ABSTRACT
Variation in signaling activity across a cell plays a crucial role in processes such as cell migration. Signaling activity specific to organelles within a cell also likely plays a key role in regulating cellular functions. To understand how such spatially confined signaling within a cell regulates cell behavior, tools that exert experimental control over subcellular signaling activity are required. Here, we discuss the advantages of using optogenetic approaches to achieve this control. We focus on a set of optical triggers that allow subcellular control over signaling through the activation of G-protein-coupled receptors (GPCRs), receptor tyrosine kinases and downstream signaling proteins, as well as those that inhibit endogenous signaling proteins. We also discuss the specific insights with regard to signaling and cell behavior that these subcellular optogenetic approaches can provide.
KEY WORDS: Optogenetics, Subcellular signaling
Introduction
The development of novel techniques and their continuing improvement has been fundamental to major advances in modern biology. Nucleic acid sequencing (Padmanabhan and Wu, 1972; Sanger et al., 1977), protein analysis using gel electrophoresis (Laemmli, 1970; Shapiro et al., 1967) and molecular cloning (Cohen et al., 1973) were crucial during the more-reductionist phase when the molecules at the basis of important cellular functions were being identified. As it became apparent that a better understanding of these molecules in the intact cell was required, reporters and sensors based on fluorescent proteins (Giepmans et al., 2006) were developed to visualize the dynamics of molecular activity at the subcellular level in live cells. These studies have shown that specific cellular outputs are mediated by signaling activity that is spatially heterogeneous across a cell.
Cells encounter gradients of extracellular stimuli and asymmetric signaling activity during processes such as migration. They also show variation in signaling activity at various subcellular sites within a cell, such as the Golgi, endoplasmic reticulum (ER) and endosomes, which likely play a key role in regulating cellular functions (Antal and Newton, 2013; Calebiro et al., 2010; Dehmelt and Bastiaens, 2010; DiPilato et al., 2004; Fivaz and Meyer, 2005; Fonseca et al., 2012; Fosbrink et al., 2010; Hewavitharana and Wedegaertner, 2012; Irannejad et al., 2013; Kreis et al., 2014; Kunkel and Newton, 2010; Machacek et al., 2009; O'Neill et al., 2012; Pertz, 2010; Saini et al., 2009; Tadevosyan et al., 2012; Zehorai et al., 2010). To obtain a better understanding of how such signaling molecules govern crucial cellular behaviors, we need to be able to exert experimental control over subcellular signaling activities.
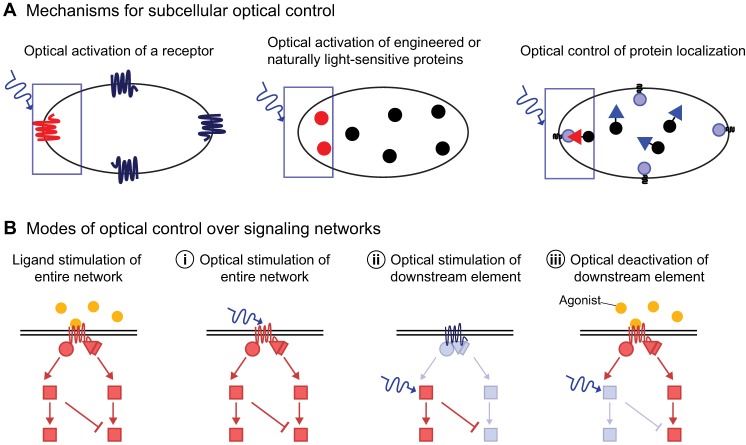
It has recently become possible to achieve such control through subcellular optogenetics. In this approach, a light signal is used to modulate the activity of a signaling protein expressed in cells. The ability to direct a light beam to a selected region of a cell allows subcellular control (Fig. 1). Signaling proteins used for subcellular optogenetic control include naturally occurring opsins, which are G-protein-coupled receptors (GPCRs), recombinant chimeras of an opsin with a ligand-binding receptor, photosensitive domains tagged to different signaling proteins, such as receptor tyrosine kinases (RTKs), small GTP-binding proteins and adenylyl cyclases, which are all described below. This type of optogenetics thus allows control over specific signaling activity in a cell and potentially the cellular behavior that is governed by the signaling activity (Fig. 1).
Fig. 1.
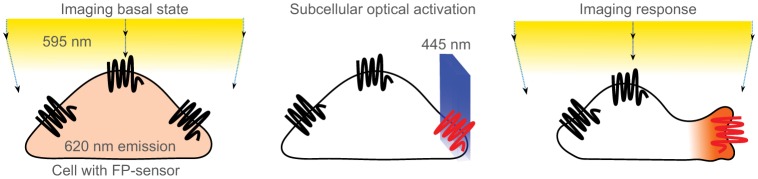
Spectral selectivity allows subcellular control over signaling while responses are imaged globally. In this diagrammatic representation, a migratory cell expressing inactive receptors (black) that are sensitive to 445 nm light is activated with a light beam on one side (blue box) resulting in asymmetric activation of the receptors (red). The cell shows a polarized response by extending lamellipodia in the direction of the optical signal. The cell also expresses a fluorescent protein (FP) sensor that is excited at 595 nm and emits at 620 nm indicating the concentration of a second messenger at the extending lamellipodia (pale orange diffuse distribution on the left and dark orange concentrated at the cellular extension on the right). The spectral tuning of a receptor to 445 nm prevents its activation during global imaging using 595 nm light and facilitates visualization response of the cell to subcellular optical activation of signaling.
In this Commentary, we describe optogenetic approaches that can be used to achieve subcellular control over signaling using the tools mentioned above. We highlight new insights that this approach can provide with regard to how dynamic intracellular signaling networks control cell behavior. Whereas the majority of optogenetics research has focused on regulating neuronal activity using light-sensitive ion channels and light-driven ion pumps (Miesenböck, 2011; Zhang et al., 2011), we focus here mainly on techniques that have been used to attain optical control over constituents of the GPCR and RTK signaling pathways. We also discuss the technical hurdles in applying these techniques widely and how they can be overcome. Finally, we outline the exciting future potential of these powerful methodologies. Overall, we focus on the development and utility of the optical tools that can be built using photosensitive pigments and not on the pigments themselves. Several comprehensive reviews focused on the pigments and the mechanistic basis of their function are available elsewhere (Imamoto and Shichida, 2014; Krauss et al., 2010; Möglich and Moffat, 2010; Pathak et al., 2013; Rockwell et al., 2006; Terakita, 2005; van der Horst and Hellingwerf, 2004; Yin and Wu, 2013; Yokoyama, 2000).
Advantages of optogenetic approaches for controlling signaling at the subcellular level
Chemical and genetic methods have been highly successful in identifying the role of signaling components involved in regulating cellular physiology. However, it is difficult to restrict diffusible factors, such as agonists, antagonists, activators and inhibitors, to selected regions within a cell or to specific cells in a tissue. The use of coated beads (Arimura and Kaibuchi, 2007) and striped substrates (Shelly et al., 2010) has been useful to identify some of the molecular mechanisms that direct neuron differentiation. Different uses of the microinjection of light-activated agents such as caged compounds into cells have been described (Ellis-Davies, 2007). However, these methods possess only limited spatial and temporal control and lack the ability to either continuously change or terminate signaling. Some of these limitations have been surmounted by using caged rapamycin (Umeda et al., 2011) to induce ruffling of membranes, and magnetic nanoparticles (Etoc et al., 2013) to induce actin remodeling. The use of micropipettes and microfluidic devices to create gradients of chemoattractants has also allowed a certain level of spatial control over signaling at the single-cell level and has been widely used to study cell migration (Croushore and Sweedler, 2013; Velve-Casquillas et al., 2010). However, there are still a number of inherent disadvantages to these approaches. For instance, rapamycin-based methods lack reversibility; magnetic nanobeads require microinjection into each cell to be examined, one cell at a time; and microfluidics can be used to generate gradients of extracellular cues, but cannot spatially confine the input signal, for example to the tip of a neurite. Therefore, with these methods, it is difficult to provide continual and variable signaling input or exert precise temporal control.
Optogenetics, which makes it possible to optically trigger signaling with temporal control in the subsecond range and spatial control that is only limited by diffraction, has several advantages over these methods. We describe below the design and optical targeting of light-sensitive proteins that can elicit specific cellular responses. Given that these optical triggers are genetically encoded, they can be continually activated, unlike chemical agents that are injected. Optical control over protein activity can be achieved by using light intensities and wavelengths that are non-toxic to cells. Furthermore, their spectral selectivity allows light-mediated activation to be combined with imaging of fluorescence-based biosensors (Fig. 1). Moreover, by tuning the wavelength of optical triggers, they can be used to exert optical control over multiple proteins within the same cell. Light signal inputs to a cell can also be applied to multiple cells simultaneously and varied continuously and almost instantaneously, with regard to location, intensity, duration of light pulses and time intervals between individual pulses. Many optical triggers demonstrate rapid reversal to their basal state in the absence of light. This allows signaling activity in the cell to be switched on and off at any time so that the crucial time periods when signaling activity is required to initiate or maintain a cellular response can be identified. The ability to apply repeated, non-invasive optical activity helps to sustain cellular responses that often occur over longer timescales of minutes or hours.
Light-sensitive proteins – a resource for engineering optogenetic constructs
Light-sensitive proteins are ubiquitous across living organisms, from bacteria to plants to human. Opsins are light-detecting GPCRs that are present in almost all metazoan species. They mediate important processes across species, such as vision and circadian rhythm in mammals (Kumbalasiri and Provencio, 2005; Lamb, 2013), and gamete release as well as vision in jellyfish (Suga et al., 2008). Light-sensing cryptochrome 2 (CRY2), phytochrome (Phy), and light-oxygen-voltage-sensing (LOV) domains govern light-dependent movement and circadian rhythms in plants, algae and fungi (Kennedy et al., 2010; Lungu et al., 2012; Shimizu-Sato et al., 2002; Wu et al., 2009). The ability of these light-sensitive proteins to modulate cellular and organismal behavior is reliant on their intrinsic molecular properties, such as their capability to respond to specific wavelengths of light, selectively activate second messengers (opsins), bind to partner proteins or undergo stereotypical conformational changes (CRY2, Phy and LOV domains).
Some light-sensitive proteins have signaling capabilities that allow them to be used without any significant modifications to control cell behaviors in heterologous cell types. For instance, a light-triggered GPCR that activates G proteins in the human retina has been used to activate native G proteins in immune cells and to optically direct their migration (Karunarathne et al., 2013b). A light-activated adenylyl cyclase from bacteria has been introduced into zebrafish pituitary cells to gain optical control over cyclic AMP (cAMP) production and glucocorticoid secretion (De Marco et al., 2013). However, most signaling proteins lack naturally occurring light-sensitive variants. Achieving optical control over their signaling activity has required the development of engineered constructs that incorporate light-sensitive domains. This approach has commonly been made use of with LOV, CRY and Phy domains as outlined further below.
Modes of optical control over signaling networks
The majority of signaling networks are regulated by cell surface receptors that respond to extracellular stimuli. Light activation of a receptor can provide activation of an entire signaling network, with the ability to continually change the location of the stimulus at selected regions on the surface of a cell (Fig. 2A). As the receptor stimulates the endogenous signaling pathway in a cell, the integrity of downstream signaling protein properties is wholly maintained and any evoked responses reflect normal cell behavior. Light can thus be used to orchestrate a cellular behavior similar to the native extracellular signal and monitor both cellular and molecular responses quantitatively. In the section below, we describe optically triggered GPCRs and RTKs as examples of this type of control over cellular signaling.
Fig. 2.
Modes of optical control. (A) Local photoactivation of a naturally light-sensitive receptor can be used to trigger signaling from a selected region at the cell surface (shown on the left). Inside the cell, signaling can be locally triggered by optical activation of naturally light-sensitive adenylyl cyclases or engineered fusions of signaling proteins with light-sensitive domains that mask or unmask their active sites in response to photoactivation (middle). Photoactivation of selected protein domains (triangles) can be used to translocate signaling proteins to specific subcellular sites, such as the plasma membrane at one side of the cell (right). (B) Optical control can be used to activate or inhibit signaling at different nodes within a signaling cascade. Optical activation of a receptor stimulates the entire signaling network (i). Alternatively, a downstream signaling protein can be directly activated independently of any receptor activity upstream (ii). Finally, a receptor is activated through its ligand, but a downstream protein is inhibited optically (iii). Activated proteins are in red and inactive proteins are in light blue. Filled blue circles are receptor-activating ligands. Wavy arrow denotes light. Activated components are shown in red and inactive in blue and black.
In contrast to receptor activation, it is also possible to optically activate a downstream signaling element in the absence of any receptor activity (Fig. 2A). We describe below how light-sensitive signaling can be engineered using a LOV domain and activated directly within a cell. Signaling activity can also be optically modulated by spatial control relying on the ability of proteins tagged with either an Arabidopsis thaliana CRY2 domain, which, on light activation, binds to a cryptochrome-binding basic helix-loop-helix protein domain (CIB1), or to phytochrome, which binds to phytochrome-interacting factors (PIFs) (Fig. 2A). We describe the design of these optical triggers and their utility below.
Dynamic networks of interacting signaling proteins govern cellular behavior. The ability to selectively control the activity of specific signaling components or nodes in a network can be valuable in gaining a better understanding of the network dynamics that executes cell behavior. The optical approaches described above can provide such control. Light-sensitive receptors can be used to activate an entire pathway (Fig. 2B) and as described below identify the role of spatial and temporal changes in a network of interacting signaling proteins in cellular behavior. Optical activation or deactivation of downstream elements in a signaling pathway using LOV, CRY2 or Phy domains can be used to identify the role of spatial and temporal changes in specific signaling proteins in controlling cell behavior (Fig. 2B). In the future, stimulation of an entire network using ligand- or light-based receptor activation could also be combined with approaches that optically modulate the activity of downstream signaling proteins (Fig. 2B).
Optical activation of GPCRs
Signaling cascades are commonly initiated by activation of the GPCR or RTK families of transmembrane receptors. Optogenetic control over GPCR signaling has advanced more rapidly than that of RTK pathways, owing to the availability of naturally occurring light-activated GPCRs. However, as discussed below, some engineered constructs that provide optical control over RTK-mediated signaling have recently been developed.
Early experiments showed that rhodopsin (Rh, encoded by RHO in humans) can activate Gαi in vitro (Kanaho et al., 1984), although, in the rod outer segments of the retina, Rh is coupled to Gαt. Subsequently, chimeric receptors were developed in which intracellular loops of Rh were replaced with those of β2 or α1 adrenergic receptors, and these were able to optically activate Gαs and Gαq and to increase cAMP and inositol trisphosphate (IP3) globally in cultured cells (Airan et al., 2009; Kim et al., 2005). Rh has also been used to optically control neuron excitability through activation of an inhibitory G protein pathway (Li et al., 2005). Furthermore, Rh-based chimeric receptors expressed in neurons can regulate behavior in mice (Airan et al., 2009). A light-activated Rh–CXCR4 chimeric receptor has been used to improve the efficacy of adoptive T-cell transfer immunotherapy to reduce tumor growth in mice by using light to increase T-cell trafficking to the tumor (Xu et al., 2014). This work highlights the therapeutic potential of optically activated GPCRs.
However, certain spectral and kinetic properties of Rh and its chimeric receptors, such as the occurrence of a prolonged active state due to slow deactivation, slow recovery (Shichida and Matsuyama, 2009), rapid bleaching (Bailes et al., 2012) and lack of spectral selectivity, make them less desirable for use in subcellular optogenetics. Rh shows single-photon sensitivity (Rieke and Baylor, 1998) and a relatively broad absorption spectrum that almost spans the visible range. Imaging of most fluorescent proteins will thus result in Rh activation.
In contrast, color opsins in the cone photoreceptors of the retina possess properties that make them particularly useful for optically controlling signaling at the subcellular level (Box 1). Although Rh and color opsins activate G proteins with comparable Km values (Imamoto et al., 2013), color opsins deactivate and recover more rapidly, and photobleach relatively slowly, allowing repeated activation. Owing to their intrinsic variation in spectral tuning, they also provide spectral selectivity (Imamoto and Shichida, 2014).
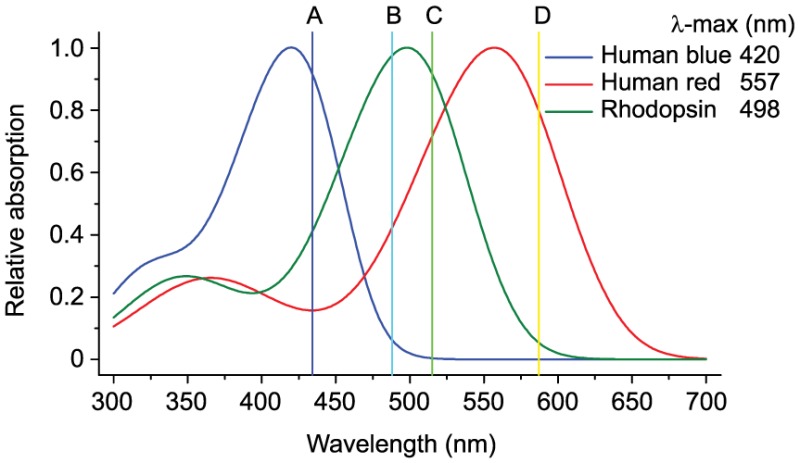
Box 1. Spectral selectivity of opsins and compatibility with fluorescence imaging.
The absorption spectra of different opsins considerably influence their utility as subcellular optogenetic signaling triggers, as shown in the figure. The vertical lines A–D indicate the wavelengths used to excite mTurquoise, GFP, YFP and mCherry. The blue opsin plot shows that absorption is minimal above ∼490 nm; this is consistent with the ability to image GFP and mCherry without activating the opsin (Karunarathne et al., 2013a). In contrast, excitation wavelengths of the commonly used fluorescent proteins all overlap with the activation spectra of rhodopsin and red opsin, indicating that these opsins are not suited for imaging the signaling responses. It might be possible to obtain a red-shifted opsin by screening naturally occurring opsins or through mutational engineering. Spectrally selective blue and red opsins that are differentially coupled to Gi, which decreases cAMP, and Gs, which increases cAMP, can be expressed in the same cell to attain further control over cell behavior. It is encouraging that recent efforts with channelrhodopsin, which shares the same chromophore as opsins, yielded a red-shifted channelrhodopsin that could be used together with a blue-sensing counterpart to achieve dual control (Klapoetke et al., 2014). Additionally, a red-shifted opsin could be used in the same cell together with optogenetic tools that are based on blue-light-activated domains such as CRY2. Thus a GPCR pathway could be activated in its entirety using the opsin, whereas a blue light-sensitive optical trigger is used to modulate specific downstream signaling elements within the same cell. This enhanced capability to interrogate signaling pathways might help to reveal new interactions and mechanisms that govern cell behaviors.
Relying on these unique properties, we have shown that the short-wavelength human cone photoreceptor opsin (blue opsin, encoded by OPN1SW) and mouse melanopsin (encoded by Opn4) can be used to optically control Gi/o and Gq signaling in subcellular regions of cells (Karunarathne et al., 2013a; Karunarathne et al., 2013b) (Fig. 3A). Blue opsin was further used to dynamically control two types of polarized cell behaviors, cell migration and neurite growth, both of which require the asymmetric activation of signaling across a cell (Karunarathne et al., 2013a; Karunarathne et al., 2013b), consistent with knowledge that these processes are regulated by Gi-coupled GPCRs (Bromberg et al., 2008; Cai and Devreotes, 2011). Furthermore, subcellular optical control with blue opsin can be combined with global imaging of biosensors that consist of fluorescent proteins that do not have spectral overlap with the opsin (Karunarathne et al., 2013a) (Fig. 1). The blue-opsin-stimulated dynamics of phosphatidylinositol (3,4,5)-trisphosphate (PtdInsP3) and β-actin, as well as the stereotypic morphological changes that are characteristic of immune cell migration and neurite growth, have been shown to closely mimic native responses (Karunarathne et al., 2013a). The unique properties of color opsins mentioned above open up the possibility of using them directly or in chimeric form to control other cellular behaviors. For instance, the ability of color opsins to be activated repeatedly has been recruited to obtain continual optical control over G protein signaling in brain slices and regulate anxiety in mice (Masseck et al., 2014).
Fig. 3.
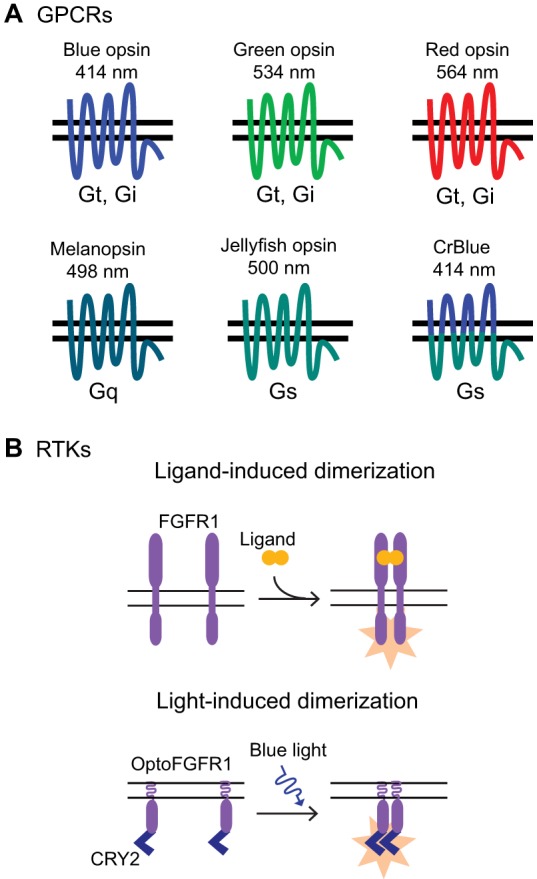
Optical control over GPCR and RTK signaling. (A) The diverse family of G-protein-coupled opsins includes receptors that are activated optimally at different wavelengths of light (colors denote wavelengths mentioned). As shown, the opsins are activated by light from across the visual spectrum. They also selectively activate different families of G proteins as indicated, which can allow optical control over distinct signaling pathways. It is possible to design chimeric opsins that combine the desired spectral tuning and G protein coupling. (B) Light-sensitive RTKs have not been found in nature, but engineered Opto-RTKs can generate the same signaling and cellular responses to those normally triggered by ligands that activate native RTKs. Opto-RTKs consist of RTKs with light-sensitive domains fused to their intracellular region. Photoactivation of these domains causes domain dimerization, resulting in receptor dimerization and activation (orange star).
To obtain subcellular optical triggers that are capable of activating different second messenger pathways, chimeric receptors that contain the light-sensing domains of blue opsin and appropriate cytosolic G-protein-coupling regions of a different receptor can be engineered. One such receptor, CrBlue, contains the Gs-coupling cytosolic regions of jellyfish opsin (Koyanagi et al., 2008) and the light-sensing domains of blue opsin (Karunarathne et al., 2013a) (Fig. 3A). CrBlue possesses spectral selectivity similar to that of blue opsin and because it is able to activate Gs and induce an increase in cAMP, can potentially be used to examine the effect of subcellular changes in cAMP on cellular behavior.
The opsin-bound chromophore retinal is enriched in photoreceptors of the retina but it also appears to be present in sufficient concentrations in neurons for both rhodopsin and color opsins to be able to modulate ion channel activity. In contrast, it is necessary to provide exogenous retinal to observe optically triggered migration of immune cells or neurite growth responses. However, opsin 3 (Opn3) homologs from mosquito (MosOpn3) and puffer fish (PufTMT) can be expressed in mammalian cells and allow continuous optical activation of G proteins without the need to add 11-cis retinal as is required for the color opsins (Koyanagi et al., 2013).
Optical activation of RTKs
Natural light-activated RTKs have not been reported, but engineered constructs termed opto-RTKs have been recently developed that allow optical control over RTK signaling (Fig. 3B). Ligand activation of RTKs often involves receptor dimerization and autophosphorylation. Optically induced dimerization of engineered RTKs has been shown to be sufficient to activate several canonical downstream responses. For example, a bacterial LOV domain that is capable of forming homodimers was fused to several different RTKs and provided optical control over their signaling pathways (Grusch et al., 2014). Similarly, fusing a CRY2 domain capable of homo-oligomerization to fibroblast growth factor receptor 1 (FGFR1) allowed optical control over receptor dimerization and signaling responses including directed cell migration (Kim et al., 2014). A similar CRY2-based approach was able to activate signaling by the tropomyosin related kinase (Trk) family of RTKs (Chang et al., 2014). These receptors are known to contribute to neurite outgrowth, and optical activation of Trk signaling has been shown to reproduce this cellular response. The mechanistic basis of LOV and CRY2 domain function is described below.
The ability of optically induced dimerization to control signaling by several different RTKs suggests that this approach can be extended to other receptors within this important family. The above experiments were performed using cultured cells, but opto-RTKs will likely be useful for studies in intact animals. For in vivo experiments it will be useful that opto-RTKs can be designed to be insensitive to their native ligand, because they no longer require the extracellular ligand-binding domains for activation (Grusch et al., 2014; Kim et al., 2014). This allows their activation to be exclusively controlled by light.
Optical activation of downstream signaling proteins
LOV domains of phototropin blue light receptors contain a flavin-based blue-light-sensing chromophore and regulate light-mediated biological processes in microbes and plants (Möglich and Moffat, 2010). In the dark state, the LOV domain interacts with a C-terminal helical region termed Jα. Light exposure leads to unwinding of the Jα helix, and this photo-sensitive conformational change has been utilized to mask and unmask signaling protein activity with light. For example, subcellular control over the small GTPase Rac has been achieved by fusing a constitutively active Rac1 mutant to a LOV2 domain that inhibits its interaction with effectors in a light-controllable manner (Wu et al., 2009). Here, light-induced unwinding of Jα relieves steric hindrance, thus allowing Rac to interact with its effectors.
Many signaling proteins rely on membrane targeting for their function, which is achieved through posttranslational lipid modifications or membrane-targeting domains. Optical control of the signaling activity of such proteins has been achieved by CRY- and Phy-based approaches that provide light-dependent membrane targeting. They rely on CRY and Phy forming heterodimers with specific partners in the presence of light. Blue light activates Arabidopsis thaliana CRY2, resulting in its binding to CIB1, and this interaction is preserved in mammalian cells (Kennedy et al., 2010). The PHR domain of CRY2 (CRY2PHR) and the N-terminal domain of CIB1 (CIBN) are sufficient for blue-light-induced binding. Similar to LOV domains, CRY domains contain a chromophore that is based on flavin adenine dinucleotide (FAD), which is present in mammalian cells at sufficient concentrations for optical control. Phytochromes can switch between red- and far-red-absorbing states (Quail, 2002). The associated conformational change regulates interaction with PIFs. Red light induces dimerization, whereas far-red light induces dissociation, allowing the interaction to be rapidly reversed. In cells expressing one member of the CRY2–CIBN or Phy–PIF pair fused to the plasma membrane, and a second member fused to a signaling protein lacking its normal membrane-targeting domain, light exposure can result in the binding of the cytosolic signaling protein to the membrane.
We have focused on the use of the LOV, CRY–CIBN and Phy–PIF because they have been used most widely in subcellular optogenetics. The existing optogenetic tools can potentially be improved through either engineering or identifying new domains from these families of light-sensitive proteins (Christie et al., 2007; Liu et al., 2013; Pathak et al., 2012; Pathak et al., 2013; Raffelberg et al., 2011; Strickland et al., 2010; Zoltowski et al., 2009). Additional approaches that have been recently developed for optical control of protein function that involve the use of unnatural amino acids, genetically encoded photosensitizers, or light-dependent association and dissociation of mutant fluorescent proteins might also prove to be widely applicable (Baker and Deiters, 2014; Bulina et al., 2006; Takemoto et al., 2013; Zhou et al., 2012).
The LOV, CRY2–CIBN and Phy–PIF approaches described above have been used to control many different intracellular signaling proteins. Here, we provide examples where engineered constructs have been used to achieve optical control over signaling proteins that are normally regulated by GPCRs, RTKs, or both. The ability to achieve optical control over their activity without the need for receptor activation can help determine which cellular responses are directly due to the activities of these downstream proteins and which responses require additional components of the signaling network to be activated by the receptor. For example, the small G protein Rac is involved in both GPCR- and RTK-mediated signaling, and subcellular optical activation has demonstrated that localized Rac activity is sufficient to drive migration independently of receptor activity in a number of cell types.
This LOV-domain-based approach has been used to address the role of different signaling proteins. It was used to control migration of border cells in the Drosophila ovary (Wang et al., 2010) and to identify the roles of Rac1 activation and deactivation in macropinocytosis (Fujii et al., 2013). It has been used to generate photoswitchable signaling peptides (Lungu et al., 2012) and to exert optical control of diaphanous-related formins (Rao et al., 2013). Optically induced heterodimerization of an LOV domain with an engineered PDZ domain has also been used to optically recruit the small G protein Cdc42, and Ste5, a scaffolding protein involved in MAPK activation, to induce polarized responses in yeast cells (Strickland et al., 2012). Here, subcellular control of a downstream element was able to evoke a response without receptor activation.
Although a chemically induced dimerization strategy (DeRose et al., 2013) has been used, optically induced dimerization strategies offer advantages, including increased spatial and temporal resolution, as well as reversibility. Several different light-induced dimerization pairs have been used in mammalian cells and they have been reviewed recently (Pathak et al., 2013). Below, we focus on strategies that use CRY2–CIBN or Phy–PIF to control signaling protein activity with subcellular resolution.
In addition to binding CIBN, photoactivated CRY2 can also form oligomers upon activation with blue light. This was exploited in light-induced clustering of CRY2-tagged low-density-lipoprotein (LDL)-receptor-related protein 6 (CRY2–LRP6) transmembrane receptors, which has been shown to activate the Wnt/β-catenin pathway (Bugaj et al., 2013). In addition, light-induced clustering of CRY2-tagged Rac1 and RhoA could initiate known responses of these small GTPases (Bugaj et al., 2013), demonstrating that clustering is sufficient to mediate their activation. This feature of CRY2 might be applicable to other transmembrane and cytosolic proteins whose activities are regulated by cluster formation.
Similar to the CRY2–CIBN system, the Phy–PIF system has been used to control signaling by light-triggered translocation to the membrane. The activity of small G proteins is controlled by a GTP hydrolysis cycle, and they can be activated by guanine-nucleotide-exchange factors (GEFs). Many GEFs require membrane targeting in order to activate small G proteins. The Phy–PIF system has been used to optically control membrane binding of several GEFs to regulate Rac, Rho, and Ras activity (Levskaya et al., 2009; Toettcher et al., 2013). The light-inducible reversibility of an optimized Phy–PIF pair is on the time scale of seconds, which provides an advantage over the CRY2–CIBN system that reverses over several minutes. A disadvantage of the Phy–PIF system is that the plant phytochrome requires a phycocyanobilin (PCB) chromophore that must be added exogenously to mammalian cells. However, a genetically encoded PCB synthesis system might be able to facilitate optogenetics using Phy–PIF (Müller et al., 2014).
The use of phytochromes in optogenetics is not limited to dimerization strategies. Bacteriophytochromes contain photosensory modules that are coupled to different output modules, such as histidine kinases, and these photosensory modules have been shown to be capable of regulating heterologous output domains. For instance, light-activated phosphodiesterases for cAMP and cGMP have been created by replacing the chemosensor of a human phosphodiesterase with the photosensor of a bacterial phytochrome (Gasser et al., 2014). Similarly, a near-infrared-light-activated adenylyl cyclase has been engineered (Ryu et al., 2014). Notably, phytochromes from bacteria use a biliverdin chromophore that is present in mammalian cells (Gasser et al., 2014; Piatkevich et al., 2013; Ryu et al., 2014). However, these approaches require a significant structural similarity between the native signaling module of the phytochrome and the new module that replaces it and might not be as broadly applicable as the translocation-based approach described above.
MAPK signaling is also triggered downstream of RTKs to control differentiation and proliferation in mammalian cells. Optically triggered activation of C-RAF, a serine/threonine kinase involved in this pathway, was achieved using CRY2 to induce light-dependent dimerization (Wend et al., 2014) and shown to produce similar downstream responses to RTK activation.
Lipid kinases and phosphatases dynamically regulate the levels of different signaling lipids within a cell, and their activities can be regulated by extracellular cues through receptor-triggered signaling pathways. CRY2–CIBN interactions have been used to obtain subcellular control over lipid kinases and phosphatases that act on phosphoinositides by optically targeting them to the plasma membrane (Idevall-Hagren et al., 2012; Kakumoto and Nakata, 2013). Such control has been useful in determining the role of plasma membrane PtdIns(4,5)P2 in recruiting extended synaptotagmins, which act as tethers between the ER and plasma membrane (Giordano et al., 2013). CRY2–CIBN has also been used to achieve subcellular control of phosphoinositide-3 kinase (PI3K) signaling at synapses and examine its role in taste-avoidance learning in Caenorhabditis elegans (Ohno et al., 2014).
Table 1 summarizes some of the crucial properties of the various optical tools described above especially in terms of their practical utility.
Table 1. Summary of crucial properties of the optical tools discussed here.
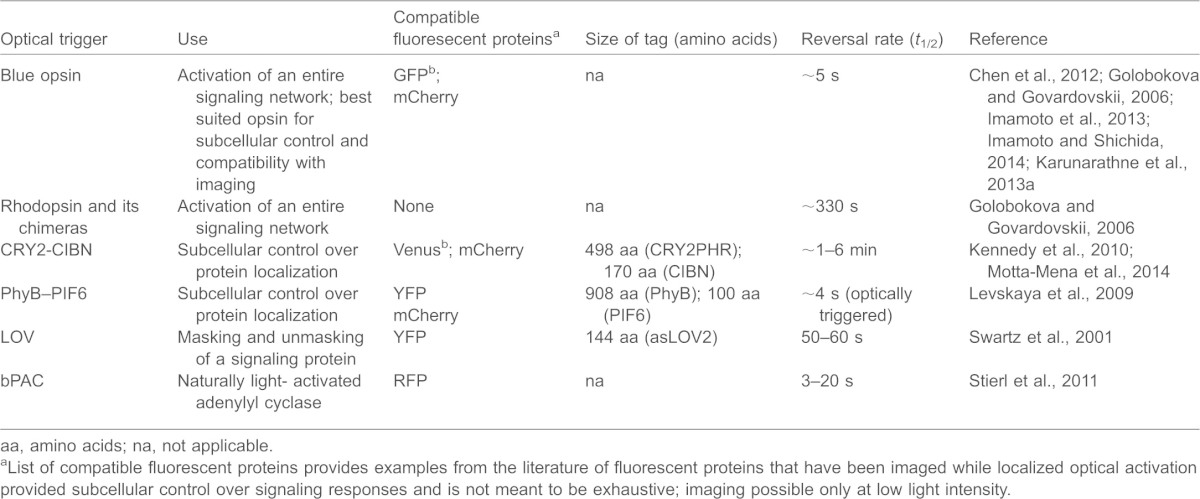
aa, amino acids; na, not applicable.
List of compatible fluorescent proteins provides examples from the literature of fluorescent proteins that have been imaged while localized optical activation provided subcellular control over signaling responses and is not meant to be exhaustive;
imaging possible only at low light intensity.
Optogenetic inhibition of endogenous signaling proteins
In classical studies of protein function, results that have been obtained using overexpression of proteins are more reliable when they are supported by appropriate knockdown studies. Similarly, results from optogenetic activation of selected signaling proteins are more valuable when they are supported by data from optically inhibiting endogenous signaling proteins. For example, experiments using optically triggered LOV–Rac were complemented with a dominant-negative mutant version that competes with endogenous Rac for binding to its effectors upon optical activation (Wu et al., 2009). Alternatively, light-triggered recruitment of signaling proteins away from their normal sites of action has been used to inhibit signaling proteins (Lee et al., 2014; Yang et al., 2013) (Fig. 4A).
Fig. 4.
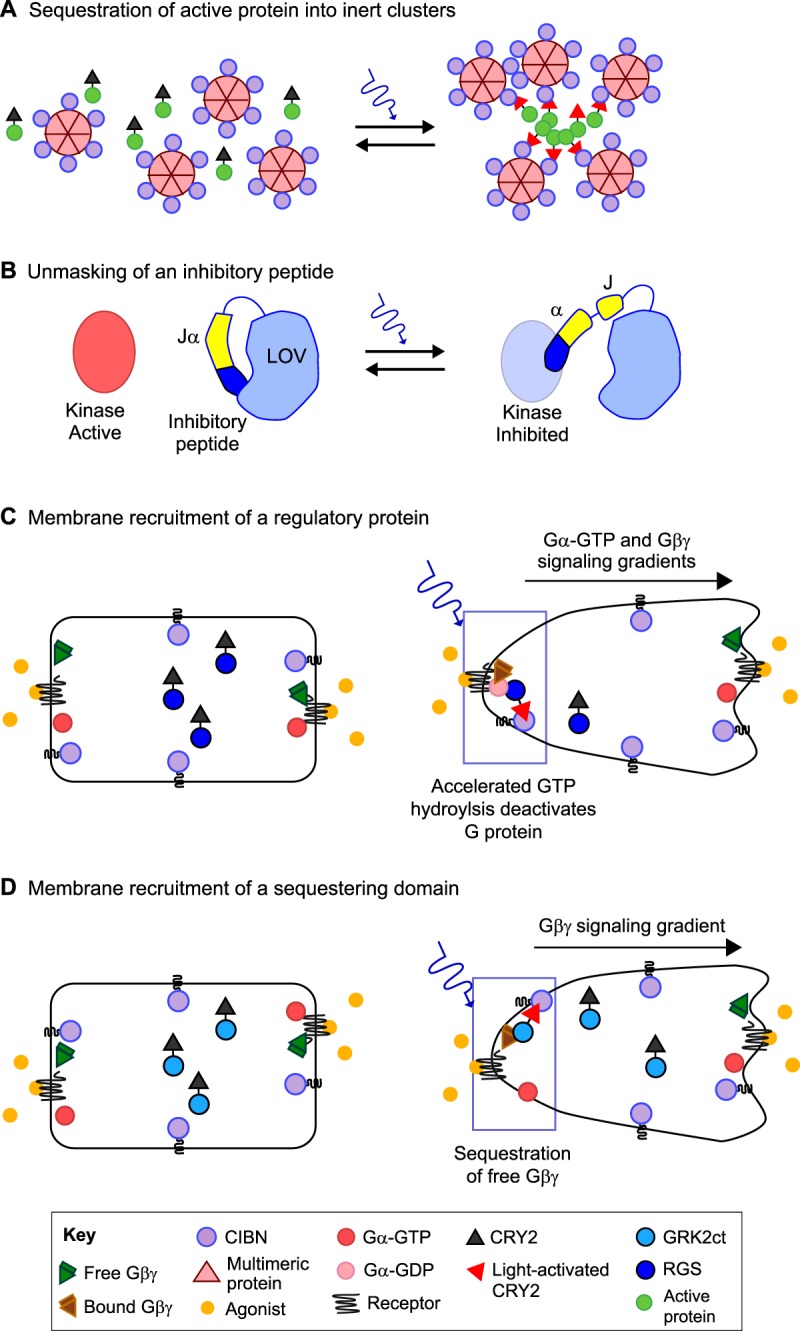
Strategies to optically inhibit signaling of endogenous proteins. (A) A protein can be prevented from interacting with its signaling partners by light-induced recruitment into large clusters. For instance, fusion of CIBN to a multimeric protein and of CRY2 to the protein of interest results in cluster formation upon light activation that effectively sequesters the protein of interest (Lee et al., 2014). (B) Fusion of inhibitory peptides to LOV domains. Steric hindrance by the LOV domain prevents the peptides from binding to their targets (e.g. a kinase as shown here) in the dark. Photoactivation unmasks the peptides, allowing them to inhibit their endogenous protein targets (Yi et al., 2014). (C) Altering the subcellular localization of a regulatory protein domain. Optically triggering the recruitment of a CRY2–RGS4 fusion to the membrane at one end of a cell can be used to induce localized G protein deactivation. When uniform agonist is applied to activate G protein signaling throughout a cell, the localized deactivation results in an intracellular gradient of Gα-GTP and Gβγ signaling (O'Neill and Gautam, 2014). (D) Optically altering the subcellular localization of a sequestering domain by recruitment to the membrane. When an agonist is applied to uniformly activate G protein signaling through a cell, optical recruitment of the G protein Gβγ-sequestering domain CRK2ct to one end of a cell results in a gradient of βγ signaling (O'Neill and Gautam, 2014).
Light-triggered unmasking of inhibitory peptides has been used to deactivate signaling mediated by protein kinase A (PKA) and myosin light chain kinase (MLCK), which was achieved by appending the inhibitory peptides for these kinases to the C-terminus of the Jα helix of LOV2 (Fig. 4B). Achieving subcellular control with this optogenetic approach might require the addition of a membrane-targeting domain to avoid rapid diffusion upon light activation.
We recently developed two optogenetic tools that inhibit G protein subunit activity to create reversible signaling gradients of endogenous heterotrimeric G protein subunits (O'Neill and Gautam, 2014). In the first tool, the native membrane-targeting domain of regulator of G protein signaling 4 (RGS4) is replaced with CRY2 to enable optical control over its membrane localization (Fig. 4C). Membrane targeting of CRY2–RGS4 results in accelerated GTP hydrolysis and deactivation of the G protein α subunit. As a result of the high affinity of inactive Gα-GDP for the G protein βγ subunit, CRY2–RGS4 also effectively inhibits βγ signaling. The subcellular control provided by this approach enabled the generation of Gα-GTP and Gβγ signaling gradients and directional control over migration of macrophage cells. The second tool was designed to specifically inhibit G protein βγ subunits by fusing CRY2 to the Gβγ-sequestering C-terminal domain of GPCR kinase 2 (GRK2ct; GRK2 is also known as ADRBK1) (Fig. 4D). When this CRY2 construct is asymmetrically activated by optical means, it generates intracellular Gβγ signaling gradients and directs polarized responses in a macrophage cell line (RAW 264.7).
What can optical control of cell behavior reveal about cell signaling mechanisms?
The properties of optical triggers described above facilitate specific experimental designs to address questions regarding the molecular basis of important cell behaviors. We outline below a few examples where such experiments have yielded useful information.
One area where the ability to control signaling at the subcellular level has helped to provide new mechanistic insights is in understanding the communication between the front and back of a migrating cell. Experiments using photoactivatable Rac (PA-Rac) have shown cross-cell coordination of protrusion and retraction by a Rac activity gradient (Wu et al., 2009). They also supported roles for PAK in mediating cell protrusion at the site of Rac activation, and for myosin in mediating cell retraction at the opposite side (Wu et al., 2009). Experiments using PA-Rac to control neutrophil migration in zebrafish revealed that Rac activity at the front of a cell controls F-actin dynamics at the back through a PI3K-dependent mechanism (Yoo et al., 2010). Two different optical approaches to trigger local retraction of cells were also found to generate responses at the distal side of the cell. Local optical activation of an inositol 5-phosphatase at one side of a cell resulted in local retraction with increases in the level of PtdIns(4,5)P2 and the amount of membrane ruffling at the opposite side (Idevall-Hagren et al., 2012). In contrast, local inhibition of Vav2, a small G protein GEF, induced local membrane retraction and generated membrane protrusions on the opposite side (Lee et al., 2014).
Subcellular optogenetics can also provide insights into signaling mechanisms underlying the differential cell responses triggered by activation of an entire signaling network versus activation of individual signaling components within the network (Fig. 2B). One example for this is comparison of migratory cell responses to optically generated gradients of GPCR activity versus gradients of G protein subunit activity. In macrophage cells, local activation of G protein signaling at one end of the cell using human blue opsin has been shown to be sufficient to induce directionally sensitive cell migration (Karunarathne et al., 2013b). Furthermore, we have shown using CRY2–RGS4 that in cells that are exposed to a spatially uniform chemoattractant, local inhibition of G protein α and βγ subunit signaling by CRY2–RGS4 is also capable of inducing migration (O'Neill and Gautam, 2014). However, the local inhibition of only Gβγ by CRY2–GRK2ct induces the formation of lamellipodia and steep PtdInsP3 gradients, but with only limited migration. These results show that gradients of G protein signaling are sufficient to drive migration independently of gradients of receptor activity and also raise the possibility that a G protein α subunit dependent pathway might be required to elicit the full migratory response.
Neurite outgrowth is another example where optical control has helped to identify differential cell responses upon activating a GPCR versus a downstream module. In primary rat hippocampal neurons, the spatially confined optical activation of Gi-coupled blue opsin at the tip of a neurite can elicit a localized increase in PtdInsP3 and induce neurite extension (Karunarathne et al., 2013a). In contrast, photoactivation of a CRY2–PI3K construct at the tip of a neurite can only elicit an increase in PtdInsP3 and the formation of filopodia and lamellipodia, but not neurite elongation (Kakumoto and Nakata, 2013). This difference suggests that Gi-mediated neurite extension involves additional pathways besides PI3K activation.
To understand how complex networks of interacting signaling proteins govern dynamic polarized cell behaviors that consist of a sequence of stereotypical changes in cell morphology, reorganization and remodeling of molecules, it is necessary to complement experiments with mathematical methods (Fivaz et al., 2008; Iglesias and Devreotes, 2012; Inagaki et al., 2011; Karunarathne et al., 2013b; Okada et al., 2013; Vilela et al., 2013; Welf and Haugh, 2011; Zhang et al., 2013). Mathematical modeling can be of value in predicting signaling mechanisms underlying dynamic cell behavior that are not obvious intuitively, so that they can then be tested experimentally. Mathematical tools can also be useful in analyzing large amounts of data and thereby arriving at unexpected insights into signaling control of a dynamic cellular process. Optical control is particularly well suited for these types of approaches because it is unique in providing quantitative information with regard to both the molecular and cellular dynamics when cell behavior is being experimentally manipulated by controlling signaling both spatially and temporally. The ability to vary the intensity of input, provide either continuous or pulsed input and vary the duration of pulses and pulse intervals allows the relationship between signal and response to be quantitatively measured. This ability was used to uncover two different classes of responses to a Ras signaling module: one that follows the input dynamics as Ras is turned on and off over a wide range of frequencies, and one that only responds to sustained pathway activation (Toettcher et al., 2013). The ability to quantitatively monitor PtdInsP3 formation in a migratory cell as a function of the number of light pulses used to activate blue opsin provides evidence for an ultrasensitive response underlying the initiation of cell migration. This finding motivated the creation of a mathematical model that predicts that there is a localized activator that is antagonistic to a diffusible inhibitor in generating the switch-like PtdInsP3 response (Karunarathne et al., 2013b).
Future perspectives
As described above, there is a requirement for experimental tools that control signaling at the subcellular level. By experimentally mimicking subcellular variations in signaling activity, the role in regulating cell behavior can be deciphered. We have discussed here the advantages of optogenetic tools for subcellular control and the specific tools that have been designed so far and used effectively. These approaches can be used to activate either entire signaling pathways or modulate signaling activity at a node within a signaling network by targeting specific proteins, as well as to optically direct signaling activity to a specific subcellular site. As mentioned above, these tools have already provided some biological insights, such as determining the roles of different types of receptor-triggered and intracellular signaling gradients in controlling polarized cell behaviors including neurite outgrowth and cell migration.
We expect that a variety of additional optogenetic tools similar to those discussed above will be developed that have new and distinct properties. Apart from regulating signaling in subcellular regions they can be used to control the behavior of individual cells in cellular clusters, such as insulin-secreting cells in islets or groups of migrating cells during morphogenesis. This can help us to understand the role that single-cell activity plays in the behavior of the population. Finally, the ability to modulate cellular behavior non-invasively with high spatiotemporal precision using light has therapeutic potential given that GPCR- and RTK-regulated pathways control the majority of crucial cellular functions.
Many photosensitive proteins from a variety of organisms have been extensively characterized (Ahmad and Cashmore, 1993; Christie et al., 1998; Frentiu and Briscoe, 2008; Hughes et al., 1997; Imamoto and Shichida, 2014; Iseki et al., 2002; Koyanagi et al., 2008; Liu et al., 2008; Ni et al., 1999; Yokoyama et al., 2008). Detailed knowledge of these molecules from such studies has led to the development of subcellular optogenetics. The characterization of additional light-sensitive proteins might reveal properties that can be harnessed for developing novel biological tools. There are over 1000 opsins that form a natural resource that can be mined for optical tools (Shichida and Matsuyama, 2009). For instance, the characterization of opsins from a colorful parrot species shows an unusual diversity of opsin types (Knott et al., 2013). This is a strong argument for curiosity-driven research and suggests that knowledge of little-understood molecular events in the extraordinarily diverse species that populate the Earth will continue to yield important, but difficult to predict, applications.
Supplementary Material
Acknowledgments
We would like to thank Matthew Toomey (Washington University School of Medicine, St Louis, MO) for discussions regarding opsin spectra and Vani Kalyanaraman (Washington University School of Medicine, St Louis, MO) for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Funding
This work was supported by National Institutes of Health [grant numbers GM069027 and GM080558 to N.G.]; and a National Research Service Award Postdoctoral Fellowship [grant number GM099351 to P.R.O.]. Deposited in PMC for release after 12 months.
References
- Ahmad M., Cashmore A. R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166 10.1038/366162a0 [DOI] [PubMed] [Google Scholar]
- Airan R. D., Thompson K. R., Fenno L. E., Bernstein H., Deisseroth K. (2009). Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 10.1038/nature07926 [DOI] [PubMed] [Google Scholar]
- Antal C. E., Newton A. C. (2013). Spatiotemporal dynamics of phosphorylation in lipid second messenger signaling. Mol. Cell Proteomics 12, 3498–3508 10.1074/mcp.R113.029819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N., Kaibuchi K. (2007). Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 8, 194–205 10.1038/nrn2056 [DOI] [PubMed] [Google Scholar]
- Bailes H. J., Zhuang L. Y., Lucas R. J. (2012). Reproducible and sustained regulation of Gαs signalling using a metazoan opsin as an optogenetic tool. PLoS ONE 7, e30774 10.1371/journal.pone.0030774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A. S., Deiters A. (2014). Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chem. Biol. 9, 1398–1407 10.1021/cb500176x [DOI] [PubMed] [Google Scholar]
- Bromberg K. D., Iyengar R., He J. C. (2008). Regulation of neurite outgrowth by G(i/o) signaling pathways. Front. Biosci. 13, 4544–4557 10.2741/3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaj L. J., Choksi A. T., Mesuda C. K., Kane R. S., Schaffer D. V. (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 10.1038/nmeth.2360 [DOI] [PubMed] [Google Scholar]
- Bulina M. E., Chudakov D. M., Britanova O. V., Yanushevich Y. G., Staroverov D. B., Chepurnykh T. V., Merzlyak E. M., Shkrob M. A., Lukyanov S., Lukyanov K. A. (2006). A genetically encoded photosensitizer. Nat. Biotechnol. 24, 95–99 10.1038/nbt1175 [DOI] [PubMed] [Google Scholar]
- Cai H., Devreotes P. N. (2011). Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Semin. Cell Dev. Biol. 22, 834–841 10.1016/j.semcdb.2011.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calebiro D., Nikolaev V. O., Lohse M. J. (2010). Imaging of persistent cAMP signaling by internalized G protein-coupled receptors. J. Mol. Endocrinol. 45, 1–8 10.1677/JME-10-0014 [DOI] [PubMed] [Google Scholar]
- Chang K. Y., Woo D., Jung H., Lee S., Kim S., Won J., Kyung T., Park H., Kim N., Yang H. W. et al. (2014). Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat. Commun. 5, 4057 10.1038/ncomms5057 [DOI] [PubMed] [Google Scholar]
- Chen M-H., Kuemmel C., Birge R. R., Knox B. E. (2012). Rapid release of retinal from a cone visual pigment following photoactivation. Biochemistry 51, 4117–4125 10.1021/bi201522h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., Briggs W. R. (1998). Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701 10.1126/science.282.5394.1698 [DOI] [PubMed] [Google Scholar]
- Christie J. M., Corchnoy S. B., Swartz T. E., Hokenson M., Han I. S., Briggs W. R., Bogomolni R. A. (2007). Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry 46, 9310–9319 10.1021/bi700852w [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. (1973). Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 70, 3240–3244 10.1073/pnas.70.11.3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croushore C. A., Sweedler J. V. (2013). Microfluidic systems for studying neurotransmitters and neurotransmission. Lab Chip 13, 1666–1676 10.1039/c3lc41334a [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco R. J., Groneberg A. H., Yeh C. M., Castillo Ramírez L. A., Ryu S. (2013). Optogenetic elevation of endogenous glucocorticoid level in larval zebrafish. Front. Neural Circuits 7, 82 10.3389/fncir.2013.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L., Bastiaens P. I. (2010). Spatial organization of intracellular communication: insights from imaging. Nat. Rev. Mol. Cell Biol. 11, 440–452 10.1038/nrm2903 [DOI] [PubMed] [Google Scholar]
- DeRose R., Miyamoto T., Inoue T. (2013). Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 465, 409–417 10.1007/s00424-012-1208-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPilato L. M., Cheng X., Zhang J. (2004). Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc. Natl. Acad. Sci. USA 101, 16513–16518 10.1073/pnas.0405973101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis-Davies G. C. (2007). Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods 4, 619–628 10.1038/nmeth1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etoc F., Lisse D., Bellaiche Y., Piehler J., Coppey M., Dahan M. (2013). Subcellular control of Rac-GTPase signalling by magnetogenetic manipulation inside living cells. Nat. Nanotechnol. 8, 193–198 10.1038/nnano.2013.23 [DOI] [PubMed] [Google Scholar]
- Fivaz M., Meyer T. (2005). Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J. Cell Biol. 170, 429–441 10.1083/jcb.200409157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivaz M., Bandara S., Inoue T., Meyer T. (2008). Robust neuronal symmetry breaking by Ras-triggered local positive feedback. Curr. Biol. 18, 44–50 10.1016/j.cub.2007.11.051 [DOI] [PubMed] [Google Scholar]
- Fonseca S. G., Urano F., Weir G. C., Gromada J., Burcin M. (2012). Wolfram syndrome 1 and adenylyl cyclase 8 interact at the plasma membrane to regulate insulin production and secretion. Nat. Cell Biol. 14, 1105–1112 10.1038/ncb2578 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fosbrink M., Aye-Han N. N., Cheong R., Levchenko A., Zhang J. (2010). Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc. Natl. Acad. Sci. USA 107, 5459–5464 10.1073/pnas.0909671107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu F. D., Briscoe A. D. (2008). A butterfly eye's view of birds. BioEssays 30, 1151–1162 10.1002/bies.20828 [DOI] [PubMed] [Google Scholar]
- Fujii M., Kawai K., Egami Y., Araki N. (2013). Dissecting the roles of Rac1 activation and deactivation in macropinocytosis using microscopic photo-manipulation. Sci. Rep. 3, 2385 10.1038/srep02385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser C., Taiber S., Yeh C. M., Wittig C. H., Hegemann P., Ryu S., Wunder F., Möglich A. (2014). Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl. Acad. Sci. USA 111, 8803–8811 10.1073/pnas.1321600111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans B. N., Adams S. R., Ellisman M. H., Tsien R. Y. (2006). The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 10.1126/science.1124618 [DOI] [PubMed] [Google Scholar]
- Giordano F., Saheki Y., Idevall-Hagren O., Colombo S. F., Pirruccello M., Milosevic I., Gracheva E. O., Bagriantsev S. N., Borgese N., De Camilli P. (2013). PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golobokova E. Y., Govardovskii V. I. (2006). Late stages of visual pigment photolysis in situ: cones vs. rods. Vision Res. 46, 2287–2297 10.1016/j.visres.2005.12.017 [DOI] [PubMed] [Google Scholar]
- Grusch M., Schelch K., Riedler R., Reichhart E., Differ C., Berger W., Inglés-Prieto Á., Janovjak H. (2014). Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 33, 1713–1726 10.15252/embj.201387695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewavitharana T., Wedegaertner P. B. (2012). Non-canonical signaling and localizations of heterotrimeric G proteins. Cell. Signal. 24, 25–34 10.1016/j.cellsig.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., Lamparter T., Mittmann F., Hartmann E., Gärtner W., Wilde A., Börner T. (1997). A prokaryotic phytochrome. Nature 386, 663 10.1038/386663a0 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O., Dickson E. J., Hille B., Toomre D. K., De Camilli P. (2012). Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA 109, E2316–E2323 10.1073/pnas.1211305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias P. A., Devreotes P. N. (2012). Biased excitable networks: how cells direct motion in response to gradients. Curr. Opin. Cell Biol. 24, 245–253 10.1016/j.ceb.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto Y., Shichida Y. (2014). Cone visual pigments. Biochim. Biophys. Acta 1837, 664–673 10.1016/j.bbabio.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Imamoto Y., Seki I., Yamashita T., Shichida Y. (2013). Efficiencies of activation of transducin by cone and rod visual pigments. Biochemistry 52, 3010–3018 10.1021/bi3015967 [DOI] [PubMed] [Google Scholar]
- Inagaki N., Toriyama M., Sakumura Y. (2011). Systems biology of symmetry breaking during neuronal polarity formation. Dev. Neurobiol. 71, 584–593 10.1002/dneu.20837 [DOI] [PubMed] [Google Scholar]
- Irannejad R., Tomshine J. C., Tomshine J. R., Chevalier M., Mahoney J. P., Steyaert J., Rasmussen S. G., Sunahara R. K., El-Samad H., Huang B. et al. (2013). Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 10.1038/nature12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki M., Matsunaga S., Murakami A., Ohno K., Shiga K., Yoshida K., Sugai M., Takahashi T., Hori T., Watanabe M. (2002). A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415, 1047–1051 10.1038/4151047a [DOI] [PubMed] [Google Scholar]
- Kakumoto T., Nakata T. (2013). Optogenetic control of PIP3: PIP3 is sufficient to induce the actin-based active part of growth cones and is regulated via endocytosis. PLoS ONE 8, e70861 10.1371/journal.pone.0070861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaho Y., Tsai S. C., Adamik R., Hewlett E. L., Moss J., Vaughan M. (1984). Rhodopsin-enhanced GTPase activity of the inhibitory GTP-binding protein of adenylate cyclase. J. Biol. Chem. 259, 7378–7381. [PubMed] [Google Scholar]
- Karunarathne W. K., Giri L., Kalyanaraman V., Gautam N. (2013a). Optically triggering spatiotemporally confined GPCR activity in a cell and programming neurite initiation and extension. Proc. Natl. Acad. Sci. USA 110, E1565–E1574 10.1073/pnas.1220697110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne W. K., Giri L., Patel A. K., Venkatesh K. V., Gautam N. (2013b). Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc. Natl. Acad. Sci. USA 110, E1575–E1583 10.1073/pnas.1220755110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. J., Hughes R. M., Peteya L. A., Schwartz J. W., Ehlers M. D., Tucker C. L. (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Hwa J., Garriga P., Reeves P. J., RajBhandary U. L., Khorana H. G. (2005). Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry 44, 2284–2292 10.1021/bi048328i [DOI] [PubMed] [Google Scholar]
- Kim N., Kim J. M., Lee M., Kim C. Y., Chang K. Y., Heo W. D. (2014). Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem. Biol. 21, 903–912 10.1016/j.chembiol.2014.05.013 [DOI] [PubMed] [Google Scholar]
- Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., Cho Y. K., Morimoto T. K., Chuong A. S., Carpenter E. J., Tian Z. et al. (2014). Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott B., Davies W. I., Carvalho L. S., Berg M. L., Buchanan K. L., Bowmaker J. K., Bennett A. T., Hunt D. M. (2013). How parrots see their colours: novelty in the visual pigments of Platycercus elegans. J. Exp. Biol. 216, 4454–4461 10.1242/jeb.094136 [DOI] [PubMed] [Google Scholar]
- Koyanagi M., Takano K., Tsukamoto H., Ohtsu K., Tokunaga F., Terakita A. (2008). Jellyfish vision starts with cAMP signaling mediated by opsin-G(s) cascade. Proc. Natl. Acad. Sci. USA 105, 15576–15580 10.1073/pnas.0806215105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M., Takada E., Nagata T., Tsukamoto H., Terakita A. (2013). Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotoreceptive tissue. Proc. Natl. Acad. Sci. USA 110, 4998–5003 10.1073/pnas.1219416110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss U., Lee J., Benkovic S. J., Jaeger K. E. (2010). LOVely enzymes - towards engineering light-controllable biocatalysts. Microb. Biotechnol. 3, 15–23 10.1111/j.1751-7915.2009.00140.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis P., Leondaritis G., Lieberam I., Eickholt B. J. (2014). Subcellular targeting and dynamic regulation of PTEN: implications for neuronal cells and neurological disorders. Front. Mol. Neurosci. 7, 23 10.3389/fnmol.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbalasiri T., Provencio I. (2005). Melanopsin and other novel mammalian opsins. Exp. Eye Res. 81, 368–375 10.1016/j.exer.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Kunkel M. T., Newton A. C. (2010). Calcium transduces plasma membrane receptor signals to produce diacylglycerol at Golgi membranes. J. Biol. Chem. 285, 22748–22752 10.1074/jbc.C110.123133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lamb T. D. (2013). Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119 10.1016/j.preteyeres.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Lee S., Park H., Kyung T., Kim N. Y., Kim S., Kim J., Heo W. D. (2014). Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods 11, 633–636 10.1038/nmeth.2940 [DOI] [PubMed] [Google Scholar]
- Levskaya A., Weiner O. D., Lim W. A., Voigt C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001 10.1038/nature08446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gutierrez D. V., Hanson M. G., Han J., Mark M. D., Chiel H., Hegemann P., Landmesser L. T., Herlitze S. (2005). Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA 102, 17816–17821 10.1073/pnas.0509030102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539 10.1126/science.1163927 [DOI] [PubMed] [Google Scholar]
- Liu Y., Li X., Li K., Liu H., Lin C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9, e1003861 10.1371/journal.pgen.1003861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lungu O. I., Hallett R. A., Choi E. J., Aiken M. J., Hahn K. M., Kuhlman B. (2012). Designing photoswitchable peptides using the AsLOV2 domain. Chem. Biol. 19, 507–517 10.1016/j.chembiol.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009). Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masseck O. A., Spoida K., Dalkara D., Maejima T., Rubelowski J. M., Wallhorn L., Deneris E. S., Herlitze S. (2014). Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron 81, 1263–1273 10.1016/j.neuron.2014.01.041 [DOI] [PubMed] [Google Scholar]
- Miesenböck G. (2011). Optogenetic control of cells and circuits. Annu. Rev. Cell Dev. Biol. 27, 731–758 10.1146/annurev-cellbio-100109-104051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A., Moffat K. (2010). Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 9, 1286–1300. [DOI] [PubMed] [Google Scholar]
- Motta-Mena L. B., Reade A., Mallory M. J., Glantz S., Weiner O. D., Lynch K. W., Gardner K. H. (2014). An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 10, 196–202 10.1038/nchembio.1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Zurbriggen M. D., Weber W. (2014). Control of gene expression using a red- and far-red light-responsive bi-stable toggle switch. Nat. Protoc. 9, 622–632 10.1038/nprot.2014.038 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J. M., Quail P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784 10.1038/23500 [DOI] [PubMed] [Google Scholar]
- O'Neill P. R., Gautam N. (2014). Subcellular optogenetic inhibition of G proteins generates signaling gradients and cell migration. Mol. Biol. Cell 25, 2305–2314 10.1091/mbc.E14-04-0870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill P. R., Karunarathne W. K. A., Kalyanaraman V., Silvius J. R., Gautam N. (2012). G-protein signaling leverages subunit-dependent membrane affinity to differentially control βγ translocation to intracellular membranes. Proc. Natl. Acad. Sci. USA 109, E3568–E3577 10.1073/pnas.1205345109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Kato S., Naito Y., Kunitomo H., Tomioka M., Iino Y. (2014). Role of synaptic phosphatidylinositol 3-kinase in a behavioral learning response in C. elegans. Science 345, 313–317 10.1126/science.1250709 [DOI] [PubMed] [Google Scholar]
- Okada S., Leda M., Hanna J., Savage N. S., Bi E., Goryachev A. B. (2013). Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell 26, 148–161 10.1016/j.devcel.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R., Wu R. (1972). Nucleotide sequence analysis of DNA. IV. Complete nucleotide sequence of the left-hand cohesive end of coliphage 186 DNA. J. Mol. Biol. 65, 447–467 10.1016/0022-2836(72)90201-X [DOI] [PubMed] [Google Scholar]
- Pathak G. P., Losi A., Gärtner W. (2012). Metagenome-based screening reveals worldwide distribution of LOV-domain proteins. Photochem. Photobiol. 88, 107–118 10.1111/j.1751-1097.2011.01024.x [DOI] [PubMed] [Google Scholar]
- Pathak G. P., Vrana J. D., Tucker C. L. (2013). Optogenetic control of cell function using engineered photoreceptors. Biol. Cell 105, 59–72 10.1111/boc.201200056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O. (2010). Spatio-temporal Rho GTPase signaling – where are we now? J. Cell Sci. 123, 1841–1850 10.1242/jcs.064345 [DOI] [PubMed] [Google Scholar]
- Piatkevich K. D., Subach F. V., Verkhusha V. V. (2013). Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 42, 3441–3452 10.1039/c3cs35458j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93 10.1038/nrm728 [DOI] [PubMed] [Google Scholar]
- Raffelberg S., Mansurova M., Gärtner W., Losi A. (2011). Modulation of the photocycle of a LOV domain photoreceptor by the hydrogen-bonding network. J. Am. Chem. Soc. 133, 5346–5356 10.1021/ja1097379 [DOI] [PubMed] [Google Scholar]
- Rao M. V., Chu P. H., Hahn K. M., Zaidel-Bar R. (2013). An optogenetic tool for the activation of endogenous diaphanous-related formins induces thickening of stress fibers without an increase in contractility. Cytoskeleton 70, 394–407 10.1002/cm.21115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieke F., Baylor D. A. (1998). Single-photon detection by rod cells of the retina. Rev. Mod. Phys. 70, 1027–1036 10.1103/RevModPhys.70.1027 [DOI] [Google Scholar]
- Rockwell N. C., Su Y. S., Lagarias J. C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 10.1146/annurev.arplant.56.032604.144208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu M. H., Kang I. H., Nelson M. D., Jensen T. M., Lyuksyutova A. I., Siltberg-Liberles J., Raizen D. M., Gomelsky M. (2014). Engineering adenylate cyclases regulated by near-infrared window light. Proc. Natl. Acad. Sci. USA 111, 10167–10172 10.1073/pnas.1324301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini D. K., Chisari M., Gautam N. (2009). Shuttling and translocation of heterotrimeric G proteins and Ras. Trends Pharmacol. Sci. 30, 278–286 10.1016/j.tips.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A. L., Viñuela E., Maizel J. V., Jr (1967). Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 28, 815–820 10.1016/0006-291X(67)90391-9 [DOI] [PubMed] [Google Scholar]
- Shelly M., Lim B. K., Cancedda L., Heilshorn S. C., Gao H., Poo M. M. (2010). Local and long-range reciprocal regulation of cAMP and cGMP in axon/dendrite formation. Science 327, 547–552 10.1126/science.1179735 [DOI] [PubMed] [Google Scholar]
- Shichida Y., Matsuyama T. (2009). Evolution of opsins and phototransduction. Philos. Trans. R. Soc. B 364, 2881–2895 10.1098/rstb.2009.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J. M., Quail P. H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044 10.1038/nbt734 [DOI] [PubMed] [Google Scholar]
- Stierl M., Stumpf P., Udwari D., Gueta R., Hagedorn R., Losi A., Gärtner W., Petereit L., Efetova M., Schwarzel M. et al. (2011). Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286, 1181–1188 10.1074/jbc.M110.185496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D., Yao X., Gawlak G., Rosen M. K., Gardner K. H., Sosnick T. R. (2010). Rationally improving LOV domain-based photoswitches. Nat. Methods 7, 623–626 10.1038/nmeth.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D., Lin Y., Wagner E., Hope C. M., Zayner J., Antoniou C., Sosnick T. R., Weiss E. L., Glotzer M. (2012). TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384 10.1038/nmeth.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H., Schmid V., Gehring W. J. (2008). Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51–55 10.1016/j.cub.2007.11.059 [DOI] [PubMed] [Google Scholar]
- Swartz T. E., Corchnoy S. B., Christie J. M., Lewis J. W., Szundi I., Briggs W. R., Bogomolni R. A. (2001). The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276, 36493–36500 10.1074/jbc.M103114200 [DOI] [PubMed] [Google Scholar]
- Tadevosyan A., Vaniotis G., Allen B. G., Hébert T. E., Nattel S. (2012). G protein-coupled receptor signalling in the cardiac nuclear membrane: evidence and possible roles in physiological and pathophysiological function. J. Physiol. 590, 1313–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K., Matsuda T., Sakai N., Fu D., Noda M., Uchiyama S., Kotera I., Arai Y., Horiuchi M., Fukui K. et al. (2013). SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci. Rep. 3, 2629 10.1038/srep02629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A. (2005). The opsins. Genome Biol. 6, 213 10.1186/gb-2005-6-3-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher J. E., Weiner O. D., Lim W. A. (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 10.1016/j.cell.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N., Ueno T., Pohlmeyer C., Nagano T., Inoue T. (2011). A photocleavable rapamycin conjugate for spatiotemporal control of small GTPase activity. J. Am. Chem. Soc. 133, 12–14 10.1021/ja108258d [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst M. A., Hellingwerf K. J. (2004). Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 37, 13–20 10.1021/ar020219d [DOI] [PubMed] [Google Scholar]
- Velve-Casquillas G., Le Berre M., Piel M., Tran P. T. (2010). Microfluidic tools for cell biological research. Nano Today 5, 28–47 10.1016/j.nantod.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela M., Halidi N., Besson S., Elliott H., Hahn K., Tytell J., Danuser G. (2013). Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. Methods Enzymol. 519, 253–276 10.1016/B978-0-12-405539-1.00009-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y. I., Hahn K. M., Montell D. J. (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591–597 10.1038/ncb2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welf E. S., Haugh J. M. (2011). Signaling pathways that control cell migration: models and analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend S., Wagner H. J., Müller K., Zurbriggen M. D., Weber W., Radziwill G. (2014). Optogenetic control of protein kinase activity in mammalian cells. ACS Synth Biol. 3, 280–285 10.1021/sb400090s [DOI] [PubMed] [Google Scholar]
- Wu Y. I., Frey D., Lungu O. I., Jaehrig A., Schlichting I., Kuhlman B., Hahn K. M. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 10.1038/nature08241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Hyun Y. M., Lim K., Lee H., Cummings R. J., Gerber S. A., Bae S., Cho T. Y., Lord E. M., Kim M. (2014). Optogenetic control of chemokine receptor signal and T-cell migration. Proc. Natl. Acad. Sci. USA 111, 6371–6376 10.1073/pnas.1319296111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Jost A. P., Weiner O. D., Tang C. (2013). A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol. Biol. Cell 24, 2419–2430 10.1091/mbc.E13-03-0126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J. J., Wang H., Vilela M., Danuser G., Hahn K. M. (2014). Manipulation of endogenous kinase activity in living cells using photoswitchable inhibitory peptides. ACS Synth. Biol [Epub ahead of print] doi: 10.1021/sb5001356 10.1021/sb5001356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., Wu Y. I. (2013). Guiding lights: recent developments in optogenetic control of biochemical signals. Pflugers Arch. 465, 397–408 10.1007/s00424-013-1244-x [DOI] [PubMed] [Google Scholar]
- Yokoyama S. (2000). Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 19, 385–419 10.1016/S1350-9462(00)00002-1 [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Yang H., Starmer W. T. (2008). Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics 179, 2037–2043 10.1534/genetics.108.090449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Deng Q., Cavnar P. J., Wu Y. I., Hahn K. M., Huttenlocher A. (2010). Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev. Cell 18, 226–236 10.1016/j.devcel.2009.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehorai E., Yao Z., Plotnikov A., Seger R. (2010). The subcellular localization of MEK and ERK – a novel nuclear translocation signal (NTS) paves a way to the nucleus. Mol. Cell. Endocrinol. 314, 213–220 10.1016/j.mce.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Zhang F., Vierock J., Yizhar O., Fenno L. E., Tsunoda S., Kianianmomeni A., Prigge M., Berndt A., Cushman J., Polle J. et al. (2011). The microbial opsin family of optogenetic tools. Cell 147, 1446–1457 10.1016/j.cell.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E. R., Wu L. F., Altschuler S. J. (2013). Envisioning migration: mathematics in both experimental analysis and modeling of cell behavior. Curr. Opin. Cell Biol. 25, 538–542 10.1016/j.ceb.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. X., Chung H. K., Lam A. J., Lin M. Z. (2012). Optical control of protein activity by fluorescent protein domains. Science 338, 810–814 10.1126/science.1226854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski B. D., Vaccaro B., Crane B. R. (2009). Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 5, 827–834 10.1038/nchembio.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.