Abstract
Biotechnology is often limited by weak interactions. We suggest that an ideal interaction between proteins would be covalent, specific, require addition of only a peptide tag to the protein of interest, and form under a wide range of conditions. Here we summarize peptide tags able to form spontaneous amide bonds, based on harnessing reactions of adhesion proteins from the bacterium Streptococcus pyogenes. These include irreversible peptide-protein interaction of SpyTag with SpyCatcher, as well as irreversible peptide-peptide interaction via SpyLigase. We describe existing applications, including polymerization to enhance cancer cell capture, assembly of living biomaterial, access to diverse protein shapes, and improved enzyme resilience. We also point to future opportunities for resisting biological force and extending the scope of protein nanotechnology.
Keywords: protein engineering, nanobiotechnology, mechanobiology, synthetic biology, supramolecular assembly, biomimetic
The challenge of protein-peptide interaction
For a generic route to extend the applications of proteins, peptide tags are ideal. Genetically fusing a peptide to a protein of interest can allow efficient purification, detection, immobilization, and derivatization. Peptide tags are characterized by a minimal chance for disrupting the function of the attached protein [1-3]. In this opinion we focus on genetically-encoded peptide-protein interactions and their optimization through spontaneous formation of isopeptide bonds (see Glossary). Protein or peptide interactions with small molecules, e.g. Ni-NTA, HaloTag, SNAP-tag, FlAsH [1], are beyond our scope.
Peptides are generally unstructured before binding their protein partner and have a limited surface area for contacts, which presents a quandary in trying to obtain stable peptide-protein interactions [4]. The most widely-used peptide-protein pairs are epitope tags (e.g. HA-, myc- and FLAG-tags) recognized by antibodies, but more recently peptide binding to streptavidin or PDZ clamps has been developed [5-7]. The limited stability and strength of peptide-protein interactions seriously hamper the use of peptide tags, especially where the peptide-protein linkage must last for a long time, link to a nanoparticle, survive high temperature, or withstand forces [5, 8-10]. Therefore, our opinion is that peptide-protein pairs forming unbreakable interactions will enable diverse new approaches for basic research and biotechnology.
Existing strong protein-peptide interactions
While advances in genetic engineering depended upon a diverse array of enzymes able to ligate, edit, or recombine DNA fragments, fewer such options exist for directing covalent reaction of proteins. Native chemical ligation (NCL) and expressed protein ligation (EPL) enable peptide fragments to be joined together, but must be at specific termini and are generally restricted to in vitro systems [11]. Split inteins can be fused to target proteins, reconstitute in solution, and splice themselves out to covalently attach the two polypeptides, although various side-reactions may occur [12, 13]. Dock-and-lock, where proteins cross-link via cysteines on terminally-attached peptide-recognition domains, also enables coupling but is not applicable in reducing environments [14].
Covalent protein coupling can also be catalyzed enzymatically. In Sortagging, an LPXTG-containing protein is covalently linked to an N-terminal pentaglycine probe via sortase catalysis [15, 16]. Transglutaminases can mediate protein cross-linking between lysine and glutamine side-chains, although are frequently promiscuous [2].
Spontaneous isopeptide bond formation for protein coupling
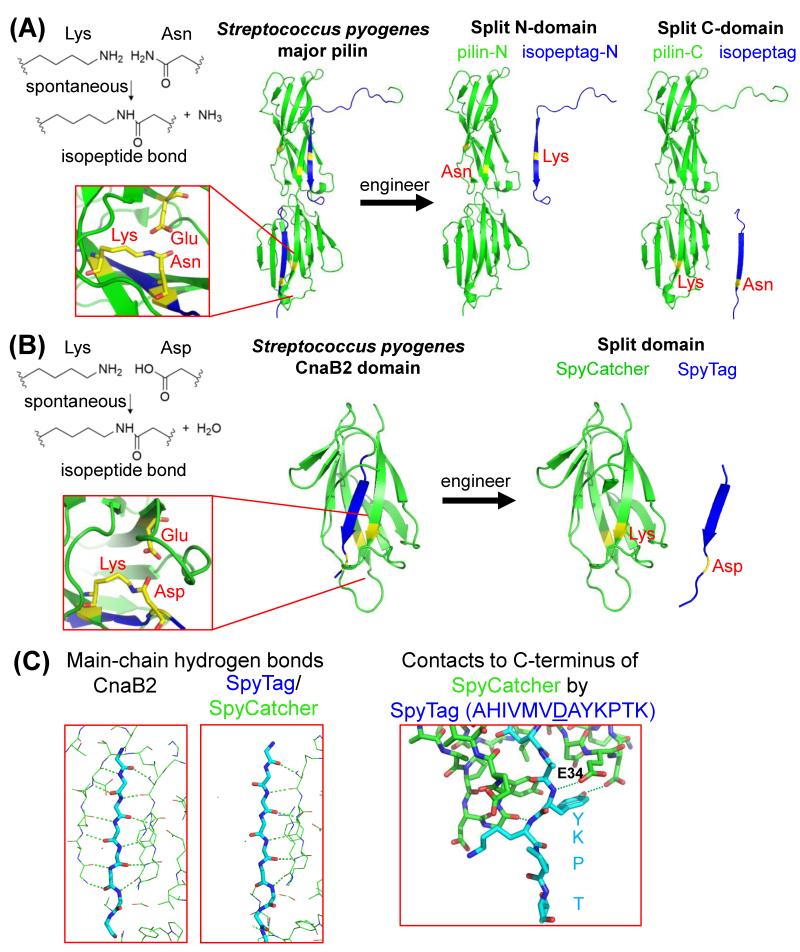
We considered that a different type of protein chemistry, spontaneous isopeptide bond formation, might be able to overcome the above challenges. An isopeptide bond is an amide bond in a protein connecting a side-chain to a side-chain, or connecting a side-chain to the protein’s main chain (Figure 1A). Important natural examples of isopeptide bonds include ATP-dependent conjugation of ubiquitin to proteins [17], and transglutaminase-catalyzed isopeptides imparting strength to hair, skin and blood vessels [18]. However, isopeptides bonds can also form autocatalytically, as first discovered in HK97, a bacteriophage discovered in Hong Kong from pig dung [19]. HK97’s capsid is composed of 420 protein subunits, which assemble non-covalently into an icosahedral particle consisting of 5- and 6-membered rings looping through each other. Spontaneous intermolecular isopeptide bond formation between adjacent subunits then locks the rings together, forming “protein chain-mail” [20].
Figure 1.
The generation of isopeptide bond-forming peptides. (A) Splitting the major pilin. Reaction between lysine and asparagine in the pilin, promoted by an apposed glutamic acid. Inset shows reactive triad in C-domain (PDB 3B2M). A cartoon is shown of splitting the pilin into two separate protein/peptide pairs: pilin-N reacts with isopeptag-N, while pilin-C reacts with isopeptag. In blue are the peptide tags or the residues from which the tags were derived. Key residues for reaction in yellow. (B) Splitting CnaB2. Reaction between lysine and aspartic acid in CnaB2, promoted by an apposed glutamic acid (PDB 2X5P). A cartoon is shown of splitting CnaB2 into the protein SpyCatcher which reacts with the SpyTag peptide. Blue marks the peptide tag and the residues from which the tag was derived. Key residues for reaction are in yellow. (C) Insights from crystal structure of SpyTag/SpyCatcher complex. Main-chain hydrogen bonds (dotted lines) form to both sides of the final β-strand in CnaB2 (left panel, PDB 2X5P) but only to one side of SpyTag (center panel, PDB 4MLI). Right panel: polar contacts made by SpyCatcher (including its E34 mutation) with the C-terminal tail of SpyTag (PDB 4MLS).
Intramolecular spontaneous isopeptide bonds were first found in 2007 by Kang et al. in Spy0128, the major pilin subunit from the human pathogen Streptococcus pyogenes (Figure 1A) [21]. Subsequently, spontaneous intramolecular isopeptide bonds were discovered in a range of surface proteins from Gram-positive bacteria. A unique biological role was shown: intramolecular isopeptides in these domains confer proteolytic, thermal and pH stability [21, 22]. Why would disulfide bonds not suffice for high stability? Many bacteria, including important human pathogens such as Staphylococci and Enterococci, lack the cellular machinery for catalyzing disulfide bond formation and often inhabit hypoxic environments [23]. Another advantage of isopeptides is that disulfides are easily cleaved by reducing agents or enzymes with reactive cysteines, whereas little is known about how lysine-asparagine or lysine-aspartic acid isopeptides are cleaved. (There is some precedent for reversal of lysine-glutamine linkages, such as transglutaminases acting in reverse [24] or destabilase from leeches hydrolyzing blood clots [25].)
The mechanism of spontaneous isopeptide bond formation has been analyzed through crystallography [21], NMR [26] and quantum mechanics/molecular mechanics (QMMM) [26, 27]. Lysine forms an isopeptide bond by nucleophilic attack on asparagine (with loss of NH3, Figure 1A) or less commonly by attack on aspartic acid (with loss of H2O, Figure 1B). A catalytic glutamic acid or aspartic acid is always positioned opposite the residue being attacked (Figure 1A/B), stabilizing the transition states via hydrogen bonding and donating/receiving a proton to facilitate reaction. The triad of residues directly involved in the reaction is located in the hydrophobic protein interior, which modulates the pKa, favoring the uncharged and more reactive forms of the residues. Interestingly, synthetic organic chemists independently developed a similar design of catalyst, for environmentally-friendly synthesis of amide bonds directly from amines and carboxylic acids, a reaction of great importance in the pharmaceutical and agrochemical industries [28].
Peptide tags forming spontaneous covalent bonds to proteins
A domain is not the minimal unit of a protein that is useful for biotechnology. Splitting up domains is a powerful way to control protein function. A polypeptide sequence can be strategically split into two fragments and, when expressed individually, the two fragments may recognize each other and reconstitute to a functional protein. Split-protein reconstitution has been achieved with a variety of scaffolds, including luciferase and fluorescent proteins [29]. Using such a strategy, we developed peptide-protein pairs that could spontaneously reconstitute and form an isopeptide bond (Figure 1A) [30]. Splitting the N-domain of the major pilin generated the peptide fragment isopeptag-N, containing the reactive lysine, and the protein fragment pilin-N containing the reactive asparagine (Figure 1A). When mixed in solution, isopeptag-N and pilin-N were able to covalently reconstitute [30]. Similarly, the C-domain of the major pilin was split, this time with the reactive asparagine on the peptide, and further engineered to develop the more efficient isopeptag (16 residues) and pilin-C (32 kDa) partners (Figure 1A). With isopeptag in two-fold excess, 98% of pilin-C reacted in 24 hours [30]. The isopeptag/pilin-C reaction was robust, proceeding with a range of temperatures (4-37°C), pH values (5-8), buffers, and detergents. Since the isopeptag/pilin-C reaction had good specificity and the partners were genetically-encodable, they served as a facile platform for covalent labeling of a specific cell-surface protein for live-cell fluorescent microscopy [30].
For a smaller and faster reacting pair, we investigated the CnaB2 domain of the fibronectin-binding protein FbaB from Streptococcus pyogenes [26, 31-33]. CnaB2 was initially split into peptide and protein partners and then engineered by removing surface-exposed hydrophobic residues and by enhancing interaction in the binding interface. This generated the optimized 13-residue SpyTag and 116-residue SpyCatcher binding partners (Figure 1B) [34]. Reconstitution between SpyTag/SpyCatcher was rapid, achieving ~50% yield in 1 minute and complete reaction of SpyCatcher with longer incubation. SpyTag/SpyCatcher still reacted together efficiently with various pH values (5-8), buffers, temperatures and detergents [33]. Reconstitution had good specificity in E. coli and mammalian cells, allowing genetically-encodable protein coupling intracellularly and extracellularly [33]. Crystallography of SpyTag/SpyCatcher revealed how the reconstituted protein had extra regions of flexibility compared to the parental domain, as well as hydrogen-bonding patterns which may explain the rapid binding of SpyTag and the role of the I34E mutation of SpyCatcher in facilitating docking (Figure 1C) [34]. This study also revealed that SpyCatcher could be truncated at each terminus, to form an 84-residue partner, without significantly reducing reaction efficiency [34].
Peptide-peptide ligation by SpyLigase
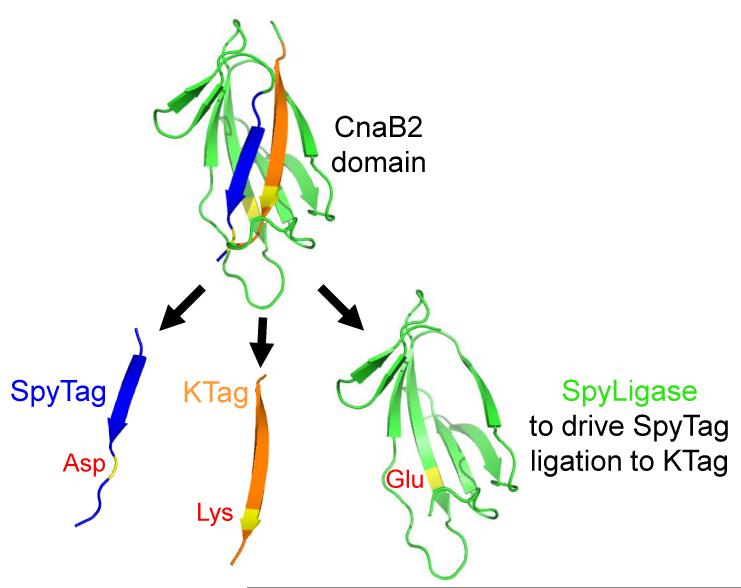
Preferably, specific connection between two proteins would be achieved simply by modifying each protein with a peptide tag. Adding SpyTag is a small modification, but adding an 84 residue SpyCatcher tag is not ideal. Therefore, we split CnaB2 into three parts, to generate a covalent peptide-peptide ligation system. SpyLigase was designed by excising from SpyCatcher the β-strand containing the reactive lysine, to obtain a new peptide tag (KTag, 10 residues) (Figure 2) [35]. SpyTag was unmodified, whereas the remaining CnaB2 scaffold, containing the catalytic glutamic acid, was circularly-permuted to give SpyLigase (107 residues) (Figure 2). SpyLigase could dock with SpyTag and KTag, reconstituting the catalytic triad and directing isopeptide formation between the two peptide tags [35].
Figure 2.
Peptide-peptide ligation by splitting CnaB2. Cartoon of the splitting strategy: β-stands were excised to obtain two peptide tags: SpyTag (blue) with the reactive aspartic acid and KTag (orange) with the reactive lysine. The remaining protein domain (SpyLigase, green) could direct isopeptide bond formation between SpyTag and KTag. Residues involved in reaction are colored yellow.
This technology is not yet optimal: SpyLigase ligation is slower and lower yielding than SpyTag/SpyCatcher, requires a specific buffer, and has not yet been demonstrated in cells [35]. Nevertheless, the unique properties for peptide-peptide linkage make this system a promising starting point for linking protein building-blocks with minimal modification.
Protein assemblies through spontaneous isopeptide bond formation
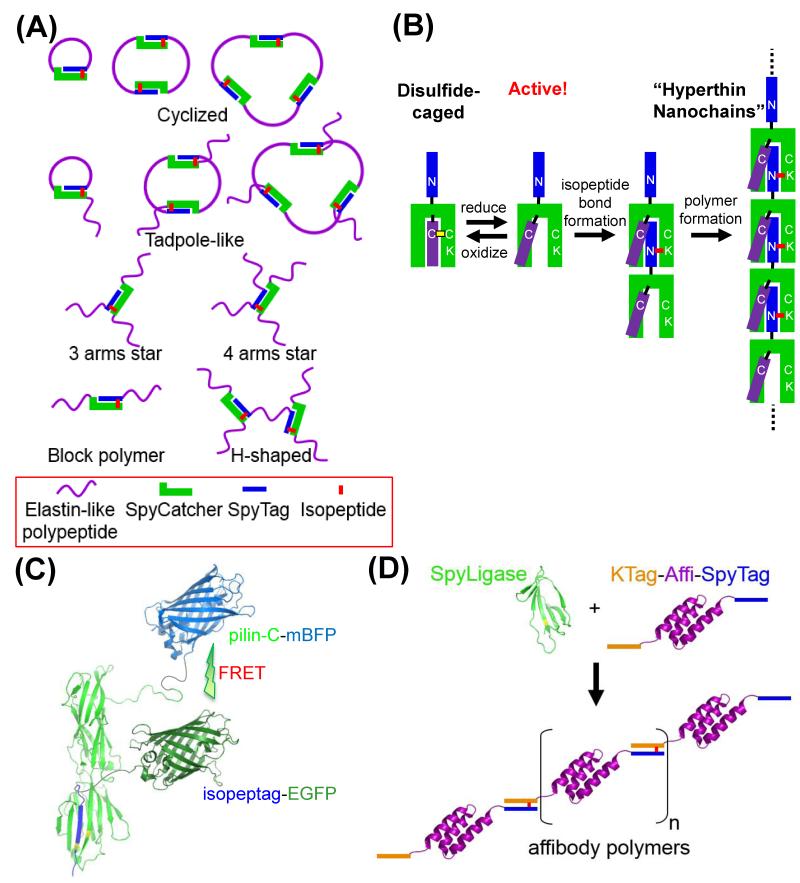
Despite the immense sophistication of multi-component machines in nature (e.g. the ribosome, replisome and spliceosome), synthetic biologists are still at an early stage in assembling artificial protein structures with desired modularity and stability [10, 14]. To generate diverse non-linear protein architectures, Zhang et al. elegantly harnessed different arrangements of SpyTag and SpyCatcher linked to elastin-like polypeptides (ELP) (Figure 3A). Cyclized, tadpole-shaped, star-shaped and H-shaped structures were produced in good yield simply upon mixing [36].
Figure 3.
Artificial assemblies created via spontaneous isopeptide bonds. (A) Diverse protein architectures from positioning SpyTag/SpyCatcher in elastin-like polypeptides. (B) Hyperthin nanochains via switchable covalent self-polymerization. Isopeptag (blue) was added to pilin-C to enable polymerization, but pilin-C reactivity was masked by a strand (purple) forming a disulfide bond (yellow). Upon reduction, isopeptag of one subunit could fit in the groove of another subunit, leading to formation of an isopeptide bond (red) and chain extension. Chains were much thinner than amyloid or collagen fibrils. Adapted from [37]. (C) Cartoon of the conjugation between pilin-C and isopeptag, each linked to different fluorescent proteins (monomeric blue fluorescent protein, mBFP, or enhanced green fluorescent protein, EGFP), enabling Förster Resonance Energy Transfer (FRET). (D) Protein tentacles: cartoon of SpyLigase locking together affibodies (purple), which were tailored with SpyTag (blue) and KTag (orange), into multivalent chains.
Linear polymers thinner than natural protein fibrils were generated by Matsunaga et al. using isopeptag/pilin C [37]. Pilin-C’s isopeptag binding pocket was engineered to introduce a redox-sensitive cap, where a disulfide inhibited binding of another isopeptag (Figure 3B). Reducing agent uncaged the isopeptag binding pocket and enabled self-polymerization, ingeniously forming pilus-like “hyperthin nanochains” in an inducible manner (Figure 3B). C- and S-shaped polymers of >200 nm were observed by atomic force microscopy (AFM) [37]. Isopeptag/pilin C were also exploited for irreversible assembly of protein conjugates with spatial control by Abe et al., enabling FRET applications between fluorescent proteins (Figure 3C) [38]. To link proteins into chains, where the protein is modified only with peptide tags, affibodies (non-immunoglobulin binding proteins) or antibodies were genetically fused to SpyTag and KTag. SpyLigase could then link these components into “protein tentacles”, over 20 units long (Figure 3D) [35]. The small size of SpyTag and its ability to react even when placed internally in proteins opens new routes for the controlled and stable assembly of proteins, reaching structural complexities not feasible with standard chemical conjugation strategies. Future applications of these assemblies include force-resistant biomaterials for tissue engineering, capsules to regulate drug delivery, and functionalizable hydrogels to control cell adhesion, growth and differentiation [36-38].
Current applications for SpyTag-based technology
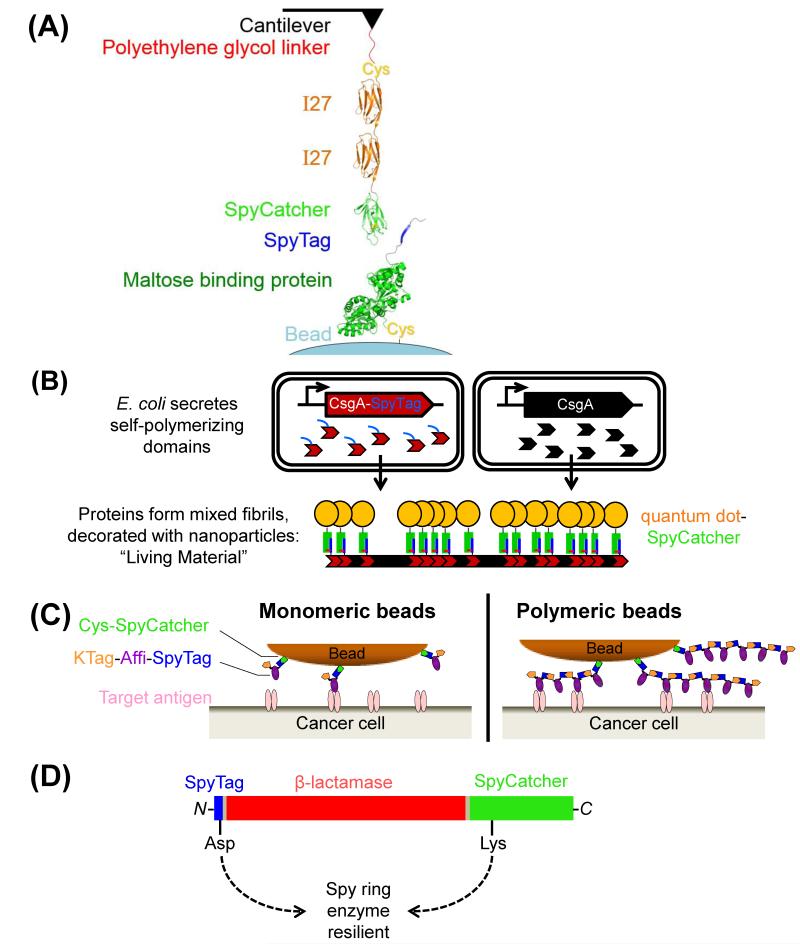
Force has widespread effects on biological function, such as regulating cell division, blood clotting and metastatic spread of cancer cells [39-41]. Single-molecule measurement of force usually employs optical tweezers or AFM. It is challenging to form specific force-resistant linkages to probe how biomolecules respond under force; often undefined hydrophobic adhesion or disulfide bonds are used. Therefore, there is great need for force-resistant, specific and genetically-encodable linkages. In fact, spontaneous isopeptide formation may have evolved to resist force [22]. AFM revealed the inextensible nature of the major pilin of S. pyogenes; the presence of isopeptides in both N- and C-terminal domains enabled the pilin to withstand forces up to 800 pN without unfolding [42]. For context, a myosin power-stroke in muscle contraction generates ~2 pN [43]. Similarly, the interaction between SpyTag and SpyCatcher has been analyzed at the single molecule level (Figure 4A) and was stable up to 1,900 pN [33], making it the strongest measured protein interaction. This mechanostability is ~20 times higher than streptavidin-biotin, one of the strongest non-covalent interactions in nature [9, 44]. Therefore, SpyTag peptide fusion has good potential for interrogating mechanical behavior of proteins, in comparison to existing approaches using less specific disulfide bonds or the larger protein fusions HaloTag or SNAP-Tag [45, 46]. SpyTag/SpyCatcher not only allows precise immobilization onto AFM tips, but should also provide the chance to study the effect of high forces on specific sites at the cell-surface. In addition, amide bonds survive higher forces than disulfides [47]. SpyTag should also be a valuable route for the formation of protein tandem repeats for AFM, to accelerate data acquisition [48].
Figure 4.
Application of isopeptide-assembled chains. (A) Cartoon of the construct to study SpyTag/SpyCatcher mechanical stability by AFM, adapted from [33]. (B) Mixed fibrils generated from E. coli CsgA (major curli subunit) and linked to nanoparticles via SpyTag/SpyCatcher. (C) SpyLigase-mediated polymerization of affibodies for enhanced cancer cell capture. Cartoon comparing the interface between a cancer cell and a magnetic bead coated with affibody monomers or SpyLigase-assembled affibody polymers. (D) Cartoon of cyclization of β-lactamase, to enhance resilience to thermal inactivation.
An application of SpyTag in synthetic biology has been to create functionalizable amyloid fibrils. CsgA is secreted from bacteria and spontaneously assembles into amyloid fibrils, facilitating bacterial surface adherence and biofilm formation. Co-culturing bacteria expressing CsgA and CsgA-SpyTag led to growth of mixed fibrils bearing surface-exposed SpyTags [49]. The fibrils could then be decorated with SpyCatcher-linked quantum dots and imaged by fluorescence microscopy (Figure 4B) [49].
The observed equilibrium constant for non-covalent binding (most commonly via antibody-antigen or streptavidin-biotin interaction) can be shifted by orders of magnitude towards dissociation when one of the partners is attached to a quantum dot or other nanoparticle [8]. Therefore, covalent linkage of biomolecules to nanoparticles will be important for going beyond labeling under gentle conditions for short time periods [50, 51]. SpyTag has been used for irreversible protein linkage to magnetic particles (Figure 4C). Magnetic microparticles or nanoparticles are the most common way to isolate rare cells. Magnetic capture of circulating tumor cells (CTCs) from blood is having a major impact on our understanding of human cancer biology and is starting to be used for cancer prognosis and personalization of cancer therapy [52]. We showed that any weak link in magnetic isolation reduces the sensitivity of cell capture, with secondary antibodies and even streptavidin-biotin interaction being limiting [9, 53]. Covalently linking SpyCatcher bearing an N-terminal cysteine (Cys-SpyCatcher) to magnetic beads enabled SpyTag-linked affibody polymers to be attached entirely through covalent bonds (Figure 4C). Affibodies ligated into covalent chains using SpyLigase enabled capture of cells bearing low levels of tumor antigen, without increasing non-specific capture [35]. These polyaffibodies or polyantibodies may find future application for increasing speed and sensitivity of capture of other rare eukaryotic cell-types (e.g. stem cells, T cell populations, Plasmodium) as well as viral or bacterial pathogens, using both magnetic and microfluidic capture.
Enhancing resilience of enzymes was also achieved via SpyTag/SpyCatcher. The instability of enzyme activity often limits diagnostic devices, biofuel production and biotransformations. Circularization of enzymes using previous approaches often only had a modest effect on thermal tolerance [54]. By fusing the termini with SpyTag and SpyCatcher, we cyclized β-lactamase and dihydrofolate reductase and found that the enzymes could be heated to 100°C and upon cooling retained high solubility and catalytic activity (Figure 4D) [55]. Differential scanning calorimetry indicated that this cyclization did not change the temperature for enzyme unfolding but increased refolding after thermal stress [55].
Regarding limitations, SpyTag is a genetically-encoded tag and therefore must be cloned onto the protein of interest case by case. Our experience is that SpyTag and SpyCatcher are well tolerated at N- or C-termini of a wide range of proteins, ideally linked through a glycine-serine spacer [33, 35, 49, 55], but there will be instances where fusion interferes with folding or protein interactions. Also, although the reaction of SpyTag with SpyCatcher can occur in minutes with high yield using micromolar protein concentrations, the reaction rate of 103 M−1s−1 for SpyTag/SpyCatcher is orders of magnitude below the diffusion limit [33]. Therefore systems such as biotin-streptavidin, with on-rate close to the diffusion limit [56], are still superior when capturing a tagged species at nanomolar concentrations or below.
Concluding remarks and future perspectives
Here we have described the emerging role of spontaneous isopeptide bonds for forming ultrastable peptide-protein interactions. Three different isopeptide-bond forming systems have been engineered, but the size and rapid reaction of SpyTag with SpyCatcher makes this the preferred system. SpyTag/SpyCatcher’s resilient interaction, forming under a wide range of conditions and able to react at any suitably-exposed site, is already enabling exciting applications. SpyTag/SpyCatcher has allowed facile assembly of diverse non-linear protein architectures, conferred resilience to boiling on enzymes, and enhanced magnetic cell capture. SpyTag/SpyCatcher also showed its mettle when expressed in cells for fluorescent microscopy and labeling biofilms. Future directions include resisting forces from molecular motors inside cells and providing unbreakable anchors between specific cellular components and surfaces or cantilevers. The modularity of SpyTag/SpyCatcher may allow proteins to be assembled irreversibly like building-blocks within living organisms, creating new opportunities for constructing evolvable and robust nanomachines.
Acknowledgements
G.V. was funded by the Medical Research Council and Merton College Oxford, B.Z. by the Clarendon Fund, Oxford University Department of Biochemistry, and St. Peter’s College Oxford, and M.H. by the European Research Council (ERC-2013-CoG 615945-PeptidePadlock). We thank Dr Jacob Orry Fierer and Mr Christopher Schoene for their stimulating comments.
Glossary
- Atomic Force Microscopy (AFM)
a high-resolution probe scanning microscopy technique, able to determine the resilience to stretching forces of biomolecules.
- HaloTag
a protein tag engineered from haloalkane dehalogenase able to react covalently to form an ester with alkyl halide ligands.
- Intein
a protein domain carrying out protein splicing. An intein excises itself to join the remaining protein’s portions.
- Isopeptag
a peptide tag (TDKDMTITFTNKKDFE) derived from C-domain of the major pilin Spy0128 able to form an isopeptide bond to pilin-C.
- Isopeptide bond
an amide bond connecting a side-chain to a side-chain or connecting a side-chain to the protein’s main chain.
- KTag
a peptide tag (ATHIKFSKRD) derived from the CnaB2 domain able to form an isopeptide bond to SpyTag in the presence of SpyLigase.
- Pilin-C
a protein derived from the major pilin Spy0128 able to form an isopeptide bond to isopeptag.
- SNAP-tag
a protein tag from engineering O6-alkylguanine-DNA alkyltransferase that forms a covalent bond with O6-benzylguanine derivatives.
- Sortase
a class of enzyme from Gram-positive bacteria that catalyzes intermolecular amide bond formation.
- SpyCatcher
a protein binding partner derived from the CnaB2 domain able to form an isopeptide bond to SpyTag.
- SpyLigase
a protein derived from the CnaB2 domain able to direct peptide-peptide ligation between SpyTag and KTag.
- SpyTag
a peptide tag (AHIVMVDAYKPTK) derived from the CnaB2 domain able to form an isopeptide bond to SpyCatcher.
Footnotes
Conflict of interest
B.Z. and M.H. are inventors on a patent application regarding isopeptide bond-forming peptides (United Kingdom Patent Application No. 1002362.0).
References
- 1.Sletten EM, Bertozzi CR. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rashidian M, et al. Enzymatic Labeling of Proteins: Techniques and Approaches. Bioconj. Chem. 2013;24:1277–1294. doi: 10.1021/bc400102w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 4.London N, et al. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Wood DW. New trends and affinity tag designs for recombinant protein purification. Curr. Op. Struct. Biol. 2014;26C:54–61. doi: 10.1016/j.sbi.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, et al. Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution. J. Mol. Biol. 2009;392:1221–1231. doi: 10.1016/j.jmb.2009.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, et al. Affinity-Guided Covalent Conjugation Reactions Based on PDZ-Peptide and SH3-Peptide Interactions. Bioconj. Chem. 2014;25:989–999. doi: 10.1021/bc500134w. [DOI] [PubMed] [Google Scholar]
- 8.Swift JL, et al. A two-photon excitation fluorescence cross-correlation assay for a model ligand-receptor binding system using quantum dots. Biophys. J. 2006;90:1396–1410. doi: 10.1529/biophysj.105.069526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chivers CE, et al. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat. Methods. 2010;7:391–393. doi: 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher JM, et al. Self-Assembling Cages from Coiled-Coil Peptide Modules. Science. 2013;340:595–599. doi: 10.1126/science.1233936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent SB. Total chemical synthesis of proteins. Chem. Soc. Rev. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 12.Shah NH, Muir TW. Inteins: Nature’s Gift to Protein Chemists. Chem. Sci. 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvajal-Vallejos P, et al. Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources. J. Biol. Chem. 2012;287:28686–28696. doi: 10.1074/jbc.M112.372680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi EA, et al. Complex and defined biostructures with the dock-and-lock method. Trends Pharm. Sci. 2012;33:474–481. doi: 10.1016/j.tips.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Mao H, et al. Sortase-mediated protein ligation: a new method for protein engineering. J. Am. Chem. Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 16.Strijbis K, et al. Protein ligation in living cells using sortase. Traffic. 2012;13:780–789. doi: 10.1111/j.1600-0854.2012.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Ann. Rev. Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 18.Klock C, Khosla C. Regulation of the activities of the mammalian transglutaminase family of enzymes. Prot. Sci. 2012;21:1781–1791. doi: 10.1002/pro.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhillon EKS, et al. Host Range, Immunity and Antigenic Properties of Lambdoid Coliphage-Hk97. J. Gen. Virol. 1980;50:217–220. doi: 10.1099/0022-1317-50-1-217. [DOI] [PubMed] [Google Scholar]
- 20.Wikoff WR, et al. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 21.Kang HJ, et al. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 22.Kang HJ, Baker EN. Intramolecular isopeptide bonds: protein crosslinks built for stress? Trends Biochem. Sci. 2011;36:229–237. doi: 10.1016/j.tibs.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Dutton RJ, et al. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamnaes J, et al. The propensity for deamidation and transamidation of peptides by transglutaminase 2 is dependent on substrate affinity and reaction conditions. Biochim. Biophys. Acta. 2008;1784:1804–1811. doi: 10.1016/j.bbapap.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavalova LL, et al. Destabilase from the medicinal leech is a representative of a novel family of lysozymes. Biochim. Biophys. Acta. 2000;1478:69–77. doi: 10.1016/s0167-4838(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 26.Hagan RM, et al. NMR spectroscopic and theoretical analysis of a spontaneously formed Lys-Asp isopeptide bond. Angew. Chem. 2010;49:8421–8425. doi: 10.1002/anie.201004340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu X, et al. Autocatalytic intramolecular isopeptide bond formation in gram-positive bacterial pili: a QM/MM simulation. J. Am. Chem. Soc. 2011;133:478–485. doi: 10.1021/ja107513t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charville H, et al. The Uncatalyzed Direct Amide Formation Reaction - Mechanism Studies and the Key Role of Carboxylic Acid H-Bonding. Eur. J. Org. Chem. 2011:5981–5990. [Google Scholar]
- 29.Shekhawat SS, Ghosh I. Split-protein systems: beyond binary protein-protein interactions. Curr. Op. Chem. Biol. 2011;15:789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zakeri B, Howarth M. Spontaneous intermolecular amide bond formation between side chains for irreversible peptide targeting. J. Am. Chem. Soc. 2010;132:4526–4527. doi: 10.1021/ja910795a. [DOI] [PubMed] [Google Scholar]
- 31.Oke M, et al. The Scottish Structural Proteomics Facility: targets, methods and outputs. J. Struct. Funct. Genom. 2010;11:167–180. doi: 10.1007/s10969-010-9090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amelung S, et al. The FbaB-type fibronectin-binding protein of Streptococcus pyogenes promotes specific invasion into endothelial cells. Cell Microbiol. 2011;13:1200–1211. doi: 10.1111/j.1462-5822.2011.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakeri B, et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E690–697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, et al. Structural analysis and optimization of the covalent association between SpyCatcher and a peptide tag. J. Mol. Biol. 2014;426:309–317. doi: 10.1016/j.jmb.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fierer JO, et al. SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1176–1181. doi: 10.1073/pnas.1315776111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang WB, et al. Controlling macromolecular topology with genetically encoded SpyTag-SpyCatcher chemistry. J. Am. Chem. Soc. 2013;135:13988–13997. doi: 10.1021/ja4076452. [DOI] [PubMed] [Google Scholar]
- 37.Matsunaga R, et al. Hyperthin nanochains composed of self-polymerizing protein shackles. Nat. Comm. 2013;4:2211. doi: 10.1038/ncomms3211. [DOI] [PubMed] [Google Scholar]
- 38.Abe H, et al. Split Spy0128 as a potent scaffold for protein cross-linking and immobilization. Bioconj. Chem. 2013;24:242–250. doi: 10.1021/bc300606b. [DOI] [PubMed] [Google Scholar]
- 39.Rago F, Cheeseman IM. Review series: The functions and consequences of force at kinetochores. J. Cell Biol. 2013;200:557–565. doi: 10.1083/jcb.201211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, et al. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stroka KM, Konstantopoulos K. Physical biology in cancer. 4.Physical cues guide tumor cell adhesion and migration. Am. J. Physiol. 2014;306:C98–C109. doi: 10.1152/ajpcell.00289.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alegre-Cebollada J, et al. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J. Biol. Chem. 2010;285:11235–11242. doi: 10.1074/jbc.M110.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore SW, et al. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev. Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong J, et al. Direct force measurements of the streptavidin-biotin interaction. Biomol. Eng. 1999;16:45–55. doi: 10.1016/s1050-3862(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 45.Popa I, et al. Nanomechanics of HaloTag Tethers. J. Am. Chem. Soc. 2013;135:12762–12771. doi: 10.1021/ja4056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kufer SK, et al. Covalent immobilization of recombinant fusion proteins with hAGT for single molecule force spectroscopy. Eur. Biophys. J. 2005;35:72–78. doi: 10.1007/s00249-005-0010-1. [DOI] [PubMed] [Google Scholar]
- 47.Beyer MK, Clausen-Schaumann H. Mechanochemistry: the mechanical activation of covalent bonds. Chem. Rev. 2005;105:2921–2948. doi: 10.1021/cr030697h. [DOI] [PubMed] [Google Scholar]
- 48.Bornschlogl T, Rief M. Single-molecule protein unfolding and refolding using atomic force microscopy. Meth. Mol. Biol. 2011;783:233–250. doi: 10.1007/978-1-61779-282-3_13. [DOI] [PubMed] [Google Scholar]
- 49.Chen AY, et al. Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Materials. 2014;13:515–523. doi: 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu DS, et al. Quantum dot targeting with lipoic acid ligase and HaloTag for single-molecule imaging on living cells. ACS Nano. 2012;6:11080–11087. doi: 10.1021/nn304793z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howarth M, et al. Monovalent, reduced-size quantum dots for single molecule imaging of receptors in living cells. Nat. Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu M, et al. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain J, et al. Cholesterol loading and ultrastable protein interactions determine the level of tumor marker required for optimal isolation of cancer cells. Canc. Res. 2013;73:2310–2321. doi: 10.1158/0008-5472.CAN-12-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwai H, Pluckthun A. Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS L. 1999;459:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 55.Schoene C, et al. SpyTag/SpyCatcher Cyclization Confers Resilience to Boiling on a Mesophilic Enzyme. Angew. Chem. 2014;53:6101–6104. doi: 10.1002/anie.201402519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hyre DE, et al. Cooperative hydrogen bond interactions in the streptavidin-biotin system. Prot. Sci. 2006;15:459–467. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]