Abstract
A well-balanced human diet includes a significant intake of non-starch polysaccharides, collectively termed “dietary fibre,” from the cell walls of diverse fruits and vegetables.1 Due to a paucity of alimentary enzymes encoded by the human genome,2 our ability to derive energy from dietary fibre depends on saccharification and fermentation of complex carbohydrates by the massive microbial community residing in our distal gut.3,4 The xyloglucans (XyGs), in particular, are a ubiquitous family of highly branched plant cell wall polysaccharides5,6 whose mechanism(s) of degradation in the human gut and consequent importance in nutrition was heretofore unknown.1,7,8 Here, we demonstrate that a single, complex gene locus in Bacteroides ovatus confers xyloglucan catabolism in this common colonic symbiont. Through targeted gene disruption, biochemical analysis of all predicted glycoside hydrolases and carbohydrate-binding proteins, and three-dimensional structural determination of the vanguard endo-xyloglucanase, we reveal the molecular mechanisms through which XyGs are hydrolysed to component monosaccharides for further metabolism. We also observe that orthologous xyloglucan utilization loci (XyGULs) serve as genetic markers of xyloglucan catabolism in Bacteroidetes, that XyGULs are restricted to a limited number of phylogenetically diverse strains, and that XyGULs are ubiquitous in surveyed human metagenomes. Our findings reveal that the metabolism of even highly abundant components of dietary fibre may be mediated by niche species, which has immediate fundamental and practical implications for gut symbiont population ecology in the context of human diet, nutrition and health.9–12
Despite our omnivory, a census of the glycoside hydrolases (GH) encoded by the human genome indicates that our inherent ability to digest carbohydrates is restricted to starch and simple saccharides, e.g. malto-oligosaccharides, sucrose and lactose.2 Consequently, the human gut microbiota and its cohort of predicted carbohydrate-active enzymes are implicated in the conversion of otherwise indigestible plant polysaccharides to short-chain fatty acids,2,7,13 which provides up to 10% of the daily caloric intake in humans14,15 and is central to colonic heath.4,9,11,16 Despite an increasing body of (meta)genomic sequence data,13,17–20 the enzymatic pathways by which the most common dietary polysaccharides are digested in the human gut have not been elucidated.7,13
XyGs are widespread in the vegetables we consume: Dicot primary cell walls, for example those of lettuce, onions and tomatoes, may contain up to 25% XyG on a dry-weight basis.1,5,6 The primary walls of commelinoid monocots, including the cereals, contain much lower (1–5%) – but still non-zero – amounts of XyGs.1,6 Seed XyGs are also widely used as food thickening agents and have been employed as drug delivery matrices in the intestine.21 This family of polysaccharides is typified by a β(1→4)-glucan main chain that is heavily substituted with pendant α(1→6)-linked xylosyl units. Depending on the species and tissue of origin, these branches may be further extended by additional monosaccharides, including galactose, fucose, and/or arabinose (Figure 1).5,22 As such, complete saccharification in the gut necessarily requires a cadre of enzymes to address the monosaccharide and linkage diversity of these complex polysaccharides.
Figure 1.
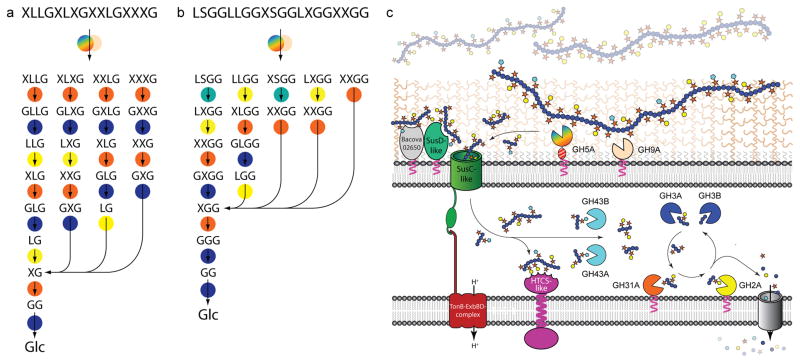
Representative structures of XXXG- and XXGG-type xyloglucans. a. XXXG-type xyloglucans, comprised of a Glc4Xyl3 repeating motif with variable branch extensions (bold residues). Tamarind seed xyloglucan and primary cell wall xyloglucans (e.g. from lettuce leaves) are distinguished by the absence of fucose in the former. b. XXGG-type xyloglucans, comprised of Glc4Xyl2 repeating motif. These xyloglucans are common to solanaceous species (e.g. tomato) and are typified by branches extended with arabinofuranosyl residues. Standard single-letter abbreviations5 for designating backbone decorations are shown.
We recently identified a polysaccharide utilization locus (PUL) in the genome of a common human gut symbiont B. ovatus, but not in the closely related model species B. thetaiotaomicron,17 that was transcriptionally up-regulated in response to growth on galactoxyloglucan.7 By homology with the archetypal starch utilization system (Sus) of B. thetaiotaomicron, this PUL was predicted to encode an outer membrane sugar-binding protein (SusD-like), a TonB-dependent sugar receptor/transporter (SusC-like), and an inner membrane hybrid two-component sensor. Further analysis strikingly revealed that this PUL was also predicted to encode eight glycoside hydrolases (GHs) from six enzyme families (Figure 2), which tantalizingly suggested a collective role in XyG utilization by B. ovatus. To establish a direct causal link for growth on XyG7,8 and outline a pathway for its degradation, we performed an in-depth molecular characterization of the PUL through reverse genetics, in vitro protein biochemistry and enzymology, and structural biology.
Figure 2.
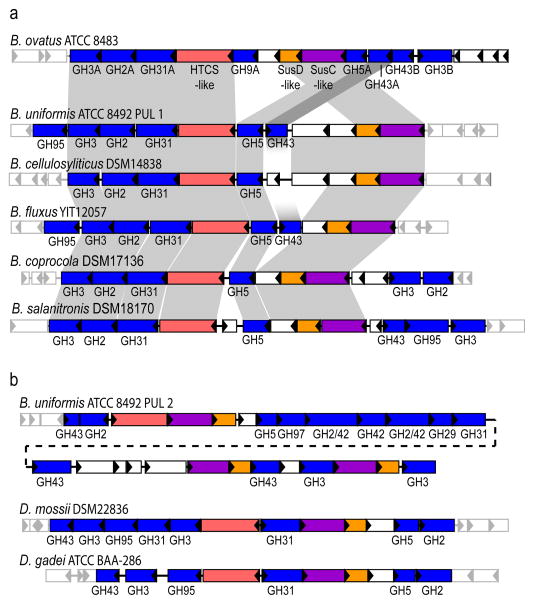
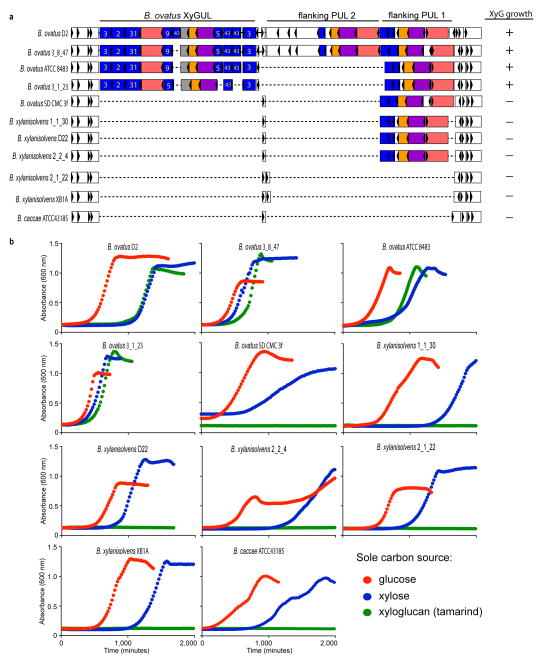
Structure of the B. ovatus xyloglucan utilization locus and evolution in the Bacteroidetes lineage. a. PULs with partial homology and synteny; homologous genes are connected by gray bars and flanking genes lacking synteny are shown as semi-transparent. b. PULs with partial homology, but lacking overall synteny. Extended Data Figure ED2 provides transcriptional evidence that each of these gene clusters is responsive to growth on XyG.
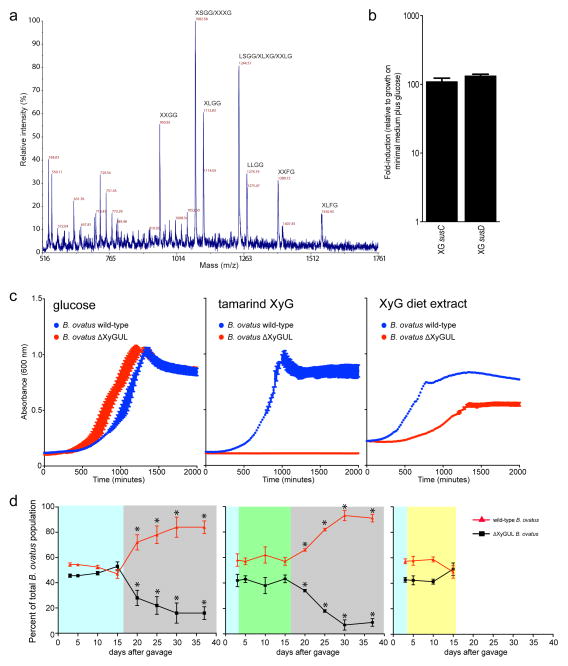
A mutant strain of B. ovatus harbouring a targeted deletion of the complete predicted xyloglucan utilization locus (XyGUL, Figure 2) was indeed completely unable to grow on tamarind XyG as the sole carbon source, but was otherwise phenotypically identical to the wild-type strain (data not shown). This indicated an absolute requirement for one or more of the corresponding gene products in XyG catabolism. Subsequently, all eight predicted GHs were produced recombinantly in E. coli and subjected to detailed enzymatic characterization to establish their substrate specificities and reaction products (Extended Data Table ED1, Supplementary Figures S1–20). All enzymes were maximally active in the pH range 6.0–7.0, which is consistent with function in the distal human gut (Supplementary Figures S1–6).
Reducing-sugar assays and mass spectrometry (MS) demonstrated that the recombinant BoGH5A and BoGH9A enzymes were versatile endo-xyloglucanases, which cleaved the backbone of the three major types of natural xyloglucans, i.e. seed galactoxyloglucan from tamarind kernel, dicot fucogalactoxyloglucan from lettuce leaves, and solanaceous arabinogalactoxyloglucan from tomato fruit, to produce xyloglucan oligosaccharides (XyGOs) based on a Glc4 backbone (Extended Data Table ED1 and Supplementary Figures S7–10, cf. Figure 1). Assays on chromogenic aryl β-glycosides of XyGOs and natural XyGOs, together with product analyses, revealed the following exo-specificities for the remaining XyGUL enzymes: BoGH2A, β-galactosidase; BoGH3A and BoGH3B, β-glucosidase; BoGH31A α-xylosidase; BoGH43A and BoGH43B, α-L-arabinofuranosidase (Extended Data Table ED1, Supplementary Data Figures S11–20). This profile of activities, together with phenotypic data from additional gene-specific knock-out strains (Extended Data Table ED2), allowed us to outline a complete model of XyG degradation by B. ovatus (Figure 3).
Figure 3.
The concerted action of XyGUL gene products in the degradation of xyloglucans. Most probable sequential pathways for the hydrolysis of (galacto)xyloglucan (a) and (arabinogalacto)xyloglucans (b) based on enzyme kinetic data, product analysis, and selected gene knock-out studies (see Fig. 1 for XyG motif abbreviations). Enzymes are represented as circles, colour-coded as in panel c: Rainbow, endo-xyloglucanase BoGH5A; tan, endo-xyloglucanase BoGH9A; orange, α-xylosidase BoGH31A; turquoise α-L-arabinofuranosides BoGH43A and/or BoGH43B; yellow β-galactosidase BoGH2; dark blue β-glucosidases BoGH3A and/or BoGH3B. c. Model of enzyme localisation by analogy with the archetypal Sus locus4 and based on inference of N-terminal lipoprotein modification from protein sequence data.
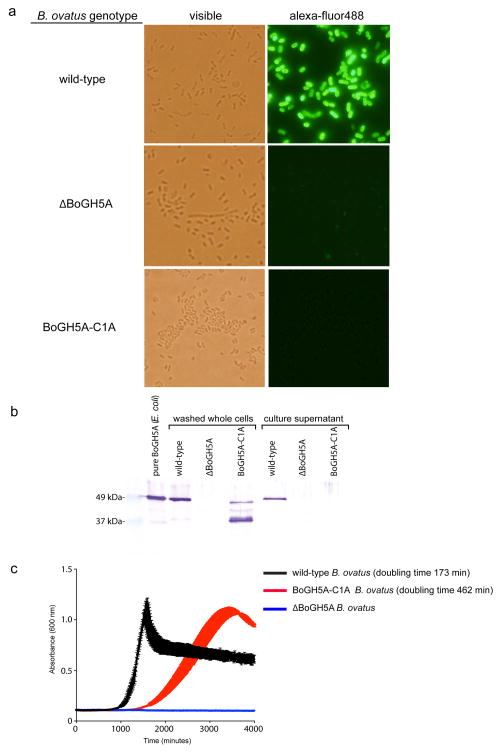
Analogous to the endo-amylase SusG of the B. thetaiotaomicron starch utilization system,4 the vanguard role in XyG utilization by B. ovatus is performed by the versatile endo-xyloglucanase BoGH5A, which generates short XyGOs for uptake (Figure 3). Indeed, a gene-specific knock-out of BoGH5A rendered B. ovatus incapable of growth on XyG polysaccharide, but this phenotype could be directly rescued by the addition of XyGOs produced exogenously by recombinant BoGH5A (Extended Data Table ED2, cf. Supplementary Figure S8). In contrast, a gene-specific knock-out of BoGH9A had no effect on growth (Extended Data Table ED2), consistent with the observation that although this enzyme produced the same limit digestion products from all XyGs as BoGH5, BoGH9A was catalytically feeble with kinetics too slow to be quantified (Extended Data Table ED1, Supplementary Figure S8–10). That a corresponding GH9 member is not found in orthologous XyGULs from other species, nor in a B. ovatus strain possessing a simpler XyGUL, which were all capable of growth on tamarind XyG (Figure 2, Extended Data Figures ED1 & ED2), suggests BoGH9A may be largely superfluous, or plays a specialised role not revealed in our analysis.
A gene-specific knock-out of the α-xylosidase BoGH31A severely reduced B. ovatus growth on tamarind XyG and completely abolished growth on XyGOs (Extended Data Table ED2). This is consistent with an essential role in removal of α(1→6)-xylosyl residues from the non-reducing-end of XyGOs (converting “X” units to “G”), to allow subsequent hydrolysis by the β-glucosidases BoGH3A and BoGH3B (removing backbone “G” units), and the β-galactosidase BoGH2A (or another β-galactosidase converting “L”→”X”, Figure 3). The two α-L-arabinofuranosidases, BoGH43A and BoGH43B, encoded by the XyGUL provide additional capacity to convert “S”→”X”, thereby enabling complete hydrolysis of solanaceous (e.g., tomato, eggplant, pepper, etc.) arabinogalactoxyloglucan. It is presently unclear if the doubling of GH3- and GH43-encoding genes in the XyGUL has any biological importance; the similar specific activities of BoGH3A and BoGH3B toward XyGO β-glucosides suggests functional redundancy, while the apparent catalytic feebleness of BoGH43B toward arabinofuranosides (Extended Data Table ED1) may suggest simple loss-of-function or evolution of a currently unresolved, orthogonal activity contrasting that of BoGH43A. Notably, the B. ovatus XyGUL does not encode an α-fucosidase, as might be anticipated for the cleavage of the “F” sidechain in dicot fucogalactoxyloglucan (Figure 1). This may reflect compensation by exogenous or endogenous α(1→2)-fucosidases2 or strain specialisation for XyGs from individual plant sources. Indeed, XyGULs from other Bacteroides species encode predicted α-fucosidases from families GH29 and GH95 (Figure 2).
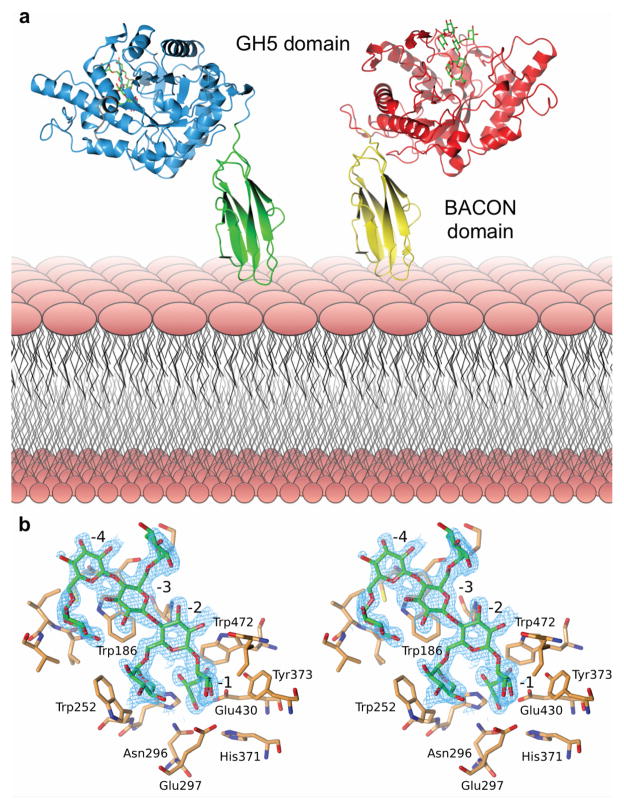
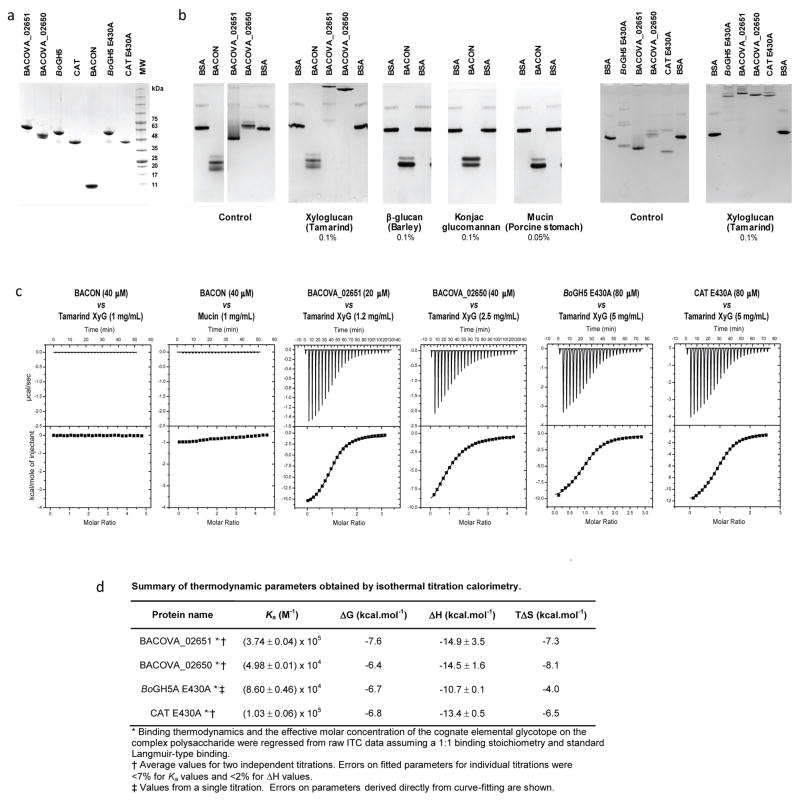
To provide further insight into XyG recognition by the keystone enzyme, we solved the three-dimensional structure of BoGH5A in complex with the heptasaccharide XXXG by X-ray crystallography (Figure 4, Extended Data Table ED3). The tertiary structure comprised a 96 residue “Bacteroidetes-Associated Carbohydrate-binding Often N-terminal (BACON)” domain23 followed by a 372 residue C-terminal GH5 domain. The BACON domain composed an 8-stranded, immunoglobulin-like β-sandwich fold and represents, to our knowledge, the first 3-D representative of this domain family. Notably, the two molecules in the asymmetric unit displayed a large difference in the relative orientations of the BACON and GH5 domains (Figure 4A). This suggested significant flexibility in the enzyme (Supplementary Video V1), which is anchored on the cell surface via N-terminal lipidation; mutation of the predicted lipidation site (Cys1->Ala) ablates surface localization and hampers growth on XyG (Extended Data Figure ED3). Notably, we were unable to find experimental evidence in vitro that the BACON domain functions in substrate binding (Extended Data Figure ED4), nor that it mediates interactions with other proteins of the XyGUL (native PAGE data not shown). In contrast, the observation of strong XyG binding (Extended Data Figure ED4) by the SusD-like protein and neighboring gene product (Figure 2, loci Bacova_02651 and Bacova_02650, respectively) indicates that, as in the archetypal Sus system,24 polysaccharide binding is mediated by independently encoded, non-catalytic proteins of the XyGUL. Thus, the broad designation of BACON domains as “carbohydrate-binding”, as inferred by bioinformatics alone, may be actively misleading.23,25 In light of current experimental data, the most parsimonious conclusion is that the primary function of the BACON domain in BoGH5A may be to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide.
Figure 4.
Structural biology of BoGH5A. a. Tertiary structure; the two conformations observed in crystallo have been oriented relative to the N-terminal, membrane-anchored BACON domain (see also Supplementary Video V1). b. Wall-eyed stereo view of the binding of XXXG in the -4 to -1 subsites (see also Supplementary Video V2). The wireframe represents an unbiased 2Fo-Fc map (contoured at 0.3 electrons per Å3) obtained using phases calculated from the best model prior to the incorporation of any ligand in refinement.
The catalytic domain has the prototypical (α/β)8 fold typical of other GH5 members and is most similar to Paenibacillus pabuli endo-xyloglucanase PpXG5 (PDB ID 2jep; r.m.s.d. 1.47 Å, 330 Cα atoms) and Clostridium cellulovorans endo-glucanase EngD (PDB ID 3icg; r.m.s.d. 1.45 Å, 345 Cα atoms) of subfamily 4 (ref. 26). The heptasaccharide XXXG was observed to extend from subsites -4 to -1 in the active site cleft in both molecules of the asymmetric unit, with binding mediated via sugar-aromatic ring interactions and hydrogen bonds to both backbone and branching substrate moieties (Figure 4B, Supplementary Video V2). The structure of this complex is well-correlated with activity on chromogenic XyGO aryl β-glycoside substrates, which indicated that the enzyme productively harnesses the Xylα(1→6) sidechains for catalysis (Extended Data Table ED1). Detailed comparison with the PpXG5:GXLG structure (PDB ID 2jeq)27 revealed that binding of the β(1→4)-glucan backbone is essentially identical in the two enzymes, while differences emerge in the subsites that accommodate the branching sugar residues (Supplementary Video V2). Regardless, it is clear that a broad active site cleft engendering binding plasticity is the key feature allowing BoGH5A to accommodate a wide range of natural xyloglucans.
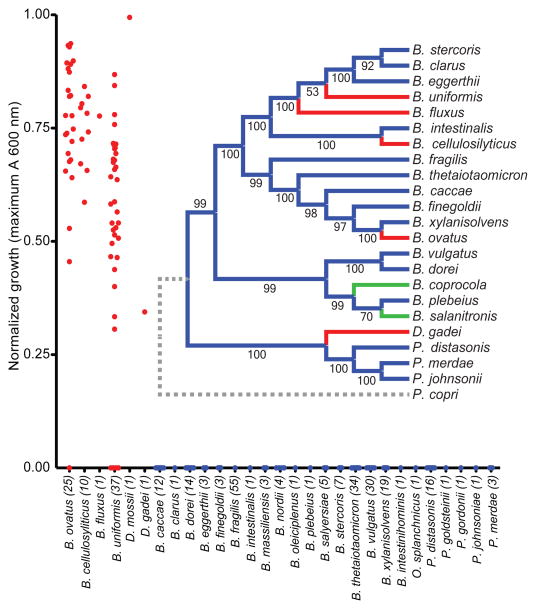
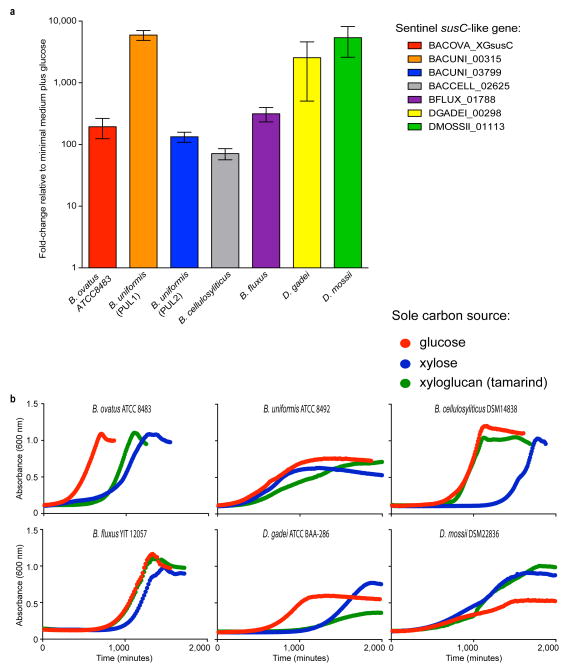
With a solid molecular understanding of the locus that confers B. ovatus with the ability to utilize XyG, we undertook a growth analysis of 292 individual Bacteroidetes strains isolated from the human gut, representing 29 different named species, to reveal the distribution of this trait in nature. XyG catabolism was rare, confined to 70 strains belonging to just 6 individual, phylogenetically dispersed species (Figure 5). Of the 25 B. ovatus strains tested, all but one grew on tamarind XyG. Interestingly, none of the 18 strains of the closely related B. xylanisolvens exhibited growth on XyG, which suggested that the XyGUL has been independently acquired by the B. ovatus lineage. This is supported by a comparative genomic analysis of the XyGUL and surrounding chromosomal region in these two species (Extended Data Figure ED1). All ten strains tested of the more distantly related B. cellulosyliticus grew very well on XyG, comparable to the less proficient B. ovatus strains. Representing a third phylogenetic clade, the single B. fluxus strain also grew to high density on XyG. Of 37 strains of B. uniformis, which is closely related to B. fluxus and among the most abundant organisms in the microbiota of Westerners,18 33 exhibited varying capacity to grow on XyG. Finally, two non-Bacteroides species, Dysgonomonas gadei and D. mossii, which are rarely observed in humans, but are abundant in termites, also possessed the ability to degrade xyloglucan, revealing that this phenotype has evolved in other lineages of Bacteroidetes.
Figure 5.
Growth of different human gut-resident Bacteroidetes on xyloglucan. Each point represents the normalized maximum growth OD600 achieved by each strain after 4 d of growth in minimal medium with tamarind XG as the sole carbon source (average of n=2 independent growth analyses per strain). The inset shows a phylogenetic tree constructed from fully sequenced strains of the species shown; those labeled in red have the ability to grow on xyloglucan as sole carbon source and those labeled in green have PULs similar to B. ovatus XyGUL but were not tested for growth on XG. Bootstrap values based on 1000 replicate trees are indicated.
A comparative analysis using genomic sequence data available for many of the strains tested showed a perfect concordance of the presence of an orthologous XyGUL and the ability to utilize XyG. Thus, we revealed putative XyGULs in B. cellulosyliticus, B. uniformis, B. fluxus, D. mossii and D. gadei, as well as in two sequenced species, B. coprocola and B. salanitronis, that were not available to directly test growth (Figures 2 & 5). These XyGULs were all activated during growth on XyG (Extended Data Figure ED2) and shared a similar overall organisation with that of B. ovatus, but did not share similar flanking regions in their genomes, suggesting that they were acquired via separate events. In notable contrast to a recent report showing that conjugative transposons are capable of mediating horizontal transfer of new PULs into Bacteroides genomes,28 there is no evidence of such a mechanism for XyGUL acquisition. Rather, the XyGUL appears to have been precisely inserted between two ancestral genes that are shared by all sequenced strains. For example, on the left side of XyGUL (Figure 2, Extended Data Figure ED1), there are only 108 bp between the 3′ end of BoGH3A (Bacova_02644) and the end of the adjacent shared region; on the right side there are only 640 bp between the end of BoGH3B (Bacova_02659) and the second adjacent shared region.
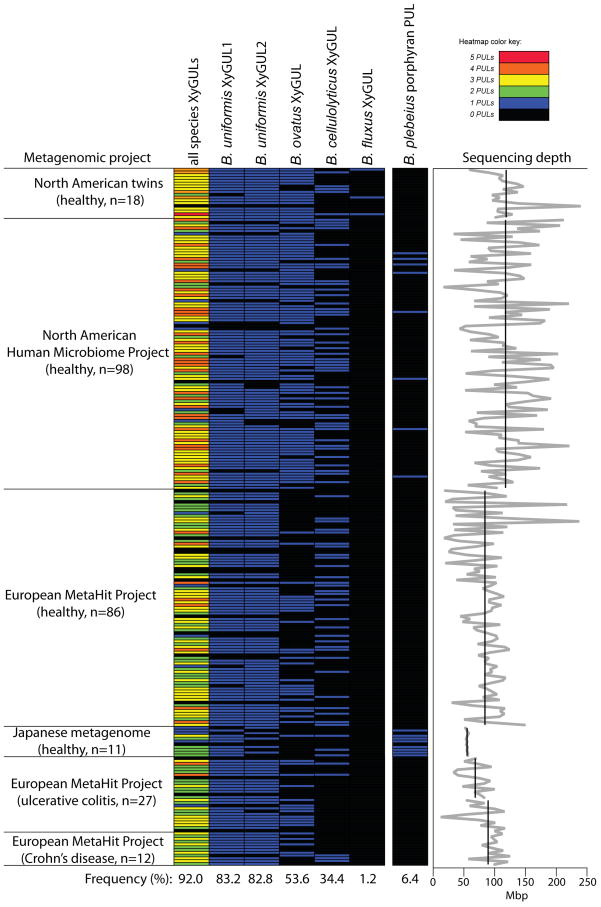
To underscore the broad importance of dietary xyloglucan metabolism, we surveyed public metagenomic data from 250 adult humans revealing that 92% harbor at least one of five different Bacteroides XyGULs identified in this study (Extended Data Figure ED5). In contrast, the presence of a PUL involved in degrading the red algal polysaccharide porphyran (a component of the food product nori)29 was exceptionally rare in the same population, and largely confined to a small cohort of Japanese subjects. To demonstrate that a metabolic advantage is associated with possessing a XyGUL, a competition experiment was performed using germfree mice co-colonized with wild-type B. ovatus and an isogenic XyGUL deletion mutant. Both strains competed equally when glucose was presented as the sole dietary carbohydrate or when XyG was presented in a mixture of polysaccharides from natural vegetable sources (a diet comprising equal amounts of cooked bell pepper, eggplant, tomato fruit and lettuce). However, when this complex diet was removed and XyG was maintained as the only exogenous polysaccharide via the drinking water, the B. ovatus XyGUL mutant exhibited a significant fitness defect over a three-week period (Extended Data Figure ED6).
The prevalence of XyGs in the human diet suggests that the mechanism by which bacteria degrade these complex polysaccharides is highly important to human energy acquisition. Moreover, the rarity of XyG metabolism (Figure 5, see also ref. 8) highlights the significance of B. ovatus and other proficient XyG-degrading Bacteroidetes as key members of the human gut microbial consortium.10 Additional work will now be required to determine if bacteria from other groups have evolved strategies to attack this ubiquitous plant cell wall polysaccharide in competition or synergy with members of the Bacteroidetes.8,30 An unexpected finding is the specific adaptation of the glycoside hydrolase complement of some XyGULs to XyGs from different plant sources; this theme that may extend to other groups of plant polysaccharides and host mucosal glycans with similar or greater degrees of fine-structural variation. Following the example presented here, defining cohorts of enzymes and binding proteins that coordinate to target other complex dietary polysaccharides will be essential to fully elucidate the systems biology of gut microbial catabolism. Such refined mechanistic understanding will be essential in designing rational intervention strategies, including prebiotics, probiotics or microbial community transplants, which aim to manipulate the membership, function and stability of this important ecosystem.4,9–12
Methods
Growth analysis of Bacteroides species & reverse genetics
The locus tags of the genes involved in the present study and the corresponding DNA primers used for knock-out, sequence alteration and qPCR studies are given in Supplementary Table S1. To construct gene deletions in B. ovatus strain ATCC 8483, a deletion of the gene encoding thymidine kinase (tdk; Bacova_03071) was first constructed using an identical strategy to that used previously for B. thetaiotaomicron.31 All subsequent gene deletions and sequence modifications were conducted in a Δtdk strain background by allelic exchange using the vector pExchange-tdk31 and primers listed in Supplementary Table S1. Wild-type Bacteroides ovatus ATCC 8483, all mutant derivatives and other Bacteroidetes species tested were grown in tryptone-yeast extract-glucose (TYG) medium, brain-heart infusion agar supplemented with 10% horse blood, or minimal media (MM) supplemented with appropriate carbohydrates as previously described.32 Antibiotics were added as needed: gentamicin (200 μg ml−1), erythromycin (25 μg ml−1), and 5-fluoro-2′-deoxyuridine (200 μg ml−1). To prepare cells for exposure to glycans, wild-type and mutant B. ovatus were grown in TYG, subcultured into MM-glucose, grown to mid-exponential phase (OD600 0.6–0.8), then washed and resuspended in 2x MM prior to addition of the appropriate glycan. Xyloglucan substrates are described below and all carbohydrate stocks were prepared at 10 mg/ml in ddH2O and sterilized by autoclaving. All quantitative growth was performed at 37°C in an anaerobic chamber (Coy Manufacturing, Grass Lake, MI; 10 % H2, 5 % CO2 and 85 % N2) in an automated plate reading device as described previously.33 In instances where bacterial growth data are either used to show growth curves or quantify differences in growth ability, strains were grown in 2–3 biological replicates. Fluorescence microscopy was performed on fixed B. ovatus cells grown to early exponential phase (A600 = 0.25–0.35) in minimal medium containing a 9:1 mixture of limit XyG oligosaccharides (XGOs) to slightly longer (average dp = 14) XGOs (see Substrates section below). These conditions circumvented the reduced growth rate of the ΔGH5 mutant on limit XGOs. Cells were fixed in formalin and stained with a polyclonal antibody raised in rabbit against purified recombinant BoGH5A (Cocalico Biologicals, Reamstown, PA), following the method previously reported.34 The same antiserum was used to probe GH5 presence via Western blot.
Construction of a Bacteroidetes phylogeny
Six Bacteroidetes core genes that encode anthranilate synthase, glucose-6-phosphate isomerase, glycerol-3-phosphate dehydrogenase, DNA-directed RNA polymerase subunit beta, chorismate synthase, 3-dehydroquinate synthase and 16S rDNA were used to construct a phylogenetic tree. For each species, nucleotide sequences were concatentated in the same order, aligned using CLUSTAL-W,35 un- or poorly aligned basepairs were trimmed out using Gblocks,36 and the resulting alignment was used to construct a maximum parsimony tree with bootstraps using MEGA5.37
Analysis of metagenomic datasets for presence of XyGULs
Human metagenomic datasets38–41 were searched by BLAST for the presence of XyGUL nucleotide sequences from B. ovatus (27.767 kbp), B. uniformis (28.433 kbp for PUL1, 59.874 kbp for PUL2), B. cellulosyliticus (26.173 kbp) and B. fluxus (25.357 kbp), plus the porphyran utilisation PUL from B. plebeius (54.476 kbp).42 Each BLAST probe was first searched against the NCBI Refseq genomes database to determine the background thresholds for Blast hits to other sequenced genomes that do not contain a XyGUL using a word size of 11; this analysis failed to reveal any hits with E-values < −20, and nucleotide identities >90% over a length >75 bp. Thus, in subsequent searches, we considered a metagenome to be positive for a particular XyGUL probe if it returned two or more hits ≥100bp in length with ≥90% identity and E-value ≤ −20. Because the most recent HMP metagenomic datasets assembled much larger contigs than previous studies (often aligning to over 20 continuous kbp of the respective XyGUL query), we considered hits in these datasets positive if they harbored just one hit that was ≥10 kbp with the same identity and E-value cut-offs listed above.
Germfree mouse experiments and diets
All animal experiments were approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan and were supervised by a veterinarian. Germfree mouse experiments were conducted in a total of 12, 6–8 week old male and female Swiss Webster mice from two litters that were born 9 days apart. Mice were randomly assigned into 3 groups of 4 animals (each was considered to be a single biological replicate) by a technician who was not familiar with the project. The investigators were not blinded to the identities of the treatment groups during the experiment. Four mice per group was chosen as the sample size based on previous studies43,44 that used similar dietary treatment regimens and found significant alterations in competitive index using 3–7 animals per group (see Extended Data Figure ED6 legend for a description of statistical tests). A custom XyG-rich diet was constructed by pureeing equal amounts by wet weight of locally purchased Roma tomatoes, Romaine lettuce leaves, common eggplant with skins, and green bell peppers with seeds. This mixture was brought to a gentle simmer for 1 hour by heating in a stainless steel vessel to simulate the cooking process for these items in human foods. The mixture was cooled and combined at a ratio of 4:1 (wet weight mixture:dry weight mouse diet) with a custom polysaccharide-free rodent diet that contained sucrose as the only additional carbohydrate (Harlan Teklad, Madison, WI USA).45 This diet mixture was dried into biscuits and autoclaved prior to feeding to germfree mice. A different custom “polysaccharide-free diet” (Harlan Teklad TD.130343; 27% protein, 44.5% glucose, 15% fat and 8% cellulose; since B. ovatus cannot grow on cellulose it is assumed that this diet does not contribute usable polysaccharides) was used for longer-term feeding studies and served as the base diet for inoculating and stabilizing the input mixture of wild-type and ΔXyGUL B. ovatus strains. The relative ratios of competing B. ovatus strains were measured from total DNA extracted from freshly voided faecal pellets as previously described.43
Recombinant protein production
Glycoside hydrolases
The genes of the glycoside hydrolases of this PUL were amplified by PCR using forward primers including Nde1 restriction sites and reverse primers including Xho1 restriction sites, except for GH43B, which had Sal1 restriction sites in the reverse primers (Supplementary Table S2). Constructs truncated to exclude predicted signal peptides and N-terminal lipidation sites46 were generated. The PCR products were digested with Nde1 and Xho1 (Nde1 and Sal1 for GH43B), and ligated into similarly digested pET21a vectors, followed by transformation into electrocompetent E. coli TOP10 cells. The cells were grown in LB overnight and plasmids were extracted using the MiniPrep kit (Qiagen) and sequenced (Eurofins MWG Operon) to identify positive clones.
Plasmids containing the PUL hydrolase genes were transformed into E. coli BL21 (DE3) cells by electroporation. The cells were grown at 37 °C with shaking in Terrific Broth containing 100 μM ampicillin to an OD600 of 0.4–0.6 when protein expression was induced by addition of 0.2 mM IPTG (isopropyl β-D-galactopyranoside) and the temperature was lowered to 25 °C. Protein expression continued for 2–3 days, after which the cells were collected by centrifugation at 4000 g for 10 minutes. The cells were resuspended in buffer A (20 mM sodium phosphate pH 7.4, 500 mM sodium chloride, 20 mM imidazole) and disrupted by either sonication or by passing twice through a French Press, followed by centrifugation at 27000 g for 45 minutes. The supernatant was loaded onto 5 ml HiTrap IMAC FF columns (GE Healthcare) using an ÄKTA FPLC (GE Healthcare) and washed thoroughly with buffer A. Each protein was purified on a separate column to eliminate the risk of cross-contamination. Tagged proteins were eluted using a linear gradient of 0–100 % buffer B (20 mM sodium phosphate pH 7.4, 500 mM sodium chloride, 500 mM imidazole) over typically 4 column volumes. Eluted proteins were concentrated using Amicon Ultra centrifugal filters (Millipore) and further purified if needed by size exclusion chromatography on a HiPrep 26/60 Sephacryl S-200 column into 50 mM sodium phosphate, pH 7.0. Liquid chromatography electrospray ionisation MS was used to verify the correct protein mass of purified proteins as described previously (data not shown).47 To improve the stability and solubility of certain of the enzymes, additives were included in the protein stock buffers, and these were present for all assays; BoGH9A was stored in 300 mM ammonium sulphate, while BoGH3A was stored in 20 % glycerol.
Non-catalytic proteins and BoGH5A truncation variants
Expression vector pET21a (Novagen) containing BoGH5A (pBoGH5) was used for subsequent cloning of the BACON domain (residues 28-116), the GH5 catalytic domain (residues 117-489), and two active-site mutants, generating pBACON, pCAT, pBoGH5-E430A and pCAT-E430A, respectively. cDNA encoding the BACON and GH5 domains were amplified by PCR using forward primers including NdeI restriction sites and reverse primers including XhoI (Supplementary Table S3). Both contained a C-terminal (His)6 tag, and a TEV-cleavage site was added before the tag at the C-terminal end of the BACON domain insert. The catalytic nucleophile mutants of full-length BoGH5A and the GH5 domain were generated using Q5® high affinity DNA polymerase (New England BioLabs) from pBoGH5 and pCAT. The gene fragments corresponding to amino acids 28-519 of SusD-like Bacova_02651 and 34-456 of Bacova_02650 gene products were amplified from B. ovatus genomic DNA by PCR using forward primers including NdeI restriction sites and reverse primers including XhoI (Supplementary Table S3). The gene products were ligated into a modified version of pET-28a (EMD Biosciences) containing a recombinant tobacco etch virus (rTEV) protease recognition site. Heterologous protein production in E. coli BL21 and purification was subsequently performed essentially as described for the glycoside hydrolases of the XyGUL; SDS-PAGE was used to confirm the purity of the proteins.
Analytical methods
Matrix-assisted laser desorption/ionisation-time-of-flight (MALDI-TOF) analysis of oligosaccharides
Xylogluco-oligosaccharides (XyGOs) products were analysed by MALDI-TOF mass spectrometry on an LT3 Plus mass spectrometer (SAI Ltd., UK) operated by the MALDI Mainframe 2, Maldi Control software (version 1.03.51, SAI Ltd., UK). A matrix of 2,5-dihydroxybenzoic acid (DHB) (10 mg ml−1 in water) was used. The number of laser pulses was varied and the resulting individual spectra were summed to optimise signal-to-noise.
Neutral sugar analysis by High Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD)
Oligo- and monosaccharides were analysed on a Dionex ICS-3000 HPLC system operated by Chromeleon software version 6.80 (Dionex) using a Dionex Carbopac PA200 column. Solvent A was water, solvent B 1 M sodium hydroxide and solvent C 1 M sodium acetate. Depending on the analytes, the following gradients were employed:
Gradient A: 0 to 5 min, 10 % B, 2 % C; 5 to 12 min, 10 % B and a linear gradient from 2–30 % C; 12 to 12.1 min, 50 % B, 50 % C; 12.1 to 13 min, an exponential gradient (curve setting 9) of B and C back to initial conditions; 13 to 17 min, initial conditions.
Gradient B: 0 to 4 min, 10 % B, 6 % C; 4 to 17 min, 10 % B and a linear gradient from 0–25 % C; 17 to 17.1 min, 50 % B, 50 % C; 17.1 to 18 min, an exponential gradient (curve setting 9) of B and C back to initial conditions; 18 to 22 min, initial conditions.
Gradient C: column pre-conditioned prior to injection by −13 to −3 min, 12 % B, 6.8 % C; −3 to 0 min, 100 % A; 0 to 25 min, 100 % A.
Substrates
The 4-nitrophenyl glycoside substrates used in this study (Glc-α-PNP, Glc-β-PNP, Gal-α-PNP, Gal-β-PNP, Xyl-α-PNP, Xyl-β-PNP, L- Araf-α-PNP, L-Araf-β-PNP) were purchased from Sigma-Aldrich. 2-chloro-4-nitrophenyl oligosaccharide substrates GGGG-β-CNP, XXXG-β-CNP, and XLLG-β-CNP were synthesised as described previously48 and L-Fuc-β-CNP was purchased from Carbosynth. Xyloglucan (tamarind seed), arabinoxylan, barley mixed linkage glucan, curdlan, galactan, gluco- and galactomannan, lichenan, carboxymethyl cellulose and hydroxyethyl cellulose, XXXG, isoprimeverose, cello-oligosaccharides, and XyGOs with a higher degree of polymerization (average degree of polymerization = 14) were purchased from Megazyme.
A mixture of oligosaccharides from tamarind xyloglucan was produced by dissolving 1 g of the polysaccharide in 100 ml water at 55 °C under vigorous stirring. The temperature was lowered to 37 °C and the endo-xyloglucanase BoGH5A (1 μM; vide infra) and phosphate buffer (to 50 mM, pH 7.0) was added, and the reaction proceeded overnight to completion, as assessed by HPAEC-PAD and LC-ESI-MS.49 The solution was passed through a Biogel-P2 column using water as eluent to remove phosphate buffer. Fractions containing XyGOs were collected, lyophilised, and analysed by HPAEC-PAD (gradient B) to determine purity. The final yield was 490 mg. XLLG was prepared from tamarind XyGO mixture as described previously.50 A similar mixture of XyGOs was also purchased from Megazyme.
GXXG was produced by dissolving XXXG in 50 mM citrate buffer, pH 6.0, and 0.4 μM of the α-xylosidase BoGH31A (vide infra) to a final substrate concentration of 40 mM. The reaction was incubated at 37 °C overnight, and MALDI-TOF was used to verify that the reaction had gone to completion. The enzyme was denatured by boiling and removed by centrifugation. Further purification of the oligosaccharide was not performed, so the GXXG prepared in this way also contained an equimolar amount of xylose.
GLLG was produced by dissolving XLLG in 20 mM citrate buffer, pH 6.0, and 0.6 μM of the α-xylosidase BoGH31A (vide infra) to a final substrate concentration of 50 mM. The reaction was incubated at 37 °C overnight, and MALDI-TOF was used to verify that the reaction had gone to completion. The enzyme was denatured by boiling and removed by centrifugation. Further purification of the oligosaccharide was not performed; therefore experiments using GLLG also contained an equimolar amount of xylose.
LLG was produced by incubating GLLG (final substrate concentration 50 mM) in 50 mM citrate buffer, pH 6.0, with 5 μM of the β-glucosidase BoGH3B (vide infra). The reaction was incubated at 37 °C overnight, and later analysed by MALDI-TOF MS; the reaction went to approximately 50 % completion. The enzyme was denatured by boiling and removed by centrifugation. As no further purification steps were taken, the LLG prepared in this way also contains an equimolar amount of glucose, as well as a significant amount of the GLLG starting material and the monosaccharide xylose from the previous preparatory step.
Extraction of tomato and lettuce xyloglucan and production of xyloglucan oligosaccharides
Using a modified and scaled-up version of the protocol outlined by Hsieh and Harris,51 xyloglucan polysaccharide was extracted from approximately 500 g freshly purchased ripe tomato fruit or iceberg lettuce leaves, obtained from a local grocery store. Fresh plant material was homogenised in 70 % aqueous ethanol using a high-speed blender, collected by filtration on Miracloth and ground to a fine powder in liquid nitrogen. Charged polysaccharides were removed by repeated 70 % ethanol wash and filtration steps. Hemicellulosic polysaccharides were extracted in 6 M sodium hydroxide, containing 1 % sodium borohydride to prevent alkaline peeling, followed by neutralisation by addition of acetic acid. The neutralised solution was filtered and the filtrate was dialysed extensively to remove salts in deionised water. The resulting hemicellulose fractions were each incubated with 135 units of the xylanase CjCBM22-GH10 (Xyn10A) and 45 units of mannanase 26A (both purchased from NZYtech) for 16 hours at 37 °C, to hydrolyse contaminating xylan and mannan, respectively. The resulting xylo- and manno-oligosaccharides were removed in subsequent ethanol washing steps. The final yield of tomato xyloglucan was 488 mg, and of lettuce xyloglucan was 560 mg.
Ethanol was added to the neutralised solution to a final concentration of 70 % to precipitate polysaccharides, followed by centrifugation at 4000 g for 30 minutes (or 24000 g for 10–15 minutes). The supernatant was discarded and the pellet washed three times with 70 % ethanol. The final pellet was dissolved in water and loaded onto a Q Sepharose column (GE Lifesciences) pre-equilibrated with 10 mM imidazole (pH 7.0). Uncharged hemicellulosic polysaccharides were washed out by addition of three column volumes of the same buffer, while charged pectins were bound to the matrix. The resulting hemicellulose containing fractions were each incubated with 150 units of the xylanase CjCBM22-GH10 (Xyn10A) and 150 units of mannanase 26A (both purchased from NZYtech) for 16 hours at 37 °C, to hydrolyse contaminating xylan and mannan, respectively. The final yield of tomato xyloglucan was 170 mg, and of lettuce xyloglucan was 180 mg.
Prior to analysis by HPAEC-PAD (gradient C), the products (1 mg) were hydrolysed by incubation with 2 M trifluoroacetic acid (TFA) for 3 hours at 120 °C (1 ml total volume). Hydrolysis products were vacuum dried and resuspended in deionised water, then filtered (0.2 μm filter). The tomato xyloglucan comprised Glc, Xyl, Gal and Ara. The lettuce xyloglucan comprised Glc, Xyl, Gal and Fuc. Both materials were found to also contain GalA and Rha, at levels indicating pectin contamination of up to 50 %. An anion exchange method adapted from Hoffman et al.52 was used to remove pectins. Dry material (84 mg lettuce xyloglucan and 63 mg tomato XyG) was redissolved in 10 mM imidazole-HCl buffer, pH 7.0. This solution was mixed thoroughly with Q-sepharose fast-flow resin (Sigma-Aldrich), to which anionic pectic polysaccharides and any proteins bound. The flow-through, containing neutral polysaccharides (xyloglucans and any trace remaining amounts of mannans and xylans), was collected by applying to a vacuum. Neutral sugar analysis showed a marked decrease in pectin contamination, to around 5 %.
One hundred mg of crude xyloglucan polysaccharide obtained from tomato or lettuce was dissolved in water (20 ml) and incubated with 0.2 μM of the BoGH5A in 50 mM phosphate buffer (pH 7.0) at 37 °C overnight. Ethanol was added to a final concentration of 70 % to precipitate remaining polysaccharides, and the soluble oligosaccharide products were recovered by filtration and lyophilisation of the filtrate. The XyGOs were analysed by HPAEC-PAD (gradient B) and MALDI-TOF. The final yield was 64 mg from the tomato xyloglucan and 58 mg from the lettuce XyG.
Enzyme assays
All assays were carried out at 37 °C, at or near the buffer (50 mM) and pH optima of the individual enzyme, unless otherwise stated. Curve fitting was performed using Origin 8 (OriginLab). For all quantitative enzyme assays at least two technical replicates were performed at each assay condition (e.g., pH value, substrate concentration, etc.).
pH dependence studies
pH optima for each enzyme were determined, where possible, using the PNP-Gly assay, described below, except for BoGH5A for which it was determined using tamarind xyloglucan as substrate and the bicinchoninic acid (BCA) reducing sugar assay.53,54 The buffers used are shown in Source Data Figures S1–S6. The concentrations of enzyme used were typically in the range 0.2 – 2 μM, unless otherwise stated.
PNP- and CNP-glycoside assays
Assays in which PNP-glycosides were used as substrates were monitored for the release of 4-nitrophenolate using A410, on a Cary 50 spectrophotometer. For discontinuous assays, samples were added to an equal volume of 200 mM Na2CO3 to terminate the reactions by raising the pH to 11.0. An extinction coefficient of 18500 M−1 cm−1 was used to calculate product concentration from absorbance values.55 For some very slow reactions, a continuous assay was used in a Cary 300 spectrophotometer; an extinction coefficient of 10500 M−1 cm−1 (pH 7.0) was used for these assays.56
Assays utilising CNP-glycosides were monitored continuously for the release of 2-chloro-4-nitrophenolate using a Cary 300 spectrophotometer (Agilent Technologies). An extinction coefficient of 12936 M−1 cm−1, determined from a standard curve, was used to calculate product concentration from A405 values.
Reducing sugar assays
Hydrolysis of polysaccharide substrates was measured by an increase in reducing sugars, using the 3,5-dinitrosalicylic acid reducing sugar assay (DNSA)57 versus a standard curve of Glc (1–6 mM). Reaction samples were added to an equal volume of DNSA reagent to terminate the reaction, and the colour was developed by boiling for 20 minutes, prior to measuring A575 on a Cary 50 spectrophotometer (Agilent Technologies).
Assays for the detection of specific monosaccharide release
A linked galactose dehydrogenase/galactose mutarotase assay (Megazyme) was used to quantify the release of galactose or arabinose from xyloglucan or XyGOs. The release of galactose or arabinose led to the stoichiometric reduction of NAD+ to NADH, giving an increase in A340 (ε 6230 M−1 cm−1 at pH 7.0),56 which was read continuously using a Cary 300 spectrophotometer.
A second linked assay (Glucose/Mannose/Fructose detection kit, Megazyme) was used to quantify the release of glucose monosaccharides. The protocol provided in the manufacturer’s instructions was modified for use as a continuous assay. The release of a glucose monosaccharide corresponds stoichiometrically with the reduction of a molecule of NADP+ to NADPH, which leads to an increase in A340 (ε 6220 M−1 cm−1),56 observed continuously using a Cary 300 spectrophotometer. Reactions were carried out in the triethylamine buffer (pH 7.6) provided with the assay kit.
Assays using HPAEC-PAD analysis
Enzymatic reactions on xylogluco- and cello-oligosaccharides were performed in 50 μl of 50 mM buffer and were stopped by addition of 2 μl of 5 M sodium hydroxide prior to HPAEC-PAD analysis. Gradient A was used for reactions with XXXG or XLLG and Gradient B was used for assays against xyloglucan polysaccharides and cello-oligosaccharides.
Affinity gel electrophoresis
Affinity gel electrophoresis was performed for 90 min at room temperature on non-denaturing 10% (w/v) polyacrylamide gels, essentially as previously described.58
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) was performed essentially as previously described59,60 using a MicroCal VP-ITC calorimeter. Proteins were dialysed into 20 mM HEPES 100 mM NaCl pH 7.0 and polysaccharides were dissolved in the dialysis buffer separately. The proteins (20–40 μM) were placed in the sample cell, and after the temperature equilibrated to 25°C, a first injection of 2 μL was performed followed by 25 subsequent injections of 10 μL of either polysaccharide (1–2.5 mg/mL). The solution was stirred at 280 rpm and the resulting heat of reaction was recorded. Data were analysed using the Origin software program.
Structural biology
BoGH5A Preparation for Crystallisation
Typically ~ 5 g of cell paste were resuspended in 5x volumes of Buffer C (50 mM HEPES pH 7.0, 0.5 M NaCl, 30 mM imidazole) and cells were lysed using three 15 s pulses of sonication at maximum amplitude using an MSE Soniprep 150 sonicator. Cell debris was removed by centrifugation at 38000 g for 30 minutes at 4°C. The supernatant was applied to a 5 ml GE Healthcare HiTrap Nickel NTA column that had been equilibrated in Buffer C at 10°C using an Äkta Xpress purification system. The column was washed with 5 column volumes of Buffer C before a 100 ml gradient of Buffer D (Buffer C + 300 mM imidazole) was applied to elute the protein. Fractions of 1.6 ml were collected along the gradient. Peak fractions containing the enzyme were pooled and concentrated using a 30 kDa molecular weight cut off Sartorius concentrator by centrifugation at 5 g to a volume of approximately 1 ml. The protein was then applied to a GE Healthcare 16/60 HiLoad Superdex 200 column, which had been equilibrated with gel filtration buffer (10 mM Tris pH 8.0, 250 mM NaCl). Following a void volume of 45 ml, 1.6 ml fractions were collected. Fractions containing BoGH5A were combined and buffer exchanged into 10 mM Tris pH 8.0, 50 mM NaCl before final concentration to 53 mg ml−1 as judged by absorption at 280 nm using an extinction coefficient of 79300 M−1 cm−1 and a molecular weight of 51674 Da.
To prepare the protein:XyGO complex 53 mg ml−1 BoGH5A was mixed with 100 mM XyGO (10 % XLXG, 90 % XXLG) to give final concentrations of 48 mg ml−1 protein and 10 mM XyGO.
Structure solution and refinement
Crystallisation trials were set up in sitting drops using a Mosquito robot (TTP Labtech). Diffraction-quality crystals were obtained in 0.1 M Bis-Tris pH 6.5, 20 % w/v polyethylene glycol monomethyl ether 5,000. Crystals were cryo-protected by soaking in mother liquor supplemented with 20 % (v/v) ethylene glycol for 30 seconds prior to flash freezing in liquid N2.
X-ray diffraction data were collected at the Diamond Synchrotron; station I03. Data were processed using XDS.61 The structure was determined by molecular replacement in PHASER62 using chain A of the Paenibacillus pabuli GH5 (PDB ID 2jep) as the search model. Two molecules were located in the asymmetric unit with log-likelihood gains of 99 and 364 for the first and second molecules respectively. The crucial step in model building was to harness 3 cycles of phase improvement, including NCS averaging, in PARROT63, prior to model construction using ARPwARP64. Subsequent model building and refinement was performed using REFMAC565 and COOT66, respectively, with the validity of the model monitored using MolProbity67 prior to deposition (PDB ID 3zmr).
Extended Data
Figure ED1.
Evolution of the genomic region containing the XyGUL and corresponding growth on XyG as a sole carbon source. a. Genomic organisation of 11 representative strains from 3 different species of Bacteroides. b. Growth of these strains on tamarind XyG with glucose and xylose as controls (average of n=2 growths per strain). The observation that one B. ovatus strain (SD CMC 3f) lacks a corresponding XyGUL, as do all B. xylanisolvens (the closest cultured relative of B. ovatus), suggests that the XyGUL entered B. ovatus after it diverged. Also note that two other unrelated flanking PULs show variable presence at this locus, suggesting that it is a ‘hotspot’ for PUL evolution.
Figure ED2.
Activation of putative XyGULs in Bacteroidetes isolates via growth on XyG. a. Sentinel (ref. 7) susC-like gene expression (n=3, expression measurements from separately grown cultures relative to a minimal medium plus glucose control). b. Growth profiles of the corresponding strains in medium containing tamarind XyG with glucose and xylose as controls (average of n=2 growths per strain). Error bars represent standard errors of the mean.
Figure ED3.
Cell-surface localisation of BoGH5A and effect of localisation on B. ovatus growth. a. staining of fixed wild-type and mutant B. ovatus strains. b. Western blot indicating that BoGH5A is still produced in the C1A lipidation site mutant, albeit in multiple degraded forms. c. Growth of wild-type (positive control), ΔBoGH5A (negative control) and BoGH5A-C1A strains on tamarind XyG. The BoGH5A-C1A mutant exhibits ~2.6-fold slower exponential growth than the wild-type. Vertical error bars on each curve indicate the standard deviation of the mean (n=3 replicates). The residual growth ability of the BoGH5A-C1A strain, despite mis-localization, is unlikely to be explained by the presence of BoGH5A enzyme accumulation in the supernatant, which was only detected by Western blot for wild-type bacteria expressing BoGH5A on the cell surface. Detection of BoGH5A in panels a and b was achieved with a rabbit polyclonal antibody raised against the recombinant protein produced in E. coli. Panels a nd b show representative data two experiments each that yielded very similar results.
Figure ED4.
Non-catalytic interaction of BoGH5A variants, SusD-like Bacova_02651 and Bacova_02650 of the XyGUL with polysaccharides. a. SDS-PAGE of recombinant proteins (representative data from at least three preparations for each protein is shown). b. Affinity gel electrophoresis (representative data from at least two gels for each experimental condition). c. Isothermal titration calorimetry (ITC); the upper graph in each pair shows the raw heat during titration, while the lower graph shows the integrated heats after correction. d. Association constants and thermodynamic parameters obtained from ITC data. Bovine serum albumin (BSA) was used as non-interacting negative control protein. Other protein names were abbreviated as follows: BACON, residues Cys1-Tyr97 corresponding to the BACON domain of BoGH5A; Bacova_02651, SusD-like XyGUL gene product; Bacova_02650, SusE-positioned XyGUL gene product; BoGH5A E430A, full-length (Cys1 to Asn470) catalytic nucleophile mutant of BoGH5A; CAT E430A, catalytic nucleophile mutant of the BoGH5A catalytic domain only (Ser98 to Asn470). Reducing-sugar assays confirmed the catalytic mutants had no detectable hydrolytic activity on xyloglucan (data not shown), while an active variant (i.e., E430) of CAT had a 2-fold higher specific activity than the full-length, wild-type BoGH5A at saturating xyloglucan concentrations (0.5 – 3 mM).
Figure ED5.
Abundance of Bacteroides XyGULs in human from a survey of metagenomic sequencing data from a total of 250 adult human samples (211 healthy, 27 ulcerative colitis, 12 Crohn’s Disease; see Methods for references). Datasets were individually queried by BLAST using the entire XyGUL nucleotide sequence from each of the four Bacteroides species listed at the top (cf. Fig. 2) and a PUL involved in degrading the red algal polysaccharide porphyran. Each horizontal line represents the presence or absence of a hit in a single individual. The leftmost column summarizes the total XyGUL content in each person (annotated according to the color key in the upper right corner). The XyGUL frequency across all 250 samples is shown at the bottom for each condition. The graph at the far right illustrates the variation in sequencing depth for each sample/study: black lines show the average depth in megabasepairs (Mbp) for each study; the light gray line shows the depth for each individual sample.
Figure ED6.
Presence of the XyGUL confers a fitness advantage to B. ovatus in the presence of dietary xyloglucan, but only when other dietary polysaccharides are eliminated. a. MALDI analysis of BoGH5A-digested alkaline extract from a custom mouse diet that contained a large amount of xyloglucan from natural vegetable sources (equal amounts of cooked bell pepper, eggplant, tomato fruit and lettuce; see Methods), indicating the presence of both solanaceous (arabinogalacto)xyloglucan and (fucogalacto)xyloglucan. b. qPCR analysis of XyGUL sentinel gene (ref. 7) expression in wild-type B. ovatus grown on extracted polysaccharides from the XyG-rich custom diet, demonstrating that it significantly activates XyGUL expression over a glucose control (error bars show the standard deviation of the mean of three biological replicates for both growth conditions). c. In vitro growth of wild-type and ΔXyGUL B. ovatus strains in the polysaccharide extract from the XyG-rich diet, including glucose and tamarind XyG as positive and negative control substrates, respectively. Compared to growth on tamarind XyG (middle panel), the incomplete growth defect of ΔXyGUL mutant on the food extract (right panel) indicates that the food contains other polysaccharides that are accessible by B. ovatus. Vertical error bars on each curve indicate the standard deviation of the mean of 3 replicates. d. In vivo competition of wild-type and ΔXyGUL B. ovatus strains in mice consuming various amounts of dietary XyG. All mice were initially fed a synthetic diet containing glucose as the sole digestible carbohydrate for 1 week and then gavaged with a 7:3 ratio of ΔXyGUL:WT (based on independent culture optical densities, total of ~108 viable B. ovatus) and the communities were allowed to equilibrate for 3 days. Despite the initial ratio biased in favour of the ΔXyGUL strain, the communities equilibrated in the range 5:5 – 4:6, but thereafter remained stable while mice were maintained on the XyG-free diet (blue boxes in three competition plots). Subsequent to community stabilization, three different dietary regimens were analysed. Left panel: Mice were maintained on the control diet (glucose only, devoid of XyG) between days 5–37, but switched to water containing 0.25% purified XyG for days 15–37 (gray box); Middle panel: Mice were switched to a xyloglucan-rich, custom diet from natural food sources while simultaneously drinking water containing 0.25% purified XyG (green box). These mice were then switched to the glucose-only, XyG-free control diet while remaining on water containing 0.25% XyG (gray box); Right panel: Mice were switched to the XyG-rich diet between days 3–15 but given normal water (yellow box), these mice were not continued further on any dietary regimen. Maintenance on either XyG food/XyG water (middle panel) or XyG food only (right panel) does not exert a measurable fitness pressure on the competing WT and ΔXyGUL strains. However, when the complex natural food polysaccharides were withheld while 0.25% XyG was maintained in water, a clear fitness pressure was observed via the significant, sequential reduction of the ΔXyGUL mutant between 15–37 days. These data suggest that although the XyGUL confers an advantage to B. ovatus by broadening its substrate range to include XyG, the presence of alternative oligo- and polysaccharides (e.g., other hemicelluloses, pectins) in a complex vegetable-based diet is nonetheless sufficient to support strains lacking this locus in vivo. Each data point is the mean abundance of the indicated strain in 4 separate mice and error bars represent one standard deviation. Measurements conformed to a normal distribution based on the observation that 67% of all assay values were within one standard deviation of their respective means. Asterisks indicate statistically significant alterations (p < 0.01; student’s t test, one-tailed, paired) in strain abundance relative to the day 15 samples, which immediately preceded the diet switch aimed at isolating XyG as the sole exogenous polysaccharide.
Table ED1.
Summary of kinetic analyses of XyGUL glycoside hydrolases.*
| Enzyme | Substrate b | kcat (s−1)† | Km (mM)‡ | kcat/Km (s−1 mM−1)‡,§ | Assay|| |
|---|---|---|---|---|---|
| BoGH2A | Gal-β-PNP | 4.0 ± 0.092 | 0.087 ± 0.0074 | 46.0 | PNP |
|
| |||||
| BoGH3A | Glc-β-PNP | Trace activity | PNP | ||
| cellobiose | Trace activity | Glc-kit | |||
| cellotetraose | 65.0 ± 3.81 | 2.49 ± 0.42 | 21.3 | Glc-kit | |
| cellohexaose | 0.62 ± 0.15 | 0.698 ± 0.52 | 0.90 | Glc-kit | |
| GLLG | n.d.¶ | n.d. | 0.15 | Glc-kit | |
| GXXG | n.d. | n.d. | 3.41 | Glc-kit | |
|
| |||||
| BoGH3B | Glc-β-PNP | 1.74 ± 0.21 | 0.155 ± 0.039 | 11.2 | PNP |
| Gal-β-PNP | Trace activity | PNP | |||
| cellobiose | 1.57 ± 0.11 | 0.68 ± 0.16 | 2.3 | Glc-kit | |
| cellotetraose | 3.99 ± 0.31 | 0.23 ± 0.060 | 17.3 | Glc-kit | |
| cellohexaose | 4.91 ± 0.71 | 0.47 ± 0.21 | 10.4 | Glc-kit | |
| GLLG | n.d. | n.d. | 0.18 | Glc-kit | |
| GXXG | n.d. | n.d. | 3.34 | Glc-kit | |
|
| |||||
| BoGH5A | XXXG-β-CNP | 10.5 ± 0.14 | 0.036 ± 0.0027 | 291.7 | CNP |
| XLLG-β-CNP | 11.1 ± 1.13 | 0.145 ± 0.024 | 76.5 | CNP | |
| GGGG-β-CNP | 0.12 ± 0.025 | 3.59 ± 1.27 | 0.034 | CNP | |
| Tamarind XyG | 435.3 ± 25.6 | 0.82 ± 0.17 mg ml−1 | 534.0 s−1 mg−1 ml | DNSA | |
| Lettuce XyG | n.d. | n.d. | 543.1 s−1 mg−1 ml | DNSA | |
| Tomato XyG | n.d. | n.d. | 501.5 s−1 mg−1 ml | DNSA | |
|
| |||||
| BoGH9A | Tomato XG | Active (MALDI-MS data) | DNSA | ||
|
| |||||
| BoGH31A | Xyl-α-PNP | 1.60 ± 0.031 | 7.7 ± 0.273 | 0.21 | PNP |
| Glc-α-PNP | 0.071 ± 0.015 | 31.8 ± 5.1 | 0.0022 | PNP | |
| XXXG | 32.6 ± 2.1 | 0.223 ± 0.046 | 146.2 | HPLC | |
| XLLG | 31.0 ± 1.9 | 0.378 ± 0.075 | 82.0 | HPLC | |
| Isoprimeverose | 1.78 ± 0.21 | 38.1 ± 7.1 | 0.047 | Glc-kit | |
|
| |||||
| BoGH43A | L-Araf-α-PNP | 0.057 ± 0.001 | 0.71 ± 0.03 | 0.081 | PNP |
| Xyl-β-PNP | 0.26 ± 0.005 | 6.58 ± 0.21 | 0.039 | PNP | |
| Tomato XyGOs | n.d. | n.d. | 0.013 s−1 mg−1 ml | GDh-kit | |
| Tomato XyG | Active | GDh-kit | |||
|
| |||||
| BoGH43B | L-Araf-α-PNP | 5.0 × 10−4 ± 8.1 × 10−5 | 6.6 ± 2.3 | 7.6 × 10−5 | Cont. assay |
| Tomato XyGOs | n.d. | n.d. | 0.0024 s−1 mg−1 ml | GDh-kit | |
| Tomato XyG | Active | GDh-kit | |||
pH-rate profiles, full kinetic data plots, and carbohydrate product analyses are available in Supplementary Figures S1–S20 for all enzymes.
Data is only given for those substrates on which a given enzyme was active. For example, BoGH5A and BoGH9A were not active on arabinoxylan, barley mixed-linkage glucan, curdlan, galactan, gluco- and galactomannan, lichenan, carboxymethyl cellulose, or hydroxyethyl cellulose. The full panel of chromogenic and natural substrates against which enzymes were tested is described in the Methods (Supplementary Information).
These units apply to all values in the column, unless otherwise stated. Standard errors of means are indicated.
For substrates for which individual kcat and Km values are not reported, kcat/Km values were obtained from linear curve fitting to initial rate data in the [S] ≪ Km(apparent) regime, where the standard Michaelis-Menten equation reduces to vo = kcat[E][S]/Km
n.d.: not determined.
Table ED2.
Growth of mutant B. ovatus strains on xyloglucan relative to wild-type B. ovatus 8384.
| Strain | Relative Growth* (%) | Relative Rate (ΔA600/Δtime) | Relative Lag (ΔtimeMutant/ΔtimeWT) | |
|---|---|---|---|---|
| ΔGH5 | tamarind xyloglucan | No Growth | ||
| dp14 tamarind xyloglucan | 139 ± 17† | 2.2 ± 0.2† | 1.2 ± 0.1 | |
| GH5-digested tamarind xyloglucan | 70 ± 9 | 0.6 ± 0.2‡ | 1.2 ± 0.2 | |
| XGOs (Megazyme) | 71 ± 6‡ | 0.6 ± 0.1† | 1.3 ± 0.3 | |
|
| ||||
| ΔGH9 | tamarind xyloglucan | 119 ± 11 | 1.2 ± 0.2 | 1.0 ± 0.1 |
| dp14 tamarind xyloglucan | 114 ± 18 | 0.9 ± 0.1 | 1.1 ± 0.1 | |
| GH5-digested tamarind xyloglucan | 105 ± 6 | 1.0 ± 0.1 | 1.3 ± 0.3 | |
| XGOs (Megazyme) | 95 ± 13 | 0.9 ± 0.1 | 1.2 ± 0.2 | |
|
| ||||
| ΔGH31 | tamarind xyloglucan | 18 ± 2† | 0.06 ± 0.05† | 3.3 ± 0.8† |
| dp14 tamarind xyloglucan | No Growth | |||
| GH5-digested tamarind xyloglucan | No Growth | |||
| XGOs (Megazyme) | No Growth | |||
Growth conditions are described in Methods. Relative growth values are average percent of wild-type B. ovatus 8384 growth density. No growth was defined as an increase of OD600 < 0.05, measured from the initial baseline to the maximum achieved. Standard errors of mean values for three biological replicates are indicated.
p value ≤0.05 as determined by unpaired t-test
p value = 0.05–0.1 as determined by unpaired t-test
Table ED3.
Data collection and refinement statistics for BoGH5A:XXXG complex.
| BoGH5A:XXXG (PDB ID 3zmr) | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 46.8, 147.2, 84.1 |
| α, β, γ (°) | 90.0, 92.8, 90.0 |
| Resolution (Å) | 72.99 - 1.43 (1.47 – 1.43)* |
| Rmerqe | 0.058 (0.575) |
| I/σI | 12.2 (2.2) |
| Completeness (%) | 99.5 (99.8) |
| Redundancy | 2.1 (2.0) |
| Refinement | |
| Resolution (Å) | 72.99 - 1.43 |
| No. reflections (Work/Free) | 197432/10458 |
| Rwork/Rfree | 0.12/0.16 |
| No. atoms | |
| Protein | 7403 |
| Ligand/ion | 226 |
| Water | 1342 |
| Avg B-factors | |
| Protein | 19 |
| Ligand/ion | 38 |
| Water | 32 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.46 |
Values in parentheses are for highest-resolution shell.
Supplementary Material
Acknowledgments
We are grateful to the following colleagues for providing materials or experimental assistance: Dr. Francisco Vilaplana (KTH Glycoscience, neutral sugar analysis of xyloglucan preparations), Dr. Gustav Sundqvist (KTH Glycoscience, protein mass spectrometry), Mr. Farid Ibatullin (KTH Glycoscience/Petersburg Nuclear Physics Institute, aryl glycoside syntheses), Ms. Sara Prexler (KTH Glycoscience, protein production and purification), Mr. Sami Tuomivaara and Dr. William York (Complex Carbohydrate Research Centre, provision of acetylated tomato xyloglucan samples), the staff at the University of Michigan Germfree Laboratory (exceptional technical assistance with gnotobiotic mouse experiments) and the staff of the Diamond Light Source (provision of data collection facilities). Work in Stockholm was supported by the Mizutani Foundation for Glycoscience, The Swedish Research Council Formas (via CarboMat – the KTH Advanced Carbohydrate Materials Centre), and the Wallenberg Wood Science Centre (salary support to O.S. and L.S.M.). Work in Vancouver was supported by faculty funding from the Michael Smith Laboratories, University of British Columbia; the Natural Sciences and Engineering Research Council of Canada (NSERC, Discovery Grant); the Canada Foundation for Innovation (CFI) and the British Columbia Knowledge Development Fund (BCKDF). Work in York was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) under Ref BB/I014802/1. Work in Ann Arbor was supported by National Institutes of Health grants DK084214 and GM099513; T.E.R. was supported in part by the Global Probiotics Council Young Investigator Grant for Probiotics Research awarded to E.C.M.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Contributions
J.L., T.E.R. and G.R.H. contributed equally to this work. J.L. performed gene cloning, recombinant production, and biochemical/enzymatic characterisation for all enzymes. T.E.R. constructed B. ovatus genetic mutants and tested mutant growth phenotypes. G.R.H. performed all protein X-ray crystallography. L.S.M. performed enzyme kinetic analyses and product analyses on select enzymes and substrates (BoGH3A & 3B, BoGH5A, BoGH9A, BoGH43A & 43B). A.S.T. performed all experiments and data analysis relating to the BACON domain and carbohydrate-binding proteins. O.S. and S.K. performed initial gene cloning and production of all enzymes, and enzymatic characterisation of BoGH5A and BoGH2A. N.A.P. performed growth profiling of various Bacteroides strains, including B. ovatus deletion mutants (ΔGH31), on xyloglucan oligo- and polysaccharides and analysed in vivo competition data by qPCR. K.U. analysed growth data from 292 Bacteroidetes isolates on xyloglucan and other substrates and assisted with metagenomic surveys. N.M.K. provided advice and assistance on XyGUL recombinant carbohydrate-binding protein production. A.L.C. and C.A.H. assisted with calorimetry and data analysis. A.G.K. assisted with comparative genomic locus identification and performed Bacteroidetes phylogenetic analysis. S.N.C. assisted with recombinant production of all enzymes and performed stability studies. E.C.M. constructed the B. ovatus Δtdk strain and N-terminal lipidation mutant, performed corresponding cellular localisation and growth studies, and performed comparative genomic analyses and metagenomic surveys. H.B., E.C.M. and G.J.D. conceived the study, directed research, analysed data, and wrote the article, including significant data analysis and writing input from J.L., T.E.R., and L.S.M.
Atomic coordinates and structure factors for the BoGH5A:XXXG complex structure have been deposited with the Protein Data Bank under accession code 3zmr.
The authors declare no competing financial interests.
References
- 1.McDougall GJ, Morrison IM, Stewart D, Hillman JR. Plant cell walls as dietary fibre: Range, structure, processing and function. J Sci Food Agric. 1996;70:133–150. [Google Scholar]
- 2.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 3.Flint H, Scott K, Duncan S, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman M, et al. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res. 2005;340:1826–1840. doi: 10.1016/j.carres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Martens EC, et al. Recognition and Degradation of Plant Cell Wall Polysaccharides by Two Human Gut Symbionts. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartemink R, VanLaere KMJ, Mertens AKC, Rombouts FM. Fermentation of xyloglucan by intestinal bacteria. Anaerobe. 1996;2:223–230. doi: 10.1006/anae.1996.0031. [DOI] [Google Scholar]
- 9.Kootte RS, et al. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes Metab. 2012;14:112–120. doi: 10.1111/j.1463-1326.2011.01483.x. [DOI] [PubMed] [Google Scholar]
- 10.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 11.van Nood E, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 12.Petrof E, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tasse L, et al. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605–1612. doi: 10.1101/gr.108332.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. Clinical Nutrition. 1997;16:3–11. doi: 10.1016/S0261-5614(97)80252-X. [DOI] [PubMed] [Google Scholar]
- 15.McNeil NI. The contribution of the large-intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 16.Smith MI, et al. Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 18.Qin JJ, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/Nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huttenhower C, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson KE, et al. A Catalog of Reference Genomes from the Human Microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamatoya K, Shirakawa M. Xyloglucan: structure, rheological properties, biological functions and enzymatic modification. Polymer Science. 2003;8:27–72. [Google Scholar]
- 22.Hsieh YSY, Harris PJ. Xyloglucans of Monocotyledons Have Diverse Structures. Molecular Plant. 2009;2:943–965. doi: 10.1093/Mp/Ssp061. [DOI] [PubMed] [Google Scholar]
- 23.Mello LV, Chen X, Rigden DJ. Mining metagenomic data for novel domains: BACON, a new carbohydrate-binding module. FEBS Lett. 2010;584:2421–2426. doi: 10.1016/j.febslet.2010.04.045. [DOI] [PubMed] [Google Scholar]
- 24.Cameron EA, et al. Multidomain Carbohydrate-binding Proteins Involved in Bacteroides thetaiotaomicron Starch Metabolism. J Biol Chem. 2012;287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakjang S, Ndeh DA, Wipat A, Bolam DN, Hirt RP. A Novel Extracellular Metallopeptidase Domain Shared by Animal Host-Associated Mutualistic and Pathogenic Microbes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aspeborg H, Coutinho PM, Wang Y, Brumer H, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC Evolutionary Biology. 2012;12:186. doi: 10.1186/1471-2148-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gloster TM, et al. Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J Biol Chem. 2007;282:19177–19189. doi: 10.1074/jbc.M700224200. [DOI] [PubMed] [Google Scholar]
- 28.Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci U S A. 2012;109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–U123. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 30.Wegmann U, et al. Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (‘Ruminococcus bicirculans’) reveals two chromosomes and a selective capacity to utilize plant glucans. Environmental microbiology. 2013 doi: 10.1111/1462-2920.12217. [DOI] [PubMed] [Google Scholar]
- 31.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martens EC, et al. Recognition and Degradation of Plant Cell Wall Polysaccharides by Two Human Gut Symbionts. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron EA, et al. Multidomain Carbohydrate-binding Proteins Involved in Bacteroides thetaiotaomicron Starch Metabolism. J Biol Chem. 2012;287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL-W - Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 37.Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin JJ, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/Nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–U487. doi: 10.1038/Nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huttenhower C, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci U S A. 2012;109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martens EC, Chiang HC, Gordon JI. Mucosal Glycan Foraging Enhances Fitness and Transmission of a Saccharolytic Human Gut Bacterial Symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonnenburg ED, et al. Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations. Cell. 2010;141:1241–U1256. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamada N, et al. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juncker AS, et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/Ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundqvist G, Stenvall M, Berglund H, Ottosson J, Brumer H. A general, robust method for the quality control of intact proteins using LC-ESI-MS. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2007;852:188–194. doi: 10.1016/j.jchromb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 48.Ibatullin FM, Baumann MJ, Greffe L, Brumer H. Kinetic Analyses of Retaining endo-(Xylo)glucanases from Plant and Microbial Sources Using New Chromogenic Xylogluco-Oligosaccharide Aryl Glycosides. Biochemistry. 2008;47:7762–7769. doi: 10.1021/bi8009168. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Fleites C, et al. Crystal structures of Clostridium thermocellum xyloglucanase, XGH74A, reveal the structural basis for xyloglucan recognition and degradation. J Biol Chem. 2006;281:24922–24933. doi: 10.1074/jbc.M603583200. [DOI] [PubMed] [Google Scholar]
- 50.Greffe L, Bessueille L, Bulone V, Brumer H. Synthesis, preliminary characterization, and application of novel surfactants from highly branched xyloglucan oligosaccharides. Glycobiology. 2005;15:437–445. doi: 10.1093/glycob/cwi013. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh YSY, Harris PJ. Xyloglucans of Monocotyledons Have Diverse Structures. Molecular Plant. 2009;2:943–965. doi: 10.1093/Mp/Ssp061. [DOI] [PubMed] [Google Scholar]
- 52.Hoffman M, et al. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res. 2005;340:1826–1840. doi: 10.1016/j.carres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 53.Mopper K, Gindler E. A new noncorrosive dye reagent for automatic sugar chromatography. Anal Biochem. 1973;56:440–442. doi: 10.1016/0003-2697(73)90210-8. [DOI] [PubMed] [Google Scholar]
- 54.Mcfeeters RF. A Manual Method for Reducing Sugar Determinations with 2,2′-Bicinchoninate Reagent. Anal Biochem. 1980;103:302–306. doi: 10.1016/0003-2697(80)90614-4. [DOI] [PubMed] [Google Scholar]
- 55.Brumer H, Sims PFG, Sinnott ML. Lignocellulose degradation by Phanerochaete chrysosporium: purification and characterization of the main alpha-galactosidase. Biochem J. 1999;339:43–53. [PMC free article] [PubMed] [Google Scholar]
- 56.Cartmell A, et al. The Structure and Function of an Arabinan-specific alpha-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases. J Biol Chem. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller GL. The use of dinitrosalicylic acid for the determination of reducing sugar. Anal Chem. 1959;31:426–428. [Google Scholar]
- 58.Freelove ACJ, Bolam DN, White P, Hazlewood GP, Gilbert HJ. A novel carbohydrate-binding protein is a component of the plant cell wall-degrading complex of Piromyces equi. J Biol Chem. 2001;276:43010–43017. doi: 10.1074/jbc.M107143200. [DOI] [PubMed] [Google Scholar]
- 59.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Schantz L, et al. Affinity maturation generates greatly improved xyloglucan-specific carbohydrate binding modules. BMC Biotechnol. 2009;9 doi: 10.1186/1472-6750-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D-Biological Crystallography. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 63.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 65.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 66.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 67.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.