1, 2–3, 4–6, or >6 T/F viruses from aggregated reports of heterosexuals (n = 174)[2–4] and unvaccinated IDUs (n = 42)[11, 12] analyzed by comparable single genome sequencing methods. The frequency of MVT in IDUs (43%) is greater than in heterosexuals (19%, P = .002). Transmission of between 1 and 3 viruses occurred in 97% heterosexuals and 93% of IDUs, P > .05. Abbreviations: HIV-1, human immunodeficiency virus type 1; IDU, injection drug user; MVT, multiple virus transmission.
Although the frequency of MVT in IDUs was approximately double that in heterosexuals, the actual number of viruses establishing productive infection was only slightly greater in IDUs than in heterosexuals (mean of 1.3 T/F viruses [range 1–6] in heterosexuals, mean of 1.7 [range 1–5] in VAX003 placebo-recipients, and mean of 2.1 [range 1–16] in all unvaccinated IDUs (n = 42), P > .05 for both comparisons, unequal-variance t-test). Thus, as shown in Figure 6, the majority of HIV-1 infections in both heterosexuals and IDUs were founded by a single virus (81% and 57%, respectively), and the vast majority of infections were founded by between 1 and 3 viruses (98% and 93%, respectively).
What might be the explanation for the surprisingly low numbers of T/F viruses in IDUs and the clinical significance of a modest difference in the proportion of cases of MVT in IDUs compared with heterosexuals? People who inject drugs have concomitant injection and sexual risks for HIV acquisition [33]. Studies in VAX003 trial participants, however, found injection practices, not sexual behaviors, correlated with HIV-1 incidence [34]. Further, we selected only trial participants reporting high-risk IDU practices for this analysis. Despite our focus on a high-injection, low sexual-risk cohort, it is probable that some subjects were infected via sexual routes. Another possible contributor to the low T/F virus numbers is that injection practices of VAX003 participants generally led to the transfer of very small amounts of blood and a low virus inoculum. Plasma-associated factors in the infected partner (eg, neutralizing antibodies[35]) or in virus-naive recipients could similarly contribute.
Epidemiological studies have consistently shown HIV transmission to be a low-probability event, with a small fraction of HIV exposures via any route causing infection [9, 36–39]. While these data are consistent with our findings, they also suggest that a small increase in the number of T/F viruses could shift the outcome of an exposure and reduce the efficacy of a marginally effective vaccine. This concept is consistent with the loss of vaccine efficacy seen in RV144 subjects with higher risks, suggesting that a higher risk challenge (eg, IDU or MSM exposures) may have overwhelmed the low-level immune responses elicited in RV144 [15, 16]. In the advent of more robust vaccines proven to be highly efficacious in heterosexual cohorts, however, the low multiplicity of infection of VAX003 IDUs shown here suggests such a vaccine is likely to be similarly successful in the majority of IDUs.
Our second finding addresses the concern for antibody-mediated enhancement of infection, which may be relevant in vaccination strategies that induce lower potency antibody responses [40]. Multiple lines of in vitro evidence demonstrate antibody-mediated enhancement of HIV infection via interactions with low potency or low concentrations of antibodies [40, 41]. More recently, this concern was raised by 3 independent nonhuman primate studies suggesting antibody-dependent enhancement by weakly- or nonneutralizing antibodies administered via passive infusion or induced by vaccination strategies similar to VAX003 and RV144 [19, 20, 42]. Because the VAX003 immunogen elicited a nonprotective response akin to in vitro and animal model experiments invoking enhancement [40, 42], we reasoned the VAX003 trial warranted assessment for vaccine-associated enhancement using T/F analyses [19, 20, 42, 43]. We found no evidence that antibodies elicited by the AIDSVAX vaccine led to enhancement of HIV-1 infection. In fact, our T/F analysis of the VAX003 trial revealed a trend, albeit statistically nonsignificant, toward a vaccine-related reduction with lower frequency of MVT and numbers of T/F viruses in vaccinees compared with the placebo arm. Further, we saw no examples of vaccine recipients with high numbers of T/F viruses, with at most 3 T/F viruses establishing infection. The lack of enhancement in virus acquisition measured by T/F analysis of VAX003 may reduce concerns that an ineffective antibody response may lead to enhancement of clinical infection.
Our last question was whether VAX003 subjects' T/F sequences had evidence of vaccine-associated selection, or a sieve effect. We chose to study T/F viruses rather than evolved virus populations represented later in infection (beyond 100 days) to focus on the earliest immune pressures present at or near the time of transmission. This strategy had an associated cost in statistical power that studying the entire VAX003 cohort may have increased. Despite the restricted sample size, the VAX003 sieve analysis demonstrated modest evidence of an atypical sieve effect at Env position 170. This signature of vaccine-associated selection corroborates and complements the RV144 analysis, which implicated typical and atypical sieve effects in the same V2 crown region, both of which were associated with increased VE and supported by biological correlates analyses [17, 32]. Atypical (ie, greater mismatch in the placebo sequences) sieve effects are in some ways counterintuitive and provocative but are supported by the associated VE shown in the RV144 sieve analysis and merit further study [18, 44].
In summary, our analyses of the T/F viruses in 50 VAX003 subjects showed a surprisingly low multiplicity of HIV-1 infection in IDUs, indicating that a successful vaccine or other prevention modality generally needs to protect against only 1 or a few viruses regardless of mode of acquisition. The stringent HIV-1 transmission bottleneck in both mucosal (sexual) and nonmucosal (IDU) transmission suggests that formidable barriers to HIV transmission lie both within and outside of the genital mucosa.
Funding
This work was supported by the National Institutes of Health Center for HIV/AIDS Vaccine Immunology (U19AI067854), the Center for HIV/AIDS Vaccine Immunology and Immunogen Development (UM1AI100645), the Bill and Melinda Gates Foundation (grant 37874), and the W. W. Smith Charitable Trust.
Supplementary Material
Acknowledgements
The authors thank Faruk Sinangil and Keith Higgins at Global Solutions for Infectious Diseases for sample coordination, Patricia Crystal for manuscript preparation, and the University of Pennsylvania Center for AIDS Research SGA Sequencing Core for sequencing support. They also acknowledge the Bangkok Vaccine Evaluation Group members and valued contributions of the VAX003 trial participants.
Conflict of Interest Statement: None of the authors of the article have any commercial or other association that would pose a conflict of interest with this work.
References
- 1.(UNAIDS) JUNPoHA. Global Report: UNAIDS report on the global AIDS epidemic 2013. 2013. In: Data WLC-i-P, ed. http://www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2013/gr/UNAIDS_Global_Report__en.pdf .
- 2.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haaland RE, Hawkins PA, Salazar-Gonzalez J, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahams MR, Anderson JA, Giorgi EE, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol. 2009;83:3556–67. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenier JL, Miller CJ, Lu D, et al. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J Virol. 2001;75:3753–65. doi: 10.1128/JVI.75.8.3753-3765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keele BF, Li H, Learn GH, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–34. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Keele BF, Li H, et al. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol. 2010;84:10406–12. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone M, Keele BF, Ma ZM, et al. A limited number of simian immunodeficiency virus (SIV) env variants are transmitted to rhesus macaques vaginally inoculated with SIVmac251. J Virol. 2010;84:7083–95. doi: 10.1128/JVI.00481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baggaley RF, Boily MC, White RG, et al. Risk of HIV-1 transmission for parenteral exposure and blood transfusion: a systematic review and meta-analysis. AIDS. 2006;20:805–12. doi: 10.1097/01.aids.0000218543.46963.6d. [DOI] [PubMed] [Google Scholar]
- 10.Powers KA, Poole C, Pettifor AE, et al. Rethinking the heterosexual infectivity of HIV-1: a systematic review and meta-analysis. Lancet Infect Dis. 2008;8:553–63. doi: 10.1016/S1473-3099(08)70156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84:6241–7. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masharsky AE, Dukhovlinova EN, Verevochkin SV, et al. A substantial transmission bottleneck among newly and recently HIV-1-infected injection drug users in St Petersburg, Russia. J Infect Dis. 2010;201:1697–702. doi: 10.1086/652702. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Blair L, Chen Y, et al. Molecular mechanisms of HIV type 1 prophylaxis failure revealed by single-genome sequencing. J Infect Dis. 2013;208:1598–603. doi: 10.1093/infdis/jit485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitisuttithum P, Gilbert P, Gurwith M, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 15.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 16.Robb ML, Rerks-Ngarm S, Nitayaphan S, et al. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis. 2012;12:531–7. doi: 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolland M, Tovanabutra S, deCamp AC, et al. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med. 2011;17:366–71. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolland M, Edlefsen PT, Larsen BB, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–20. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton DR, Hessell AJ, Keele BF, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci USA. 2011;108:11181–6. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sholukh AM, Byrareddy SN, Shanmuganathan V, et al. Passive immunization of macaques with polyclonal anti-SHIV IgG against a heterologous tier 2 SHIV: outcome depends on IgG dose. Retrovirology. 2014;11:8. doi: 10.1186/1742-4690-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Bar KJ, Wang S, et al. High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 24.Lee HY, Giorgi EE, Keele BF, et al. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol. 2009;261:341–60. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolland M, Larsen BB, Edlefsen PT. Sequence Analysis of HIV-1 Breakthrough Infections in the RV144 Trial. AIDS Vaccine. 2011 Bangkok, Thailand. Page 46 Abstract S07.02. [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Stat Methodol. 1995;57:289–300. [Google Scholar]
- 27.Giorgi EE, Funkhouser B, Athreya G, et al. Estimating time since infection in early homogeneous HIV-1 samples using a poisson model. BMC Bioinformatics. 2010;11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar KJ, Tsao CY, Iyer SS, et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–72. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salazar-Gonzalez JF, Salazar MG, Keele BF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–89. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert PB, Wu C, Jobes DV. Genome scanning tests for comparing amino acid sequences between groups. Biometrics. 2008;64:198–207. doi: 10.1111/j.1541-0420.2007.00845.x. [DOI] [PubMed] [Google Scholar]
- 32.Haynes BF, Gilbert PB, McElrath MJ, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–86. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kral AH, Bluthenthal RN, Lorvick J, et al. Sexual transmission of HIV-1 among injection drug users in San Francisco, USA: risk-factor analysis. Lancet. 2001;357:1397–401. doi: 10.1016/S0140-6736(00)04562-1. [DOI] [PubMed] [Google Scholar]
- 34.Martin M, Vanichseni S, Suntharasamai P, et al. Drug use and the risk of HIV infection amongst injection drug users participating in an HIV vaccine trial in Bangkok, 1999–2003. Int J Drug Policy. 2010;21:296–301. doi: 10.1016/j.drugpo.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Ma ZM, Stone M, Piatak M, Jr., et al. High specific infectivity of plasma virus from the pre-ramp-up and ramp-up stages of acute simian immunodeficiency virus infection. J Virol. 2009;83:3288–97. doi: 10.1128/JVI.02423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baggaley RF, White RG, Boily MC. HIV transmission risk through anal intercourse: systematic review, meta-analysis and implications for HIV prevention. Int J Epidemiol. 2010;39:1048–63. doi: 10.1093/ije/dyq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell DM. Occupational risk of human immunodeficiency virus infection in healthcare workers: An overview. Am J Med. 1997;102:9–15. doi: 10.1016/s0002-9343(97)89441-7. [DOI] [PubMed] [Google Scholar]
- 38.Wawer MJ, Gray RH, Sewankambo NK, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–9. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 39.Jin F, Jansson J, Law M, et al. Per-contact probability of HIV transmission in homosexual men in Sydney in the era of HAART. AIDS. 2010;24:907–13. doi: 10.1097/QAD.0b013e3283372d90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorlani A, Forthal DN. Antibody-dependent enhancement and the risk of HIV infection. Curr HIV Res. 2013;11:421–6. doi: 10.2174/1570162x113116660062. [DOI] [PubMed] [Google Scholar]
- 41.Robinson WE, Jr., Montefiori DC, Mitchell WM. A human immunodeficiency virus type 1 (HIV-1) infection-enhancing factor in seropositive sera. Biochem Biophys Res Commun. 1987;149:693–9. doi: 10.1016/0006-291x(87)90423-2. [DOI] [PubMed] [Google Scholar]
- 42.Pegu P, Vaccari M, Gordon S, et al. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol. 2013;87:1708–19. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kijak GH, Kim JH. Timing, adherence, resistance, and … persistence? New insight into the mechanisms of failure of HIV type 1 postexposure prophylaxis. J Infect Dis. 2013;208:1542–4. doi: 10.1093/infdis/jit486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edlefsen PT, Gilbert PB, Rolland M. Sieve analysis in HIV-1 vaccine efficacy trials. Curr Opin HIV AIDS. 2013;8:432–6. doi: 10.1097/COH.0b013e328362db2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.