Abstract
The X-box binding protein 1 (XBP1) is not only an important component of the unfolded protein response (UPR), but also an important nuclear transcription factor. Upon endoplasmic reticulum stress, XBP1 is spliced by inositol-requiring enzyme 1 (IRE1), thereby generating functional spliced XBP1 (XBP1s). XBP1s functions by translocating into the nucleus to initiate transcriptional programs that regulate a subset of UPR- and non-UPR-associated genes involved in the pathophysiological processes of various diseases. Recent reports have implicated XBP1 in metabolic diseases. This review summarizes the effects of XBP1-mediated regulation on lipid metabolism, glucose metabolism, obesity, and atherosclerosis. Additionally, for the first time, we present XBP1s-dependent transcriptional reprogramming in metabolic diseases under different conditions, including pathology and physiology. Understanding the function of XBP1 in metabolic diseases may provide a basic knowledge for the development of novel therapeutic targets for ameliorating these diseases.
Introduction
The endoplasmic reticulum (ER) is the primary organelle for secretory and membrane protein synthesis, protein folding and secretion. Additionally, the ER is also the primary intracellular calcium reservoir, and many rate-limiting enzymes of lipid biosynthesis are located in the ER membrane. Some stimuli, including oxidative stress, ischemic insult and overexpression of normal and/or misfolded proteins, can lead to ER stress. The unfolded protein response (UPR) is then triggered to maintain ER homeostasis. In mammals, the UPR is induced by stress sensors on the ER membrane, namely, activating transcription factor 6 (ATF6) (Haze et al., 1999), PKR-like ER kinase (PERK) (Harding et al., 1999) and inositol-requiring enzyme 1α (IRE1α). The UPR and other signaling pathways, including metabolic signaling, have interactive effects, and such profound reprogramming is essential for maintaining proper ER function. Furthermore, a growing number of studies have shown that UPR signaling is involved in pathophysiological and metabolic alterations (Park and Ozcan, 2013; Lee and Ozcan, 2014). The X-box binding protein 1 (XBP1), which is downstream of IRE1α, is an important component of the UPR and is a nuclear transcription factor. In this review, we discuss the current knowledge regarding the involvement of the IRE1α-XBP1 pathway and spliced XBP1 (XBP1s)-dependent transcriptional reprogramming in lipid metabolism, glucose metabolism, obesity, and vascular diseases, which may provide a basis for future studies to expand our understanding of XBP1 function in the pathophysiology of metabolic diseases.
XBP1 and the IRE1α-XBP1 Pathway
XBP1 was first obtained by cloning in 1990 and was discovered as a unique basic-region leucine zipper (bZIP) protein possessing the capability of binding to the cis-acting X box present in the promoter regions of human major histocompatibility complex class II genes (Liou et al., 1990). More than a decade later, several studies confirmed that XBP1 is a stress-inducible transcription factor in the UPR that exists in both invertebrate and vertebrate cells and is crucial for cell survival under stress conditions (Shen et al., 2001; Yoshida et al., 2001; Romero-Ramirez et al., 2004). The UPR is one type of ER stress response. Of the three branches of the mammalian UPR, IRE1α can be activated upon ER stress. Activated IRE1α triggers unconventional cytoplasmic splicing of XBP1 mRNA. IRE1α splices 26 nucleotides from the unspliced XBP1 (XBP1u) mRNA, thereby leading to a frameshift and the generation of XBP1s containing a C-terminal transactivation domain that is absent from XBP1u (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2002). Because of the extremely short half-life of XBP1u, little is known regarding its biological function (Navon et al., 2010). Recently, Zhao et al. (2013) reported that XBP1u binds to Forkhead box O1 (FoxO1), which is dependent on XBP1u phosphorylation by ERK, thereby recruiting FoxO1 to the 20S proteasome for degradation. Additionally, XBP1u could control the termination of UPR responses through mediating the proteasomal degradation of XBP1s in the cytosol upon prolonged ER stress, which is involved in the interaction between these molecules (Yoshida et al., 2006). In addition, studies have found that the C-terminus of XBP1u is the primary domain for interacting with 20S proteasomes (Yoshida et al., 2006, 2009). These findings indicate that XBP1u functions in promoting protein degradation. Benhamron et al. (2014) confirmed that XBP1u also plays an important role in controlling IRE1 expression. Moreover, numerous studies of XBP1u have demonstrated XBP1's involvement in splicing XBP1 mRNA (Yanagitani et al., 2009, 2011; Majumder et al., 2012). With the development and improvement of measurement techniques, we propose that many biological functions of XBP1u will be found. However, in contrast to XBP1u, XBP1s has multiple functions and is involved in both classical UPR and non-UPR pathways. During the UPR, the XBP1s protein translocates into the nucleus to initiate transcriptional programs that upregulate a broad spectrum of UPR-associated genes involved in protein entry into the ER, protein folding, ER-associated degradation (ERAD), and ER biogenesis. For example, one study showed that XBP1 was required for the terminal differentiation of B lymphocytes into plasma cells (Reimold et al., 2001). Although XBP1-deficient B cells exhibited normal proliferation and activation, they expressed decreased levels of J chain, a component required for Ig assembly, resulting in decreased Ig production (Reimold et al., 2001). However, restoration of XBP1s expression rescued Ig production (Reimold et al., 2001). Further study also showed that XBP1 was required for IgM synthesis and secretion (Tirosh et al., 2005). These findings suggest that the IRE1-XBP1 pathway is required for stimulated B cells to secrete antibodies (Aragon et al., 2012). XBP1s can also contribute to the differentiation of end-stage effector CD8+ T cells (Kamimura and Bevan, 2008). Additionally, XBP1 is required for Paneth cell function, gastric zymogenic cell function, exocrine pancreas function, and salivary gland function (Lee et al., 2005; Kaser et al., 2008; Huh et al., 2010). In addition, the IRE1α-XBP1 pathway is involved in insulin and glucagon secretion through the regulation of pancreatic β- and α-cell functions (Lee et al., 2011a; Akiyama et al., 2013). Finally, XBP1s can regulate triglyceride (TG)-rich very-low-density lipoprotein (VLDL) assembly and secretion in mice with a hepatocyte-specific deletion of IRE1α (Wang et al., 2012). Concerning the non-UPR-related genes associated with these functions, some reports have certified that XBP1s is involved in the regulation of lipid metabolism by directly regulating the expression of a subset of lipogenic genes (Lee et al., 2008; Ning et al., 2011; Fang et al., 2013). Additionally, XBP1 enhances ERα-dependent transcriptional activity in a ligand-independent manner in breast tumors, and this involves the interaction between XBP1 and ERα, which functions in estrogen receptors signal transduction (Ding et al., 2003). Further study found that XBP1-regulated, large-scale chromatin unfolding may be responsible for the enhancement of ERα transcriptional activity (Fang et al., 2004). Sengupta et al. (2010) reported that ERα along with the coactivators, steroid receptor coactivator-3 (SRC-3) and steroid receptor coactivator-1 (SRC-1), accumulate at the promoter and enhancer regions of the XBP1 gene. These findings suggest that XBP1 may regulate the expression of ERs at the transcriptional level. In addition, XBP1s can regulate a group of inflammatory cytokines (Gargalovic et al., 2006; Martinon et al., 2010; Li et al., 2011). Finally, XBP1s triggers autophagy in endothelial cells (ECs) by regulating BECLIN-1 expression, which promotes ECs proliferation by inhibiting VE-cadherin transcription, and regulates human coronary artery smooth muscle cell (HCSMC) calcification through Runx2 (Zeng et al., 2009; Liberman et al., 2011; Margariti et al., 2013).
IRE1 is a type I transmembrane glycoprotein of the ER that possesses Ser/Thr protein kinase and endoribonuclease (endoRNase) functions. Under normal circumstances, IRE1 binds to BiP (Grp78) in the ER. However, ER stress disturbs interactions between IRE1 and BiP, leading to IRE1 autophosphorylation and, eventually, the activation of its endoRNase activity. In addition to mediating XBP1 mRNA splicing, overactive IRE1α initiates the regulated IRE1-dependent decay (RIDD) of cytosolic mRNAs through an XBP1 deficiency-triggered feedback-activated mechanism (So et al., 2012). RIDD then regulates many physiological processes, including degradation of mRNAs encoding a subset of ER or secretory proteins prone to misfolding (Hollien and Weissman, 2006; Han et al., 2009; Hollien et al., 2009; Oikawa et al., 2010) and the regulation of lipid metabolism genes (So et al., 2012) and fatty acid transport proteins (Coelho et al., 2013). Intriguingly, RIDD can also cause rapid decay of select microRNAs (miRs-17, 34a, 96, 125b) that normally repress translation of caspase-2, which controls the induction of apoptosis upon continued ER stress (Upton et al., 2012). However, a recent report claimed that ER stress did not cause the upregulation and activation of caspase-2 to initiate apoptosis (Sandow et al., 2014). Thus, the precise mechanisms of these actions need to be further investigated. One study demonstrated that CD59 mRNA is a cleavage target of IRE1α (Oikawa et al., 2010). Additionally, this study identified 13 novel mRNAs as candidate IRE1α cleavage targets (Oikawa et al., 2010). Importantly, these mRNAs as well as the XBP1 mRNA were confirmed to have a consensus sequence and a stem-loop structure (Oikawa et al., 2010). These findings provide potential molecular mechanisms for RIDD. However, although both IRE1α activity and ER stress are required for RIDD, XBP1 splicing can be artificially induced by the ATP analogue 1NM-PP1 (4-amino-1-tert-butyl-3-(1′-naphthyl methyl)pyrazolo(3,4-d) pyrimidine) in the absence of ER stress, suggesting that the two functions of IRE1α are dependent on different conditions (Hollien et al., 2009).
The IRE1α-XBP1 signaling pathway is highly conserved from yeast to humans. This signaling pathway can be regulated at the level of XBP1 and IRE1α. These findings have been widely described in many reviews (He et al., 2010; Hetz et al., 2011). However, the mechanism for the unconventional cytoplasmic splicing of XBP1 through IRE1α remains obscure. Recently, Yanagitani et al. (2009) found that nascent XBP1u polypeptide recruited its own mRNA to the ER membrane through a hydrophobic region within XBP1u, pulling the XBP1 mRNA close to IRE1α as a substrate for splicing. Subsequently, this author also found that an evolutionarily conserved peptide module at the C-terminus of XBP1u that results in translational pausing also results in higher splicing efficiency (Yanagitani et al., 2011). After splicing, the generated 2′,-3′-cyclic phosphate structure at the cleavage site is enzymatically joined to form the mature XBP1s mRNA (Shinya et al., 2011). The small amount of generated XBP1s mRNA is rapidly degraded under physiological conditions. In the early UPR, eukaryotic initiation factor-2α (eIF2α) phosphorylation by PERK inhibits XBP1s mRNA translation (Majumder et al., 2012). Then, both the number of XBP1s mRNAs generated by splicing and the suppression of translation result in XBP1s mRNA stabilization (Majumder et al., 2012). Along with the progression of the UPR, eIF2α is dephosphorylated by GADD34, an ER-associated phosphatase regulatory subunit, and translation is then reinitiated (Majumder et al., 2012). Finally, synthesized XBP1s protein, in turn, stimulates the transcription of XBP1 and its target genes, suggesting that a positive feedback mechanism regulates in this process through the induction of phosphorylated and dephosphorylated eIF2α (Majumder et al., 2012). In addition, XBP1u may interact with XBP1s to mediate its proteasomal degradation in the cytosol (Fig. 1).
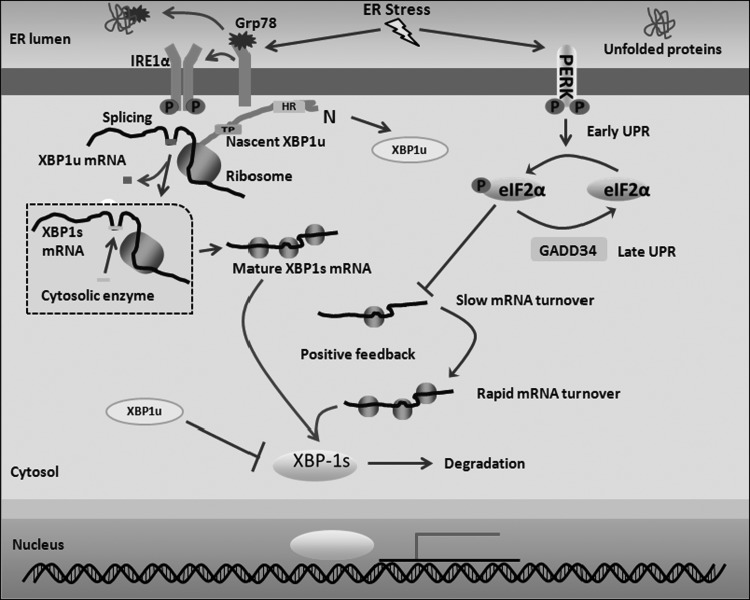
FIG. 1.
The schematic diagram of XBP1s production. Under normal circumstances, IRE1α is located on the ER membrane and binds to Grp78. Upon ER stress, IRE1α separates from Grp78 and oligomerizes, resulting in its autophosphorylation. Nascent XBP1u polypeptide recruits its own mRNA to the ER membrane through a hydrophobic region (HR) within XBP1u, facilitating the activated IRE1α to splice 26 nucleotides from the XBP1u mRNA. In addition, the C-terminus of XBP1u results in translational pausing (TP), which increases the efficiency of this process. After splicing, the generated 2′,-3′-cyclic phosphate structure at the cleavage site is enzymatically joined to form the mature XBP1s mRNA. During the early UPR, eIF2α phosphorylation by PERK inhibits XBP1s mRNA translation. Then, both the number of XBP1s mRNAs generated by splicing and translation suppression result in the stabilization and slow turnover of XBP1s mRNA. During the progression of the UPR, eIF2α is dephosphorylated by GADD34, an ER-associated phosphatase regulatory subunit, and translation is then reinitiated, resulting in rapid XBP1s mRNA turnover. Finally, synthesized XBP1s protein stimulates XBP1 transcription and its nuclear translocation to mediate the transcription of additional factors through a positive feedback mechanism. In addition, XBP1u may interact with XBP1s to mediate its proteasomal degradation in the cytosol. eIF2α, eukaryotic initiation factor-2α; ER, endoplasmic reticulum; IRE1, inositol-requiring enzyme 1; PERK, PKR-like ER kinase; UPR, unfolded protein response; XBP1, X-box binding protein 1; XBP1s, spliced XBP1; XBP1u, unspliced XBP1.
The IRE1α–XBP1 Pathway in Lipid Metabolism
XBP1 plays an important role in membrane lipid synthesis in the ER. Recently, XBP1 was also implicated in lipogenesis. One study reported that XBP1 deficiency in the liver resulted in profound compromise of de novo hepatic lipid synthesis, thereby leading to decreased serum TG, cholesterol, and free fatty acids (FFAs); however, this decrease occurred without causing hepatic steatosis, and defective assembly and secretion of VLDL particles were not observed (Lee et al., 2008). This XBP1 function is primarily mediated through two different pathways: a) XBP1s directly regulates the expression of a subset of lipogenic genes in a carbohydrate response element-binding protein (ChREBP)- and sterol regulatory element-binding protein (SREBP)-independent manner (Lee et al., 2008; So et al., 2012); and b) XBP1 ablation activates IRE1α because of the feedback regulation of IRE1α activity by the abundance of its substrate, XBP1s (Lee et al., 2008), and hyperactivated IRE1α then downregulates the expression of a group of lipid metabolism genes, including Angptl3 and the carboxylesterase 1 (Ces1) gene family, by RIDD (So et al., 2012). However, another study also reported that XBP1s could regulate TG-rich VLDL assembly and secretion in mice with a hepatocyte-specific deletion of IRE1α without affecting TG synthesis, de novo lipogenesis, or the synthesis or secretion of apolipoprotein B (apoB) under physiological conditions, which affected microsomal triglyceride transfer protein (MTP) activity by regulating protein disulfide isomerase (PDI) levels (Wang et al., 2012). Although XBP1 is an important transcription factor, this study does not illustrate how PDI is regulated. Moreover, different effects on hepatic lipid metabolism were clearly observed in the two studies. We speculate that this phenomenon may be involved in two functions. XBP1 deletion causes IRE1α overactivation through a feedback-modulated mechanism (Lee et al., 2008) that can either activate downstream signaling pathways or mediate mRNA degradation by RIDD to further complicate the phenotype observed in XBP1 knockout mice (Zhang, and Kaufman, 2008). In contrast, one recent study reported that XBP1u expression was observed in IRE1α deletion mice; however, XBP1u expression was not observed in XBP1-deletion mice. Consequently, existing XBP1u may affect hepatic lipid metabolism through an uncharacterized mechanism. Surprisingly, although XBP1 deletion in ApoE-deficient mice could significantly decrease plasma cholesterol, atherosclerotic lesion formation did not significantly decrease (So et al., 2012). Nevertheless, XBP1 clearly participates in lipogenesis in the liver. However, the precise mechanism behind this function must be defined. Additionally, XBP1s could interact with the promoter of the SREBP-1c gene following insulin treatment, and XBP1-mediated lipogenesis required SREBP-1c (Ning et al., 2011; Fang et al., 2013). In addition, XBP1s overexpression stimulated transcription by the FAS promoter (Ning et al., 2011). However, the mechanism by which XBP1 regulates lipogenesis through this pathway remains unclear. In addition, some studies have shown that mammalian target of rapamycin complex-1 (mTORC1) participates in the regulation of the lipogenic program in the liver (Porstmann et al., 2008, 2009). Another study found that the postprandial environment (i.e., rats fed a high-carbohydrate diet) promotes the activation of the IRE1-XBP1 branch of the UPR in the liver, which is partially mediated by mTORC1 (Pfaffenbach et al., 2010).
Interestingly, one study demonstrated that a circadian clock-dependent rhythmic activation of the IRE1α-XBP1 pathway in the liver with a 12-h period could influence hepatic lipid metabolism (Cretenet et al., 2010). Animals lacking a functional circadian clock exhibited constitutive activation of the IRE1α-XBP1 pathway, which might explain the perturbed lipid metabolism and TG accumulation in the liver in these animals. First, XBP1 can directly bind to associated lipid metabolism genes. Second, the activities of ER-resident enzymes, such as HMGCR and SCD1, may be modified because of the activation of the IRE1α-XBP1 pathway. Third, the author also reported that SREBP target gene expression is altered (Cretenet et al., 2010). Thus, IRE1α-XBP1 may disturb lipid metabolism through the SREBP pathway. Fourth, blood glucose can be affected by circadian clock variations. Accordingly, activation of the IRE1α-XBP1 pathway disturbs hepatic lipid metabolism as a result of the increased glucose concentration. Fifth, a recent study demonstrated the circadian-dependent involvement of histone deacetylase 3 (HDAC3) in regulating hepatic lipid metabolism (Feng et al., 2011). However, Tao et al. (2011) demonstrated that XBP1 physically and genetically interacts with the histone deacetylase Rpd3 complex. It will be interesting to determine whether a functional correlation between XBP1s and HDAC3 exists. In summary, many XBP1-mediated pathways under circadian clock conditions may participate in lipogenesis in the liver.
In addition, Sha et al. (2009) also reported that XBP1 was indispensable for adipogenesis in mouse embryonic fibroblasts and preadipocytes. Interestingly, this author found that XBP1 could also mediate adipocyte differentiation. The underlying mechanism involved CCAAT/enhancer-binding protein β (C/EBPβ), a key early adipogenic factor that could regulate XBP1 expression by directly binding to its proximal promoter region and, in turn, activating the master adipogenic factor C/EBPα during adipogenesis (Hollien and Weissman, 2006). Additionally, XBP1 regulated C/EBPα expression by binding to its promoter (Sha et al., 2009). This mechanism is consistent with other studies. For example, fat deposits were absent in XBP1-deficient neonates (Lee et al., 2005), and XBP1 was highly expressed in both embryonic adipose deposits (Clauss et al., 1993) and white adipose cells (Kajimura et al., 2008). XBP1 inhibition in preadipocytes resulted in incomplete adipogenesis in vitro (Basseri et al., 2009). However, a recent study reported that IRE1α activation or deletion did not alter adipocyte differentiation in preadipocytes (Han et al., 2013). Thus, future studies are required to delineate the function of the IRE1α-XBP1 pathway in adipocyte differentiation. Finally, IRE1β limited chylomicron production through MTP mRNA degradation in intestinal epithelial cells (Iqbal et al., 2008), suggesting a physiological role for IRE1β in lipid metabolism.
Taken together, although these studies suggest that the IRE1α-XBP1 signaling pathway participates in lipid metabolism under different conditions, several questions must be answered. The essential mechanism by which XBP1s induces the transcription of genes encoding lipogenic enzymes remains unknown (Fig. 2).
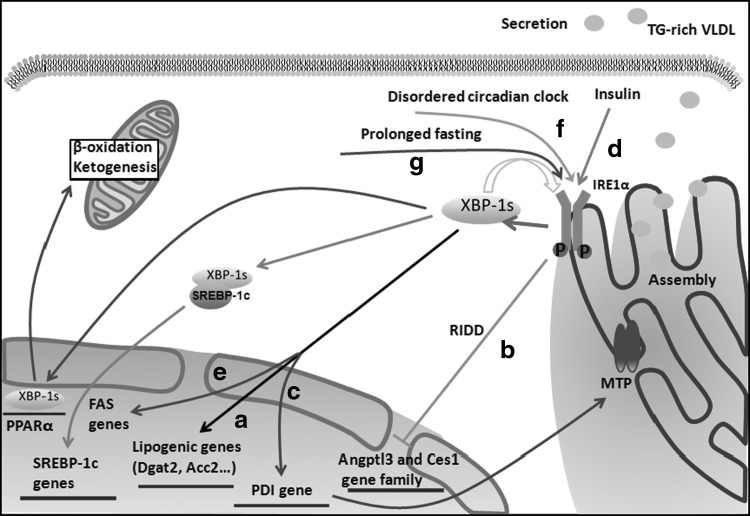
FIG. 2.
The IRE1α-XBP1 pathway in hepatic lipogenesis. (a) XBP1s directly regulates the expression of a subset of lipogenic genes. (b) XBP1 ablation activates IRE1α because of feedback regulation of IRE1α activity caused by the abundance of its substrate XBP1s. Hyperactivated IRE1α then downregulates the expression of a group of lipid metabolism genes, including Angptl3 and the carboxylesterase1 (Ces1) gene family, by regulated IRE1-dependent decay (RIDD). (c) XBP1s regulates TG-rich VLDL assembly and secretion in hepatocyte-specific IRE1α deletion mice under physiological conditions, which affects MTP activity by regulating PDI at the transcriptional level. (d) XBP1s can interact with the promoter of the SREBP-1c gene following insulin treatment, thereby mediating lipogenesis. (e) XBP1s overexpression stimulates transcription by the FAS promoter. (f) A disordered circadian clock results in the constitutive activation of the IRE1α-XBP1 pathway, thus influencing hepatic lipid metabolism. (g) Hepatic IRE1α-XBP1 signaling is activated by prolonged fasting. Then, XBP1s can directly bind to the endogenous PPARα promoter and upregulate PPARα expression, thereby modulating mitochondrial β-oxidation and ketogenesis. MTP, microsomal triglyceride transfer protein; PPARα, peroxisome proliferator activator receptor α; PDI, protein disulfide isomerase; TG, triglyceride; VLDL, very-low-density lipoprotein.
The IRE1α-XBP1 Pathway in Glucose Metabolism and Obesity
A previously published article reported that the IRE1α-XBP1 pathway is involved in obesity, insulin resistance, and type 2 diabetes through the IRE1α-c-Jun N-terminal kinase (JNK) signaling axis (Sha et al., 2011). Nonetheless, the roles of JNK1 in both obesity and insulin resistance are tissue- and cell type-dependent. For example, global JNK1 deficiency in obese mice improved insulin sensitivity and enhanced insulin receptor signaling (Hirosumi et al., 2002). Additionally, JNK1 in hematopoietic cells of adipose tissue increased insulin resistance (Solinas et al., 2007; Sabio et al., 2008). However, JNK1 was required to maintain insulin sensitivity and prevent hepatic steatosis in hepatocytes (Sabio et al., 2009). Furthermore, a recent study demonstrated that lipid (particularly diacylglycerol (DAG)) accumulation, but not JNK activation, was required for ER stress to cause hepatic insulin resistance and glucose intolerance when on a high-fructose diet (Chan et al., 2013). Thus, the underlying molecular mechanisms of the role of the IRE1α-JNK signaling pathway in insulin resistance and obesity remain unclear and require further studies.
Adiponectin is an important insulin-sensitizing hormone, and its serum level negatively correlates with insulin resistance and obesity (Turer and Scherer, 2012). Nuclear XBP1s can improve insulin sensitivity through regulating the expression of UPR target genes involved in adiponectin multimerization (Sha et al., 2014). Indeed, a severe defect in the capacity of XBP1s to translocate into the nucleus has been observed under obesity conditions, and this defect is crucial for the development of ER stress and type 2 diabetes in obesity (Park et al., 2010). When XBP1s was reactivated in the liver by forced ectopic expression in severely obese and diabetic mice, the blood glucose levels were reduced to euglycemia (Zhou et al., 2011). Additionally, deletion of XBP1 in mouse adipocytes resulted in obesity during lactation (Gregor et al., 2013). In addition, XBP1s overexpression in adipocytes also could improve glucose tolerance and insulin sensitivity in both lean and obese mice (Sha et al., 2014). Recently, several studies reported that p38 mitogen-activated protein kinase (p38 MAPK), a stress-activated protein kinase, and PI3K, an essential mediator of the metabolic actions of insulin that is composed of catalytic (p110 or p110) and regulatory (p85, p85, or p55) subunits, have also been implicated in modulating the UPR by regulating the nuclear translocation of XBP1s (Park et al., 2010; Winnay et al., 2010; Lee et al., 2011b). For example, it has been shown that p38 MAPK phosphorylates XBP1s on its Thr48 and Ser61 residues, greatly enhancing its nuclear migration in mice (Lee et al., 2011b). Insulin promoted p85α and p85β association with XBP-1s by disrupting their heterodimerization, which subsequently facilitated XBP1s nuclear translocation independent of PI3K catalytic activity (Park et al., 2010). Most recently, a study further confirmed that bromodomain-containing protein 7 (BRD7), a subunit of the polybromo-associated BRG1-associated factor (PBAF) complex, interacted with p85a/p85b/XBP1s and that insulin increased the formation of the BRD7-p85-XBP1s complex, which subsequently increased the nuclear translocation and activity of XBP1s (Park et al., 2014). Intriguingly, researchers also found that both p38 MAPK activity and the interaction between p85 and XBP1s were markedly reduced in obese mice. Thus, these findings indicate that obesity actually creates a relative XBP1s-deficient condition through decreasing the nuclear translocation of XBP1s by affecting p38 MAPK activity and the interaction between p85 and XBP1s. Further research should investigate whether other insulin signaling- or UPR-related molecules function through similar mechanisms to mediate glucose homeostasis. Expression of UPR target genes should be downregulated upon obesity, leading to relative XBP1 deficiency. Accordingly, this downregulation should increase insulin resistance. However, a recent study reported that XBP1 knockout mice were protected from hepatic insulin resistance despite increased hepatic ER stress and JNK activation (Jurczak et al., 2012). This finding is paradoxical. As mentioned, XBP1 deficiency results in profound compromise of de novo hepatic lipid synthesis. Additionally, XBP1s deletion led to IRE1α overactivation, thereby downregulating the expression of a group of lipid metabolism genes by RIDD. This downregulation will lead to a decrease in lipids. Insulin resistance has been attributed to both increased ER stress and lipid accumulation. Thus, we propose that IRE1α-XBP1 pathway-mediated lipogenesis is dominant in models of ER stress. In addition, nuclear XBP1s expression is increased in the livers of obese mice (Kammoun et al., 2009). Mice with a liver-specific deletion of p85α exhibited improved hepatic and peripheral insulin sensitivities (Taniguchi et al., 2006). p85α was also required for JNK activation in insulin resistance states in high-fat diet-induced obese mice (Taniguchi et al., 2007). These reports are inconsistent with the above-mentioned reports. Thus, it is necessary to continue examining the crosstalk between the IRE1α-XBP1 pathway and insulin signaling (Fig. 3).
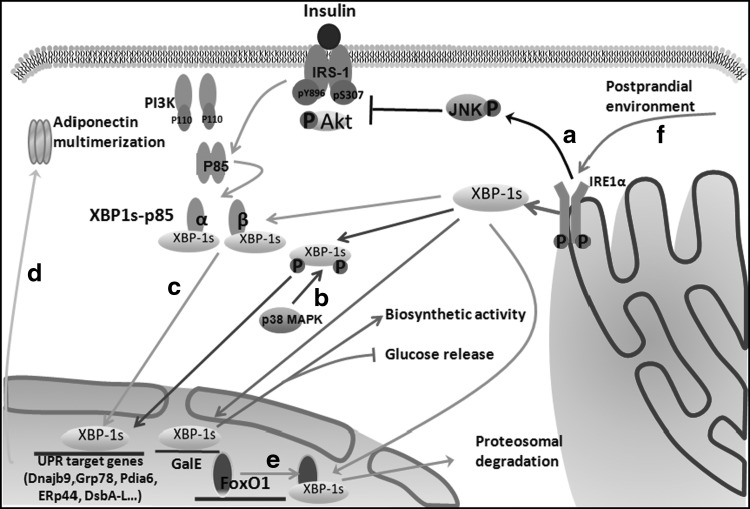
FIG. 3.
The IRE1α-XBP1 pathway in glucose homeostasis. (a) IRE1α-JNK signaling is activated by ER stress, which reduces IRS1 (pY896) tyrosine phosphorylation and Akt phosphorylation whereas enhancing IRS1 (pS307) serine phosphorylation, thereby increasing insulin resistance. (b) p38 MAPK phosphorylates the spliced form of XBP1 on residues Thr48 and Ser61 and greatly enhances its nuclear migration. Then, nuclear XBP1s induces the expression of UPR target genes. (c) Insulin promotes p85α and p85β (subunits of PI3K) association with XBP1s by disrupting their heterodimerization, which subsequently facilitates XBP1s nuclear translocation independent of PI3K catalytic activity. Nuclear XBP1s then induces the expression of UPR target genes. (d) During the UPR, nuclear XBP1s can improve insulin sensitivity by regulating the expression of UPR target genes that are involved in adiponectin multimerization in adipocytes. (e) In the nucleus, XBP1s also directly interacts with FoxO1, a transcription factor involved in gluconeogenesis resulting in its proteasomal degradation and thereby promoting glucose tolerance in the liver. (f) IRE1α-XBP1 signaling is activated by the postprandial environment. XBP1s regulates UDP-galactose-4-epimerase (GalE) expression at the transcriptional level, thereby increasing its biosynthetic activity and reducing hepatic glucose release. FoxO1, Forkhead box O1; JNK, c-Jun N-terminal kinase; p38 MAPK, p38 mitogen-activated protein kinase.
Interestingly, XBP1s also directly interacted with the FoxO1 transcription factor in obese mice, thereby regulating glucose homeostasis independent of its effects on ER folding capacity and insulin signaling (Zhou et al., 2011). For example, modest hepatic XBP1s overexpression improved serum glucose concentrations without improving insulin signaling or ER folding capacity in insulin deficiency or insulin resistance mouse models through proteasome-mediated degradation of FoxO1 (Zhou et al., 2011). Additionally, an XBP1s mutant unable to bind DNA could reduce serum glucose concentrations and increase glucose tolerance in severely insulin-resistant obese mice due to FoxO1 accumulation (Zhou et al., 2011). In addition, overexpression of XBP1s in pancreatic α cells treated with insulin decreased the nuclear FoxO1 level (Akiyama et al., 2013). These studies reveal an unexpected function of XBP1s in improving glucose homeostasis in addition to regulating gene expression as a transcription factor and, hence, raise the question of whether this function also exists in other important metabolic organs, such as adipose tissue. A previous study reported that XBP1u interacted with FoxO1 in the cytosol to activate autophagy in cancer cells (Zhao et al., 2013). Thus, XBP1u and XBP1s interact with FoxO1 in the cytosol and in the nucleus, respectively, resulting in different effects on various cells. In summary, XBP1 interactions with FoxO1 may be cell type dependent (Fig. 3).
The IRE1α-XBP1 pathway is also involved in insulin and glucagon secretion, thus regulating glucose homeostasis. XBP1 deletion in a mouse model markedly decreased the amount of insulin granules in β-cells, increased the serum proinsulin:insulin ratio, impaired proinsulin processing, inhibited cell proliferation, and inhibited glucose-stimulated insulin secretion, thereby leading to glucose intolerance and modest hyperglycemia (Lee et al., 2011a). Mechanistically, XBP1 deficiency not only weakened the ER stress response in β-cells, but also caused constitutive IRE1α hyperactivation, thus degrading a subset of mRNAs encoding proinsulin processing enzymes by RIDD (Lee et al., 2011a). The total effect of XBP1 deficiency manifested as β-cell dysfunction. In addition, one study found that XBP1 deficiency in α-cells resulted in altered insulin signaling and dysfunctional glucagon secretion in vivo and in vitro (Akiyama et al., 2013). α-Cell-specific XBP1 knockout mice were not able to inhibit glucagon secretion after treatment with glucose and exhibited mild insulin resistance and glucose intolerance (Akiyama et al., 2013). XBP1 knockdown in α-cells led to the activation of the IRE1α-XBP1 signaling pathway, a reduction of tyrosine IRS1 and the phosphorylation of Akt, whereas enhancing IRS1 serine phosphorylation reduced glucagon secretion inhibition after insulin treatment under high-glucose conditions (Akiyama et al., 2013). Intriguingly, rat β-cells were sensitized to interleukin-1 beta (IL-1β) in insulin resistance and lipid accumulation related with obesity, generating a severe inflammatory response through IRE1α-XBP1 activation contributing to the pathogenesis of type 1 diabetes (Miani et al., 2012). Additionally, dominant-negative hepatic nuclear factor 1α (DN HNF1α) expression sensitized the β-cells to ER stress by directly downregulating XBP1 transcription (Kirkpatrick et al., 2011). These findings suggest that XBP1 is important for the regulation of pancreatic β- and α-cell functions.
The IRE1α-XBP1 Pathway in Vascular Diseases
Atherosclerosis
Atherosclerosis is a complication caused by metabolic disorders. The pathophysiology of atherosclerosis is involved in the inflammatory response/apoptosis and autophagy in ECs, the inflammatory response/cell death in macrophages and foam cell formation. These pathological processes are prevalent in atherosclerotic cardiovascular disease. XBP1, particularly XBP1s, is abundantly found at branch points and areas of atherosclerotic lesions in the arteries of ApoE−/− mice (Zeng et al., 2009). Sustained activation of XBP1s induces atherosclerosis in an aortic isograft model (Zeng et al., 2009).
Recently, some studies have reported that the IRE1α-XBP1 pathway is involved in macrophage cell death. For example, Martinet et al. (2007) demonstrated that XBP1 participated in NO-induced ER stress in both macrophages and smooth muscle cells (SMCs), most likely through the inhibition of protein synthesis, but only induced macrophage cell death without affecting SMC viability. Additionally, aging also promoted ER stress-induced apoptosis in macrophages, which involved alterations in the IRE1α-XBP1 signaling pathway (Song et al., 2013). CD36-mediated oxidized low-density lipoprotein (ox-LDL) uptake in macrophages triggered the ER stress response, which, in turn, played a critical role in CD36 upregulation, thus enhancing foam cell formation by taking up more ox-LDL (Yao et al., 2014). IRE1, PERK, XBP1, and Grp-78 expression levels were upregulated in the process. Yao et al. (2010) found that minimally modified LDL induced intimal foam cell formation, which was promoted by ER stress. Another study demonstrated that the IRE1α-XBP1 pathway participated in this process, which was mediated by toll-like receptor 4 (TLR4) (Yao et al., 2012). However, Yao et al. (2013) also demonstrated that ER stress-related proteins, particularly ATF6 and its downstream molecule, CHOP, were involved in ox-LDL-induced cholesterol accumulation and apoptosis in macrophages. Although these findings are discrepant, there is no doubt that the IRE1α-XBP1 pathway participates in foam cell formation. In addition, the IRE1α-XBP1 signaling pathway is also involved in the inflammatory response in macrophages. TLR2 and TLR4 activation through lipopolysaccharide engaged the IRE1α-XBP1 pathway in macrophages, and this activation was required for the optimal, sustained expression of a subset of inflammatory cytokines, including interleukin-6 (IL-6) and tumor necrosis factor alpha (TNFα) (Martinon et al., 2010).
In addition, XBP1 deletion led to increased expression of vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) in retinal vascular ECs (Li et al., 2011). Conversely, forced expression of XBP1s abated the TNFα-induced phosphorylation of IκBα, IKK, and NF-κB p65, accompanied by decreased NF-κB activity and reduced adhesion molecule expression (Li et al., 2011). These findings suggest that XBP1 may have an anti-inflammatory role in ECs. Recently, disturbed flow was found to cause XBP1s overexpression in human umbilical vein ECs (Zeng et al., 2009). XBP1s overexpression induced EC apoptosis, which was involved in VE-cadherin downregulation (Zeng et al., 2009). Another study found that forced expression of XBP1s mediated VE-cadherin downregulation through transcriptional suppression and matrix metalloproteinase-mediated degradation (Zeng et al., 2009). Finally, the VE-cadherin decrease-mediated EC apoptosis may be dependent on caspase activation (Zeng et al., 2009). Accordingly, XBP1 splicing and sustained activation through disturbed flow leads to EC apoptosis. Most recently, a study reported that endostatin-induced XBP1 mRNA splicing triggered an autophagic response in ECs, which was involved in autophagic vesicle formation, leading to EC survival or apoptosis (Margariti et al., 2013). XBP1s could regulate BECLIN-1 expression by binding directly to its promoter at the region from −537 to −755 nt, and activated BECLIN-1 then led to microtubule-associated protein 1 light chain 3-β (LC3-βII) activation and, in turn, induced the autophagic response (Margariti et al., 2013). Additionally, activation of IRE1α-XBP1 signaling could regulate autophagy upon ER stress in human neuroglioma cell lines (Pehar et al., 2012). During this process, IRE1α-XBP1 signaling first induces an acetyl-CoA influx into the ER lumen through the membrane transporter AT-1; the acetyl-CoA levels in the lumen of the ER determine the acetylation status of Atg9A, which serves as the last signal for the induction of autophagy-dependent ERAD (Pehar et al., 2012). However, Vidal et al. (2012) found that XBP1deficiency in cells led to autophagy through FoxO1 upregulation, which may contribute to autophagy-mediated clearance of mutant huntingtin in Huntington's disease, consistent with a previous finding showing the negative regulation FoxO1, the transcription factor of XBP1s, in hepatic cells (Zhou et al., 2011). Thus, these findings indicate that both upregulation and downregulation of XBP1s induce autophagy, depending on the distinct signal pathways active in different cell lines. In an article recently published in Cell Research, XBP1u reportedly interacted with FoxO1 to trigger autophagy in cancer cells (Zhao et al., 2013). Although XBP1 can induce autophagy through different pathways in cells, these pathways require further study to uncover their existence in ECs or other atherosclerosis-related cells. In summary, XBP1s, which plays different roles in diverse cellular events, is involved in atherosclerotic initiation and progression (Fig. 4).
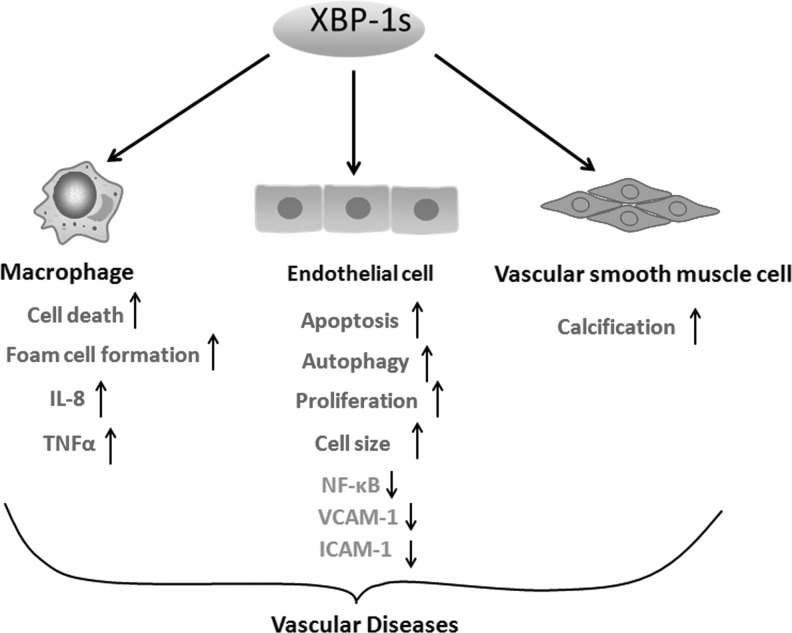
FIG. 4.
XBP1s in vascular diseases. XBP1s mediates macrophage cell death, foam cell formation, and IL-8 and TNFα up-expression. XBP1s attenuates NF-κB, VCAM-1, and ICAM-1 expression in retinal vascular endothelial cells. XBP1s induces endothelial cell apoptosis, autophagy and proliferation, and increases cell size. XBP1s results in smooth muscle cell calcification. These different cellular events are involved in vascular diseases, including atherosclerotic initiation and progression and tissue and organ ischemia. ICAM-1, intercellular adhesion molecule-1; IL-8, interleukin-8; TNFα, tumor necrosis factor alpha; VCAM-1, vascular cell adhesion molecule-1.
Ischemia
XBP1 can also ameliorate ischemic injury by regulating endothelial proliferation and angiogenesis. Zeng et al. (2009) found that transient activation of XBP1 splicing may increase EC proliferation. Another study confirmed that transient, XBP1s following treatment with vascular endothelial growth factor, which is involved in the kinase insert domain receptor (KDR)/XBP1u/IRE1α interaction, regulated EC proliferation in a PI3K/Akt/GSK3β/β-catenin/E2F2-dependent manner (Zeng et al., 2013). In animal models, global knockout of XBP1 reduced vessel formation during embryonic development (Zeng et al., 2013). Early stage retinal vasculogenesis and angiogenesis in ischemic muscles were affected in EC-specific XBP1 knockout mice (Zeng et al., 2013). These findings are consistent with previous reports showing that XBP1 activation can ameliorate cerebral ischemia/reperfusion injury (Urban et al., 2009; Nakka et al., 2010; Ibuki et al., 2012). In addition, transient XBP1activation resulted in an EC cell size increase, but not in an Akt/GSK/β-catenin/E2F2-dependent manner (Zeng et al., 2013). How XBP1 regulates EC cell size will be the focus of a future investigation. In addition, fewer vascular smooth muscle cells are present in ischemic tissues in XBP1 knockout mice, which may be due to decreased SMC recruitment (Zeng et al., 2013). Runx2, which is a master transcription factor essential for osteoblast and chondrocyte differentiation, can regulate the expression of bone-related proteins that are important for calcification (Ducy et al., 1997; Komori et al., 1997; Inada et al., 1999). XBP1s can bind to the Runx2 promoter at −596 to −591 in bone morphogenetic protein-2 (BMP-2)-treated HCSMCs, thus increasing Runx2 expression to modulate HCSMC calcification (Liberman et al., 2011) (Fig. 4).
Heart failure
Several studies have confirmed the existence of XBP1s in patients with heart failure (Okada et al., 2004; Dally et al., 2009; Sawada et al., 2010). Additionally, XBP1s expression is significantly increased in a heart failure mouse model (Jiao et al., 2012). In rat neonatal cultured cardiomyocytes, hypoxia increases XBP1 mRNA splicing (Thuerauf et al., 2006). However, XBP1 downregulation augments hypoxia/reoxygenation-induced apoptosis. In addition, XBP1s can regulate the expression of brain natriuretic peptide, which is a non-UPR-target gene, in cardiomyocytes by binding to a novel AP1/CRE-like element (Sawada et al., 2010). These findings suggest that XBP1 may be involved in the initiation and progression of heart failure.
XBP1s-Dependent Transcriptional Reprogramming Involving Metabolic Diseases
As described above, XBP1s translocates into the nucleus to initiate transcriptional programs that regulate a subset of UPR- and non-UPR-associated genes involved in metabolic diseases. In the inflammatory responses of macrophages and EC, XBP1s can regulate a group of inflammatory cytokines, including IL-6, TNFα, TNFβ, IL-8, MCP1, CXCL3, VCAM-1, and ICAM-1. Additionally, XBP1s triggers autophagy in ECs by regulating BECLIN-1 expression, thereby determining EC fate. In addition, XBP1s inhibits VE-cadherin transcription, promoting EC proliferation. Finally, Runx2 is modulated by XBP1s, regulating HCSMC calcification. Microarray analyses indicated that various lipid metabolism genes, such as SCD1, Fdps, Cyp51, Sqle, Pmvk, Mvk, Idi1, Sc4mol, Fdft1, and Hsd17b7, are regulated by XBP1s (Lee et al., 2008; So et al., 2012). Experimental studies confirmed that SREBP-1c, FAS, C/EBPα, PDI, and SCD1 are XBP1s target genes. Furthermore, XBP1s protein directly regulates the expression of several ER chaperones, including Grp78, DsbA-L, ERp44, and Pdia6, contributing to the amelioration of insulin resistance (Sha et al., 2014). One study reported that XBP1 drove plasma cell differentiation by regulating many genes that encode secretory pathway components (Shaffer et al., 2004). Using genome-wide approaches, another study found that a core group of XBP1s target genes is involved in the constitutive maintenance of ER function (Acosta-Alvear et al., 2007). Hence, XBP1s may regulate almost all of the genes involved in the physiological function of the ER. XBP1s also serves as a regulator of liver metabolic reprogramming under physiological conditions. When transforming the postprandial environment into prolonged fasting states, the liver experiences an extensive metabolic reprogramming that is required for the switch from anabolism to catabolism. The postprandial environment, which is characterized by an increase in protein synthesis and a switch from glucose production to glucose assimilation, induces XBP1 splicing. Using a liver-specific XBP1s overexpression mouse model, researchers found that XBP1s was sufficient to provoke a metabolic switch characteristic of the postprandial state, namely, increased biosynthetic activity, reduced hepatic glucose release, and enhanced glucose assimilation (Deng et al., 2013). Another study identified UDP-galactose-4-epimerase (GalE) as a direct transcriptional target of XBP1s and as the key mediator of this effect (Deng et al., 2013). Similarly, enhanced glucagon and FFAs mediated the activation of the hepatic IRE1α-XBP1 pathway in a prolonged fasting state. Then, XBP1s could directly bind to the endogenous peroxisome proliferator activator receptor α (PPARα) promoter and upregulate PPARα expression, thereby modulating the mitochondrial β-oxidation and ketogenesis programs to generate energy under a fasting state (Shao et al., 2014). Interestingly, hepatic IRE1α can respond to both anabolic and catabolic states, which are involved in IRE1α-Xbp1s-GalE and IRE1α-XBP1s-PPARα axis signaling. These findings may provide an unacknowledged mechanism that underlies the pathological progression of metabolic disorders.
Taken together, many genes involved in metabolic diseases and physiological conditions are modulated by XBP1s. We propose that many more genes will be identified as XBP1s targets in the future. Further studies are also required to identify the stimuli in pathological and physiological conditions that modulate these targets, thereby providing researchers with a precise mechanism of XBP1s function in metabolic diseases.
Conclusions
The recognition that metabolic disorders are associated with the IRE1α-XBP1 pathway, which regulates the pathogenesis of lipid metabolism, glucose metabolism, obesity, and atherosclerosis, has extended our appreciation of the etiology of metabolic syndromes. XBP1s has been shown to be an important nuclear transcription factor and regulates non-UPR and UPR genes involved in metabolic disorders, revealing crucial links between ER stress and different components of metabolic syndromes at the transcriptional level. Thus, there are two primary approaches to targeting IRE1α-XBP1 for the treatment of metabolic disorders. The first approach involves regulating the IRE1α-XBP1 pathway at multiple levels, including IRE1α activation by its interacting proteins, RIDD, the IRE1α-JNK signaling axis, transcriptional XBP1 induction, XBP1s nuclear translocation, and XBP1s posttranslational modifications. Several kinase inhibitors, including sunitinib, can directly activate IRE1α and lead to XBP1 splicing (Korennykh et al., 2009). The second approach is to intervene in XBP1-dependent transcriptional reprogramming, including non-UPR and UPR XBP1s regulatory genes. Although targeting the IRE1α-XBP1 pathway seems to be a promising potential therapy for metabolic diseases, there are several limitations of our knowledge: (1) How is the IRE1α-XBP1 pathway activated under physiological or pathological conditions? (2) What is the XBP1s binding site in the promoter regions of these lipid metabolism genes? (3) What is the physiological function of XBP1u? (4) How can we deliver the agent to the targeted tissues? Consequently, only a better understanding of the mechanisms that underlie IRE1α-XBP1 metabolic disorders may provide novel therapeutic targets for metabolic diseases.
Acknowledgments
The authors gratefully acknowledge the financial support from the National Natural Sciences Foundation of China (No. 30570958, 81270360) and the Key Project of Department of Hunan Province Science and Technology Plan (No. 2014FJ2012).
Disclosure Statement
No competing financial interests exist.
References
- Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N.H., Arias C., Lennon C.J., Kluger Y., and Dynlacht B.D. (2007). XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell 27,53–66 [DOI] [PubMed] [Google Scholar]
- Akiyama M., Liew C.W., Lu S., Hu J., Martinez R., Hambro B., Kennedy R.T., and Kulkarni R.N. (2013). X-box binding protein 1 is essential for insulin regulation of pancreatic alpha-cell function. Diabetes 62,2439–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon I.V., Barrington R.A., Jackowski S., Mori K., and Brewer J.W. (2012). The specialized unfolded protein response of B lymphocytes: ATF6alpha-independent development of antibody-secreting B cells. Mol Immunol 51,347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basseri S., Lhotak S., Sharma A.M., and Austin R.C. (2009). The chemical chaperone 4-phenylbutyrate inhibits adipogenesis by modulating the unfolded protein response. J Lipid Res 50,2486–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamron S., Hadar R., Iwawaky T., So J.S., Lee A.H., and Tirosh B. (2014). Regulated IRE1-dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur J Immunol 44,867–876 [DOI] [PubMed] [Google Scholar]
- Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., and Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415,92–96 [DOI] [PubMed] [Google Scholar]
- Chan S.M., Sun R.Q., Zeng X.Y., Choong Z.H., Wang H., Watt M.J., and Ye J.M. (2013). Activation of PPARalpha ameliorates hepatic insulin resistance and steatosis in high fructose-fed mice despite increased endoplasmic reticulum stress. Diabetes 62,2095–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss I.M., Gravallese E.M., Darling J.M., Shapiro F., Glimcher M.J., and Glimcher L.H. (1993). In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev Dyn 197,146–156 [DOI] [PubMed] [Google Scholar]
- Coelho D.S., Cairrao F., Zeng X., Pires E., Coelho A.V., Ron D., Ryoo H.D., and Domingos P.M. (2013). Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep 5,791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet G., Le Clech M., and Gachon F. (2010). Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab 11,47–57 [DOI] [PubMed] [Google Scholar]
- Dally S., Monceau V., Corvazier E., Bredoux R., Raies A., Bobe R., del Monte F., and Enouf J. (2009). Compartmentalized expression of three novel sarco/endoplasmic reticulum Ca2+ATPase 3 isoforms including the switch to ER stress, SERCA3f, in non-failing and failing human heart. Cell Calcium 45,144–154 [DOI] [PubMed] [Google Scholar]
- Deng Y., Wang Z.V., Tao C., Gao N., Holland W.L., Ferdous A., Repa J.J., Liang G., Ye J., Lehrman M.A., Hill J.A., Horton J.D., and Scherer P.E. (2013). The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest 123,455–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Yan J., Zhu J., Zhong H., Lu Q., Wang Z., Huang C., and Ye Q. (2003). Ligand-independent activation of estrogen receptor alpha by XBP-1. Nucleic Acids Res 31,5266–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V., Ridall A.L., and Karsenty G. (1997). Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89,747–754 [DOI] [PubMed] [Google Scholar]
- Fang D.L., Wan Y., Shen W., Cao J., Sun Z.X., Yu H.H., Zhang Q., Cheng W.H., Chen J., and Ning B. (2013). Endoplasmic reticulum stress leads to lipid accumulation through upregulation of SREBP-1c in normal hepatic and hepatoma cells. Mol Cell Biochem 381,127–137 [DOI] [PubMed] [Google Scholar]
- Fang Y., Yan J., Ding L., Liu Y., Zhu J., Huang C., Zhao H., Lu Q., Zhang X., Yang X., and Ye Q. (2004). XBP-1 increases ERalpha transcriptional activity through regulation of large-scale chromatin unfolding. Biochem Biophys Res Commun 323,269–274 [DOI] [PubMed] [Google Scholar]
- Feng D., Liu T., Sun Z., Bugge A., Mullican S.E., Alenghat T., Liu X.S., and Lazar M.A. (2011). A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331,1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargalovic P.S., Gharavi N.M., Clark M.J., Pagnon J., Yang W.P., He A., Truong A., Baruch-Oren T., Berliner J.A., Kirchgessner T.G., and Lusis A.J. (2006). The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol 26,2490–2496 [DOI] [PubMed] [Google Scholar]
- Gregor M.F., Misch E.S., Yang L., Hummasti S., Inouye K.E., Lee A.H., Bierie B., and Hotamisligil G.S. (2013). The role of adipocyte XBP1 in metabolic regulation during lactation. Cell Rep 3,1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Lerner A.G., Vande Walle L., Upton J.P., Xu W., Hagen A., Backes B.J., Oakes S.A., and Papa F.R. (2009). IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138,562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Murthy R., Wood B., Song B., Wang S., Sun B., Malhi H., and Kaufman R.J. (2013). ER stress signalling through eIF2alpha and CHOP, but not IRE1alpha, attenuates adipogenesis in mice. Diabetologia 56,911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., and Ron D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397,271–274 [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., and Mori K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10,3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Sun S., Sha H., Liu Z., Yang L., Xue Z., Chen H., and Qi L. (2010). Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr 15,13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Martinon F., Rodriguez D., and Glimcher L.H. (2011). The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev 91,1219–1243 [DOI] [PubMed] [Google Scholar]
- Hirosumi J., Tuncman G., Chang L., Gorgun C.Z., Uysal K.T., Maeda K., Karin M., and Hotamisligil G.S. (2002). A central role for JNK in obesity and insulin resistance. Nature 420,333–336 [DOI] [PubMed] [Google Scholar]
- Hollien J., Lin J.H., Li H., Stevens N., Walter P., and Weissman J.S. (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186,323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J., and Weissman J.S. (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313,104–107 [DOI] [PubMed] [Google Scholar]
- Huh W.J., Esen E., Geahlen J.H., Bredemeyer A.J., Lee A.H., Shi G., Konieczny S.F., Glimcher L.H., and Mills J.C. (2010). XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology 139,2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibuki T., Yamasaki Y., Mizuguchi H., and Sokabe M. (2012). Protective effects of XBP1 against oxygen and glucose deprivation/reoxygenation injury in rat primary hippocampal neurons. Neurosci Lett 518,45–48 [DOI] [PubMed] [Google Scholar]
- Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., Ochi T., Endo N., Kitamura Y., Kishimoto T., and Komori T. (1999). Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn 214,279–290 [DOI] [PubMed] [Google Scholar]
- Iqbal J., Dai K., Seimon T., Jungreis R., Oyadomari M., Kuriakose G., Ron D., Tabas I., and Hussain M.M. (2008). IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab 7,445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Q., Takeshima H., Ishikawa Y., and Minamisawa S. (2012). Sarcalumenin plays a critical role in age-related cardiac dysfunction due to decreases in SERCA2a expression and activity. Cell Calcium 51,31–39 [DOI] [PubMed] [Google Scholar]
- Jurczak M.J., Lee A.H., Jornayvaz F.R., Lee H.Y., Birkenfeld A.L., Guigni B.A., Kahn M., Samuel V.T., Glimcher L.H., and Shulman G.I. (2012). Dissociation of inositol-requiring enzyme (IRE1alpha)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem 287,2558–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S., Seale P., Tomaru T., Erdjument-Bromage H., Cooper M.P., Ruas J.L., Chin S., Tempst P., Lazar M.A., and Spiegelman B.M. (2008). Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev 22,1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura D., and Bevan M.J. (2008). Endoplasmic reticulum stress regulator XBP-1 contributes to effector CD8+ T cell differentiation during acute infection. J Immunol 181,5433–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammoun H.L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., Ferre P., and Foufelle F. (2009). GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest 119,1201–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A., Lee A.H., Franke A., Glickman J.N., Zeissig S., Tilg H., Nieuwenhuis E.E., Higgins D.E., Schreiber S., Glimcher L.H., and Blumberg R.S. (2008). XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134,743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick C.L., Wiederkehr A., Baquie M., Akhmedov D., Wang H., Gauthier B.R., Akerman I., Ishihara H., Ferrer J., and Wollheim C.B. (2011). Hepatic nuclear factor 1alpha (HNF1alpha) dysfunction down-regulates X-box-binding protein 1 (XBP1) and sensitizes beta-cells to endoplasmic reticulum stress. J Biol Chem 286,32300–32312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., and Kishimoto T. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89,755–764 [DOI] [PubMed] [Google Scholar]
- Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., Stroud R.M., and Walter P. (2009). The unfolded protein response signals through high-order assembly of Ire1. Nature 457,687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Chu G.C., Iwakoshi N.N., and Glimcher L.H. (2005). XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24,4368–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Heidtman K., Hotamisligil G.S., and Glimcher L.H. (2011a). Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci U S A 108,8885–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Scapa E.F., Cohen D.E., and Glimcher L.H. (2008). Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320,1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., and Ozcan U. (2014). Unfolded protein response signaling and metabolic diseases. J Biol Chem 289,1203–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Sun C., Zhou Y., Lee J., Gokalp D., Herrema H., Park S.W., Davis R.J., and Ozcan U. (2011b). p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat Med 17,1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., and Kaufman R.J. (2002). IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16,452–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J.J., and Zhang S.X. (2011). Preconditioning with endoplasmic reticulum stress mitigates retinal endothelial inflammation via activation of X-box binding protein 1. J Biol Chem 286,4912–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman M., Johnson R.C., Handy D.E., Loscalzo J., and Leopold J.A. (2011). Bone morphogenetic protein-2 activates NADPH oxidase to increase endoplasmic reticulum stress and human coronary artery smooth muscle cell calcification. Biochem Biophys Res Commun 413,436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou H.C., Boothby M.R., Finn P.W., Davidon R., Nabavi N., Zeleznik-Le N.J., Ting J.P., and Glimcher L.H. (1990). A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science 247,1581–1584 [DOI] [PubMed] [Google Scholar]
- Majumder M., Huang C., Snider M.D., Komar A.A., Tanaka J., Kaufman R.J., Krokowski D., and Hatzoglou M. (2012). A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation. Mol Cell Biol 32,992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margariti A., Li H., Chen T., Martin D., Vizcay-Barrena G., Alam S., Karamariti E., Xiao Q., Zampetaki A., Zhang Z., Wang W., Jiang Z., Gao C., Ma B., Chen Y.G., Cockerill G., Hu Y., Xu Q., and Zeng L. (2013). XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem 288,859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet W., Croons V., Timmermans J.P., Herman A.G., and De Meyer G.R. (2007). Nitric oxide selectively depletes macrophages in atherosclerotic plaques via induction of endoplasmic reticulum stress. Br J Pharmacol 152,493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Chen X., Lee A.H., and Glimcher L.H. (2010). TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 11,411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miani M., Colli M.L., Ladriere L., Cnop M., and Eizirik D.L. (2012). Mild endoplasmic reticulum stress augments the proinflammatory effect of IL-1beta in pancreatic rat beta-cells via the IRE1alpha/XBP1s pathway. Endocrinology 153,3017–3028 [DOI] [PubMed] [Google Scholar]
- Nakka V.P., Gusain A., and Raghubir R. (2010). Endoplasmic reticulum stress plays critical role in brain damage after cerebral ischemia/reperfusion in rats. Neurotox Res 17,189–202 [DOI] [PubMed] [Google Scholar]
- Navon A., Gatushkin A., Zelcbuch L., Shteingart S., Farago M., Hadar R., and Tirosh B. (2010). Direct proteasome binding and subsequent degradation of unspliced XBP-1 prevent its intracellular aggregation. FEBS Lett 584,67–73 [DOI] [PubMed] [Google Scholar]
- Ning J., Hong T., Ward A., Pi J., Liu Z., Liu H.Y., and Cao W. (2011). Constitutive role for IRE1alpha-XBP1 signaling pathway in the insulin-mediated hepatic lipogenic program. Endocrinology 152,2247–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D., Tokuda M., Hosoda A., and Iwawaki T. (2010). Identification of a consensus element recognized and cleaved by IRE1 alpha. Nucleic Acids Res 38,6265–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Minamino T., Tsukamoto Y., Liao Y., Tsukamoto O., Takashima S., Hirata A., Fujita M., Nagamachi Y., Nakatani T., Yutani C., Ozawa K., Ogawa S., Tomoike H., Hori M., and Kitakaze M. (2004). Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation 110,705–712 [DOI] [PubMed] [Google Scholar]
- Park S.W., Herrema H., Salazar M., Cakir I., Cabi S., Basibuyuk Sahin F., Chiu Y.H., Cantley L.C., and Ozcan U. (2014). BRD7 regulates XBP1s' activity and glucose homeostasis through its interaction with the regulatory subunits of PI3K. Cell Metab 20,73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., and Ozcan U. (2013). Potential for therapeutic manipulation of the UPR in disease. Semin Immunopathol 35,351–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., Zhou Y., Lee J., Lu A., Sun C., Chung J., Ueki K., and Ozcan U. (2010). The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med 16,429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehar M., Jonas M.C., Hare T.M., and Puglielli L. (2012). SLC33A1/AT-1 protein regulates the induction of autophagy downstream of IRE1/XBP1 pathway. J Biol Chem 287,29921–29930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenbach K.T., Nivala A.M., Reese L., Ellis F., Wang D., Wei Y., and Pagliassotti M.J. (2010). Rapamycin inhibits postprandial-mediated X-box-binding protein-1 splicing in rat liver. J Nutr 140,879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T., Santos C.R., Griffiths B., Cully M., Wu M., Leevers S., Griffiths J.R., Chung Y.L., and Schulze A. (2008). SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 8,224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T., Santos C.R., Lewis C., Griffiths B., and Schulze A. (2009). A new player in the orchestra of cell growth: SREBP activity is regulated by mTORC1 and contributes to the regulation of cell and organ size. Biochem Soc Trans 37,278–283 [DOI] [PubMed] [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., and Glimcher L.H. (2001). Plasma cell differentiation requires the transcription factor XBP-1. Nature 412,300–307 [DOI] [PubMed] [Google Scholar]
- Romero-Ramirez L., Cao H., Nelson D., Hammond E., Lee A.H., Yoshida H., Mori K., Glimcher L.H., Denko N.C., Giaccia A.J., Le Q.T., and Koong A.C. (2004). XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res 64,5943–5947 [DOI] [PubMed] [Google Scholar]
- Sabio G., Cavanagh-Kyros J., Ko H.J., Jung D.Y., Gray S., Jun J.Y., Barrett T., Mora A., Kim J.K., and Davis R.J. (2009). Prevention of steatosis by hepatic JNK1. Cell Metab 10,491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabio G., Das M., Mora A., Zhang Z., Jun J.Y., Ko H.J., Barrett T., Kim J.K., and Davis R.J. (2008). A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science 322,1539–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandow J.J., Dorstyn L., O'Reilly L.A., Tailler M., Kumar S., Strasser A., and Ekert P.G. (2014). ER stress does not cause upregulation and activation of caspase-2 to initiate apoptosis. Cell Death Differ 21,475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T., Minamino T., Fu H.Y., Asai M., Okuda K., Isomura T., Yamazaki S., Asano Y., Okada K., Tsukamoto O., Sanada S., Asanuma H., Asakura M., Takashima S., Kitakaze M., and Komuro I. (2010). X-box binding protein 1 regulates brain natriuretic peptide through a novel AP1/CRE-like element in cardiomyocytes. J Mol Cell Cardiol 48,1280–1289 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Sharma C.G., and Jordan V.C. (2010). Estrogen regulation of X-box binding protein-1 and its role in estrogen induced growth of breast and endometrial cancer cells. Horm Mol Biol Clin Invest 2,235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H., He Y., Chen H., Wang C., Zenno A., Shi H., Yang X., Zhang X., and Qi L. (2009). The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab 9,556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H., He Y., Yang L., and Qi L. (2011). Stressed out about obesity: IRE1alpha-XBP1 in metabolic disorders. Trends Endocrinol Metab 22,374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H., Yang L., Liu M., Xia S., Liu Y., Liu F., Kersten S., and Qi L. (2014). Adipocyte spliced form of x-box-binding protein 1 promotes adiponectin multimerization and systemic glucose homeostasis. Diabetes 63,867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., Hurt E.M., Petroulakis E., Sonenberg N., Yewdell J.W., Calame K., Glimcher L.H., and Staudt L.M. (2004). XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21,81–93 [DOI] [PubMed] [Google Scholar]
- Shao M., Shan B., Liu Y., Deng Y., Yan C., Wu Y., Mao T., Qiu Y., Zhou Y., Jiang S., Jia W., Li J., Li J., Rui L., Yang L., and Liu Y. (2014). Hepatic IRE1alpha regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARalpha axis signalling. Nat Commun 5,3528. [DOI] [PubMed] [Google Scholar]
- Shen X., Ellis R.E., Lee K., Liu C.Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D.M., Mori K., and Kaufman R.J. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107,893–903 [DOI] [PubMed] [Google Scholar]
- Shinya S., Kadokura H., Imagawa Y., Inoue M., Yanagitani K., and Kohno K. (2011). Reconstitution and characterization of the unconventional splicing of XBP1u mRNA in vitro. Nucleic Acids Res 39,5245–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J.S., Hur K.Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A.H., Iwawaki T., Glimcher L.H., and Lee A.H. (2012). Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab 16,487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G., Vilcu C., Neels J.G., Bandyopadhyay G.K., Luo J.L., Naugler W., Grivennikov S., Wynshaw-Boris A., Scadeng M., Olefsky J.M., and Karin M. (2007). JNK1 in hematopoietically derived cells contributes to diet-induced inflammation and insulin resistance without affecting obesity. Cell Metab 6,386–397 [DOI] [PubMed] [Google Scholar]
- Song Y., Shen H., Du W., and Goldstein D.R. (2013). Inhibition of x-box binding protein 1 reduces tunicamycin-induced apoptosis in aged murine macrophages. Aging Cell 12,794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi C.M., Aleman J.O., Ueki K., Luo J., Asano T., Kaneto H., Stephanopoulos G., Cantley L.C., and Kahn C.R. (2007). The p85alpha regulatory subunit of phosphoinositide 3-kinase potentiates c-Jun N-terminal kinase-mediated insulin resistance. Mol Cell Biol 27,2830–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi C.M., Tran T.T., Kondo T., Luo J., Ueki K., Cantley L.C., and Kahn C.R. (2006). Phosphoinositide 3-kinase regulatory subunit p85alpha suppresses insulin action via positive regulation of PTEN. Proc Natl Acad Sci U S A 103,12093–12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R., Chen H., Gao C., Xue P., Yang F., Han J.D., Zhou B., and Chen Y.G. (2011). Xbp1-mediated histone H4 deacetylation contributes to DNA double-strand break repair in yeast. Cell Res 21,1619–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuerauf D.J., Marcinko M., Gude N., Rubio M., Sussman M.A., and Glembotski C.C. (2006). Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res 99,275–282 [DOI] [PubMed] [Google Scholar]
- Tirosh B., Iwakoshi N.N., Glimcher L.H., and Ploegh H.L. (2005). XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J Exp Med 202,505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turer A.T., and Scherer P.E. (2012). Adiponectin: mechanistic insights and clinical implications. Diabetologia 55,2319–2326 [DOI] [PubMed] [Google Scholar]
- Upton J.P., Wang L., Han D., Wang E.S., Huskey N.E., Lim L., Truitt M., McManus M.T., Ruggero D., Goga A., Papa F.R., and Oakes S.A. (2012). IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 338,818–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P., Pavlikova M., Sivonova M., Kaplan P., Tatarkova Z., Kaminska B., and Lehotsky J. (2009). Molecular analysis of endoplasmic reticulum stress response after global forebrain ischemia/reperfusion in rats: effect of neuroprotectant simvastatin. Cell Mol Neurobiol 29,181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal R.L., Figueroa A., Court F.A., Thielen P., Molina C., Wirth C., Caballero B., Kiffin R., Segura-Aguilar J., Cuervo A.M., Glimcher L.H., and Hetz C. (2012). Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum Mol Genet 21,2245–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chen Z., Lam V., Han J., Hassler J., Finck B.N., Davidson N.O., and Kaufman R.J. (2012). IRE1alpha-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab 16,473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnay J.N., Boucher J., Mori M.A., Ueki K., and Kahn C.R. (2010). A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat Med 16,438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K., Imagawa Y., Iwawaki T., Hosoda A., Saito M., Kimata Y., and Kohno K. (2009). Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell 34,191–200 [DOI] [PubMed] [Google Scholar]
- Yanagitani K., Kimata Y., Kadokura H., and Kohno K. (2011). Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331,586–589 [DOI] [PubMed] [Google Scholar]
- Yao S., Miao C., Tian H., Sang H., Yang N., Jiao P., Han J., Zong C., and Qin S. (2014). Endoplasmic reticulum stress promotes macrophage-derived foam cell formation by up-regulating cluster of differentiation 36 (CD36) expression. J Biol Chem 289,4032–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Yang N., Song G., Sang H., Tian H., Miao C., Zhang Y., and Qin S. (2012). Minimally modified low-density lipoprotein induces macrophage endoplasmic reticulum stress via toll-like receptor 4. Biochim Biophys Acta 1821,954–963 [DOI] [PubMed] [Google Scholar]
- Yao S., Zong C., Zhang Y., Sang H., Yang M., Jiao P., Fang Y., Yang N., Song G., and Qin S. (2013). Activating transcription factor 6 mediates oxidized LDL-induced cholesterol accumulation and apoptosis in macrophages by up-regulating CHOP expression. J Atheroscler Thromb 20,94–107 [DOI] [PubMed] [Google Scholar]
- Yao S.T., Sang H., Yang N.N., Kang L., Tian H., Zhang Y., Song G.H., and Qin S.C. (2010). [Oxidized low density lipoprotein induces macrophage endoplasmic reticulum stress via CD36.]. Sheng Li Xue Bao 62,433–440 [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107,881–891 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Oku M., Suzuki M., and Mori K. (2006). pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol 172,565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Uemura A., and Mori K. (2009). pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Struct Funct 34,1–10 [DOI] [PubMed] [Google Scholar]
- Zeng L., Xiao Q., Chen M., Margariti A., Martin D., Ivetic A., Xu H., Mason J., Wang W., Cockerill G., Mori K., Li J.Y., Chien S., Hu Y., and Xu Q. (2013). Vascular endothelial cell growth-activated XBP1 splicing in endothelial cells is crucial for angiogenesis. Circulation 127,1712–1722 [DOI] [PubMed] [Google Scholar]
- Zeng L., Zampetaki A., Margariti A., Pepe A.E., Alam S., Martin D., Xiao Q., Wang W., Jin Z.G., Cockerill G., Mori K., Li Y.S., Hu Y., Chien S., and Xu Q. (2009). Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc Natl Acad Sci U S A 106,8326–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., and Kaufman R.J. (2008). From endoplasmic-reticulum stress to the inflammatory response. Nature 454,455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Li X., Cai M.Y., Ma K., Yang J., Zhou J., Fu W., Wei F.Z., Wang L., Xie D., and Zhu W.G. (2013). XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res 23,491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lee J., Reno C.M., Sun C., Park S.W., Chung J., Lee J., Fisher S.J., White M.F., Biddinger S.B., and Ozcan U. (2011). Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat Med 17,356–365 [DOI] [PMC free article] [PubMed] [Google Scholar]