Abstract
Significance: Venous leg ulcers (VLUs) are susceptible to microbial invasion, and serious complications can result without the timely control of infection. Diagnosis of wound infection is primarily based on subjective clinical characteristics and patient-reported symptoms, and the treatment with antimicrobials has not consistently shown improvement in healing outcomes. This is a review of studies using bacterial cultures and/or new molecular-based methods associating microbial bioburden with healing outcomes in VLU patients, with the goal of guiding future studies to better determine significant patterns of microbial involvement in chronic wounds.
Recent Advances: Studies reviewed here use cultivation-based identification of bacteria and next-generation sequencing of the bacterial 16S rRNA gene to gain insight into microbial bioburden in VLUs. Further application of sophisticated DNA sequencing and bioinformatic analyses has the potential to revolutionize our ability to further discern, with high resolution, complex microbial communities in chronic wounds.
Critical Issues: Few previous studies of microbial bioburden in VLUs have incorporated the knowledge of clinical treatments, which includes close monitoring of patients' symptoms and responses to therapy. Thus, wound care practitioners are currently without evidence-based guidance for the diagnosis and treatment of wound infections.
Future Directions: Clinically relevant breakthroughs are possible by combining advanced microbial detection techniques with improved study designs that reflect clinical practices. Well-designed longitudinal studies have great potential to lead to better evidence-based diagnosis of chronic wounds. A greater understanding of microbial bioburden in chronic wounds is likely to lead to better therapies that speed healing and prevent wound infection without risking the development of antimicrobial resistance.

Marie S. Tuttle, MD
Scope and Significance
Venous leg ulcers (VLUs) are the most common cause of chronic leg wounds with distinct pathogenesis and unique susceptibilities to microbial invasion. Lack of timely control of microbial invasion can lead to serious complications, including delayed healing, cellulitis, and sepsis. This is a review of the evidence regarding the effect of microbial bioburden on healing outcomes in VLUs, including clinical studies using bacterial culture and advanced molecular methods. The combination of new microbial detection techniques with improved study designs learned from the strengths and weaknesses of previous studies has great potential to lead to improved diagnostics and therapeutics for chronic wounds.
Translational Relevance
This review compares the study designs and results from clinical investigations of microbial bioburden in VLUs with associated patient information, including wound parameters and healing outcomes. Methods used for microbial detection in these studies range from cultivation-based methods to molecular-based methods, including DNA sequencing of the bacterial 16S rRNA gene for microbiome analysis. Next-generation DNA sequencing has revolutionized our ability to discern complex microbial communities with high resolution. However, clinically relevant breakthroughs are only possible if further applications of these technologies are guided by studies that incorporate clinical practice to determine significant patterns of pathogenic microbial involvement in VLUs.
Clinical Relevance
Current clinical practice for wound infection includes the use of subjective clinical signs and symptoms for diagnosis, then antimicrobial treatment with guidance by conventional bacterial culture. However, as reviewed in this study, there are very few studies of wound microbial bioburden that include patients' wound parameters and healing outcomes, and no studies that examine changes in microbial communities with antimicrobial therapy. Additionally, there is little evidence that the use of antimicrobials leads to improvements in clinical outcomes. We will discuss the lessons learned from previous studies to guide future investigations to better reflect clinical practice and improve the prevention and treatment of wound infections.
Background
VLUs are a common and costly problem, accounting for up to 70% of all chronic leg ulcers.1,2 Among people aged 65 years and older, the annual prevalence of VLUs is 1.7%.3 Of all types of chronic ulcers, VLUs have the worst long-term healing prognosis and are more likely to be recurrent.2,4 In fact, half of the patients diagnosed with VLUs have an ulcer duration greater than 1 year and recurrence has been estimated at >75% of those affected.1,2 With such long ulcer duration and high recurrence rate, patients presenting for wound care can be at any point along this continuum, and often have an extensive medical history with several comorbidities and complex medication regimens. Thus, thorough patient assessment at initial and follow-up visits is essential to determine optimal treatment strategies.
An essential element of thorough assessment of VLU patients includes frequent and regular monitoring for infection. For the evaluation and treatment of bacterial infection of chronic wounds, most wound care practitioners rely on subjective clinical signs and patient-reported symptoms.5 This reliance on subjective findings is necessitated by the lack of objective diagnostics that can guide clinical decisions to improve patient outcomes. Conventional bacterial culture is the most widely available tool for the diagnosis of bacterial infection, but has been shown consistently to underestimate the complexity of wound bacterial burden as revealed by DNA-based molecular methods.6–10 Bacteria in chronic wounds grow in biofilms, which include many anaerobes and other species not identified by cultivation-based methods.7,10–12 This complexity of the microbial flora in chronic wounds may explain the lack of improved clinical outcomes observed in many studies of antimicrobial use targeting bacteria identified by cultivation-based methods.13 Thus, rather than aiming to eradicate microbes with an antimicrobial, the enhanced view of wound microbial bioburden revealed by molecular methods supports a more comprehensive approach, including further consideration of patient and wound factors that may contribute to increased susceptibility to microbial invasion.
VLUs occur in people with chronic venous insufficiency, which has many sequelae that can increase susceptibility to microbial invasion. These include edema, lipodermatosclerosis, iron overload and hemosiderin deposition, dermatitis, atrophie blanche, and persistent proinflammatory immune responses (Fig. 1).14–17 VLU patients also have propensities for neuropathy, limited ankle mobility, deep vein thrombosis, thrombophlebitis, and frequent occurrence of comorbidities such as obesity, arterial insufficiency, diabetes, and autoimmune diseases.1,2,14,17,18 Most studies of VLU microbial bioburden either exclude or do not account for these characteristics, making deduction of significant patterns difficult. In this review, we will examine the studies of microbial bioburden in VLUs that have included information on patient wound parameters and outcomes, with the goal of guiding future studies to improve the objective assessment of bacterial infection and clarify the role of antimicrobial treatment in VLUs.
Figure 1.
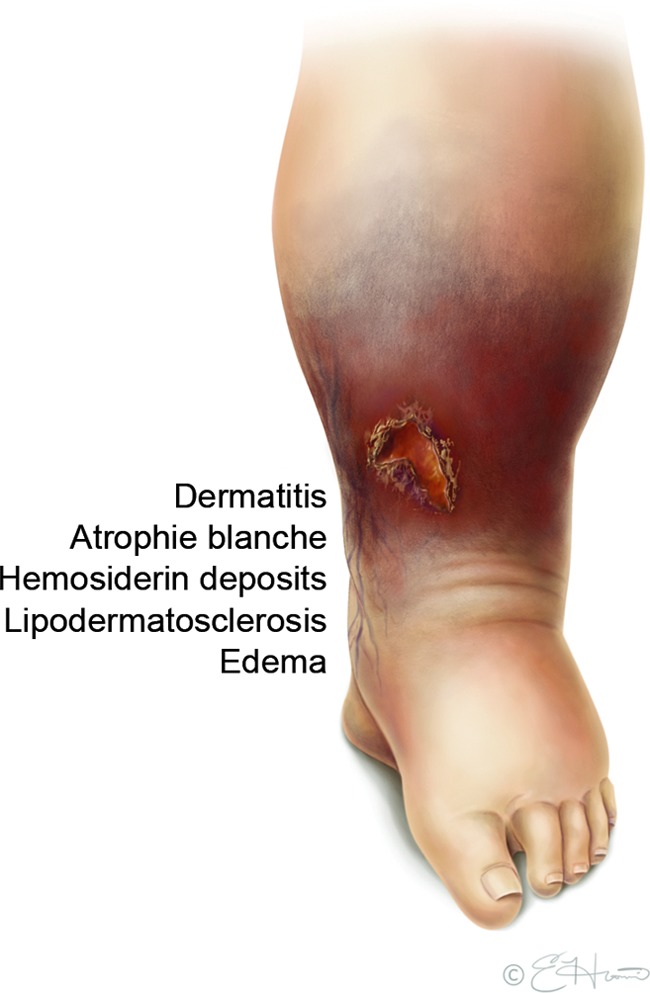
Venous leg ulcer (VLU) characteristics that can increase susceptibility to microbial invasion. The underlying etiology of VLUs, chronic venous insufficiency, has many manifestations that can alter normal barriers to microbial invasion as illustrated. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Discussion
Microbial bioburden and VLU healing outcomes
The concept of increased wound microbial bioburden has been proposed as an important predictor of poor healing outcomes, including three dimensions of microbiology: (1) microbial load, (2) presence of pathogenic organisms, and (3) microbial diversity.19 We will further examine the current evidence behind these dimensions of microbial bioburden in studies that have included VLU patient healing outcomes.
Microbial load
Microbial load of >105 colony-forming units (CFU)/g is often held as the reference standard definition of clinical infection, although this figure is primarily based upon early studies of heterogeneous patient populations, including few VLU patients healing by secondary intention (healing without intervention with skin grafting, delayed surgical closure, or other surgical procedures).20–22 These studies included 50 skin graft patients with a mix of VLUs, traumatic, and burn wounds,20 40 patients requiring delayed surgical closures (types of wounds not otherwise described),22 and 95 surgical incisions requiring delayed surgical closure.21 All of these studies demonstrated >90% graft survival or successful delayed surgical closure only when bacterial colonization was ≤105 CFU/g and <20% successful closure when bacterial colonization was >105 CFU/g. Unfortunately, the results have not been as clear in studies of wounds healing by secondary intention without further surgical intervention.23,24
Most studies of quantitative bacterial cultures in chronic wounds have not associated microbial load with healing outcomes.25–27 Two exceptions include Davies et al.23 and Lantis et al.24 Davies et al. prospectively performed quantitative tissue biopsies and surface swabs of 66 patients with clinically noninfected chronic VLUs at baseline and monitored healing outcomes. They showed that neither quantitative tissue biopsy (CFU/g) nor surface swabs (CFU/mL) of the wound at baseline predicted healing at 6 months in a logistic regression model.23 The investigators state that the 7-log–fold variation in bacterial numbers suggests that the microbial load is not directly related to the healing outcome in clinically noninfected VLUs. However, they did report that increased bacterial load in the tissue biopsies was significantly correlated with a longer ulcer duration at the time of sampling. There was also a significant relationship between healing at 4 weeks (as measured in percent reduction in surface area) and bacterial load in the biopsies and swabs, suggesting that microbial load may be applicable to healing rates within a month of sampling.
Lantis et al.24 analyzed the impact of microbial load and other measured variables in 227 patients in a prospective randomized VLU trial of HP802-247, an allogeneic bioformulation of neonatal keratinocytes and fibroblasts administered through pump spray.28 Similar to Davies et al.,23 they found that increased bacterial load in quantitative tissue biopsies was significantly greater in wounds of longer duration, although there was no association between bacterial load and healing. However, an overall negative effect on healing was seen by regression analysis for wounds harboring ≥1×104 CFU/g of select bacterial species termed “inhibitory bacteria” (species associated with a lower than average proportion of wounds healed), supporting the multidimensional concept of microbial bioburden requiring integration of both the load and species of bacterial involvement.
Both studies of VLU patients suggest that increased bacterial load in tissue biopsies is associated with ulcers of longer duration, but cannot be used to predict the overall healing outcomes. Further associations of bacterial load and healing outcomes may be difficult to determine due to the underestimation of CFU by cultivation-based quantitation compared with molecular methods, such as quantitative PCR, as demonstrated in diabetic foot ulcers.7 Thus, application of molecular methods in future studies of VLUs may help illuminate the potential correlation between bacterial load and healing rates further.
Presence of pathogenic organisms
Although there have been many investigations aimed at identifying the effect of different bacteria on the size, duration, and healing outcomes of VLUs, there are few clear or consistent associations for any single species implicated (summarized in Tables 1–3). As summarized in Table 1, several studies have shown no association between any specific bacterial species and healing outcomes.16,29–33 Among studies that show associations, there are conflicting conclusions on the relevance of many of the commonly isolated species, such as Staphylococcus aureus and Pseudomonas species, as well as anaerobes (Table 2). Most studies of S. aureus show no association with healing delay or increased ulcer size.24,34,35 However, there are studies that associate S. aureus both with delayed healing14,36 and smaller, healing ulcers.23,37 Similarly, Pseudomonas species have been associated both with larger ulcers and delayed healing,24,34–36,38 whereas other (although fewer) studies have found that ulcers harboring Pseudomonas were more likely to heal normally.10,14 Fewer studies comment on anaerobes and the rigor of anaerobic cultivation methods vary widely, but even these studies disagree on the association of anaerobes with healing.24,34,35,37 Results for Streptococcus species are most consistent, with most studies showing association with ulcers of increased size, longer duration, and healing delay.24,34–36
Table 1.
Studies that conclude bacteria are unlikely associated with healing outcomes of venous leg ulcers
| Source | No. of Patients (Ulcers) | Patient Details | Sampling Time Points | Sampling Method | Report of Healing Outcome | Summary of Study Conclusions |
|---|---|---|---|---|---|---|
| Eriksson et al.29 | 53 | VI by clinical judgment Excluded patients with clinical signs of infection, diabetes, or tendon/bone involvement Concurrent study of dextranomer beads, a freeze-dried porcine skin, an aluminium dressing, and double-layer bandaging |
Baseline, then every 1–2 weeks | 2 swabs of ulcer (for aerobic and anaerobic culture) Swab of intact skin on contralateral leg Agar bacteriological imprint of ulcer Combined findings from methods for conclusions |
10 weeks | No difference between occurrence of various bacterial species or semiquantitative levels initially or throughout the study irrespective of treatment or outcome Noted that “patients tended to keep the initial bacterial flora throughout the whole study” Intact skin grew lower levels of bacteria overall, mainly S. epidermidis |
| Gilchrist and Reed30 | 18 (20) |
VI by clinical judgment Patient resistant to two previous treatments, no antibiotic treatment in prior 2 weeks, and without significant abnormalities in AI, blood counts, blood sugar level, liver, renal, or thyroid function Concurrent study of hydrocolloid dressing under compression stockings |
Baseline, then weekly | Swab 1 mL exudate upon dressing removal for anaerobic culture Combined findings from methods for conclusions |
12 weeks | Ulcers treated with hydrocolloid dressing maintain a fairly stable microbial flora, except for appearance of Streptococcus faecalis in 10/20 ulcers, and consistent decline and loss of Pseudomonas Anaerobic bacteria cultured from 12/20 ulcers, with presence in 6/9 ulcers that healed completely |
| Skene et al.16 | 140 | VI confirmed by US and PPG Extensive characterization of patients for evaluation of prognostic factors Excluded patients with clinical signs of infection, diabetes, rheumatoid arthritis, AI with ABPI <0.75, and neurological disease Concurrent study of hydrocolloid dressing vs. tulle and iodine dressing under compression stockings |
Baseline | Bacteriological growth not otherwise detailed | 16 weeks | No association between bacteria present at baseline and healing Observed that bacteria present changed from month to month, although data not shown |
| Hansson et al.31 | 58 | VI by clinical judgment Excluded patients with clinical signs of infection, significant AI Several different dressings were used under compression stockings |
Baseline, then monthly | 10 mm sampling discs pressed against ulcer surface for 1 min In 10 patients, discs compared with swab, showing same result regardless of method |
16 weeks | No single microbial strain was correlated to ulcer size changes, with a multiple linear regression analysis showing that the presence of different species could at most explain 1/5th of variability of ulcer size changes 57/58 ulcers had one or more of the same species present in all but one of the samples, termed “resident flora” Also, no association between microbial quantities and change in ulcer size |
| Moore et al.32 | 176 | VI by clinical judgment Excluded patients with AI with ABPI <0.8, history of malignancy within 3 years, and history of use of immunosuppressants, chemotherapy, or topcial corticosteroids within 1 month Did not exclude patients with clinical signs of infection Standard treatment, including compression bandage therapy |
Weekly | Swab in a zigzag pattern over the whole ulcer surface | 12 weeks | No significant association with any strains identified by aerobic and anaerobic culture, total number of anaerobes, number of species identified, or four or more taxonomic groups 54% of patients had clinical signs of infection by week 12, although no mention of bacteria associated with these signs or use of antibiotics Clostridium striatum and Pseudomonas aeruginosa were present with greater frequency in ulcers with a poor prognosis (area >10 cm2, duration >12 months) Noted that “weekly sampling demonstrated the dynamic nature of wound bacterial populations, with some organisms only detected at isolated time points” |
ABPI, ankle brachial pressure index; AI, arterial insufficiency; US, ultrasonography; PPG, photoplethysmography; VI, venous incompetence.
Table 2.
Studies that conclude bacteria are likely associated with healing outcomes of venous leg ulcers
| Source | No. of Patients | Patient Details | Sampling Time Points | Sampling Method | Report of Healing Outcome | Summary of Study Conclusions |
|---|---|---|---|---|---|---|
| Lookingbill et al.33 | 9 (13) |
VI by clinical judgment No signs of infection noted throughout the study Concurrent study of benzoyl peroxide 10% lotion |
All ulcers: baseline and 2 weeks Only three ulcers: 6 weeks |
Swab 1 cm2 of ulcer Biopsy 3 mm of same area |
16 weeks | Eight ulcers with >105 did not heal Of five ulcers with <105, three healed, one almost healed, one had little healing Qualitative differences between ulcers described, no differences noted Good correlation noted between sampling methods, although swabs had higher quantitative and qualitative recovery rate than biopsies |
| Halbert et al.34 | 82 (100) |
VI confirmed by PPG Excluded patients with other causes of ulceration by performing testing for blood glucose, urea and electrolytes, rheumatoid serology, autoantibody screen, and AI with ABPI <0.9 Did not exclude patients with clinical signs of infection Concurrent dressing trial for a zinc oxide impregnated paste bandage and a calcium alginate fiber dressing |
Baseline | Swab not otherwise specified | 24 weeks | Compared with ulcers with no bacterial growth, ulcers were significantly larger and of longer duration if colonized with anaerobes, beta-hemolytic streptococci, P. aeruginosa alone, and coliforms (not otherwise specified) Ulcer healing time was prolonged when colonized with most organisms, although not significantly prolonged with Staphylococcus aureus+P. aeruginosa, P. aeruginosa alone, and normal skin flora Two patients developed cellulitis and were treated with antibiotics; cultures showed S. aureus and mixed coliforms from one and anaerobes and mixed coliforms from the other |
| Madsen et al.36 | 59 | VI confirmed by US Excluded patients with AI with toe pressures <60 mmHg and diabetes Did not exclude patients with clinical signs of infection Concurrent study of patients undergoing ligation of incompetent perforators and wide debridement |
Weekly×4 weeks, then monthly | Swab samples from proximal ulcer edge | 24 weeks |
P. aeruginosa, S. aureus, and hemolytic streptococci were associated with healing delay P. aeruginosa was associated with ulcer enlargement Forty-one percent of patients had clinical signs of cellulitis on one to three occasions; cultures showed P. aeruginosa in seven, hemolytic streptococci in four, and others not noted Ulcers with P. aeruginosa and cellulitis were not significantly larger than without cellulitis Healing delay occurred despite antibiotic treatment when cellulitis occurred No mention of changes in bacterial species throughout healing |
| Trengove et al.35 | 52 | VI confirmed by PPG Included patients with AI, although all had ABPI >0.7 Did not exclude patients with clinical signs of infection Concurrent study of different compression therapies |
Weekly | Two swabs of ulcer for aerobic and anaerobic cultures | 24 weeks | A significantly greater number of ulcers with four or more bacteria failed to heal The presence of any one specific bacterial group was not associated with healing The bacterial flora changes as the ulcers heal, although the changes were not related to healing No mention of clinical signs of infection or antibiotic use |
| Gjodsbol et al.38 | 36 | VI confirmed by US Patient had ABPI >0.6, no diabetes or other diseases that could influence the ulcer No patients had antibiotics within 14 days of inclusion, or antibiotics during the study |
Swab Baseline only Biopsy Baseline and after 4 weeks Filter paper Baseline and every 2 weeks |
Swab Biopsy 4 mm from ulcer center Sterile 10 mm2 filter paper pad placed in ulcer until saturated Combined findings from methods for conclusions |
8 weeks | All the ulcers had resident flora (defined as one or more of the same species reisolated from all or all but one of the series of samples from the ulcer) P. aeruginosa was present as resident bacteria in all ulcers that increased in size, but nonresident in all ulcers that reduced in size Ulcers with S. aureus were significantly smaller No significant difference in ulcer size was associated with the presence of anaerobic bacteria |
| Davies et al.23 | 66 | VI confirmed with US Excluded patients with AI with ABPI <0.08, diabetes, immunosuppression, or antimicrobials within 1 month |
Baseline | Swab of 1 cm2 of central ulcer Biopsy 6 mm of same area of ulcer Differentiated swabs and biopsies in conclusions |
24 weeks | No association between individual bacteria or bacterial groups present at baseline and overall healing The presence of ≥4 bacterial genera in the ulcer was associated with delayed healing Ulcer duration correlated with bacterial load in the biopsy, but not the swab Significant relationship between reduction in surface area at 4 weeks and bacterial load in the biopsies and swabs |
| Moffat et al.14 | 113 | VI confirmed with US and PPG AI assessed with ABPI, but not excluded Did not exclude patients with clinical signs of infection, clinically characterizing cellulitis by pain, odor, swelling, and erythema Extensive characterization of patients for evaluation of clinical predictors of healing |
Not stated | Swabs not otherwise specified | Up to 48 weeks | Clinical cellulitis, S. aureus, hemolytic streptococcus were associated with reduced healing, but did not approach statistical significance The presence of Pseudomonas was associated with an increase in healing The number of species in an ulcer did not show a direct relationship with healing, although the presence of at least on bacterium reduced the chance of healing |
| Lantis et al.24 | 227 | VI confirmed by US Excluded patients with clinical signs of infection, elevated HbA1c or prealbumin, thyroid disease, systemic lupus, or elevated anti-DNA Concurrent, prospective randomized trial of HP802-247, an allogeneic bioformulation of neonatal keratinocytes and fibroblasts |
Baseline | Biopsy 4 mm from ulcer center | 12 weeks | Bacteria divided into inhibitory (lower than average [60%] proportion healed) vs. noninhibitory bacteria Inhibitory bacteria, all facultative anaerobes, with the exception of Pseudomonas species Regression analysis showed a significantly negative effect on healing associated with the inhibitory bioburden index, which was the number of inhibitory bacteria at ≥104 CFU/g No significant association between bacterial load and healing |
CFU, colony-forming unit.
Table 3.
Studies that use molecular-based techniques for microbial detection and include healing outcomes of venous leg ulcers
| Source | No. of Patients | Patient Details | Sampling Time Points | Sampling Method | Report of Healing Outcome | Summary of Study Conclusions |
|---|---|---|---|---|---|---|
| Davies et al.6 | 18 | VI confirmed by US Excluded patients with AI (by ABPI ≥0.8), diabetes, all antibiotic use×1 month |
Baseline | Swab of 1 cm2 of central ulcer for aerobic and anaerobic culture Biopsy 6 mm of same area of ulcer bisected for culture and molecular analysis |
24 weeks | More micrococcus and streptococcus in nonhealers, although not significant due to small numbers No difference between anaerobes between healers and nonhealers |
| Tuttle et al.10 | 10 | VI by clinical judgment Did not exclude patients with clinical signs of infection or use of antibiotics |
Baseline | Curette 3 mm from proximal and distal ulcer edge and center Swab of ulcer base in areas curetted Combined findings from methods for conclusions |
24 weeks | More diverse bacterial populations in nonhealed ulcers Differences between nonhealed and healed wounds shown with 16S sequencing methods, including more Actinomycetales in nonhealed wounds, and more Pseudomonadaceae in healed wounds No differences revealed by culture or other molecular diagnostic methods studied (Ibis) |
Interestingly, the studies that did not find associations between bacterial populations and healing were more likely to exclude patients with clinical signs of infection in the study design (Table 1 vs. Table 2). In all of the studies examined, clinical signs of infection were not defined, except for the report by Moffatt et al., which listed pain, odor, swelling, erythema, and confirmation by microbiological results without further definition.14
As shown in Table 3, there have been two studies using molecular-based methods for identification of bacteria that also include VLU healing outcome measures, Davies et al.6 and Tuttle et al.10 To compare and contrast these studies, both studies compared aerobic and anaerobic bacterial cultures to sequencing of the bacterial 16S rRNA gene, which can be used for the identification of bacteria by matching the unique 16S rRNA gene sequence to the taxonomic classification based on large reference databases such as the Ribosomal Database Project (RDP), SILVA, and Greengenes. Before the dissemination of the next-generation sequencing technology, Davies et al. selected 16S rRNA genes for sequencing that were unique from culture isolates on the basis of profiling by denaturing gradient gel electrophoresis.6 In contrast, Tuttle et al. utilized the next-generation (454 FLX titanium series) 16S rRNA sequencing as well as the Ibis T5000 universal biosensor, which uses microbial DNA for taxonomic identification based on the molecular sizes of amplicons generated by using multiple PCR primer pairs.10
Overall, the reports from both Davies et al. and Tuttle et al. demonstrate that sequencing methods identify significantly more bacteria than cultivation methods, with increased detection of even known culturable bacteria. Increased detection of culturable microbes could be, in part, due to the ability of molecular methods to detect DNA from nonviable or viable, but unculturable microbes in the ulcer. Whereas Davies et al. did not identify significant differences between microbial communities from ulcers that were either healed or unhealed at the 6-month follow-up, Tuttle et al. showed a significantly greater bacterial diversity in unhealed ulcers, which incorporates information on the type and relative proportion of the bacteria, as discussed further in the microbial diversity section.6,10
Microbial diversity
Another component of microbial bioburden of chronic ulcers that has been implicated in delayed healing is bacterial diversity, which is defined differently depending on the data used for analysis. For the cultivation-based studies reviewed here, diversity is defined as the number of different species of bacteria cultured from an ulcer sample. As summarized in Tables 1 and 2, some studies have concluded that nonhealing ulcers were more likely to have a higher microbial diversity,23,35 but others have shown that diversity could not be used to predict healing.14,32 For studies using DNA sequencing data, more specific ecological concepts of diversity can be used. Tuttle et al.10 used the Shannon diversity index, which takes into account the relative proportions of each species, showing that diversity was significantly higher in VLUs that were not healed by the 6-month follow-up. This diversity within each sample is referred to as “alpha diversity” or “species richness.”
The concept of diversity also includes variation in bacterial communities between different samples, which is termed “beta diversity.” Evaluation of beta diversity could include comparison of (1) different sampling time points of the same ulcer, (2) different sampling methods (e.g., swab vs. curette/biopsy), (3) different geographic regions within the same ulcer (intrawound variation), (4) similar time points in different ulcers (interwound variation), and (5) between wounds and intact skin. Of the studies reviewed here, only the cultivation-based studies included sampling schemes that could be used to determine beta diversity. Unfortunately, few conclusions can be drawn, mostly from the descriptive culture data, due to insufficient details in the results. For example, of the studies with multiple sampling time points, some concluded that the bacterial communities remained relatively the same,29–31,38 whereas others reported wide variations with time;16,32,35 however, there is no objective measure of the relative extent of variation in the different studies, making these conclusions subjective. Also, these studies included minimal information on changes in patient/ulcer characteristics (such as clinical signs and symptoms of infection) that could aid in further interpretation of differences between samples at various time points.
There have not yet been studies using molecular methods to further determine the different aspects of beta diversity in VLUs that also incorporated patient/ulcer characteristics or healing outcome data. Therefore, this is an important future direction for research. To guide this research, we can learn from studies that have been done examining the beta diversity in chronic wounds of mixed etiology using cultivation and/or molecular-based methods. These studies have aimed to determine variation in microbial composition in (1) different sampling methods (e.g., biopsy vs. swab) and (2) intra- versus interwound variation (using similar sampling methods), as detailed in the following sections. Although these studies do not directly contribute to determining the influence of microbes on healing, they can help understand how differences in the sampling method or location may obscure patterns in investigations of microbial bioburden.
Differences between sampling methods
The studies comparing different wound sampling methods—including tissue biopsies versus less invasive techniques such as swabbing and wound fluid collection—have primarily used cultivation-based methods in chronic wounds arising from multiple etiologies (as reviewed by Reddy et al.39). Overall, cultivation methods have been shown to be comparable between biopsies and swabs for aerobes, although there are more discrepancies for anaerobes and overall quantities of bacteria, suggesting uneven vertical distribution of at least some species as discussed in the Intra- and Interwound Variation section.23,25,37,40 Molecular-based studies performed to date have predominantly used sharp debridement samples without comparison of different sampling methods or indication of the depth of the wound sampled.
Intra- and interwound variation
Although cultivation-based methods are unlikely to be sensitive enough to determine intrawound variation in bacterial composition of ulcers, quantitative culture studies have suggested that bacterial concentration is increased superficially in wounds.35,40 Sophisticated imaging techniques such as fluorescent in situ hybridization and confocal laser scanning microscopy have confirmed this result, further displaying wound bacteria in both mixed- and single-species biofilms, as well as scattered individual cells.41–43 In general, S. aureus has been visualized more toward the surface of the ulcers and biofilms, particularly compared with Pseudomonas aeruginosa, potentially explaining why S. aureus is most commonly isolated from swab samples of chronic wounds and P. aeruginosa is likely frequently underestimated.41,43,44
Studies that have used bacterial 16S rRNA sequencing to investigate intrawound variation in chronic wounds include Price et al.,45 and Wolcott et al.46 In both studies, curette samples were collected from different geographic regions of chronic wounds, including the leading edge, opposing leading edge, and/or center of chronic wounds. Wolcott et al. concluded there was substantial variation across the wound surface without further statistical analysis.46 Price et al. further compared intrawound variation to bacterial communities from wounds on different individuals (interwound variation), showing that the intrawound variation is significantly smaller than the interwound variation.45 They, therefore, argue that studies that do not control for sampling site within the wound can still be valid for comparing interwound variation, although controlling for sampling site likely improves the overall quality of the study. It is important to note that these were cross-sectional studies of patients without further characterization of the healing trajectory. To date, no other studies have compared different time points, patient characteristics, wound parameters, or healing outcomes.
Clinical signs and symptoms of VLU infection
Studies to date have regarded patient clinical signs and symptoms as subjective and secondary to more objective measures, most often using >105 CFU/g tissue as the reference standard for the diagnosis of infection. However, as discussed above, this one-dimensional reference standard does not fully capture the extent of microbial bioburden, of which microbial pathogenicity and diversity are also important aspects. Regardless of whether quantitative tissue cultures are an adequate reference standard of infection, they are generally considered impractical for routine use in clinical practice due to the variability in cultivation methods in different clinical laboratories and frequency of refusal by patients to undergo biopsy. Therefore, in reality, most wound care practitioners rely solely on clinical signs and symptoms to identify wound infection.5 Once infection is suspected, topical antiseptics (for superficial infection) and/or systemic antibiotics (for deep infection) are recommended, and conventional culture may or may not be performed to guide antimicrobial therapy.47
Because studies on the accuracy of clinical signs and symptoms of infection use >105 CFU/g as a reference standard and include minimal information on healing outcomes, there is currently no evidence to assess how well clinical signs and symptoms predict infection-related complications. As reviewed by Reddy et al.,39 no sign or symptom was consistently evaluated in more than two studies, with the likelihood of infection being increased when the ulcer causes increased pain, and being decreased in wounds that had no serous exudates or were healing rapidly.48 Other clinical signs and symptoms evaluated include nonhealing status, increase in wound exudate (more than 50% of the dressing stained with exudate), red tissue at the wound base that bleeds easily, the presence of slough or nonviable tissue at the wound base, and unpleasant odor for a superficial infection, for which a topical antiseptic would be recommended.49 Wounds with a deeper tissue infection, for which a systemic antibiotic would be recommended, have additional signs and symptoms, including increased size, surrounding areas of tissue breakdown, and increased periwound temperature, redness, and swelling.49 Of note, these studies have included leg and foot ulcers of various etiologies, not only VLUs; therefore, some signs and symptoms may be more or less relevant for VLUs.
Summary
Thorough assessment of VLU patients includes regular monitoring for signs of infection. Failure to do so may lead to serious complications, including further wound breakdown, cellulitis, and sepsis. As this review demonstrates, attempts to define wound infection using a one-dimensional measure have been largely unsuccessful; rather, a full evaluation of microbial bioburden—including microbial load, pathogenicity, and diversity—as well as a defined set of clinical parameters and patient-reported symptoms is likely required, although studies have not been designed to support this practice. Previous studies linking microbial bioburden to wound parameters and healing outcomes have focused primarily on clinically noninfected wounds. Few cultivation-based studies and no molecular-based studies have analyzed changes in wound microbial bioburden over time. Even rarer are the studies that link results on microbial bioburden with the clinical status of the patient or use of antimicrobials or other therapies.
Therefore, recommendations for future studies to better support clinical practice include the following:
(1) Further use of molecular methods for a more comprehensive evaluation of microbial burden;
(2) Prospective determination of patients' signs and symptoms of infection and wound parameters to better link changes in microbial bioburden with patients' stages of healing and responses to treatments, including wound exacerbation, stalled healing, and resolution; and
(3) Inclusion of different sampling methods as well as serial sampling to compare intrawound variation throughout the course of healing.
Implementation of these study design changes would enhance and corroborate with the clinical decision-making process, in which modifications to therapy are made at frequent and regular intervals based on the changes in patients' clinical signs and symptoms.47 These studies would better elucidate the responses of microbial bioburden to therapeutics, clarifying the impact of antimicrobials versus other treatments such as compression therapy. Results from these studies would likely support a more comprehensive approach to infection control in chronic wounds that would include controlling patient factors that increase susceptibility to microbial invasion (illustrated in Fig. 1).
Overall, a combination of improved study design and the use of higher resolution molecular methods to assess wound microbial bioburden has great potential to lead to better evidence-based diagnosis of chronic wounds. In addition to taxonomic information, the future of molecular methods also includes further discernment of functional differences between bacterial communities, potentially leading to more targeted therapies that speed healing and prevent wound infection without risking the development of antimicrobial resistance.
Take-Home Messages.
• There is no established reference standard for diagnosis of wound infection in VLUs.
• To better support clinical practice, further studies are needed that include evaluation of all aspects of microbial bioburden—including microbial load, pathogenicity, and diversity—correlated with patient signs and symptoms of infection and healing outcomes.
• Molecular methods identify significantly more bacteria than cultivation methods, with increased detection of even culturable bacteria.
• Therefore, a combination of improved study design and use of molecular methods has the potential to lead to improved diagnostics and therapeutics to speed healing of chronic wounds.
Abbreviations and Acronyms
- ABPI
ankle brachial pressure index
- AI
arterial insufficiency
- CFU
colony-forming unit
- PPG
photoplethysmography
- RDP
Ribosomal Database Project
- US
ultrasonography
- VI
venous incompetence
- VLU
venous leg ulcer
Acknowledgments and Funding Sources
This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland, KL2TR000440 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Thank you to Christine Ammons, Jeremy Honaker, Daniel Sprockett, and Tom McCormick for critical review of the article.
Author Disclosure and Ghostwriting
No competing financial interests exist. The content of this article was expressly written by the author listed. No ghostwriters were used to write this article.
About the Author
Marie Tuttle, MD, is an Assistant Professor in the Department of Dermatology at Case Western Reserve University/University Hospitals Case Medical Center and Louis Stokes Cleveland Veterans Affairs Medical Center. She is a dermatologist-scientist with a research focus on factors that influence susceptibility to microbial invasion in nonhealing cutaneous wounds.
References
- 1.Baker SR, Stacey MC, Jopp-McKay AG, Hoskin SE, Thompson PJ. Epidemiology of chronic venous ulcers. Br J Surg 1991;78:864–867 [DOI] [PubMed] [Google Scholar]
- 2.Nelzen O, Bergqvist D, Lindhagen A. Venous and non-venous leg ulcers: clinical history and appearance in a population study. Br J Surg 1994;81:182–187 [DOI] [PubMed] [Google Scholar]
- 3.Margolis DJ, Bilker W, Santanna J, Baumgarten M. Venous leg ulcer: incidence and prevalence in the elderly. J Am Acad Dermatol 2002;46:381–386 [DOI] [PubMed] [Google Scholar]
- 4.Nelzen O, Bergqvist D, Lindhagen A. Long-term prognosis for patients with chronic leg ulcers: a prospective cohort study. Eur J Vasc Endovasc Surg 1997;13:500–508 [DOI] [PubMed] [Google Scholar]
- 5.Bamberg R, Sullivan PK, Conner-Kerr T. Diagnosis of wound infections: current culturing practices of U.S. wound care professionals. Wounds 2002;14:314–328 [Google Scholar]
- 6.Davies CE, Hill KE, Wilson MJ, et al. . Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J Clin Microbiol 2004;42:3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013;62:923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melendez JH, Frankel YM, An AT, et al. . Real-time PCR assays compared to culture-based approaches for identification of aerobic bacteria in chronic wounds. Clin Microbiol Infect 2009;16:1762–1769 [DOI] [PubMed] [Google Scholar]
- 9.Rhoads DD, Wolcott RD, Sun Y, Dowd SE. Comparison of culture and molecular identification of bacteria in chronic wounds. Int J Mol Sci 2012;13:2535–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuttle MS, Mostow E, Mukherjee P, et al. . Characterization of bacterial communities in venous insufficiency wounds by use of conventional culture and molecular diagnostic methods. J Clin Microbiol 2011;49:3812–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowd SE, Sun Y, Secor PR, et al. . Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 2008;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price LB, Liu CM, Melendez JH, et al. . Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One 2009;4:e6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Meara S, Al-Kurdi D, Ologun Y, Ovington LG. Antibiotics and antiseptics for venous leg ulcers. Cochrane Database Syst Rev CD003557, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Moffatt CJ, Doherty DC, Smithdale R, Franks PJ. Clinical predictors of leg ulcer healing. Br J Dermatol 2010;162:51–58 [DOI] [PubMed] [Google Scholar]
- 15.Sindrilaru A, Peters T, Wieschalka S, et al. . An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest 2011;121:985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skene AI, Smith JM, Dore CJ, Charlett A, Lewis JD. Venous leg ulcers: a prognostic index to predict time to healing. BMJ 1992;305:1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milic DJ, Zivic SS, Bogdanovic DC, Karanovic ND, Golubovic ZV. Risk factors related to the failure of venous leg ulcers to heal with compression treatment. J Vasc Surg 2009;49:1242–1247 [DOI] [PubMed] [Google Scholar]
- 18.Yim E, Vivas A, Maderal A, Kirsner RS. Neuropathy and ankle mobility abnormalities in patients with chronic venous disease. JAMA Dermatol 2014;150:385–389 [DOI] [PubMed] [Google Scholar]
- 19.Grice EA, Segre JA. Interaction of the microbiome with the innate immune response in chronic wounds. Adv Exp Med Biol 2012;946:55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krizek TJ, Robson MC, Kho E. Bacterial growth and skin graft survival. Surg Forum 1967;18:518 [Google Scholar]
- 21.Robson MC, Heggers JP. Delayed wound closure based on bacterial counts. J Surg Oncol 1970;2:379–383 [DOI] [PubMed] [Google Scholar]
- 22.Robson MC, Lea CE, Dalton JB, Heggers JP. Quantitative bacteriology and delayed wound closure. Surg Forum 1968;19:501. [PubMed] [Google Scholar]
- 23.Davies CE, Hill KE, Newcombe RG, et al. . A prospective study of the microbiology of chronic venous leg ulcers to reevaluate the clinical predictive value of tissue biopsies and swabs. Wound Repair Regen 2007;15:17–22 [DOI] [PubMed] [Google Scholar]
- 24.Lantis JC, 2nd, Marston WA, Farber A, et al. . The influence of patient and wound variables on healing of venous leg ulcers in a randomized controlled trial of growth-arrested allogeneic keratinocytes and fibroblasts. J Vasc Surg 2013;58:433–439 [DOI] [PubMed] [Google Scholar]
- 25.Gardner SE, Frantz RA, Saltzman CL, Hillis SL, Park H, Scherubel M. Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen 2006;14:548–557 [DOI] [PubMed] [Google Scholar]
- 26.Levine NS, Lindberg RB, Mason AD, Jr., Pruitt BA., Jr.The quantitative swab culture and smear: a quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma 1976;16:89–94 [PubMed] [Google Scholar]
- 27.Bill TJ, Ratliff CR, Donovan AM, Knox LK, Morgan RF, Rodeheaver GT. Quantitative swab culture versus tissue biopsy: a comparison in chronic wounds. Ostomy Wound Manage 2001;47:34–37 [PubMed] [Google Scholar]
- 28.Kirsner RS, Marston WA, Snyder RJ, Lee TD, Cargill DI, Slade HB. Spray-applied cell therapy with human allogeneic fibroblasts and keratinocytes for the treatment of chronic venous leg ulcers: a phase 2, multicentre, double-blind, randomised, placebo-controlled trial. Lancet 2012;380:977–985 [DOI] [PubMed] [Google Scholar]
- 29.Eriksson G, Eklund AE, Kallings LO. The clinical significance of bacterial growth in venous leg ulcers. Scand J Infect Dis 1984;16:175–180 [DOI] [PubMed] [Google Scholar]
- 30.Gilchrist B, Reed C. The bacteriology of chronic venous ulcers treated with occlusive hydrocolloid dressings. Br J Dermatol 1989;121:337–344 [DOI] [PubMed] [Google Scholar]
- 31.Hansson C, Hoborn J, Moller A, Swanbeck G. The microbial flora in venous leg ulcers without clinical signs of infection. Repeated culture using a validated standardised microbiological technique. Acta Derm Venereol 1995;75:24–30 [DOI] [PubMed] [Google Scholar]
- 32.Moore K, Hall V, Paull A, et al. . Surface bacteriology of venous leg ulcers and healing outcome. J Clin Pathol 2010;63:830–834 [DOI] [PubMed] [Google Scholar]
- 33.Lookingbill DP, Miller SH, Knowles RC. Bacteriology of chronic leg ulcers. Arch Dermatol 1978;114:1765–1768 [PubMed] [Google Scholar]
- 34.Halbert AR, Stacey MC, Rohr JB, Jopp-McKay A. The effect of bacterial colonization on venous ulcer healing. Aust J Dermatol 1992;33:75–80 [DOI] [PubMed] [Google Scholar]
- 35.Trengove NJ, Stacey MC, McGechie DF, Mata S. Qualitative bacteriology and leg ulcer healing. J Wound Care 1996;5:277–280 [DOI] [PubMed] [Google Scholar]
- 36.Madsen SM, Westh H, Danielsen L, Rosdahl VT. Bacterial colonization and healing of venous leg ulcers. APMIS 1996;104:895–899 [DOI] [PubMed] [Google Scholar]
- 37.Gjodsbol K, Skindersoe ME, Christensen JJ, et al. . No need for biopsies: comparison of three sample techniques for wound microbiota determination. Int Wound J 2012;9:295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 2006;3:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy M, Gill SS, Wu W, Kalkar SR, Rochon PA. Does this patient have an infection of a chronic wound? JAMA 2012;307:605–611 [DOI] [PubMed] [Google Scholar]
- 40.Cooper RA, Ameen H, Price P, McCulloch DA, Harding KG. A clinical investigation into the microbiological status of ‘locally infected’ leg ulcers. Int Wound J 2009;6:453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fazli M, Bjarnsholt T, Kirketerp-Moller K, et al. . Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 2009;47:4084–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han A, Zenilman JM, Melendez JH, et al. . The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011;19:532–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malic S, Hill KE, Hayes A, Percival SL, Thomas DW, Williams DW. Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiology 2009;155:2603–2611 [DOI] [PubMed] [Google Scholar]
- 44.Kirketerp-Moller K, Jensen PO, Fazli M, et al. . Distribution, organization, and ecology of bacteria in chronic wounds. J Clin Microbiol 2008;46:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price LB, Liu CM, Frankel YM, et al. . Macroscale spatial variation in chronic wound microbiota: a cross-sectional study. Wound Repair Regen 2011;19:80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolcott RD, Gontcharova V, Sun Y, Dowd SE. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol 2009;9:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sibbald RG, Goodman L, Woo KY, et al. . Special considerations in wound bed preparation 2011: an update(c). Adv Skin Wound Care 2011;24:415–436; quiz 437–418. [DOI] [PubMed] [Google Scholar]
- 48.Gardner SE, Frantz RA, Doebbeling BN. The validity of the clinical signs and symptoms used to identify localized chronic wound infection. Wound Repair Regen 2001;9:178–186 [DOI] [PubMed] [Google Scholar]
- 49.Woo KY, Sibbald RG. A cross-sectional validation study of using NERDS and STONEES to assess bacterial burden. Ostomy Wound Manage 2009;55:40–48 [PubMed] [Google Scholar]


