Abstract
Infectious diseases and increasing antibiotic resistance among diverse classes of microbes are global health concerns and a prime focus of omics systems science applications in novel drug discovery. Plumbagin is a plant-derived naphthoquinone, a natural product that exhibits antibacterial activity against gram-positive bacteria. In the present study, we investigated the antimicrobial effects of plumbagin against Bacillus subtilis using two complementary proteomics techniques: two-dimensional electrophoresis (2-DE) and isobaric tag for relative and absolute quantification (iTRAQ). Comparative quantitative proteomics analysis of plumbagin treated and untreated control samples identified differential expression of 230 proteins (1% FDR, 1.5 fold-change and ≥2 peptides) in B. subtilis after plumbagin treatment. Pathway analysis involving the differentially expressed proteins suggested that plumbagin effectively increases heme and protein biosynthesis, whereas fatty acid synthesis was significantly reduced. Gene expression and metabolic activity assays further corroborated the proteomics findings. We anticipate that plumbagin blocks the cell division by altering the membrane permeability required for energy generation. This is the first report, to the best of our knowledge, offering new insights, at proteome level, for the putative mode(s) of action of plumbagin and attendant cellular targets in B. subtilis. The findings also suggest new ways forward for the modern omics-guided drug target discovery, building on traditional plant medicine.
Introduction
Infectious diseases are vastly challenging healthcare professionals worldwide. They are major cause of disease-related deaths in developing countries (Christian et al., 2013; Hay et al., 2013a, 2013b). Ability of different classes of bacteria to survive in the presence of many of the commonly used antibiotics and emergence of multidrug resistant species have been observed over the past 2 decades. At the same time, the limited availability of novel antibiotics and drug targets has intensified the need for new approaches to antibiotic research. Natural products often exhibit antimicrobial activity against a wide range of pathogenic microorganisms. Recent advanced high-throughput multi-omics approaches integrating genomics, proteomics, and metabolomics have effectively accelerated the growth of antimicrobial research (Karaosmanoglu et al., 2014; Plichta et al., 2012; Wecke and Mascher, 2011). In addition, several research groups have reported that bacterial cell division process is one of the exciting targets for the development of next-generation antibiotics due to its conserved nature among the prokaryotes. In recent years, many natural compounds, which can perturb the bacterial cell division, have been studied extensively as potential microbial agents (Jaiswal et al., 2007; Rai et al., 2008; Sun et al., 2014).
Plumbagin is a plant-derived naphthoquinone, a yellow natural compound extracted from the root of Plumbago zeylanica L. This medicinal shrub had a role in ancient medicine in many countries including India, China, and Ceylon. It has a wide range of activities, including anti-cancer (Acharya et al., 2008; Gomathinayagam et al., 2008), anti-malarial (Krungkrai et al., 2002), anti-fungal (Curreli, et al., 2001), anti-inflammatory (Luo et al., 2010), and anti-mutagenic activity (Edenharder and Tang, 1997). It also possesses anti-bacterial activity by generating reactive oxygen species, affecting respiratory process (Imlay and Fridovich, 1992), inhibiting NADH dehydrogenase (Imlay and Fridovich, 1992), and lactose carriers (Neuhaus and Wright, 1983). Recent biophysical studies have revealed that plumbagin treatment causes perturbation of bacterial cell division by elongating the cell length with multiple nucleoids (Bhattacharya et al., 2013).
Bacillus subtilis is sensitive, whereas Escherichia coli is resistant to plumbagin (Bhattacharya et al., 2013; de Paiva et al., 2003). Previous proteomics analysis investigating the effect of plumbagin on E. coli showed upregulation of multiple proteins (mar/sox regulon) involved in detoxification of reactive oxygen species (Chen et al., 2006; Lin et al., 2010). Here we report the first comprehensive proteomics analysis of plumbagin effects on a gram-positive bacterium: B. subtilis. The present study aimed to explore the proteome alterations of B. subtilis owing to plumbagin treatment by using two-dimensional electrophoresis (2-DE) and isobaric tag for relative and absolute quantification (iTRAQ)-based quantitative proteomics (LTQ-Orbitrap and Q-TOF). With these complementary proteomics technologies, we identified 18 and 230 differentially expressed proteins in 2-DE (p≤0.05) and iTRAQ-based quantitative proteomic analysis respectively [1% false discovery rate (FDR)]. In silico analysis of the differentially expressed proteins indicated modulation of TCA, heme biosynthesis, fatty acid synthesis, and ribosomes. Further, the metabolic activity assay using resazurin and gene expression analysis using RT-PCR were performed to validate the findings obtained from the discovery-phase proteomics analysis. In summary, this is the first comprehensive study on B. subtilis at the proteome level, to investigate the putative mode(s) of action of plumbagin and attendant cellular targets. Modern multi omics-guided drug target discovery and other biophysical studies are necessary to take traditional medicine forward.
Materials and Methods
Microscopic analysis of drug treatment
Bacillus subtilis AH75 strain containing spectinomycin resistance marker (plasmid having the spectinomycin antibiotic marker gene) obtained from Prof. R. Losick (Harvard University, Cambridge MA) was used to study the effect of plumbagin (Sigma, USA) (Handler et al., 2008). Two different concentrations of plumbagin: IC50 (5 μM) and 2×IC50 (10 μM) were used for morphological analysis (Bhattacharya et al., 2013). Cultures were grown for 2 h in the presence of the drug, and the same culture was used to monitor the morphological changes. DAPI (1 μg/mL) staining of nucleic acid was performed for 20 min in dark after fixing with 2.8% formaldehyde and 0.04% glutaraldehyde at 370C for 30 min. Fluorescence microscopic images (Eclipse TE-2000 U microscope; Nikon) at 40X magnification were captured for morphological observations.
Whole cell protein extraction
Global proteomic analysis was performed after treating the cultures with IC50 of plumbagin which can lead to filamentation, but allows sufficient cell viability for protein extraction. The in-house standardized protein extraction protocol was followed for this study (Reddy et al., 2013). In brief, plumbagin was added to the freshly growing B. subtilis cultures at OD 0.2 and grown further for 2 h to reach mid-exponential phase, whereas DMSO was added to the control. Cultures were harvested and washed with PBS buffer for 4 times to remove the media components. Cell lysis was done with lysozyme (1 mg/mL) treatment for 30 min and sonication (2 sec pulse, 2 sec gap till 2.5 min at 30% amplitude).
Classical two-dimensional electrophoresis and data analysis
600 μg of protein samples (control and 120 min plumbagin treated) were used for passive rehydration of 24 cm IPG strips (linear pH 4–7: GE Healthcare) for overnight. IEF was carried out with the same parameters as reported previously (Reddy et al., 2013). Focused IPG strips were equilibrated with DTT and IAA for 15 min each. 12.5% SDS-PAGE was used to perform the second-dimension separation using an Ettan DALTsix instrument (GE Healthcare) to minimize the technical artifacts. After electrophoretic separation, the gels were stained with PhastGel™ Blue R stain (GE Healthcare) for visualization of the protein spots. After staining and de-staining, the gels were scanned using LabScan 4.0 software (GE Healthcare) and imported into ImageMaster 2D Platinum 7.0 software (GE Healthcare) for differential proteomics analysis. Triplicates of control and plumbagin-treated gels were analyzed using automatic detection and matching tools. Afterwards, each spot was manually verified to remove the artifacts. Finally, the statistical analysis was performed using t-test (p≤0.05) for evaluation of the significance of differential expression.
In-gel digestion and protein identification
Differentially expressed (plumbagin treated vs. control) statistically significant (p≤0.05) protein spots were excised from the 2-DE gels and in-gel digestion was performed following the same protocol reported previously (Shevchenko et al., 2006; Reddy et al., 2013; Rao et al., 2014). In brief, the gel pieces were washed with 25 mm ABC and Sol-A (1:2 of 25 mM ABC and acetonitrile), followed by reduction (10 mM DTT in 100 mM ABC) for 1 h at 65°C and alkylation (50 mM IAA in 100 mM ABC) for 30 min in dark. Trypsin (Trypsin Gold; Promega, Madison, WI) was added and the gel pieces were incubated at 37°C for 16 h. The digested peptides were extracted using a gradient of ACN from 50%–80% and 0.1% TFA in the presence of mild sonication. Peptide extracts were further processed by using C-18 Zip-tips (Millipore, USA) and spotted on a MALDI plate along with CHCA matrix (5 mg/mL CHCA in 50% ACN/0.1% TFA) for co-crystallization. 4800 MALDI-TOF/TOF mass spectrometer (AB Sciex, Framingham, MA) linked to 4000 series explorer software (v.3.5.3) was used for protein identification with the mass range of 800–4000 Da and the laser used was Nd:YAG 355 nm after calibration. GPS™ Explorer software version 3.6 (AB Sciex) was used to generate the peak list and imported into MASCOT version 2.1 (http://www.matrixscience.com) search engine for protein identification using the following parameters: taxonomy, B. subtilis; database, Swiss-Prot; enzyme used for digestion, trypsin with single missed cleavage; fixed modification, carbamidomethylation of cysteine; variable modification, oxidation of methionine; mass tolerance,75 ppm for MS and 0.4 Da for MS/MS.
iTRAQ labeling, SCX fractionation, and OFFGEL fractionation
Two independent biological pools (each pool has three controls and three plumbagin-treated samples) were used for duplex iTRAQ labeling. 60 μg of proteins from each sample was subjected to in-solution digestion after treating with 2 μL of reducing reagent (TCEP) at 60°C for 1 h and 1 μL of cysteine blocking reagent (MMTS) for 10 min in dark. After in-solution digestion with trypsin (protein: trypsin 20:1) for 16 h at 37°C, iTRAQ labeling (Applied Biosystems Inc., Forster City, CA) of the digested peptides was performed with control-114 and plumbagin-117 for 1 h at RT, followed by quenching with milli Q water for 30 min. iTRAQ-labeled samples were divided into two aliquots and pre-fractionation was performed using strong cation exchange (SCX) chromatography and OFFGEL fractionation. Strong cation exchange (SCX) was performed using an Agilent 1100 series LC system connected with Poly-SULFOETHYL A column (PolyLC, Columbia, MD) (100×2.1 mm, 5 μm particles with 300 Å pores). A total of 96 fractions were collected using solution-A (5 mM KH2PO4 pH 2.7, 30% ACN) and solution-B (5 mM KH2PO4 pH 2.7, 30% ACN, 350 mM KCl) with a gradient of 0%–100% solution-B with a flow rate of 0.25 mL/min for 50 min. The second aliquot of labeled samples were pooled and fractionated using OFFGEL fractionation on a high resolution IPG strip (24 cm, pH 3–10) with default settings following the manufacture's instructions for peptide fractionation. A total of 24 fractions were collected and processed with C18 STAGE tips before being subjected to mass spectrometry.
LTQ-Orbitrap and QTOF mass spectrometry analysis
The SCX fractions were cleaned with C18 STAGE tips and injected into LTQ- Orbitrap Velos mass spectrometer (Thermo Fischer Scientific, Bermen, Germany) equipped with Proxeon Easy nLC liquid chromatography having magic C18 AQ reversed phase material (Michrom Bioresources, 5 μm,100 Å). Peptides were enriched with a trap column (75 mm×62 cm) at a flow rate of 3 μL/min and resolved with an analytical column (75 mm×10 cm) at a flow rate of 350 nL/min. In the analytical column, the peptides were eluted with linear gradient of 7%–35% ACN for 60 min. Full scan was performed with 60,000 resolution at 400 m/z and top 20 peaks were considered for MS/MS at 15,000 resolution at 400 m/z with 40% normalized collision energy using LTQ-Orbitrap mass analyzer having polydimethylcyclosiloxane (m/z, 445.1200025) for internal calibration.
The processed OFFGEL fractions were subjected to a 6550 ESI Q-TOF iFunnel instrument (Agilent Technology, Santa Clara, CA) coupled with 1260 Infinity HPLC-nano-chip. A polaris C18A chip (150 mm×0.075 mm) along with 160 nL trap column was used for peptide separation. A total run time for each fraction was 90 min with linear gradient of 7%–35% of acetonitrile for 60 min and 95% to 90 min. The flow rate was 2 μL/min in the capillary pump and 200 nL/min for the nano pump. Online MS and MS/MS data acquisition was performed with m/z range from 100–3200 with the speed of 6 spectra/sec in MS mode, and 3 spectra/sec in MS/MS mode and top 15 peaks were selected for MS/MS analysis.
Mass spectrometry data analysis
Data obtained from both LTQ-Orbitrap and Q-TOF were analyzed with Proteome Discoverer 1.3 (Thermo Fischer Scientific, Bermen, Germany) configured with SEQUEST (SCM build 59). Following search parameters: database, B. subtilis UniProtKB database having 4227 sequences; MS and MS/MS tolerance, 20 ppm and 0.1 Da, enzyme- trypsin with single missed cleavage; modifications, iTRAQ on N-terminal, lysine and alkylation as a fixed modifications and oxidation of methionine as a variable modification were employed for protein identification. Protein quantitation was performed by measuring the reporter ion intensity and data were validated using a validator node for FDR calculations. Data were normalized with ‘normalize on protein median’ with minimum protein count as 20 proteins.
Functional annotation and pathway analysis
DAVID (Database for Annotation, Visualization and Integrated Discovery) database version 6.7 (http://david.abcc.ncifcrf.gov/home.jsp) was used for functional annotation of the differentially expressed proteins obtained from our proteomics analysis. The list of UniProt accession IDs were uploaded into the DAVID database as a tab limited text and mapped against a B. subtilis dataset as a reference for pathway analysis with the default parameters (Huang et al., 2009a; 2009b). Pathway analysis was also performed by using KOBAS, which can identify the statistically enriched pathways (Xie et al., 2011).
Resazurin microtiter assay for metabolic activity
Inoculum was prepared from untreated control, IC50 (5 μM) and 2×IC50 (10 μM) plumbagin-treated B. subtilis samples with four different dilutions having cell numbers in the range of 108 to 106 cells/mL. Cell pellets were washed with PBS buffer for three times to remove the media components. Resazurin dye was added to each culture with a concentration of 10 μg/mL and the fluorescence intensity of resorufin was measured using a RT-PCR (MyiQ2 system, BioRad, Hercules, CA) machine for next 30 min with 15 sec intervals at 590 nm. Resazurin is the tracer dye, which was used to estimate the aerobic respiration based on metabolic activity. Actively growing cells were able to reduce the resazurin into fluorescence resorufin, which absorbs light at 590 nm.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from the control and plumbagin treated (5 μm) B. subtilis (20 mL culture each) using TRIzol LS reagent (Invitrogen, Carlsbad, CA) following manufacturer's protocol, and quality was checked by agarose gel electrophoresis. 4 μg of RNA was used for cDNA synthesis using RevertAid™ first strand kit (Fermentas, Europe) following the manufacturer's instructions. Eco Real-Time PCR system (Eco Real-Time PCR, Illumina, USA) was used for gene expression analysis of target genes containing 1×Maxima™ SYBR Green qPCR Master Mix (Fermentas), primers (Table 1) and cDNA template with the following settings: initial denaturing at 95°C for 5 min, 40 cycles of 95°C for 15 sec, annealing at 46°C for 15 sec, and extension at 72°C for 30 sec, followed by a melting curve for 15 sec at 95°C, 15 sec at 46°C, and finally again at 95°C for 15 sec. Data were normalized using 16S rRNA as an internal control and relative gene expression analysis was performed in triplicate.
Table 1.
Primers Used for Quantitative RT-PCR Analysis for Selected Genes
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Annealing temperature (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| ClpX | 5′CAGGTTCGTAAGCTTGTAG3′ | 5′GTGGTTATACACAGCAACAG3′ | 46 | 213 |
| MurAA | 5′GTACAGGTCATGCAAGAGT3′ | 5′TTCTCTGTAGCTCCTACACT3′ | 46 | 196 |
| Hit | 5′CTTGATATCAGCCAAGTGAC3′ | 5′GTAGTGGAACACAGATTGTC3′ | 46 | 210 |
| 16S rRNA | 5′GATCTTAGTTGCCAGCATTC-3′ | 5′TTACTAGCGATTCCAGCTTC-3′ | 46 | 233 |
3D structure modeling and molecular docking
3D structures of NADH dehydrogenase from both B. subtilis and E. coli were modeled using the Ab initio algorithm of I-TASSER server (Roy et al., 2010). The server predicted the accuracy with an average error of 0.90±0.06 TM score and 4.2±2.8 Å RMSD for E. coli and 0.99±0.04 TM score and 3.1±2.2 Å RMSD for B. subtilis. 3D structure of plumbagin was retrieved from PubChem compound database (CID 10205). Docking was performed using AutoDock 4.0 (Huey et al., 2007) and the protein molecules and the drug were converted in .pdbqt format using ADT tools. Blind docking was performed with plumbagin for NADH dehydrogenase from both B. subtilis and E. coli separately with default settings; 100 runs for each protein were executed to get the least docking energy and inhibition constant. The docked structures were visualized with DS visualiser v2.0. Top five docking sites were considered based on ΔG values.
Results
Morphological changes in B. subtilis
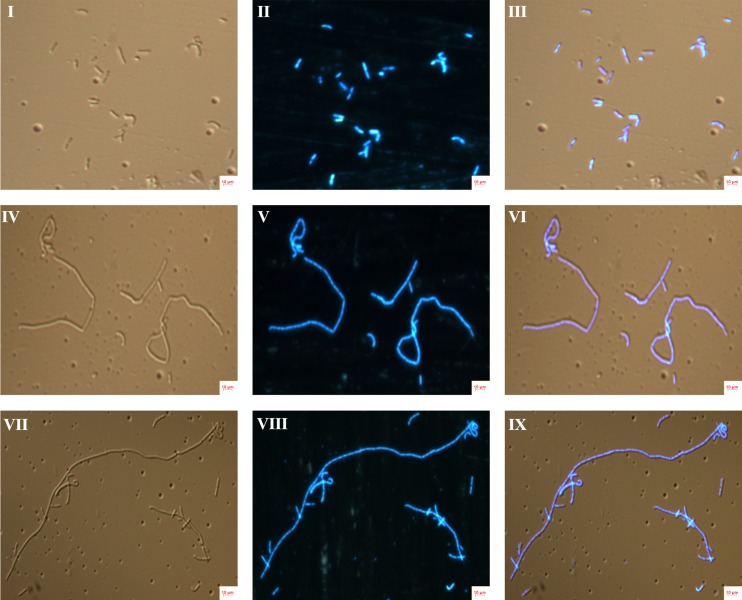
Morphological analysis of control, IC50 (5 μM) and 2×IC50 (10 μM) plumbagin-treated B. subtilis cultures were performed using fluorescence microscopy. Microscopic analysis indicated that control cells exhibit typical normal cell length with single or double nucleoids per cell, but in the case of the IC50 and 2×IC50 plumbagin treatments, the cells were found to be elongated with multi-nucleoid per cell. Both IC50 and 2×IC50 plumbagin-treated cells exhibited elongated cell morphology, but there was no significant difference between the cell length and viability (Fig. 1). IC50 plumbagin-treated samples were used for proteome analysis because we need to have sufficient biomass for protein extraction and IC50 is considered as a standard for studying the proteome level alterations after drug treatment.
FIG. 1.
Effect of plumbagin treatment on the B. subtilis cell morphology. B. subtilis AH75 strains were grown in the presence of IC50 (5 μM), 2×IC50 (10 μM) and absence (control) of plumbagin for 2 h; nuclear materials were stained using 1 μg/μL DAPI for 20 min. The fluorescence microscopic images were captured with both DAPI and DIC filters. I, II, and III are the DIA, DAPI, and overlaid images of control; IV, V, and VI are the DIA, DAPI, and overlaid images of IC50 plumbagin treated samples; VII, VIII, and IX indicating the DIA, DAPI, and overlaid images of 2×IC50 plumbagin treated samples.
Proteome analysis of plumbagin treated B. subtilis using 2-DE
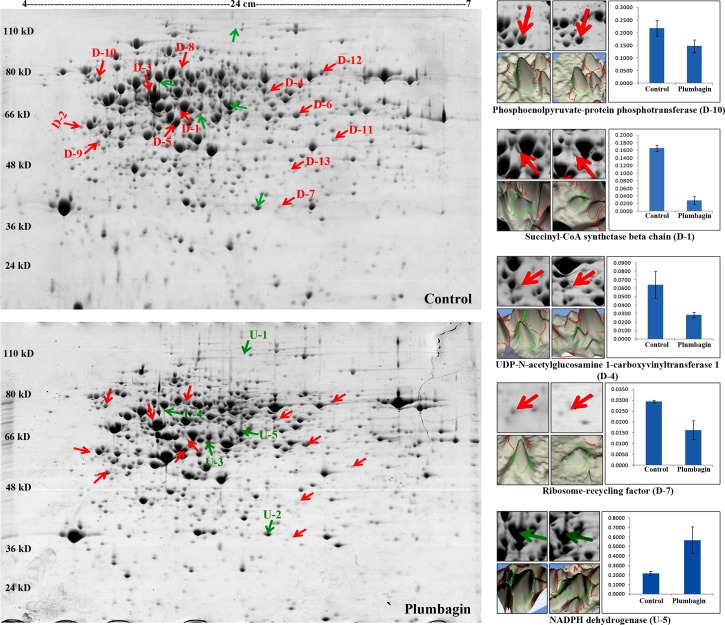
Proteomic analysis of untreated control and plumbagin-treated (IC50) B. subtilis showed more than 1000 spots on the CBB stained 2-DE gels. Comparative analysis of triplicates of control and plumbagin-treated samples indicated the differential expression of 18 protein spots with statistical significance (p≤0.05). Among the differentially expressed spots, 13 were down regulated and 5 were up regulated with respect to the expression levels observed in controls. Representative 2-DE images for control and plumbagin-treated samples with selected differentially expressed protein spots, and their 3D views and histograms are displayed in Figure 2 and Supplementary Figure S1 (supplementary material is available on line at www.liebertpub.com/omi). Identification of all the significantly differentially expressed spots were performed by MALDI-TOF/TOF analysis. Only succinyl-CoA synthetase beta chain was identified as two different spots on the 2-DE gels, most probably due to the protein degradation or presence of multiple isoforms (Table 2 and Supplementary Table S1). Most of the 2-DE identified proteins were also observed in quantitative iTRAQ analysis with a similar trend.
FIG. 2.
Representative 2-DE gel images of B. subtilis proteome in response to the plumbagin treatment (IC50). 3D views, histogram and gel position views for selected differentially expressed protein spots are displayed.
Table 2.
Significant Proteins Identified on 2-DE and its Comparison with iTRAQ Data
| Trend | Name of the protein | M.wt | 2-DE | iTRAQ | Biological function | Molecular component |
|---|---|---|---|---|---|---|
| D-1 | Succinyl-CoA synthetase beta chain | 41.34 | −5.79 | −2.59 | TCA | Ligase |
| D-2 | Acetoin:2,6-dichlorophenolindophenol oxidoreductase subunit beta | 36.69 | −3.67 | −4.31 | Acetoin catabolism | Oxidoreductase |
| D-3 | Elongation factor Tu | 43.56 | −2.40 | −1.79 | Protein biosynthesis | Elongation factor |
| D-4 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | 46.67 | −2.24 | −1.06 | Cell division, cell wall biogenesis | Transferase |
| D-5 | Succinyl-CoA ligase [ADP-forming] subunit beta | 41.34 | −2.06 | −2.59 | TCA | Ligase |
| D-6 | Probable butyrate kinase | 39.73 | −2.05 | −5.41 | NA | Kinase |
| D-7 | Ribosome-recycling factor | 20.62 | −1.81 | −1.01 | Protein biosynthesis | NA |
| D-8 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | 47.37 | −1.69 | −1.04 | Glycolysis | Transferase |
| D-9 | Manganese-dependent inorganic pyrophosphatase | 33.96 | −1.57 | −1.08 | NA | Hydrolase |
| D-10 | Phosphoenolpyruvate-protein phosphotransferase | 63.03 | −1.48 | −1.41 | Transport | Kinase, Transferase |
| D-11 | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2 | 35.40 | −1.46 | −1.58 | Lipid biosynthesis, fatty acid biosynthesis | Transferase |
| D-12 | D-3-phosphoglycerate dehydrogenase | 57.07 | −1.41 | −1.72 | Serine biosynthesis | Oxidoreductase |
| D-13 | Transcriptional regulatory protein degU | 25.85 | −1.13 | −1.59 | Transcription, two component system | Activator/repressor |
| U-1 | Isoleucyl-tRNA synthetase | 104.7 | 1.78 | 1.27 | Protein synthesis | Ligase |
| U-2 | Uncharacterized protein yvyD | 21.96 | 2.11 | 1.70 | Response | NA |
| U-3 | Glyceraldehyde-3-phosphate dehydrogenase 1 | 35.67 | 2.14 | 2.31 | Glycolysis | Oxidoreductase |
| U-4 | ATP-dependent Clp protease ATP-binding subunit ClpX | 46.37 | 2.23 | −1.09 | Stress response | Chaperone |
| U-5 | NADPH dehydrogenase | 55.12 | 2.63 | 3.56 | Detoxification, Stress response | Oxidoreductase |
NA=not available.
Modulation in protein expressions after plumbagin treatment identified by quantitative proteome analysis using LTQ-orbitrap and Q-TOF
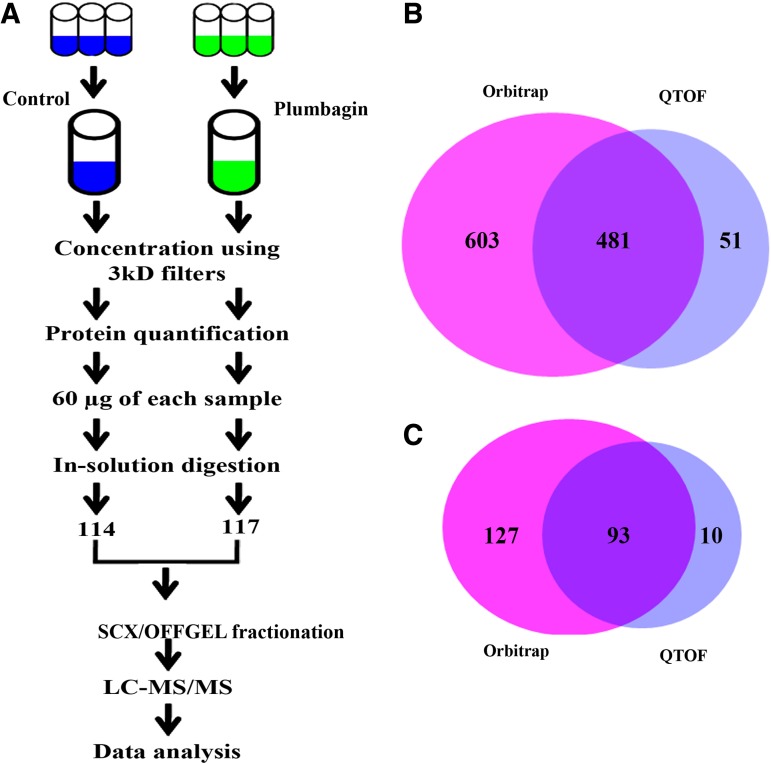
Quantitative iTRAQ analysis of two independent pooled biological replicates (each biological pool contained control (n=3) and plumbagin (n=3)) samples were performed using LTQ-Orbitrap and Q-TOF (Fig. 3A). Using proteome discoverer search engine, 1084 and 532 proteins were identified in LTQ-Orbitrap and Q-TOF, respectively (1% FDR). The comparative analysis of the proteins identified in LTQ-Orbitrap and Q-TOF indicated that 481 proteins were common between the two techniques, whereas 603 proteins were unique to LTQ-Orbitrap and 51 proteins were unique to Q-TOF (Fig. 3B). Further analysis of LTQ-Orbitrap and Q-TOF identified proteins indicated that 230 proteins (115 up regulated and 115 down regulated) were differentially expressed (1% FDR, 1.5-fold change and ≥2 peptides) either with similar trend in LTQ-Orbitrap and Q-TOF analysis or unique to one of the MS analyses. Among the 230 proteins, 93 proteins were common between LTQ-Orbitrap and Q-TOF, whereas 127 proteins were unique to LTQ-Orbitrap and 10 proteins were unique to Q-TOF (Fig. 3C). The quantitative information of the differential expressed proteins, peptides information, scores, sequence coverage, accession numbers and reporter ion intensity are presented in Table 3 and Supplementary Table S2. In addition, comparison of 2-DE and iTRAQ data (LTQ-orbitrap) showed a similar trend of differential expression for all the 18 proteins (5 up regulated and 13 down regulated) identified in 2-DE.
FIG. 3.
(A) Schematic representation of the iTRAQ-based quantitative proteomics workflow used in the present study. (B) Venn diagram representing the comparative analysis of the total identified proteins from LTQ-Orbitrap and Q-TOF analysis. (C) Venn diagram representing the differentially expressed proteins identified in LTQ-Orbitrap and Q-TOF (1.5-fold change, and identified with ≥2 peptides) analysis.
Table 3.
Partial List of Differentially Expressed Proteins in B. subtilis after Plumbagin Treatment Obtained from iTRAQ-Based Quantitative Proteome Analysisa
| Orbitrap | Q-TOF | |||||||
|---|---|---|---|---|---|---|---|---|
| Accession | Name of the protein | Gene name | Coverage | Unique peptides | Plumbagin/control | Coverage | Unique peptides | Plumbagin/control |
| TCA cycle | ||||||||
| P09124 | Glyceraldehyde-3-phosphate dehydrogenase 1 | GapA | 42.39 | 12 | 2.31 | 56.72 | 13 | 2.647 |
| Q03224 | Fructose-1,6-bisphosphatase class 2 | glpX | 22.43 | 6 | 0.464 | – | – | – |
| P54533 | Dihydrolipoyl dehydrogenase | BfmBC | 19.20 | 8 | 0.377 | 15.61 | 5 | 0.520 |
| P80886 | Succinyl-CoA ligase [ADP-forming] subunit beta | SucC | 47.53 | 16 | 0.386 | 62.08 | 22 | 0.495 |
| P80865 | Succinyl-CoA ligase [ADP-forming] subunit alpha | sucD | 29.00 | 5 | 0.465 | – | – | – |
| P08066 | Succinate dehydrogenase iron-sulfur subunit | sdhB | 18.97 | 4 | 0.313 | – | – | – |
| P39126 | Isocitrate dehydrogenase [NADP] OS | Icd | 36.17 | 15 | 0.581 | 55.32 | 21 | 0.488 |
| P09339 | Aconitate hydratase OS | CitB | 23.54 | 17 | 0.317 | 24.31 | 16 | 0.427 |
| P39120 | Citrate synthase 2 OS | CitZ | 27.15 | 7 | 0.454 | 37.90 | 12 | 0.471 |
| Electron transport | ||||||||
| P94551 | Electron transfer flavoprotein subunit alpha | etfA | 9.54 | 2 | 0.478 | – | ||
| P80861 | NADH dehydrogenase-like protein YjlD | yjlD | 30.10 | 8 | 0.335 | 51.53 | 16 | 0.499 |
| O32117 | NADH dehydrogenase-like protein YutJ | yutJ | 12.11 | 3 | 0.552 | – | – | – |
| P42974 | NADH dehydrogenase OS | AhpF | 25.15 | 12 | 1.57 | 35.36 | 13 | 1.592 |
| P54524 | Probable NADH-dependent flavin oxidoreductase YqiG OS | yqiG | 18.82 | 5 | 3.926 | 24.73 | 6 | 1.240 |
| P42175 | Nitrate reductase alpha chain | narG | 4.72 | 4 | 1.650 | – | – | – |
| P42176 | Nitrate reductase beta chain | narH | 5.95 | 2 | 2.469 | – | – | – |
| P39605 | FMN reductase (NADPH) | nfrA1 | 24.10 | 4 | 2.774 | 34.54 | 5 | 1.262 |
| P94424 | FMN reductase [NAD(P)H] | nfrA2 | 20.88 | 4 | 7.150 | 16.06 | 3 | 6.020 |
| Fatty acid biosynthesis | ||||||||
| P49786 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase OS | accB | 28.93 | 2 | 0.464 | 33.33 | 3 | 0.698 |
| P49787 | Biotin carboxylase 1 OS | accC1 | 18.22 | 6 | 0.462 | 26.44 | 8 | 0.707 |
| P94549 | Probable enoyl-CoA hydratase OS | fadB | 10.85 | 2 | 0.581 | |||
| P94584 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ OS | fabZ | 9.93 | 2 | 1.799 | 5.67 | 1 | 3.105 |
| O07600 | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2 OS | fabHB | 7.08 | 2 | 0.634 | – | – | – |
| P54616 | Enoyl-[acyl-carrier-protein] reductase [NADH] FabI OS | fabI | 39.53 | 9 | 0.440 | 27.91 | 6 | 0.595 |
| Cell wall and cell division | ||||||||
| P28264 | Cell division protein FtsA OS | FtsA | 11.36 | 4 | 0.529 | |||
| P28015 | Putative septation protein SpoVG OS | SpoVG | 40.21 | 3 | 0.475 | |||
| P26497 | Stage 0 sporulation protein J OS | Spo0J | 32.27 | 7 | 0.563 | |||
| P45693 | Stage V sporulation protein S OS | SpoVS | 38.37 | 2 | 0.617 | |||
| Q02114 | N-acetylmuramoyl-L-alanine amidase LytC | lytC | 5.24 | 2 | 0.221 | – | – | – |
| P54421 | Probable peptidoglycan endopeptidase LytE | lytE | 8.08 | 2 | 0.266 | – | – | – |
| P96612 | D-alanine-D-alanine ligase OS | ddl | 5.93 | 2 | 0.644 | – | – | – |
| P80698 | Trigger factor OS | Tig | 36.08 | 13 | 3.354 | 59.67 | 20 | 1.664 |
| P19670 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase 2 OS | MurAB | 11.66 | 3 | 2.115 | 10.96 | 3 | 2.120 |
| P39131 | UDP-N-acetylglucosamine 2-epimerase | MnaA | 11.32 | 3 | 1.639 | – | – | – |
| P42976 | 4-hydroxy-tetrahydrodipicolinate reductase | dapB | 14.98 | 2 | 1.799 | 40.07 | 7 | 2.647 |
| Heme biosynthesis | ||||||||
| P30950 | Delta-aminolevulinic acid dehydratase | HemB | 34.26 | 7 | 2.42 | 64.20 | 15 | 1.944 |
| P32396 | Ferrochelatase OS | HemH | 22.58 | 6 | 2.09 | 26.13 | 6 | 1.369 |
| P30949 | Glutamate-1-semialdehyde 2,1-aminomutase | HemL | 21.86 | 6 | 2.06 | 34.88 | 9 | 2.022 |
| P16616 | Porphobilinogen deaminase OS | HemC | 27.07 | 6 | 1.86 | 35.03 | 8 | 2.116 |
| Protein synthesis | ||||||||
| P21473 | 30S ribosomal protein S15 OS | rpsO | 19.10 | 4 | 4.689 | 41.57 | 3 | 1.300 |
| P21474 | 30S ribosomal protein S16 OS | rpsP | 55.56 | 4 | 3.141 | 63.33 | 5 | 2.220 |
| P21476 | 30S ribosomal protein S19 OS | rpsS | 50.00 | 6 | 3.357 | 48.91 | 5 | 1.735 |
| P21477 | 30S ribosomal protein S20 OS | rpsT | 23.86 | 2 | 15.518 | 26.14 | 3 | 1.220 |
| P21470 | 30S ribosomal protein S9 OS | rpsI | 49.23 | 4 | 2.190 | 49.23 | 4 | 1.698 |
| Q06796 | 50S ribosomal protein L11 OS | rplK | 44.68 | 6 | 1.814 | 66.67 | 8 | 1.430 |
| P19946 | 50S ribosomal protein L15 OS | rplO | 40.41 | 5 | 1.908 | 52.05 | 7 | 1.684 |
| P20277 | 50S ribosomal protein L17 OS | rplQ | 45.00 | 5 | 1.975 | 39.17 | 5 | 1.273 |
| P05657 | 50S ribosomal protein L27 OS | rpmA | 43.62 | 5 | 6.451 | 43.62 | 5 | 3.351 |
| P42920 | 50S ribosomal protein L3 OS | rplC | 39.23 | 6 | 1.599 | 48.33 | 7 | 2.472 |
| P19947 | 50S ribosomal protein L30 OS | rpmD | 52.54 | 4 | 6.322 | 42.37 | 4 | 1.251 |
| O34967 | 50S ribosomal protein L31 type B OS | rpmE2 | 30.49 | 2 | 2.998 | – | – | – |
| P55874 | 50S ribosomal protein L35 OS | rpmI | – | – | – | 45.45 | 4 | 0.434 |
Full list of the proteins are described in Supplementary Table ST2.
Alterations in different physiological pathways due to plumbagin treatment
DAVID and KOBAS were used for pathway analysis using the 230 differentially expressed proteins identified in the discovery phase proteomic analysis. Pathway analysis indicated significant alteration of citric acid cycle, ribosome, heme biosynthesis, and fatty acid biosynthesis in B. subtilis due to plumbagin treatment. The complete information of the modulated pathways is provided in Supplementary Table S3.
Metabolic activity assays for validation of respiratory arrest
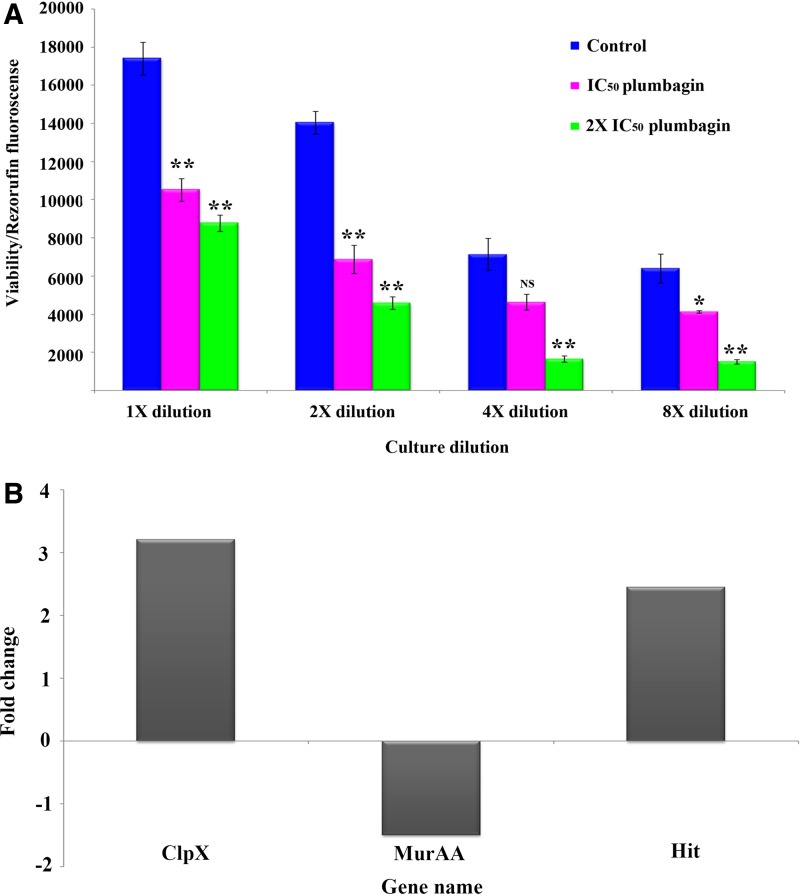
Cell viability/metabolic activity assay was performed with resazurin reagent, which is a nonfluorescent dye converted into pink resorufin by using intracellular redox equivalents. Measurement of fluorescent resorufin showed that the control (untreated) samples had relatively higher intensity of resorufin as compared to the IC50 (5 μM) and 2×IC50 (10 μM) plumbagin-treated samples, which could be due to the existence of active metabolism in controls compared to the plumbagin-treated samples (Fig. 4A).
FIG. 4.
(A) Resazurin assay for metabolic activity. Control, IC50, and 2×IC50 plumbagin treated cultures were used for assay with 10 μg/mL of resazurin, and the product (resorufin) formed in active cells was measured at 590 nm; NS indicates p>0.05; *indicates p<0.05, ** indicates p<0.005 in a paired t-test analysis. (B) The relative gene expression patterns of ClpX, MurAA and Hit calculated by taking the mean Ct values from triplicate runs (* indicates p<0.05 in a paired t-test analysis).
Gene expression analysis for validation of proteomics data
Gene expression analysis of selected candidates under plumbagin treatment was performed to correlate their gene (mRNA) and protein expression levels. UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurAA), Protein hit (Hit) and ATP-dependent Clp protease ATP-binding subunit ClpX (ClpX) were selected for quantitative real-time PCR analysis based on their fold-change of differential expression and physiological roles. Expression analysis of all the above mentioned genes were performed in triplicates after total RNA extraction with TRIzol. Alterations in expression patterns of three genes ClpX, MurAA, and Hit under plumbagin treatment were consistent with the proteome level findings (Fig. 4B). MurAA involved in cell wall biosynthesis has showed 1.5-fold down regulation, whereas Hit and ClpX have showed 2.5- and 3.2-fold up regulation at mRNA level, respectively.
Molecular docking for interaction analysis
NADH dehydrogenase protein was modeled with I-TASSER from both B. subtilis and E. coli and was docked with plumbagin. Blind docking was performed with AutoDock 4 program with default settings and 100 runs. The top five docking sites were considered for the binding energies, inhibition constant, and other parameters. The top hit model is displayed in Supplementary Figure S2. Docking results indicate that in the case of B. subtilis, NADH dehydrogenase has two hydrogen bonds with plumbagin, whereas E. coli NADH dehydrogenase does not form any hydrogen bond with the drug. Consequently, B. subtilis NADH dehydrogenase has comparatively higher affinity to plumbagin than E. coli NADH dehydrogenase (Supplementary Table S4).
Discussion
Quantitative proteomics analysis is frequently used to study the effect of various drugs and environmental stress on diverse organisms to understand the physiological responses against those adverse conditions. Previous reports on plumbagin have suggested that respiratory chain (Imlay and Fridovich, 1992), NADH dehydrogenase (Imlay and Fridovich, 1992), and FtsZ are the potential targets (Bhattacharya et al., 2013) of the drug; however, its obvious mechanism of action is still obscure. For the first time, we have used two complementary quantitative proteomics techniques (2-DE and iTRAQ) to investigate the physiological response and possible mechanism of action of plumbagin on a gram-positive organism B. subtilis.
Aerobic respiration is a fundamental process for cellular energy production via glycolysis, citric acid cycle (TCA), and electron transport chain. TCA is the prime pathway to generate the majority of reducing equivalents (i.e., NADH in cell) and generates the proton motive force or membrane potential across the membrane essential for ATP synthesis (Möbius et al., 2010). Our results indicate that plumbagin treatment significantly repressed TCA and the electron transport chain, which indicates the blockage in energy generation. Alternatively, glycolysis, which can provide the energy via substrate level phosphorylation, anaerobic respiration or fermentative process to support the growth, was found to be induced due to plumbagin treatment (Cruz Ramos et al., 2000; Kohler et al., 2003) (Fig. 5).
FIG. 5.
Modulation of the central metabolism and fatty acid biosynthesis due to plumbagin treatment. This pathway is generated based on the information obtained from KOBAS and DAVID analysis.
Moreover, the induction of nitrate reductase alpha and nitrate reductase beta chain strongly indicates the anaerobic respiration (Cortial et al., 2010; Kohler et al., 2003; Ye et al., 2000) and induction of anaerobic marker protein YwfI suggests that the anaerobic respiration was dominated under plumbagin treatment (Marino et al., 2000). In addition, nitroreductases involved in oxidation of NADH and scavengers of peroxides were induced. Consequently, metabolic activity using the resazurin assay also showed that respiration process was inhibited significantly after plumbagin treatment. A study by Imlay et al. (1992) on E. coli demonstrated that plumbagin reversibly inhibits the NADH dehydrogenase activity (Km=22 μM) and blocks respiration, but interestingly in our study, NADH dehydrogenase was slightly induced (1.5-fold up). Our molecular docking analysis indicated that the inhibition constant of NADH dehydrogenase (Ki=28.14 μM) was close to the experimental results (Km=22 μM) in E. coli, (Imlay et al., 1992) whereas B. subtilis NADH dehydrogenase exhibited slightly more affinity (Ki=22.7 μM); however, no experimental data are available in literature to support this observation. We presume that plumbagin might inhibit the NADH dehydrogenase activity in B. subtilis at molecular level similar to E. coli NADH dehydrogenase (Supplementary Fig. S2).
Heme is the major component of the electron transport chain, which can transport electrons from NADH to molecular oxygen for energy generation. Interestingly, most of the enzymes involved in heme synthesis coupled to electron transport were found to be induced significantly after plumbagin treatment. The enhanced heme biosynthesis is probably an indication of the reprogramming of the normal growth via anaerobic respiration (Ye et al., 2000). Besides, plumbagin is believed to be an antimicrobial agent by generating the reactive oxygen species. As expected, catalase and superoxide dismutase were induced significantly to protect the cell from the reactive oxygen species (Imlay and Fridovich, 1992).
Recent findings highlighted that many cell division targeting drugs disturb the membrane potential or membrane permeability, which is not only essential for energy production via respiration, but also inevitable for the cell division process (Foss et al., 2012; Strahl and Hamoen, 2008). Similar results were reported with lantibiotics, indole, and daptomycin, which can disturb the membrane potential or PMF and lead to mislocalization of the cell division proteins (Chimerel et al., 2012; Pogliano et al., 2012; Wenzel et al., 2012). In addition, fatty acid synthesis, which is crucial for phospholipid biosynthesis to maintain membrane permeability during vegetative growth, was repressed significantly after plumbagin treatment (Fig. 5). We anticipate that plumbagin treatment may lead to the loss of membrane permeability, which is essential for ATP synthesis and localization of essential cell division proteins (Paoletti et al., 2007; Pogliano et al., 2012; Strahl and Hamoen, 2010).
On the other hand, crucial proteins involved in bacterial cell division were found to be altered under plumbagin treatment. Cell division protein FtsA plays very important roles in both vegetative and asymmetric cell division along with FtsZ protein. FtsA is the membrane anchored protein and provides strength to FtsZ polymers during cell division by physically interacting with FtsZ. SpoVG also plays a role in asymmetric cell division (Feucht et al., 2001; Matsuno et al., 1999). In our study, expression levels of both FtsA and SpoVG were found to be repressed by plumbagin treatment. Quite a few proteins such as N-acetylmuramoyl-L-alanine amidase LytC, probable peptidoglycan endopeptidase LytE, and D-alanine-D-alanine ligase involved in cell separation during division, cell wall synthesis, and motility were also found to be significantly reduced. MurAA, which is one of the ClpX target and involved in the first committed step in cell wall biosynthesis (Kock et al., 2004), was repressed at both protein and RNA levels, whereas MurAB involved in cell wall synthesis along with MurAA was induced. In addition, ATP-dependent Clp protease ATP-binding subunit ClpX, which is the major protease family protein that regulates the cellular protein quality, including cell division protein FtsZ and cell wall biosynthesis, was found to be induced (Camberg et al., 2011; Haeusser et al., 2009; Krüger et al., 2000). From these findings we anticipate that MurAA was repressed slightly due to the induction of ClpX, which can degrade MurAA. In addition, expression levels of quite a few of the ribosomal (both 30S and 50S subunits) proteins involved in protein synthesis were significantly induced after plumbagin treatment.
Conclusion
This is the first report, to the best of our knowledge, offering new insights, at a proteome level, for the putative mode(s) of action of plumbagin and attendant cellular targets in B. subtilis. The findings also suggest new ways forward for omics-guided drug target discovery building on traditional plant medicine. Respiratory arrest due to plumbagin treatment is an interesting finding, which was demonstrated by the alteration of primary dehydrogenases, electron transporter proteins linked to cell division process. In addition, expression levels of multiple proteins linked to cell division process, including FtsA, SpoVG, MurAA, and ClpX, were directly affected. Notably, we found that plumbagin can also inhibit the bacterial cell division machinery in B. subtilis by elongating the cell length to several folds. We anticipate that plumbagin blocks cell division by altering the membrane permeability required for energy generation. Further in-depth analysis using multi-omics, medicinal chemistry, microbiology, and biophysical approaches together will be useful to completely decipher the molecular targets and mechanism of action of the drug.
Supplementary Material
Acknowledgments
This research was supported by a start-up grant 09IRCC007 from IIT Bombay to SS and a grant from Department of Science and Technology, Government of India, to DP. We thank the Center for Research in Nanotechnology and Science (CRNTS), Indian Institute of Technology Bombay, for providing the fluorescence activated cell sorting (FACS) and LC-MS/MS facilities and MALDI-TOF/TOF central facility, BSBE, IITB. We sincerely thank Dr. Richard Losick (Harvard University, Cambridge, MA) for providing us the B. subtilis AH75 strain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Acharya BR, Bhattacharyya B, and Chakrabarti G. (2008). The natural naphthoquinone plumbagin exhibits antiproliferative activity and disrupts the microtubule network through tubulin binding. Biochemistry 47, 7838–7845 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Jindal B, Singh P, Datta A, and Panda D. (2013). Plumbagin inhibits cytokinesis in Bacillus subtilis by inhibiting FtsZ assembly: A mechanistic study of its antibacterial activity. FEBS J 280, 4585–4599 [DOI] [PubMed] [Google Scholar]
- Camberg JL, Hoskins JR, and Wickner S. (2011). The interplay of ClpXP with the cell division machinery in Escherichia coli. J Bacteriol 193, 1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JW, Sun CM, Sheng WL, Wang YC, and Syu WJ. (2006). Expression analysis of up-regulated genes responding to plumbagin in Escherichia coli. J Bacteriol 188, 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimerel C, Field CM, Piñero-Fernandez S, Keyser UF, and Summers DK. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim Biophys Acta 1818, 1590–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian KA, Ijaz K, Dowell SF, et al. (2013). What we are watching—five top global infectious disease threats, 2012: A perspective from CDC's Global Disease Detection Operations Center. Emerg Health Threats J 6, 20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortial S, Chaignon P, Iorga BI, et al. (2010). NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: Insight into its biological role. FEBS Lett 584, 3916–3922 [DOI] [PubMed] [Google Scholar]
- Cruz Ramos H, Hoffmann T, Marino M, et al. (2000). Fermentative metabolism of Bacillus subtilis: Physiology and regulation of gene expression. J Bacteriol 182, 3072–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curreli N, Sollai F, Massa L, et al. (2001). Effects of plant-derived naphthoquinones on the growth of Pleurotus sajor-caju and degradation of the compounds by fungal cultures. J Basic Microbiol 41, 253–259 [DOI] [PubMed] [Google Scholar]
- de Paiva SR, Figueiredo MR, Aragão TV, and Kaplan MA. (2003). Antimicrobial activity in vitro of plumbagin isolated from Plumbago species. Mem Inst Oswaldo Cruz 98, 959–691 [DOI] [PubMed] [Google Scholar]
- Edenharder R, and Tang X. (1997). Inhibition of the mutagenicity of 2-nitrofluorene, 3-nitrofluoranthene and 1-nitropyrene by flavonoids, coumarins, quinones and other phenolic compounds. Food Chem Toxicol 35, 357–372 [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, and Errington J. (2001). Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol 40, 115–125 [DOI] [PubMed] [Google Scholar]
- Foss MH, Eun YJ, Grove CI, et al. (2013). Inhibitors of bacterial tubulin target bacterial membranes in vivo. Med Chem Comm 4, 112–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomathinayagam R, Sowmyalakshmi S, Mardhatillah F, Kumar R, Akbarsha MA, and Damodaran C. (2008). Anticancer mechanism of plumbagin, a natural compound, on non-small cell lung cancer cells. Anticancer Res 28, 785–792 [PubMed] [Google Scholar]
- Haeusser DP, Lee AH, Weart RB, and Levin PA. (2009). ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J Bacteriol 191, 1986–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AA, Lim JE, and Losick R. (2008). Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol 68, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay SI, Battle KE, Pigott DM, et al. (2013). Global mapping of infectious disease. Philos Trans R Soc Lond B Biol Sci 368, 20120250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay SI, George DB, Moyes CL, and Brownstein JS. (2013). Big data opportunities for global infectious disease surveillance. PLoS Med 10, e1001413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA. (2009a). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA. (2009b). Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc 4, 44–57 [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, and Goodsell DS. (2007). A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28, 1145–1152 [DOI] [PubMed] [Google Scholar]
- Imlay J, and Fridovich I. (1992). Exogenous quinones directly inhibit the respiratory NADH dehydrogenase in Escherichia coli. Arch Biochem Biophys 296, 337–346 [DOI] [PubMed] [Google Scholar]
- Jaiswal R, Beuria TK, Mohan R, Mahajan SK, and Panda D. (2007). Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry 46, 4211–4220 [DOI] [PubMed] [Google Scholar]
- Karaosmanoglu K, Sayar NA, Kurnaz IA, and Akbulut BS. (2014). Assessment of berberine as a multi-target antimicrobial: A multi-omics study for drug discovery and repositioning. OMICS 18, 42–53 [DOI] [PubMed] [Google Scholar]
- Kock H, Gerth U, and Hecker M. (2004). MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol Microbiol 51, 1087–1102 [DOI] [PubMed] [Google Scholar]
- Kohler C, von Eiff C, Peters G, Proctor RA, Hecker M, and Engelmann S. (2003). Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J Bacteriol 185, 6928–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger E, Witt E, Ohlmeier S, Hanschke R, and Hecker M. (2000). The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol 182, 3259–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krungkrai J, Kanchanarithisak R, Krungkrai SR, and Rochanakij S. (2002). Mitochondrial NADH dehydrogenase from Plasmodium falciparum and Plasmodium berghei. Exp Parasitol 100, 54-961 [DOI] [PubMed] [Google Scholar]
- Lin CN, Syu WJ, Sun WS, et al. (2010). A role of ygfZ in the Escherichia coli response to plumbagin challenge. J Biomed Sci 17, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Wong YF, Ge L, et al. (2010). Anti-inflammatory and analgesic effect of plumbagin through inhibition of nuclear factor-κB activation. J Pharmacol Exp Ther 335, 735–742 [DOI] [PubMed] [Google Scholar]
- Marino M, Hoffmann T, Schmid R, Möbitz H, and Jahn D. (2000). Changes in protein synthesis during the adaptation of Bacillus subtilis to anaerobic growth conditions. Microbiology 146, 97–105 [DOI] [PubMed] [Google Scholar]
- Matsuno K, and Sonenshein AL. (1999). Role of SpoVG in asymmetric septation in Bacillus subtilis. J Bacteriol 181, 3392–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möbius K, Arias-Cartin R, Breckau D, et al. (2010). Heme biosynthesis is coupled to electron transport chains for energy generation. Proc Natl Acad Sci USA 107, 10436–41041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus JM, and Wright JK. (1983). Chemical modification of the lactose carrier of Escherichia coli by plumbagin, phenylarsinoxide or diethylpyrocarbonate affects the binding of galactoside. Eur J 137, 615–621 [DOI] [PubMed] [Google Scholar]
- Paoletti L, Lu YJ, Schujman GE, de Mendoza D, and Rock CO. (2007). Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol 189, 5816–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plichta JK, Nienhouse V, and Radek KA. (2012). Integrating “omics” technologies to conceptualize dynamic antimicrobial peptide responses. Front Immunol 2012 3, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Pogliano N, and Silverman JA. (2012). Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 194, 4494–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Singh JK, Roy N, and Panda D. (2008). Curcumin inhibits FtsZ assembly: An attractive mechanism for its antibacterial activity. Biochem J 410, 147–155 [DOI] [PubMed] [Google Scholar]
- Rao AA, Patkari M, Reddy PJ, et al. (2014). Proteomic analysis of Streptomyces coelicolor in response to ciprofloxacin challenge. J Proteomics 97, 222–234 [DOI] [PubMed] [Google Scholar]
- Reddy PJ, Rao AA, Malhotra D, et al. (2013). A simple protein extraction method for proteomic analysis of diverse samples. Curr Proteomics 10, 298–311 [Google Scholar]
- Roy A, Kucukural A, and Zhang Y. (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protoc 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, and Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- Strahl H, and Hamoen LW. (2010). Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA 107,12281–12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Chan FY, Lu YJ, et al. (2014). Rational design of berberine-based FtsZ inhibitors with broad-spectrum antibacterial activity. PLoS One 9, e97514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecke T, and Mascher T. (2011). Antibiotic research in the age of omics: From expression profiles to interspecies communication. J Antimicrob Chemother 66, 2689–2704 [DOI] [PubMed] [Google Scholar]
- Wenzel M, Kohl B, Münch D, et al. (2012). Proteomic response of Bacillus subtilis to lantibiotics reflects differences in interaction with the cytoplasmic membrane. Antimicrob Agents Chemother 56, 5749–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, et al. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39(Web Server issue),W316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye RW, Tao W, Bedzyk L, Young T, Chen M, and Li L. (2000). Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol 182, 4458–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.