Abstract
Early detection is vital to improve the overall survival rate of bladder cancer (BCa) patients, yet there is a lack of a reliable urine-based assay for early detection of BCa. Urine metabolites represented a potential rich source of biomarkers for BCa. This study aimed to develop a metabolomics approach for high coverage discovery and identification of metabolites in urine samples. Urine samples from 23 early stage BCa patients and 21 healthy volunteers with minimum sample preparations were analyzed by a short 30 min UPLC-HRMS method. We detected and quantified over 9000 unique UPLC-HRMS features, which is more than four times than about 2000 features detected in previous urine metabolomic studies. Furthermore, multivariate OPLS-DA classification models were established to differentiate urine samples from bladder cancer cohort and normal health cohort. We identified three BCa-upregulated metabolites: nicotinuric acid, trehalose, AspAspGlyTrp, and three BCa-downregulated metabolites: inosinic acid, ureidosuccinic acid, GlyCysAlaLys. Finally, analysis of six post-surgery BCa urine samples showed that these BCa-metabolomic features reverted to normal state after tumor removal, suggesting that they reflected metabolomic features associated with BCa. ROC analyses using two linear regression models to combine the identified markers showed a high diagnostic performance for detecting BCa with AUC (area under the ROC curve) values of 0.919 to 0.934. In summary, we developed a high coverage metabolomic approach that has potential for biomarker discovery in cancers.
Introduction
Bladder cancer (BCa) is one of the most prevalent malignancies of the urinary system, and there is a trend of increasing incidence and mortality of BCa worldwide in recent decades (Kaufman et al., 2009, Lozano et al., 2012, Siegel et al., 2012). Early detection is vital to the overall survival rate of BCa patients. The 5-year survival rate for patients diagnosed at Stage I of the BCa can reach 94% (Kaufman et al., 2009). However, conventional detection methods including voided urinary cytology, cystoscopy imaging, transurethral bladder biopsy are highly invasive, expensive, inconvenient, and are not sensitive to detect early onset of BCa (Tetu, 2009). Typically, when BCa is diagnosed, transurethral resection of the bladder tumor (TURBT) is recommended as the standard of treatment. However, BCa is known to have high rate of recurrence, which calls for extensive long-term surveillance program following TURBT treatment. Ideally, a convenient biomarker assay with minimal cost shall be employed routinely to monitor BCa relapse before more invasive diagnostic options are recommended. Employing fast and cost-effective bioassay-based monitoring strategy can help to lower the mental and financial burden for the patients, which will lead to better patient compliance, thus in turn may increase detection rate for BCa relapse. Despite years of attempts to develop such a vital screening assay, there is still a dearth of suitable candidates with sufficient sensitivity and specificity as compared to routine cytology or cystoscopy-based methods.
Malignant cells or tissue usually display a wide range of metabolic abnormalities that can be reflected by concentration changes of specific metabolite species compared to normal cells or tissue. Therefore, measuring these metabolic products with abnormal levels allows differentiation of cancerous from normal samples. Of the various analytical strategies, the high throughput metabolomic survey provides a nonbiased, quantitative methodology for a large portion of metabolites in a given biological system. For the choice of biofluid samples, urine seems to be the ideal reservoir from which BCa diagnostic marker can be derived, as it is in direct contact with the cancer lesion on the bladder transitional epithelium. Based on this rationale, urinary metabolomics studies for BCa diagnosis has been carried out previously with various analytical platforms, including GC-MS, NMR and LC-MS (Huang et al., 2011, Issaq et al., 2008, Jobu et al., 2012, Pasikanti et al., 2010, Srivastava et al., 2010).
However, the catalog of the urinary metabolome has recently reached near 4000 species according to the ever-growing human metabolome project (www.urinemetabolome.ca), yet such a diverse urinary metabolite spectrum was nonetheless under-represented in most previous studies that only partially covered fewer than 2000 features per study (Huang et al., 2011, Issaq et al., 2008, Jobu et al., 2012, Pasikanti et al., 2010, Srivastava et al., 2010). As the result, none of the markers identified so far have shown validated clinical values, particularly for early BCa detections. This is probably because only small fraction of abundant urinary components, which are unlikely to reflect the local lesion at early stage, were surveyed due to the low sensitivity of previous analytical platforms. Thanks to the latest development of ultra performance liquid chromatography (UPLC) and electrospray ionization mass spectrometry (ESI-MS) instruments with significant improvement in detection sensitivity, investigation is possible of low abundant metabolites that were never been detected before to achieve deep coverage of urine metabolome. Additionally, the higher mass resolution of TOF-HRMS now helps to narrow down each acquired analyte to a small list of plausible molecular formulas with high identification confidence.
In this study, we specifically aimed to develop multivariate models using time-of-fly high resolution mass spectrometry (TOF-HRMS) technology providing high metabolomics coverage to differentiate samples from BCa cohort and normal health cohort. Based on this model, we further aimed to develop urinary metabolite markers for early stage BCa detection.
Methods
Subjects
The study cohort consists of 23 patients with BCa at early stages and 21 healthy controls without a history of BCa or any suggestive BCa symptoms. All patients in the control group were also confirmed by abdominal ultrasound examination. Patients with benign urinary conditions were excluded from this study. All BCa patients were treated with transurethral resection of bladder tumor (TURBT). Microscopic histology and CT imaging were used for pathologic staging of BCa according the American Joint Committee on Cancer (AJCC) TNM staging system. Characteristics of all enrolled cases are summarized in Table 1. Informed consent was obtained from each patient and approval was granted by Institutional Review Board of Shaoxing People's Hospital and Clinical Research Ethics Committee of Sir Run Run Shaw Hospital of Zhejiang University before commencing the study.
Table 1.
Characteristics of Enrolled Patients
| Groups | BCa | Normal | Post-surgery |
|---|---|---|---|
| No. patients | 23 | 21 | 6 |
| Average age | 65.14±13.27 | 53.76±19.47 | 66.83±6.91 |
| Sex ratio, M:F | 18:5 | 12:9 | 4:2 |
| Tumor Stage | |||
| Ta | 4 | 0 | 0 |
| T1 | 18 | 0 | 6 |
| T2 | 1 | 0 | 0 |
| T3 | 0 | 0 | 0 |
Sample collection and processing
Voided urine samples were collected in the morning before breakfast from all subjects at Shaoxing People's Hospital (Shaoxing, China) and Sir Run Run Shaw Hospital (Hangzhou, China). For BCa patients, all samples were collected a day before TURBT operation. In addition, post-surgery urine samples were collected at 7 days after TURBT treatment from 6 BCa patients. The collected urine samples were centrifuged at 3000 g for 10 min at 4°C to remove suspended debris, and the resulting supernatants were aliquoted and immediately stored at −80°C without any preservatives. Prior to UPLC-MS analysis, urine samples were diluted 1:1 with HPLC water and were distributed randomly into 96-well injector trays kept at 4°C.
Metabolomics analysis
Urine metabolites fingerprinting was performed on a UPLC-HRMS system consisting of a Agilent 1290 UPLC hyphened with a Agilent 6230 TOF-MS. Urine, treated as described above, was injected at a volume of 5 μL into a reversed-phase column (ACQUITY 100×2.1 mm, 1.7 μm, Waters, Milford, MA, USA) at 50°C. The system was operated at a flow rate of 0.3 mL/min of mobile phase consisting of solvent A: water with 0.1% formic acid (FA, Sigma-Aldrich, St. Louis, MO, USA), and organic solvent B: acetonitrile (ACN, Sigma-Aldrich) with 0.1% FA. The total analysis time per sample was 25 min. The gradient started with 3% of B and increased to 60% in 12 min, then reached 100% in 3 min, and maintained at 100% for 1 min. The gradient was ended by return to 3% B in 1 min and kept at the re-equilibration condition for 8 min until next injection.
Samples were analyzed in both positive and negative ESI modes in separate runs on a TOF mass analyzer operated in full scan mode from m/z 100 to 1000 Th. The capillary voltage was 4000 V; the nebulizer nitrogen flow rate was 11 L/min maintained at 310 kPa; and the temperature at 325°C. During the analyses, reference masses (m/z 121.0509, m/z 922.0098 for positive ESI and m/z 112.9856, m/z 1033.9891 for negative ESI) were continuously infused to allow constant mass correction. During the analyses, samples were kept in the LC auto-sampler maintained at 4°C.
The LC-MS raw data were obtained by MassHunter (Agilent, Santa Clara, CA) in centroid mode and converted to mzData format processed in MZmine 2.10 (mzmine.sourceforge.net) software environment. Briefly, the LC-MS profiles were first cleaned of background noise. Individual compounds within the 15 min effective LC gradient window from each sample were recognized by unique m/z and retention time (RT) values. Isotopic peaks of each compound were grouped. Chromatograms of each compound were then aligned across samples/injections for comparison. Metabolite features were then tabulated with unique m/z, RT, and peak areas from each sample.
Statistical analysis and multivariate modeling
To build sound statistical models, only features presented in 90% of all samples across the whole experiment were chosen. The filtered features were then gap filled by Peak Finder module in MZmine 2.0 to avoid missing data points. Instrumental signal variations were corrected against the peak area of the total ion chromatogram of each LC-MS run. The corrected data were imported to SIMCA-P v12 (Umetrics AB, Sweden) for multivariate data analysis. Data were Ln transformed and Pareto scaled before principal component analysis (PCA) or orthogonal partial least squares-discriminant analysis (OPLS-DA) to model the difference between the BCa patients and healthy controls. Potential biomarkers were selected based on OPLS-DA model by S-plot and Variable Importance in the Project (VIP>2) value. Student's t-tests were employed to further ensure the selected species significantly differentially expressed (p<0.01) between the BCa patients and the controls. In addition, only species with stable LC profile (RSD% of RT<0.5%) across the whole experiment were considered as final biomarker candidates.
Metabolite identification
For structure elucidation of potential biomarker candidates, CID assisted MS/MS experiments were performed on a Premier Q-TOF mass spectrometer (Waters, Milford, MA, USA) with the same UPLC system configuration and chromatographic conditions. The capillary voltage was set at 3.0 kV, and the sampling cone voltage was set at 50.0 V for positive ESI or 40.0 V for negative ESI. The nebulizer nitrogen flow rate was 7.5 L/min maintained at 120°C. During the analyses, reference masses (m/z 556.2771 for positive ESI and m/z 554.2615 for negative ESI) were continuously infused via lock mass spray. For each candidate on separate LC-MS/MS runs, the quadruple was set to isolate candidate precursor m/z during the whole UPLC gradient, while the TOF mass analyzer operated in full scan mode from m/z 50 to 1000 Th. Argon was used as the collision gas with different collision energies ranging from 30 to 80 V, according to the respective chemical stability. The UPLC-MSMS data were collected by MassLynx v4.1 software (Waters Co.). Tandem spectra of each metabolite candidate were exported in mgf format. To identify potential biomarkers, both HMDB (http://www.hmdb.ca/), METLIN (http://metlin.scripps.edu/) were searched using MS/MS peak list with 10 ppm mass error and precursor mass with 5 ppm mass error.
Results
UPLC-HRMS profiling of urine metabolome
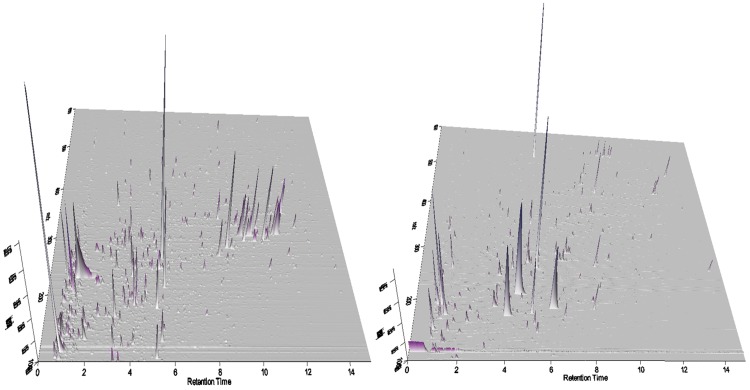
Fifty urine samples (23 BCa, 21 normal, and 6 post-surgery) were analyzed using UPLC-HRMS nontargeting profiling. The urine metabolites were detected in both positive and negative electrospray modes. The typical UPLC-HRMS chromatograms in both ESI modes are shown in 3D format as in Figure 1. The successful alignment of metabolomics profiles between samples depends on the stability of the UPLC performance. To assure this, 14 injections of QC samples made from a mixture of all samples were used throughout both ESI positive and negative mode experiments.
FIG. 1.
Representative 3D profiles of urinary metabolome from both positive (left) and negative (right) mode UPLC-HRMS analyses.
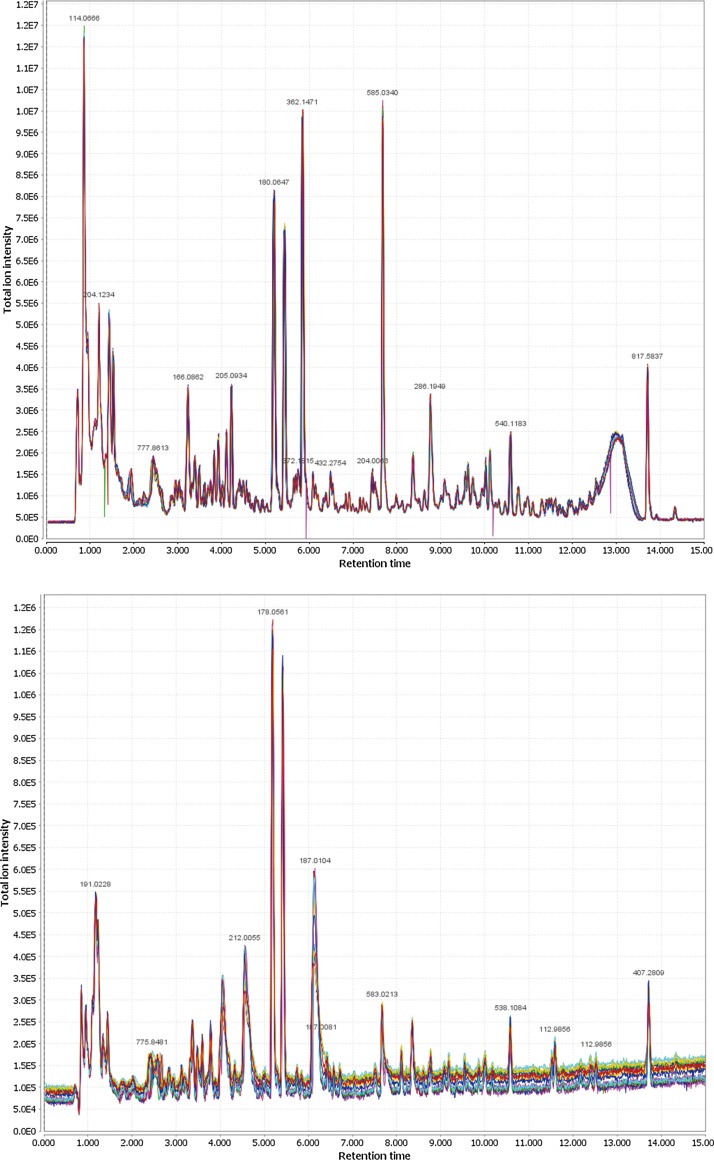
The median RSD% of the retention time of the 1000 most intensive peaks in the 14 positive QC runs is 0.342%, whereas that of the 500 most intensive peaks in the 14 negative QC runs is 0.247% (Fig. 2). For the same selection of peaks in QC runs, the variations of m/z values were less than 10 ppm in both positive and negative modes. These results demonstrated the robust stability and reproducibility of the UPLC separation and TOF mass measurement. Using the minimum intensity threshold of 200 counts, a total of 9110 and 530 unique entities (defined by a unique pair of RT and M/Z value) in positive and negative ESI mode, respectively, were consistently detected in at least 90% (n=45) urine samples.
FIG. 2.
Superposed chromatograms from 14 QC runs (color-coded) in positive (upper) and negative (lower) mode UPLC-HRMS analyses without any post-run RT correction, alignment, or background subtraction.
Multivariate models establishment
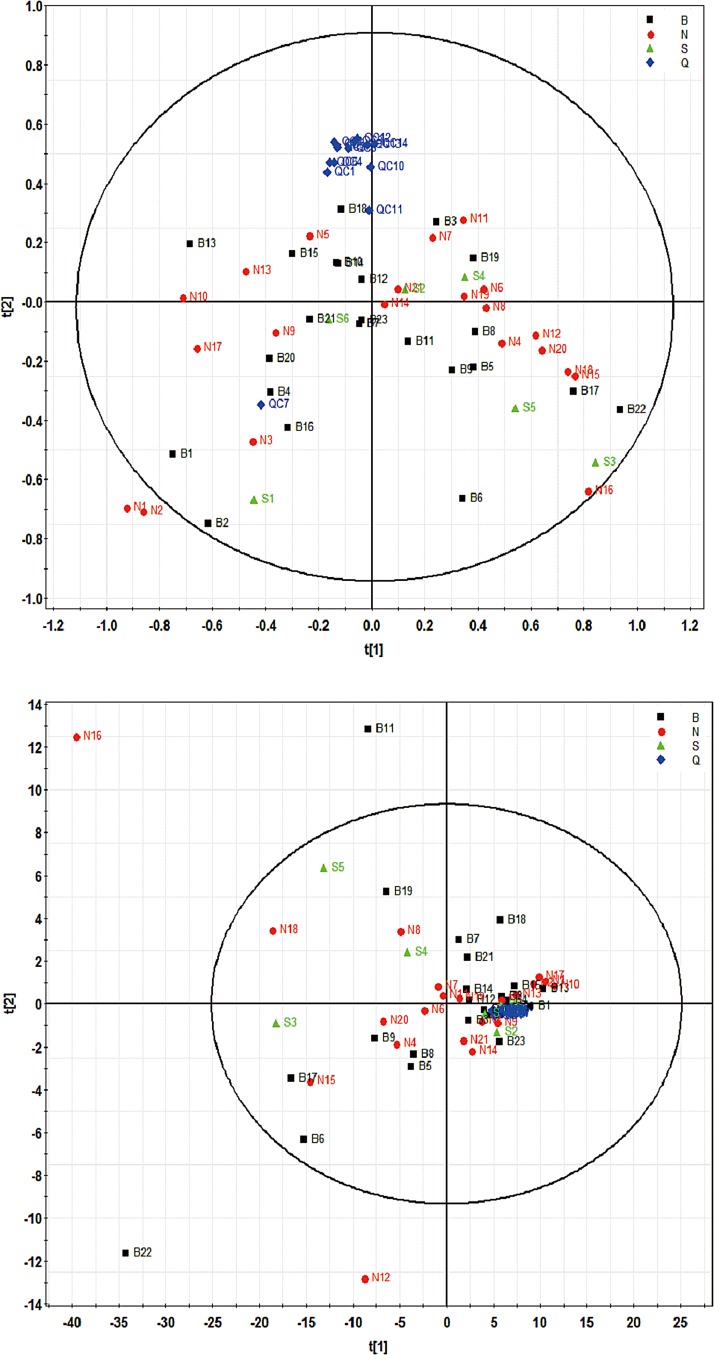
After data normalization, log transformation, and Pareto scaling, unsupervised PCA was performed to assess the overall intergroup separation as shown in Figure 3. According to the PCA score plots in both positive (R2Xcum=0.445, Q2cum=0.19) and negative ESI mode (R2Xcum=0.761, Q2cum=0.404), there were no distinctive differences between normal and cancer groups. Considering the complex and dynamic nature of human urine metabolome, this is possible that negligible individual differences comprised mostly by BCa-irrelevant variations make it unlikely to separate groups without a supervised based approach. However, it is notable that all QC data clustered closely together in both positive and negative ESI modes, indicating the satisfactory stability of our UPLC-HRMS method.
FIG. 3.
PCA score plots of ESI-positive (upper) and ESI-negative (lower) metabolome from all samples. The black square, red dot, green triangle, and blue diamond markers represent BCa, normal, post-surgery, and QC samples.
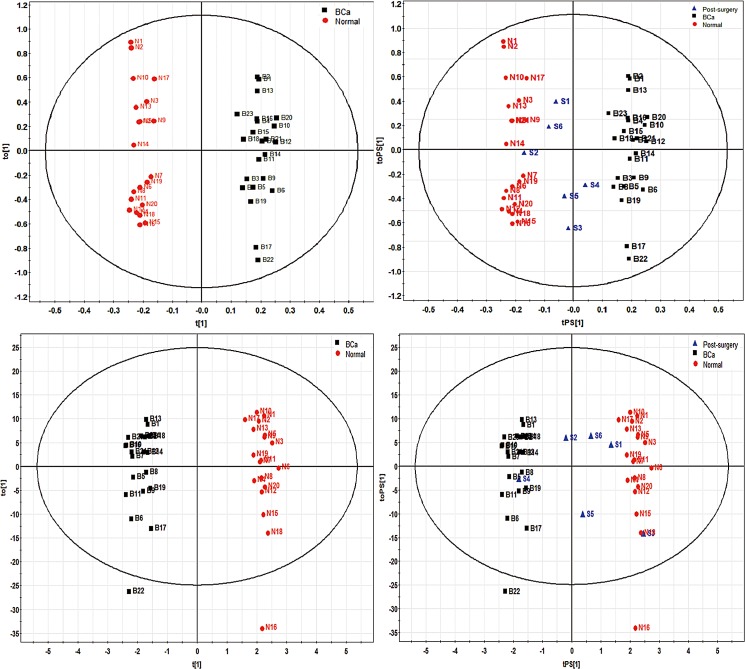
To specify BCa-related metabolomic alterations, supervised orthogonal OPLS-DA models were constructed using pre-surgery BCa and normal control datasets collected from both ESI modes (Fig. 4). The OPLS-DA model for positive ESI mode dataset showed satisfactory predictive ability with 1 predictive component and 2 orthogonal components (R2Xcum=0.238, R2Ycum=0.976, Q2cum=0.33). The OPLS-DA model for negative ESI mode dataset consisted of 1 predictive component and 5 orthogonal components (R2Xcum=0.699, R2Ycum=0.982, Q2cum=0.648) showed even better predictive ability. To guard against model overfitting, analysis of variance testing of cross-validated predictive residuals (CV-ANOVA) (Erikssona et al., 2008) was applied, and statistical significance (p=0.014 for positive ESI datasets and p=0.001 for negative ESI datasets) was achieved. In both OPLS-DA models, cancer samples were completely discriminated from the control samples in the predictive component. Our modeling results suggested potential value of using urinary metabolomic profile for early diagnosis of BCa.
FIG. 4.
OPLS-DA score plots for ESI-positive (top panel) and ESI-negative (bottom panel) models on the left, and the prediction of post-surgery samples in both models on the right.
Additionally, we further analyzed the urine metabolome from post-surgery groups (n=6). Except for sample S4, which was clearly deemed as cancer sample, other post-surgery samples have shown overall return trends of the metabolomic trajectory toward to the normal state, hence indicated a recovering event as a result of TURBT operation. These metabolomic shifts demonstrated the potential use of metabolomics data to assist the evaluation of BCa treatment outcome. Interestingly, patient S4 was the only case reported BCa relapse within the study cohorts. This patient was diagnosed with BCa and has received TURBT 3 years ago. The sample S4 was collected after the recent second TURBT treatment. Therefore, future study shall investigate if the metabolomics models developed in this study can be use for BCa recurrence monitoring.
Metabolite markers associated with bladder cancer
Based on the established OPLS-DA models, potential biomarkers contributed to the discrimination of cancer and control groups were selected according to the S-plot (Fig. 5) and the threshold of VIP>2. To further discard possible spurious markers due to peak mismatches and to pave foundation for robust assay development, only species with average MS intensity >1000 counts, LC retention time RSD% <0.5%, and significant differences in raw MS intensity (t-test p<0.01) were selected. This narrowed down the targets to a final list (Table 1) of 21 and 3 metabolites detected in positive and negative ESI mode, respectively.
FIG. 5.
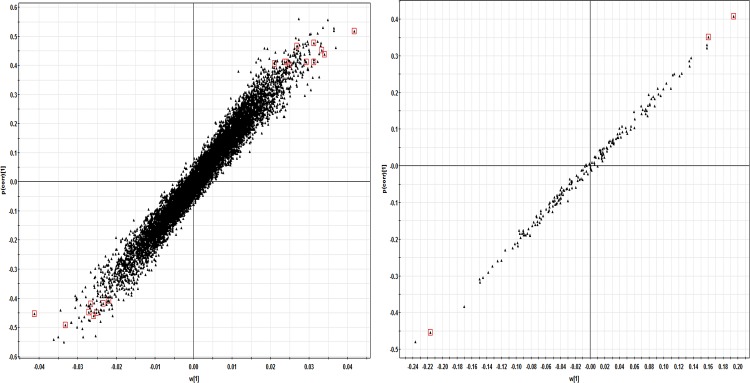
The loading S-plots showing the contribution of individual feature to the OPLS-DA models derived from ESI-positive (left) and -negative (right) metabolomic datasets comparing early BCa samples and normal samples. Features approaching top-right corner in S-plots are significantly upregulated in BCa samples, whereas those on the bottom-left corner are significantly downregulated in BCa samples. The final list of potential markers filtered with additional intensity, RT-RSD%, and t-test significance criteria are marked with red squares.
The pooled urine QC samples were analyzed again by UPLC-QTOF to obtain the CID fragmentation pattern for each of the 24 plausible metabolites. Corresponding MS/MS peak lists were searched against the HMDB or Metlin database. Some putative biomarkers were identified and are summarized in Table 2. Three upregulated metabolites: nicotinuric acid (NTUA), tetrapeptide AspAspGlyTrp (DDGW), trehalose (TRHL), and three downregulated metabolites: tetrapeptide GlyCysAlaLys (GCAK), inosinic acid (ISNA), and ureidosuccinic acid (UDSA) were identified. The relative intensities of these SIX metabolites all show significant differences (p<0.01) between healthy and cancerous samples as displayed in Figure 6, which also includes their relative intensities in urine samples collected from six post-surgery BCa patients. To our expectation, all six markers showed a reversing tendency toward the normal level from BCa state after TURBT operation, with three of them (GCAK, NTUA, ISNA) showing significant changes compared to cancerous samples. Such recovering alterations indicated a direct link between these metabolites markers and the physical existence of the BCa tumor.
Table 2.
Statistically Significant Urinary Metabolites Differentiating BCa from Healthy Cohort
| MZ (Th) | Ion | RT (min) | Identification/ion | VIP | FC | T test |
|---|---|---|---|---|---|---|
| 288.2836 | + | 14.3 | Unknown species (possible formula C17H37NO2 [M+H]) | 3.98 | 7.24 | 0.0040 |
| 274.2688 | + | 13.8 | Unknown species (possible formula C16H35NO2 [M+H]) | 3.93 | 0.39 | 0.0028 |
| 378.1805 | + | 11.2 | GlyCysAlaLys [M+H] | 3.35 | 0.31 | 0.0008 |
| 288.2837 | + | 14.8 | Unknown species (possible formula C17H37NO2 [M+H]) | 3.24 | 5.52 | 0.0012 |
| 244.2583 | + | 14.9 | Unknown species | 3.18 | 4.87 | 0.0048 |
| 449.1437 | + | 13.9 | Unknown species (possible formula C22H24O10 [M+H]) | 3.16 | 0.20 | 0.0026 |
| 181.0784 | + | 9.9 | Nicotinuric acid [M+H] | 2.98 | 3.10 | 0.0021 |
| 271.1583 | + | 9.1 | Unknown species | 2.98 | 3.42 | 0.0020 |
| 203.1208 | + | 6.5 | Unknown species | 2.80 | 1.81 | 0.0053 |
| 773.4909 | + | 14.3 | Unknown species (possible formula C48H68O8 [M+H]) | 2.59 | 0.37 | 0.0058 |
| 317.1144 | + | 4.9 | Unknown species | 2.56 | 2.63 | 0.0035 |
| 393.217 | + | 11.5 | Unknown species (possible formula C25H28O4 [M+H]) | 2.54 | 0.25 | 0.0085 |
| 399.1325 | + | 13.6 | Unknown species (possible formula C22H22O7 [M+H]) | 2.47 | 0.38 | 8.80E-05 |
| 365.1478 | + | 13.9 | Unknown species (possible formula C22H20O5 [M+H]) | 2.40 | 0.42 | 0.0004 |
| 609.3597 | + | 13.1 | Unknown species | 2.38 | 4.23 | 0.0055 |
| 426.9059 | + | 0.7 | Unknown species | 2.30 | 2.27 | 0.0096 |
| 195.0689 | + | 3.9 | Unknown species | 2.26 | 1.58 | 0.0061 |
| 143.0008 | + | 0.7 | Unknown species | 2.23 | 0.47 | 0.0016 |
| 304.2046 | + | 7.5 | Unknown species | 2.09 | 0.38 | 0.0024 |
| 492.1783 | + | 3.8 | AspAspGlyTrp [M+H] | 2.01 | 2.66 | 0.0054 |
| 349.1164 | + | 12.0 | Inosinic acid [M+H] | 2.00 | 0.50 | 0.0023 |
| 341.0293 | - | 1.1 | Trehalose [M-H] | 3.02 | 1.98 | 0.0006 |
| 174.9552 | - | 0.7 | Ureidosuccinic acid [M-H] | 2.71 | 0.68 | 0.0035 |
| 304.9121 | - | 0.7 | Unknown species | 2.24 | 0.53 | 0.0069 |
Metabolites identified by LC-MSMS are shown in bold.
FIG. 6.
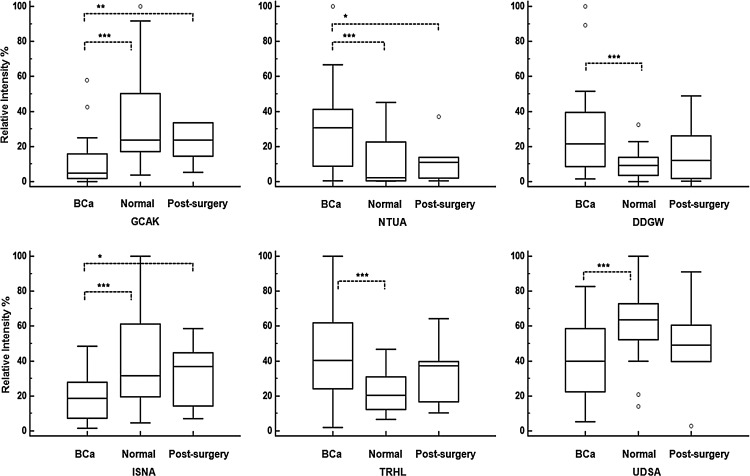
Variations of relative MS intensity of 6 identified metabolite markers in urine from BCa, healthy, and post-surgery BCa patients. GCAK, NTUA, DDGW, ISNA, TRHL, UDSA represent GlyCysAlaLys, nicotinuric acid, AspAspGlyTrp, inosinic acid, trehalose, and ureidosuccinic acid, respectively. The boxes were drawn from the 25th to 75th percentiles in the intensity distribution. The median (50th percentile) is represented by the horizontal line inside the box. Variation significance between groups are indicated by ***p<0.01, **p<0.05, *p<0.1.
Receiver operating characteristic curve analysis and linear regression models
To better assess the clinical utility potential of the markers, receiver operating characteristic (ROC) curve analyses were performed for individual markers and possible marker combinations (Table 3). The sensitivity and specificity of each marker was calculated at the best cut-off value.
Table 3.
Diagnostic Characteristics of Metabolite Marker Combinations
| Marker | AUC | SE | Cut-off | 95% CI | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| GlyCysAlaLys | 0.834 | 0.061 | <=18.34 | 0.716–0.953 | 82.61 | 76.19 |
| Nicotinuric acid | 0.774 | 0.071 | >4.75 | 0.635–0.913 | 82.61 | 66.67 |
| Asp Asp Gly Trp | 0.743 | 0.076 | >=14.04 | 0.595–0.891 | 65.22 | 80.95 |
| Inosinic acid | 0.720 | 0.078 | <=43.29 | 0.567–0.874 | 95.65 | 42.86 |
| Trehalose | 0.776 | 0.071 | >38.79 | 0.637–0.916 | 60.87 | 90.48 |
| Ureidosuccinic acid | 0.752 | 0.076 | <=42.68 | 0.603–0.900 | 60.87 | 85.71 |
| MixModel 1 | 0.934 | 0.0344 | >0.3998 | 0.866 to 1.000 | 91.30 | 80.95 |
| MixModel 2 | 0.919 | 0.0397 | >0.5651 | 0.841 to 0.997 | 82.61 | 90.48 |
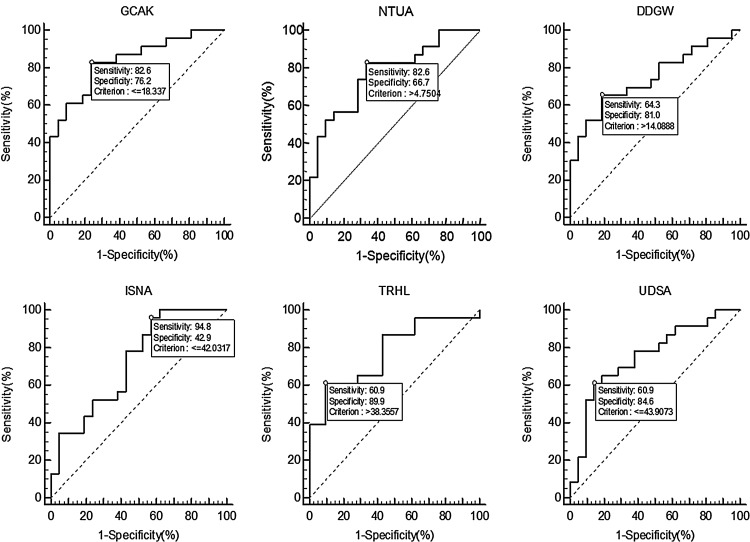
When used alone (Fig. 7), the tetrapeptide GlyCysAlaLys showed the best combination of sensitivity (82.61%) and specificity (76.19%) of all, with AUC of 0.834. The inosinic acid (AUC=0.720) although showing the best sensitivity (95.65%), had the worst specificity (42.86%) of all. On the contrary, trehalose (AUC=0.776) displayed a mediocre sensitivity at 60.87%, but nonetheless had the best specificity (90.48%) among all others.
FIG. 7.
ROC characterization of six identified metabolite markers in urine to differentiate BCa and healthy cohorts. GCAK, NTUA, DDGW, ISNA, TRHL, UDSA represent GlyCysAlaLys, nicotinuric acid, AspAspGlyTrp, inosinic acid, trehalose, and ureidosuccinic acid, respectively.
Such distinctive diagnostic characteristics of different markers identified in this dataset prompted us to try marker combinations with higher sensitivity and specificity. We further constructed two multivariate linear regression models using combinations of the relative intensity of various metabolites for early BCa determination (Fig. 8). In the first model, all six variables were used, and an optimum R2 can be achieved by:
FIG. 8.
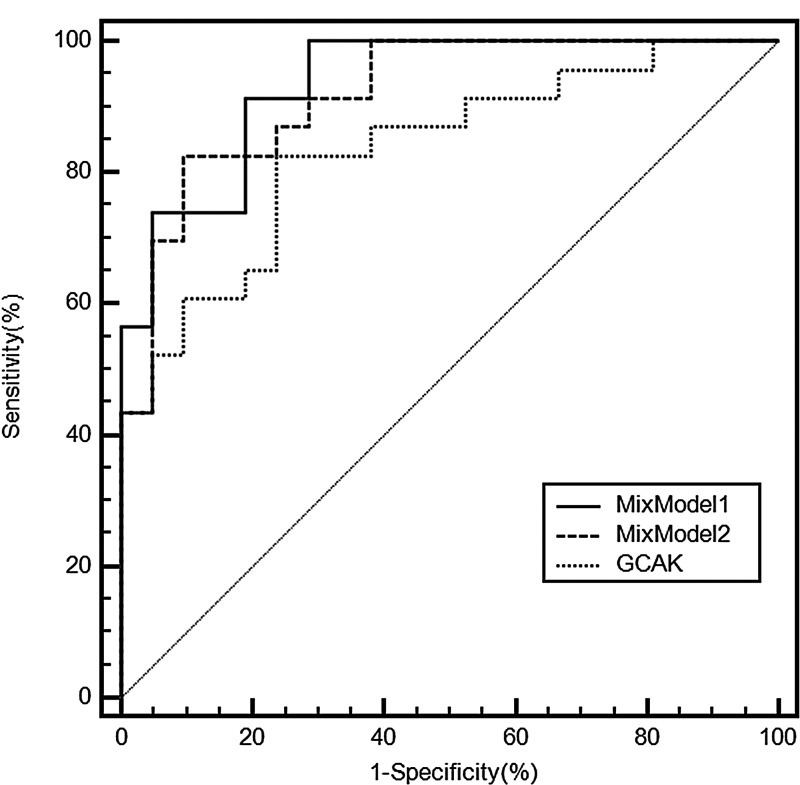
ROC characterization of two multivariate MixModels to differentiate BCa and healthy cohorts, compared to the single best metabolite candidate GCAK.
MixModel1=0.7793+0.005921×DDGW - 0.006758×GCAK 0.001266×ISNA+0.004530×NTUA+0.001669×TRHL - 0.007907×UDSA.
The ROC analysis of MixModel1 showed significant improvement of sensitivity (91.30%) and specificity (80.95%) with AUC reaching 0.934. Another much simplified model can be derived from three variables with significant association (p<0.01) in the previous full model:
MixModel2=0.9444+0.008692×DDGW - 0.007243×GCAK -0.008314×UDSA.
This combination model exhibited mildly compromised sensitivity at 82.61% but with better specificity at 90.48%, and the overall AUC reached 0.919. Other linear combinations of the different variables did not have significant improvement over sensitivity and specificity.
Discussion
In recent years, the clinical urology field has seen a surge in the development of urine-based noninvasive bladder cancer markers as alternatives or as additions to the conventional cystoscopic approaches (Grossfeld et al., 2001, Tetu, 2009). Despite a wealth of attempts to develop new diagnostic markers for BCa, none of the currently available USFDA-approved assays, namely BTA Stat, BTA Trak, NMP22, ImmunoCyt/uCyt, FDP, and UroVysion (Shariat et al., 2008, Sullivan et al., 2010) is widely adopted in clinical use. As suggested by recent meta-analyses, none of them is sensitive or robust enough to replace conventional methods (Budman et al., 2008, Giannopoulos et al., 2001, Mahnert et al., 2003, Shariat et al., 2008, Sullivan et al., 2010, Urquidi et al., 2012, Vrooman and Witjes, 2008).
Urine samples were chosen for its direct physical contact with bladder tissue and for the purpose of developing a convenient noninvasive assay. Several other studies have employed nontargeted urinary metabolomics approaches to detect BCa (Huang et al., 2011, Issaq et al., 2008, Jobu et al., 2012, Pasikanti et al., 2010, Putluri et al., 2011, Srivastava et al., 2010). However, many previous reports, particularly those performed on NMR (Srivastava et al., 2010) or GC-MS (Jobu et al., 2012, Pasikanti et al., 2010) resulted in insufficient metabolome coverage, and LC-coupled MS studies did not improve the coverage significantly due to the limited sensitivity of old model mass spectrometries (Issaq et al., 2008, Putluri et al., 2011). In comparison, despite only 2.5 μL of urine used in this study, the detection sensitivity has not been sacrificed, as over 9000 unique UPLC-HRMS features (present in 90% of all samples) were quantified in one single analysis, surpassing most previous reports. In addition, the sample preparation was extremely simple and did not require extra steps of precipitation, extraction, or chemical derivatization, leaving the technical variations as low as possible. Sample analysis can be finished within 30 minutes and the decision can be made rather quickly and objectively without the expertise of experienced pathologists. In addition, the tiny volume of urine required by our metabolomics approach allows it to be easily and routinely integrated with other urine-based tests.
Regarding the diagnostic performance of our metabolomics method, the multivariate OPLS-DA models showed 100% discrimination power to separate BCa patients from healthy donors. One of the most promising markers that contributed significantly to the OPLS-DA model, the tetrapeptide GlyCysAlaLys, achieved a specificity of 82.61% with a sensitivity of 76.19% that results an AUC at 0.834. A linear combination discriminant model using all five markers can further reach the AUC at 0.934, while a simplified combination of GlyCysAlaLys, AspAspGlyTrp and ureidosuccinic acid also have an improved AUC of 0.919. Given these merits, this urinary metabolomics-based approach showed great application potentials as alternative or supplement diagnostic procedure to cystoscopic tests. Future large-scale retrospective or prospective studies are needed to test the true clinical utility of these metabolites as bladder cancer biomarkers.
The present study indicated that the cancer group has elevated levels of urinary nicotinuric acid and trehalose. Nicotinuric acid (NTUA or acyl glycines) is an endogenous end product of nicotinate and nicotinamide metabolism and is also a minor metabolite of fatty acid beta-oxidation. NTUA has been detected at very low level in urine samples (Gronwald et al., 2011). Our report is the first to show elevated levels of NTUA in urine samples from BCa patients. Trehalose is a nonreducing sugar with antioxidant property that is usually found in extracellular space. However, very little was known about its connection to cancer pathology. Our dataset also found inosinic acid and ureidosuccinic acid were downregulated in urine samples from BCa patients. Inosinic acid (or inosine 5'-phosphate, IMP) is involved in purine metabolism. It is converted by inosine-5'-monophosphate dehydrogenases (IMPDHs) in a rate-limiting step for the de novo biosynthesis of guanine nucleotides. IMPDHs have been shown to play vital roles in the development of malignancy such as myeloma, neuroblastoma, colorectal cancer, prostate cancer, and have long been considered as an attractive target for anticancer intervention (Chen and Pankiewicz, 2007).
However, the possible alterations of the substrates of IMPDHs were never investigated, and there has been no report of possible association of IMP or IMPDHs to BCa yet. Ureidosuccinic acid (or carbamoylaspartic acid) is an intermediary product in both aspartate and pyrimidine synthesis. Ureidosuccinic acid can be detected in urine (van Kuilenburg et al., 2004) and was previously reported to associated with prostate cancer progression (Sreekumar et al., 2009). Our data also found concentration alteration of two tetrapeptides (GlyCysAlaLys and AspAspGlyTrp) in urine of BCa patients. Many small oligopeptides, including tetrapeptides, are bioactive molecules often showing affinity to a wealth of binding partners involving in intra- and extracellular signaling. Unfortunately, the identities of these two tetrapeptides were never reported before, and the source of these two small peptides cannot be specified at this point, as suggested by Blastp search which retrieved multiple proteins IDs that contains these two short sequences. Therefore, further studies are needed to interpret the biological links between those compounds and BCa pathogenesis and progression.
Conclusion
This work aims to discover urinary metabolite constituents from early bladder cancer patients that have potential to be used as sensitive and specific BCa diagnostic biomarkers. Here we first described an integrated UPLC-HRMS workflow to characterize urinary metabolome. Over 9000 unique UPLC-HRMS features were identified and quantified, making this study one of the most comprehensive survey of the human urinary metabolome compared to other state-of-art urinary metabolomics reports. Multivariate OPLS-DA classification models were built to successfully differentiate BCa patients from healthy cohorts. Given such a rich source of urinary metabolites, we further identified many BCa specific markers and identified six unique metabolites that have not been reported previously as diagnostic biomarkers for bladder cancer. Linear regression models were also built using combinations of identified markers to further improve diagnostic performance for detecting BCa.
Acknowledgments
This work was supported by the grant from the Department of Science and Technology, Zhejiang Province (2012C33SA600020), National Natural Science Foundation of China (81400589), Chinese High Tech Research and Development Programs (2012AA022705).
Author Disclosure Statement
The authors declare no competing financial interest.
References
- Budman LI, Kassouf W, and Steinberg JR. (2008). Biomarkers for detection and surveillance of bladder cancer. Can Urol Assoc J 2, 212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, and Pankiewicz KW. (2007). Recent development of IMP dehydrogenase inhibitors for the treatment of cancer. Curr Opin Drug Discov Devel 10, 403–412 [PubMed] [Google Scholar]
- Erikssona L, Trygg J, and Wold S. (2008). CV-ANOVA for significance testing of PLS and OPLS models. J Chemometrics 22, 594–600 [Google Scholar]
- Giannopoulos A, Manousakas T, Gounari A, Constantinides C, Choremi-Papadopoulou H, and Dimopoulos C. (2001). Comparative evaluation of the diagnostic performance of the BTA stat test, NMP22 and urinary bladder cancer antigen for primary and recurrent bladder tumors. J Urol 166, 470–475 [PubMed] [Google Scholar]
- Gronwald W, Klein MS, Zeltner R, et al. (2011). Detection of autosomal dominant polycystic kidney disease by NMR spectroscopic fingerprinting of urine. Kidney Int 79, 1244–1253 [DOI] [PubMed] [Google Scholar]
- Grossfeld GD, Litwin MS, Wolf JS, et al. (2001). Evaluation of asymptomatic microscopic hematuria in adults: The American Urological Association best practice policy–part I: Definition, detection, prevalence, and etiology. Urology 57, 599–603 [DOI] [PubMed] [Google Scholar]
- Huang Z, Lin L, Gao Y, et al. (2011). Bladder cancer determination via two urinary metabolites: A biomarker pattern approach. Mol Cell Proteomics 10, M111 007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaq HJ, Nativ O, Waybright T, et al. (2008). Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J Urol 179, 2422–2426 [DOI] [PubMed] [Google Scholar]
- Jobu K, Sun C, Yoshioka S, et al. (2012). Metabolomics study on the biochemical profiles of odor elements in urine of human with bladder cancer. Biol Pharm Bull 35, 639–642 [DOI] [PubMed] [Google Scholar]
- Kaufman DS, Shipley WU, and Feldman AS. (2009). Bladder cancer. Lancet 374, 239–249 [DOI] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, et al. (2012). Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnert B, Tauber S, Kriegmair M, et al. (2003). Measurements of complement factor H-related protein (BTA-TRAK assay) and nuclear matrix protein (NMP22 assay)—Useful diagnostic tools in the diagnosis of urinary bladder cancer? Clin Chem Lab Med 41, 104–110 [DOI] [PubMed] [Google Scholar]
- Pasikanti KK, Esuvaranathan K, Ho PC, et al. (2010). Noninvasive urinary metabonomic diagnosis of human bladder cancer. J Proteome Res 9, 2988–2995 [DOI] [PubMed] [Google Scholar]
- Putluri N, Shojaie A, Vasu VT, et al. (2011). Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res 71, 7376–7386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat SF, Karam JA, Lotan Y, and Karakiewizc PI. (2008). Critical evaluation of urinary markers for bladder cancer detection and monitoring. Rev Urol 10, 120–135 [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, and Jemal A. (2012). Cancer statistics, 2012. CA Cancer J Clin 62, 10–29 [DOI] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. (2009). Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457, 910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Srivastava S, Roy R, Singh S, et al. (2010). Taurine: A possible fingerprint biomarker in non-muscle invasive bladder cancer: A pilot study by 1H NMR spectroscopy. Cancer Biomark 6, 11–20 [DOI] [PubMed] [Google Scholar]
- Sullivan PS, Chan JB, Levin MR, and Rao J. (2010). Urine cytology and adjunct markers for detection and surveillance of bladder cancer. Am J Transl Res 2, 412–440 [PMC free article] [PubMed] [Google Scholar]
- Tetu B. (2009). Diagnosis of urothelial carcinoma from urine. Mod Pathol 22, S53–59 [DOI] [PubMed] [Google Scholar]
- Urquidi V, Rosser CJ, and Goodison S. (2012). Molecular diagnostic trends in urological cancer: Biomarkers for non-invasive diagnosis. Curr Med Chem 19, 3653–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuilenburg AB, van Lenthe H, Loffler M, and van Gennip AH. (2004). Analysis of pyrimidine synthesis “de novo” intermediates in urine and dried urine filter- paper strips with HPLC-electrospray tandem mass spectrometry. Clin Chem 50, 2117–2124 [DOI] [PubMed] [Google Scholar]
- Vrooman OP, and Witjes JA. (2008). Urinary markers in bladder cancer. Eur Urol 53, 909–916 [DOI] [PubMed] [Google Scholar]