Abstract
Microbial pathogens are known to express molecules that interact with host proteins, leading to invasion and colonization. For example, some pathogenic microorganisms express proteins that bind to and enhance the activity of plasminogen. In this way, pathogens utilize the host fibrinolytic system to promote invasion. We found that triosephosphate isomerase (TPI), a glycolytic enzyme produced by Staphylococcus aureus, bound to mannooligosaccharides from the pathogenic capsulated fungus Cryptococcus neoformans and human plasminogen, suggesting that TPI is a moonlighting protein. Several C. neoformans surface proteins are thought to be plasminogen-binding proteins. Here, we examined the ability of surface polymers (including polysaccharides) to bind plasminogen. Heat-killed C. neoformans cells transformed plasminogen into plasmin in a dose-dependent manner in the presence of tissue plasminogen activator. Soluble polysaccharides were found to bind plasminogen based on surface plasmon resonance (SPR) analysis. Neutral polysaccharides fractionated using DEAE column chromatography bound and activated plasminogen. However, the fraction containing glucuronoxylomannan (the primary component of the capsule) did not activate plasminogen. In addition, binding between glucuronoxylomannan and plasminogen was weak. Components of the neutral polysaccharides were identified as mannose, galactose, glucose and xylose. In conclusion, neutral polysaccharides that may affect fibrinolysis were detected on the surface of C. neoformans.
Keywords: Cryptococcus neoformans, plasminogen, capsule, glucuronoxylomannan
Introduction
Microbial pathogens are known to express molecules that interact with host proteins, leading to invasion and colonization. For example, some pathogenic microorganisms express proteins that bind to and enhance the activity of plasminogen (Bhattacharya et al., 2012; Sanderson-Smith et al., 2012; Fulde et al., 2013; Godier & Hunt, 2013; Magalhaes et al., 2013). Thus, pathogens utilize the host fibrinolytic system to promote invasion. We found that triosephosphate isomerase (TPI), a glycolytic enzyme produced by Staphylococcus aureus, interacted with mannooligosaccharides from the pathogenic capsulated fungus Cryptococcus neoformans (Ikeda et al., 2007; Furuya & Ikeda, 2009) and human plasminogen (Furuya & Ikeda, 2011), suggesting that TPI is a moonlighting protein (Henderson & Martin, 2011). Several C. neoformans surface proteins are thought to be plasminogen-binding proteins (Stie et al., 2009). Furthermore, plasmin may play a role during blood–brain barrier (BBB) invasion (Stie & Fox, 2012). Here, we examined the ability of surface polymers (including polysaccharides) to bind plasminogen.
Materials and methods
Preparation of the polysaccharide fraction
Cryptococcus neoformans B-3501 (serotype D) was cultured at 37 °C for 5 days in yeast nitrogen base broth with 2% glucose, 1% casamino acids, and 0.01% streptomycin. Soluble polysaccharides were obtained from the supernatant by ethanol precipitation followed by protease treatment, as described previously (Ikeda et al., 1991; Ikeda & Maeda, 2004). Polysaccharides were dissolved in water and fractionated using anion-exchange column chromatography (IEC DEAE-2025, 20φ × 150 mm). Bound polysaccharides were washed with 0.01 M sodium phosphate buffer (PB), pH 7.4, and eluted with a linear gradient of 0–0.4 M NaCl in PB. Total carbohydrate contents of the effluent were measured using the phenol-H2SO4 method.
Plasminogen activation by tissue plasminogen activator (t-PA)
To examine the effects of polysaccharides on plasminogen activation, plasminogen was activated to plasmin using t-PA. Reagents were diluted in 50 mM Tris-HCl buffer, pH 7.4. First, 40 μL of 500 nM plasminogen and 10 μL of polysaccharide at various concentrations were incubated for 30 min at 37 °C on a microplate. Next, 10 μL of t-PA (recombinant human; Technoclone GmbH; 10 μg mL−1) was added and incubated for 10 min at 37 °C, followed by addition of the chromogenic substrate S-2251 (40 μL of 0.5 mM solution; Chromogenix, Chapel Hill, NC). The absorbance at 405 nm (which depended on the release of p-nitroaniline from the substrate by plasmin activity) was monitored for 3 h at 10-min intervals.
Effect of intact C. neoformans cells on plasminogen activation
To examine the effects of intact C. neoformans on plasminogen activation, cells were added to the above plasminogen activation assay systems. Cell suspensions (10 μL) at various concentrations (108, 107, 106, and 105 cells mL−1) were incubated with 40 μL of 500 nM plasminogen for 30 min at 37 °C. After incubation with t-PA for 10 min at 37 °C, substrate S-2251 was added and the absorbance at 405 nm was recorded.
Interaction between polysaccharide fractions and plasminogen
To examine the interaction between polysaccharide fractions and plasminogen, SPR analysis was performed using a Biacore 3000 (GE Healthcare, Milwaukee, WI).
Plasminogen (human; Enzyme Research Laboratories, South Bend, IN) was diluted with 10 mM sodium acetate buffer (pH 5.0) to a concentration of c. 20 μg mL−1 and immobilized on a standard sensor chip (CM 5) using an amine-coupling kit according to the manufacturer's instructions. Running buffer containing 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, and 0.005% surfactant P20 was used. The flow rate was maintained at 10 μL min−1 for immobilization and 20 μL min−1 for analysis.
Results
Plasminogen activation by C. neoformans cells
To determine whether molecules on the surface of C. neoformans interacted with plasminogen, C. neoformans cells were added to the plasminogen activation system. Heat-killed cells were used to examine carbohydrate heat stability. As shown in Fig.1, the cells facilitated plasminogen activation to plasmin in the presence of t-PA in a dose-dependent manner. When cells were used at a final concentration of 107 mL−1, the reactions were significantly accelerated. At cell concentrations of 104 mL−1, reactions proceeded similarly to the control without C. neoformans. These results suggested that heat-resistant molecules on C. neoformans play a role in plasminogen activation.
Fig 1.
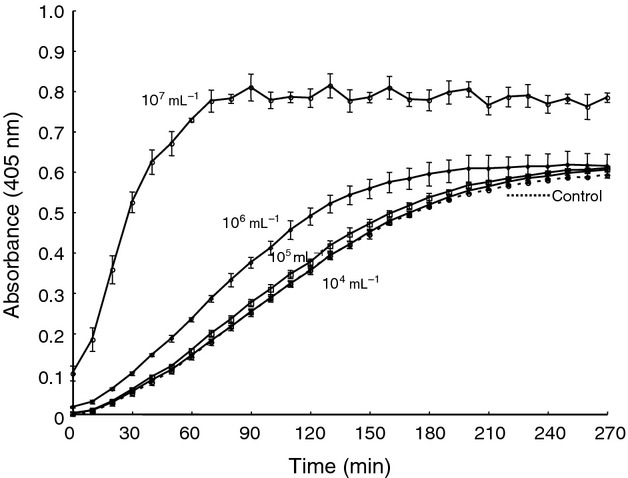
Effect of heat-killed Cryptococcus neoformans cells on plasminogen activation by t-PA. Plasminogen was pre-incubated with C. neoformans cells for 30 min at 37 °C, after which t-PA was added. After incubation for 10 min, substrate S-2251 was added. The absorbance at 405 nm was monitored for 4.5 h at 10-min intervals.
Interaction between cryptococcal soluble polysaccharides and plasminogen
To explore the role of polysaccharides located on the cell surface in plasminogen activation, soluble polysaccharides were examined using SPR. For these experiments, plasminogen was immobilized on the sensor tip as the ligand. Soluble polysaccharides at concentrations of 125, 250, 500, 1000, and 2000 μg mL−1 responded to plasminogen in a dose-dependent manner (Fig.2). SPR sensorgrams showed rapid binding and dissociation between cryptococcal molecules and plasminogen. Based on these results, the molecules that activate plasminogen are likely carbohydrates, and not proteins.
Fig 2.

Interaction between soluble polysaccharides from Cryptococcus neoformans and plasminogen based on SPR. Human plasminogen was used as the ligand, and analytes were cryptococcal polysaccharides (125–2000 μg mL−1).
Interaction between glucuronoxylomannan and plasminogen
Capsule is a representative surface structure of C. neoformans, and the primary component of the capsule is glucuronoxylomannan. Therefore, the glucuronoxylomannan fraction isolated from soluble molecules was applied to SPR. However, the binding of glucuronoxylomannan to plasminogen decreased compared with crude polysaccharides (Fig.3). To assess whether glucuronoxylomannan affected plasmin activation, glucuronoxylomannan was added to the plasminogen activation system. As shown in Fig.4, no significant enhancement of plasminogen activation by glucuronoxylomannan (250, 500 and 1000 μg mL−1) was observed (P > 0.05).
Fig 3.
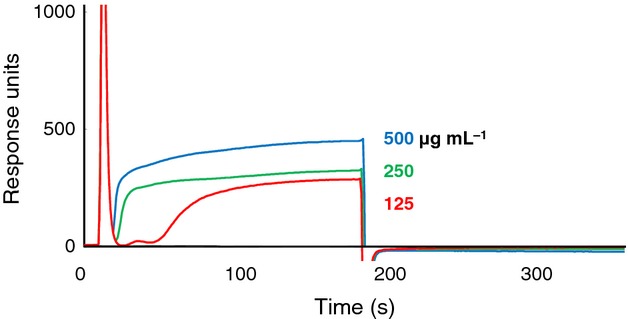
Binding between glucuronoxylomannan and plasminogen based on SPR. Human plasminogen was used as the ligand, and the analyte was the glucuronoxylomannan fraction (125–500 μg mL−1).
Fig 4.
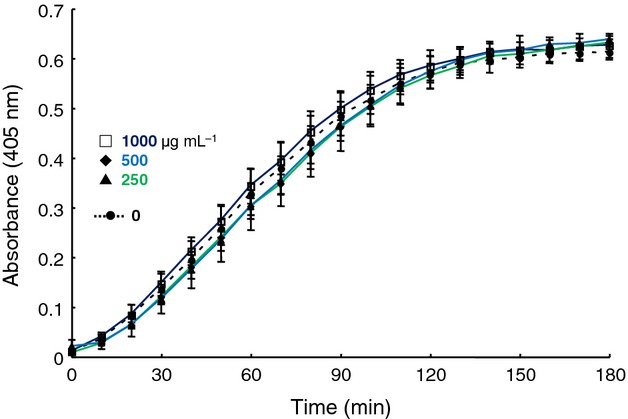
Effect of glucuronoxylomannan (250–1000 μg mL−1) on plasminogen activation in the presence of t-PA. Plasminogen was pre-incubated with glucuronoxylomannan. Closed circle; 0, closed triangle; 250, closed diamond; 500, open square; 1000 μg mL−1 for 30 min at 37 °C, after which t-PA was added. After incubation for 10 min, substrate S-2251 was added. The absorbance at 405 nm was monitored for 3 h at 10-min intervals.
Effects of the neutral fraction of polysaccharides on the interaction and activation of plasminogen
We next examined neutral polysaccharides fractionated using DEAE column chromatography. SPR showed binding between the neutral fraction and plasminogen in a dose-dependent manner (Fig.5a). Furthermore, the neutral components activated plasminogen to plasmin in the presence of t-PA (Fig.6).
Fig 5.
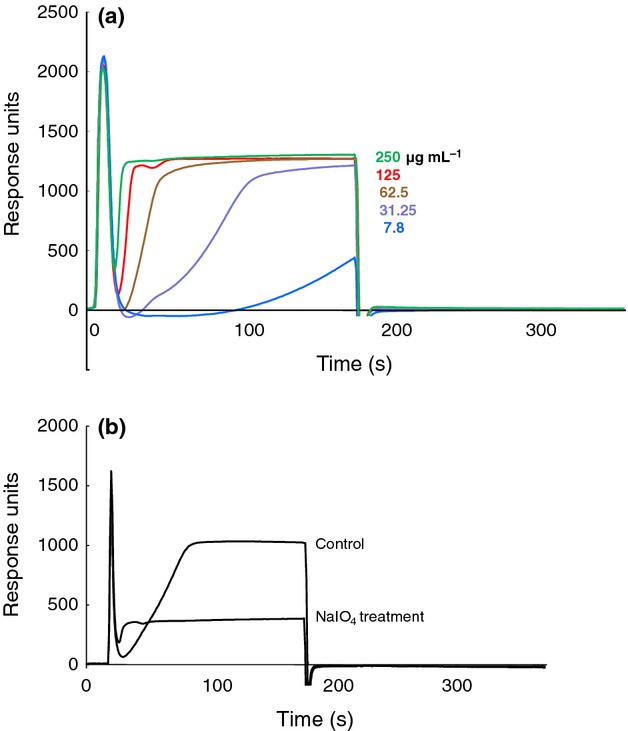
Sensorgrams from SPR analysis of polysaccharides. Human plasminogen was used as the ligand. (a) Binding between the polysaccharide fraction eluted from the DEAE column and plasminogen. The analytes were the unbound fraction (7.8–250 μg mL−1). (b) Comparison of sensorgrams between plasminogen and the neutral fraction untreated and treated with NaIO4. Polysaccharide (10 μg mL−1) before and after treatment was used as the analyte.
Fig 6.
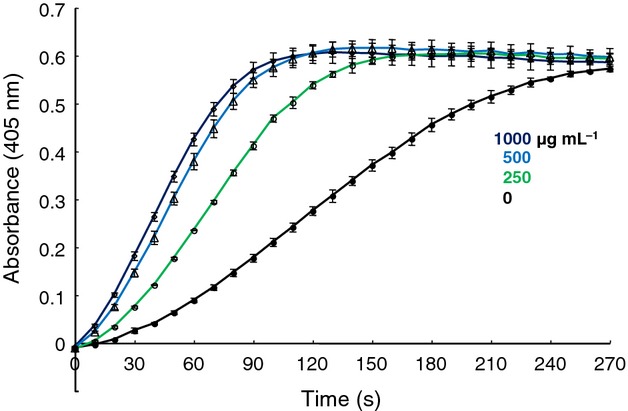
Activation of plasminogen by the unbound fraction (250–1000 μg mL−1) in the presence of t-PA. Plasminogen was pre-incubated with polysaccharide for 30 min at 37 °C, after which t-PA was added. After incubation for 10 min, substrate S-2251 was added. The absorbance at 405 nm was monitored for 4.5 h at 10-min intervals.
Contribution of carbohydrates to plasminogen activation
The neutral polysaccharide fraction eluted with water using DEAE column chromatography was acid hydrolyzed (0.5 M sulfuric acid, 100 °C, 18 h) and neutralized with barium carbonate. The sugar components were identified by HPLC after labeling with 4-aminobenzoic acid ethyl ester as mannose, galactose, glucose, and xylose at ratios of 3.42 : 1.09 : 1.00 : 0.26, respectively. To explore the role of carbohydrates in the interaction with plasminogen, polysaccharides were oxidized by sodium metaperiodate (0.02 M, 4 °C, 120 h) and the interaction with plasminogen was compared with native polysaccharides using SPR. As shown in Fig.5b, the interaction was sensitive to periodate oxidation, suggesting that carbohydrates play a role in the interaction with plasminogen. To determine whether the fraction contained proteins, SDS-PAGE analysis of the polysaccharide fraction followed by silver staining was also performed. However, no protein band was detected (data not shown). Furthermore, proteins extracted from the cell surface using 3 M LiCl hardly reacted with plasminogen according to dot immunobinding assays and SPR (data not shown).
Discussion
Microbial pathogens utilize host-derived molecules to invade target organs. Several bacteria bind extracellular matrix proteins including collagen, fibronectin, fibrin, and laminin (Singh et al., 2012). Coagulation and fibrinolytic systems are recognized by microbial proteins, such as enolase and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, which are glycolytic enzymes) in Staphylococcus and Streptococcus, M protein of Streptococcus, plasminogen-binding proteins (PgbA/PgbB) in Helicobacter pylori, and flagella in Escherichia coli (Bhattacharya et al., 2012). In our previous study, TPI, a glycolytic enzyme, was shown to interact plasminogen (Furuya & Ikeda, 2011). TPI also interacted with α-1, 3-linked mannooligosaccharide from C. neoformans and contributed to the adherence of C. neoformans and S. aureus cells (Ikeda et al., 2007; Furuya & Ikeda, 2009). Based on these results, TPI was suggested to be a moonlighting protein.
Cryptococcus neoformans is an encapsulated yeast that causes life-threatening meningitis. The polysaccharide capsule is mainly composed of glucuronoxylomannan (Bhattacharjee et al., 1992) and is considered a pathogenic factor. However, specific virulence factors (such as exotoxins produced by several bacteria) have not been identified. During C. neoformans infection, molecules derived from C. neoformans can ‘hijack’ host factors during dissemination and interact with plasminogen on the cell surface to accelerate invasion. Several proteins from C. neoformans have been identified as candidate plasminogen-binding proteins (Stie et al., 2009) and could play a role in crossing the BBB (Stie & Fox, 2012). Proteomic research showed molecular and cellular changes induced by C. neoformans in brain endothelial cells (Vu et al., 2013). The role of the carbohydrate moiety in recognition and interaction between molecules has been reported previously (Liu & Pedersen, 2007; Springer & Gagneux, 2013). However, the association between carbohydrate and plasminogen remains unknown. Here, we examined the role of carbohydrates in the interaction with plasminogen and suggested that polysaccharide (rather than glucuronoxylomannan) interacts with and enhances plasminogen activation in the presence of t-PA.
Heat-killed C. neoformans accelerated plasminogen activation in the presence of t-PA, suggesting that molecules (rather than heat-labile proteins) play an important role in this process. Plasminogen activation activity was observed in the fraction containing polysaccharides composed of neutral sugars, including mannose, galactose, glucose, and xylose. Mannose accounted for the majority of the fraction (59.3%). Glucuronoxylomannan, the primary component of the cryptococcal capsule, did not interact with or activate plasminogen. The plasminogen-activating molecules were sensitive to periodate oxidation, which suggested that carbohydrates were involved in this process. However, after treatment with sodium metaperiodate, one-third of the plasminogen-activating activity was maintained. The neutral polysaccharide fraction contained some periodate-resistant moieties that played a role in plasminogen binding. Glucuronoxylomannan contained peroxidation resistant α-1, 3-linked mannan in the backbone. Based on this information, mannose residues may play a role in plasminogen activation. Although the definitive chemical structures of the active substances have not been obtained, molecules that may function as plasminogen receptors on microorganisms could be predicted.
To determine whether our findings were limited to the strain used in the current study, we tested the interactions between plasminogen and fractions from C. neoformans CDC 551 (serotype A) and NIH 52 (serotype D) (Ikeda et al., 1982) obtained by DEAE column chromatography. We confirmed that the unbound fraction responded clearly; however, fractions containing glucuronoxylomannan hardly reacted (data not shown). To confirm that the plasminogen receptor(s) is the virulence factor, further studies will be required using more virulent strains (e.g., H99) and/or clinical isolates.
In conclusion, on the surface of C. neoformans, neutral polysaccharides (excluding glucuronoxylomannan) were detected that affect fibrinolysis. Many molecules on the surfaces of microorganisms, as well as tumor cells (Andreasen et al., 2000; Plow et al., 2012; Godier & Hunt, 2013), have been identified as plasminogen receptors. Thus, microbial pathogens may utilize host plasminogen for colonization and dissemination using protein and carbohydrate components on the cell surface.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 23590155.
References
- Andreasen PA, Egelund R. Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57:25–40. doi: 10.1007/s000180050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee AK, Kwon-Chung KJ. Glaudemans CP. The major capsular polysaccharide of Cryptococcus neoformans serotype B. Carbohydr Res. 1992;233:271–272. doi: 10.1016/s0008-6215(00)90942-x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Ploplis VA. Castellino FJ. Bacterial plasminogen receptors utilize host plasminogen system for effective invasion and dissemination. J Biomed Biotechnol. 2012;2012:482096. doi: 10.1155/2012/482096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulde M, Rohde M, Polok A, Preissner KT, Chhatwal GS. Bergmann S. Cooperative plasminogen recruitment to the surface of Streptococcus canis via M protein and enolase enhances bacterial survival. MBio. 2013;4:00629. doi: 10.1128/mBio.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya H. Ikeda R. Interaction of triosephosphate isomerase from the cell surface of Staphylococcus aureus and alpha-(1→3)-mannooligosaccharides derived from glucuronoxylomannan of Cryptococcus neoformans. Microbiology. 2009;155:2707–2713. doi: 10.1099/mic.0.028068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya H. Ikeda R. Interaction of triosephosphate isomerase from Staphylococcus aureus with plasminogen. Microbiol Immunol. 2011;55:855–862. doi: 10.1111/j.1348-0421.2011.00392.x. [DOI] [PubMed] [Google Scholar]
- Godier A. Hunt BJ. Plasminogen receptors and their role in the pathogenesis of inflammatory, autoimmune and malignant disease. J Thromb Haemost. 2013;11:26–34. doi: 10.1111/jth.12064. [DOI] [PubMed] [Google Scholar]
- Henderson B. Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun. 2011;79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R. Maeda T. Structural studies of the capsular polysaccharide of a non-neoformans Cryptococcus species identified as C. laurentii, which was reclassified as Cryptococcus flavescens, from a patient with AIDS. Carbohydr Res. 2004;339:503–509. doi: 10.1016/j.carres.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Shinoda T, Fukazawa Y. Kaufman L. Antigenic characterization of Cryptococcus neoformans serotypes and its application to serotyping of clinical isolates. J Clin Microbiol. 1982;16:22–29. doi: 10.1128/jcm.16.1.22-29.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Matsuyama H, Nishikawa A, Shinoda T. Fukazawa Y. Comparison of serological and chemical characteristics of capsular polysaccharides of Cryptococcus neoformans var. neoformans serotype A and Cryptococcus albidus var. albidus. Microbiol Immunol. 1991;35:125–138. doi: 10.1111/j.1348-0421.1991.tb01540.x. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Saito F, Matsuo M, Kurokawa K, Sekimizu K, Yamaguchi M. Kawamoto S. Contribution of the mannan backbone of cryptococcal glucuronoxylomannan and a glycolytic enzyme of Staphylococcus aureus to contact-mediated killing of Cryptococcus neoformans. J Bacteriol. 2007;189:4815–4826. doi: 10.1128/JB.00412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Pedersen LC. Anticoagulant heparan sulfate: structural specificity and biosynthesis. Appl Microbiol Biotechnol. 2007;74:263–272. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes V, Andrade EB, Alves J, Ribeiro A, Kim KS, Lima M, Trieu-Cuot P. Ferreira P. Group B streptococcus hijacks the host plasminogen system to promote brain endothelial cell invasion. PLoS One. 2013;8:e63244. doi: 10.1371/journal.pone.0063244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plow EF, Doeuvre L. Das R. So many plasminogen receptors: why? J Biomed Biotechnol. 2012;2012:141806. doi: 10.1155/2012/141806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson-Smith ML, De Oliveira DM, Ranson M. McArthur JD. Bacterial plasminogen receptors: mediators of a multifaceted relationship. J Biomed Biotechnol. 2012;2012:272148. doi: 10.1155/2012/272148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Fleury C, Jalalvand F. Riesbeck K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev. 2012;36:1122–1180. doi: 10.1111/j.1574-6976.2012.00340.x. [DOI] [PubMed] [Google Scholar]
- Springer SA. Gagneux P. Glycan evolution in response to collaboration, conflict, and constraint. J Biol Chem. 2013;288:6904–6911. doi: 10.1074/jbc.R112.424523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stie J. Fox D. Blood-brain barrier invasion by Cryptococcus neoformans is enhanced by functional interactions with plasmin. Microbiology. 2012;158:240–258. doi: 10.1099/mic.0.051524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stie J, Bruni G. Fox D. Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion. PLoS One. 2009;4:e5780. doi: 10.1371/journal.pone.0005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu K, Eigenheer RA, Phinney BS. Gelli A. Cryptococcus neoformans promotes its transmigration into the central nervous system by inducing molecular and cellular changes in brain endothelial cells. Infect Immun. 2013;81:3139–3147. doi: 10.1128/IAI.00554-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


