SUMMARY
Reprogramming to iPSCs resets the epigenome of somatic cells, including the reversal of X chromosome inactivation. We sought to gain insight into the steps underlying the reprogramming process by examining the means by which reprogramming leads to X chromosome reactivation (XCR). Analyzing single cells in situ, we found that hallmarks of the inactive X (Xi) change sequentially, providing a direct readout of reprogramming progression. Several epigenetic changes on the Xi occur in the inverse order of developmental X inactivation, whereas others are uncoupled from this sequence. Among the latter, DNA methylation has an extraordinary long persistence on the Xi during reprogramming, and, like Xist expression, is erased only after pluripotency genes are activated. Mechanistically, XCR requires both DNA demethylation and Xist silencing, ensuring that only cells undergoing faithful reprogramming initiate XCR. Our study defines the epigenetic state of multiple sequential reprogramming intermediates and establishes a paradigm for studying cell fate transitions during reprogramming.
INTRODUCTION
Understanding the mechanisms by which the identity of a cell is established and maintained is a key goal of contemporary biology. Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) through transcription factor expression (Takahashi and Yamanaka, 2006). This process entails profound changes in genome organization, histone modifications, DNA methylation, and gene expression (reviewed in Apostolou and Hochedlinger, 2013). Questions of outstanding interest are whether reprogramming proceeds through specific stages that can be defined based on epigenetic features and how and in what order the epigenetic features gradually acquired during differentiation are reversed during reprogramming. One approach to address these questions is to focus on events for which the sequence of epigenetic changes that occur during differentiation is well defined and to ask how it is reversed during reprogramming to iPSCs.
X chromosome inactivation (XCI) is induced upon differentiation of female mouse pluripotent cells and leads to the inactivation of one of the two X chromosomes (reviewed in Lee and Bartolomei, 2013; Barakat and Gribnau, 2010; Chow and Heard, 2009). The sequence of epigenetic events accompanying the silencing of the X chromosome during differentiation has been examined extensively (Chow and Heard, 2009). These events include an initiation phase characterized by the coating of the future inactive X chromosome (Xi) by the large noncoding RNA Xist, which creates a nuclear compartment devoid of RNA polymerase II (Chaumeil et al., 2006) and leads to transient recruitment of the Polycomb Repressive Complex 2 (PRC2) and the deposition of the repressive histone mark H3K27me3 by its catalytic subunit EZH2 (Plath et al., 2003; Silva et al., 2003), closely followed by gene silencing. Later in differentiation, these events are followed by incorporation of the repressive histone variant macroH2A1 and DNA methylation, stabilizing the silenced state (Gendrel et al., 2012; Mermoud et al., 1999). Thus, once established, the Xi is extraordinary stable and is only reversed in a process termed X chromosome reactivation (XCR), which, in embryos, is limited to the inner cell mass and to germ cells (Lee and Bartolomei, 2013). XCR results in erasure of Xi-heterochromatin marks, and, importantly, can also be induced experimentally by reprogramming of female mouse somatic cells to iPSCs and somatic cell nuclear transfer (SCNT) (Maherali et al., 2007; Eggan et al., 2000). It is known that XCR is a late event during reprogramming to iPSCs (Payer et al., 2013; Stadtfeld et al., 2008), but the exact dynamics of XCR and how the epigenetic hallmarks of the Xi change in this process have remained unclear.
Most insight into the molecular events of reprogramming to iPSCs have been gained from gene expression studies of populations of cells undergoing reprogramming and of subpopulations isolated using cell surface markers (O’Malley et al., 2013; Golipour et al., 2012; Polo et al., 2012; Samavarchi-Tehrani et al., 2010; Stadtfeld et al., 2008; Mikkelsen et al., 2008; reviewed in Buganim et al., 2013). These studies indicated that reprogramming is a multistep process with two predominant “waves” of gene expression changes: an early wave marked by enhanced proliferation and a mesenchymal-to-epithelial transition (MET), characterized by Cdh1 (E-cadherin) expression (Polo et al., 2012; Samavarchi-Tehrani et al., 2010; Li et al., 2010), and a late wave, characterized by reactivation of pluripotency genes such as Nanog (O’Malley et al., 2013; Buganim et al., 2012; Golipour et al., 2012; Polo et al., 2012). The variable latency and relatively low efficiency by which individual cells reprogram have also encouraged gene expression measurements at the single-cell level at various stages of reprogramming and in clonal late intermediates. These experiments have argued for a sequence of stochastic transcriptional changes early in reprogramming, where expression programs vary dramatically between individual cells, eventually leading to hierarchical activation of pluripotency genes during the final phase, which, however, may occur through multiple paths (Buganim et al., 2012; Polo et al., 2012; Parchem et al., 2014).
Despite these advances, further molecular insight into the reprogramming path and a continuous view of the molecular events and stages leading to pluripotency would benefit from alternative approaches. In situ temporal analyses that integrate the position of cells within their native reprogramming environment, as well as the level of proteins and chromatin marks and their subcellular localization, may be particularly useful. Given that reprogramming to iPSCs is associated with XCR, and in light of the detailed characterization of sequential steps of XCI during differentiation, the reprogramming process provides an unprecedented opportunity to study XCR. In turn, the Xi provides an exceptional possibility to characterize the dynamics of the reversal of epigenetic marks during reprogramming.
Here, we followed epigenetic changes on the Xi during reprogramming to iPSCs in individual cells using detailed, high-resolution in situ time course analyses to address the question of whether XCR and somatic cell reprogramming follow a precise sequence of epigenetic changes. Due to the sheer size of the X chromosome, this analysis can be done at the single-cell level using immunofluorescence and RNA FISH approaches, allowing for the identification of reprogramming stages that have been elusive in transcriptional and chromatin studies to date. Our work demonstrates that the epigenetic state of the Xi changes sequentially throughout reprogramming, along with global changes in chromatin character. To shed light on the mechanisms by which XCR takes place, we used genetically manipulated somatic cells and examined the role played by Cdh1, Nanog, Xist, Tsix, Tet1, Tet2, and DNA methylation. The highly reproducible sequence of epigenetic steps leading to XCR and induced pluripotency provides a simple readout of reprogramming progression and a basis for studying cell fate transitions during reprogramming.
RESULTS
Reprogramming Steps Defined by the Dynamics of Xi Chromatin Marks
To define epigenetic steps of XCR and reprogramming, we determined the dynamics of Xi hallmarks during the establishment of pluripotency in mouse embryonic fibroblasts (MEFs). We induced female MEFs to reprogram with retroviruses encoding Oct4, Sox2, and Klf4 and analyzed single cells in their native reprogramming environment throughout detailed time courses every other day for 1–3 weeks using multicolor immunostaining (Figure 1A). This allowed us to assess the state of the Xi and of evolving global epigenetic states in any cell of the reprogramming cultures and to delineate the sequence of epigenetic events during reprogramming relative to other markers.
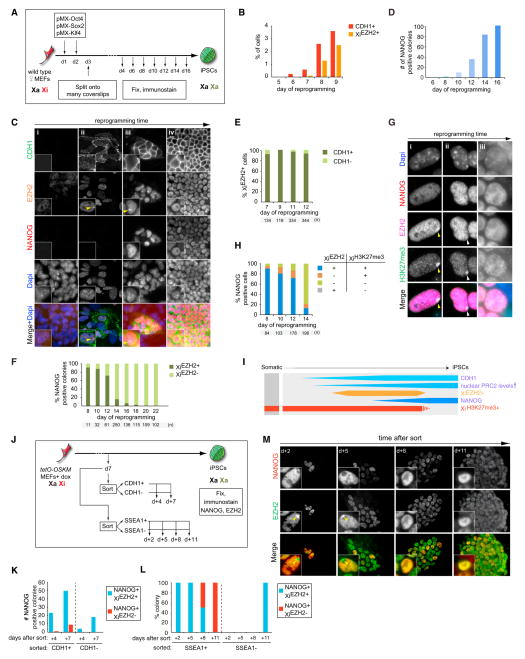
Figure 1. Time Course Analysis of XiEZH2+ during Reprograming to Pluripotency.
(A) Diagram of reprogramming time course experiments. In all experiments, results for female cells are displayed except when stated otherwise, and the time points and number (n) of cells or colonies counted are given in each subfigure.
(B) Quantitation of the proportion of CDH1+ or XiEZH2+ cells at indicated reprogramming time points. 100 cells in three randomly chosen microscopic fields were counted per time point.
(C) Multicolor immunostaining for CDH1 (green in merge), EZH2 (orange), and NANOG (red) at different stages of reprogramming. Dapi staining (blue) marks nuclei. Unlike MEFs (i), cells with elevated nuclear levels of EZH2 and XiEZH2+ (arrowhead) are seen within CDH1+ cells during reprogramming starting around day 7 of reprogramming (ii). (iii) NANOG+ colonies are first marked by XiEZH2+ and elevated EZH2 levels in the nucleus (image from day 9). (iv) Later, NANOG+ colonies become larger and are characterized by high nuclear EZH2 levels without XiEZH2+ (image from day 14).
(D) Number of NANOG+ colonies throughout reprogramming (a colony is defined as four or more closely localized cells).
(E) Proportion of XiEZH2+ cells with and without CDH1 expression during reprogramming.
(F) Proportion of NANOG+ colonies with or without XiEZH2+ at indicated time points. All NANOG+ colonies present in the reprogramming cultures were counted up to day 14 and only a subset thereafter.
(G) Multicolor immunostaining for EZH2 (magenta in merge) and NANOG (red) in combination with H3K27me3 (green). The images depict various states of XiEZH2+ and XiH3K27me3+ in NANOG+ cells quantified in (H). During reprogramming, (i) NANOG+/XiEZH2+ cells are initially XiH3K27me3+ (ii) and, at a later time point, become XiEZH2−/XiH3K27me3+ for a very short time and subsequently become (iii) XiEZH2−/XiH3K27me3−. Yellow and white arrowheads indicate XiEZH2+/XiH3K27me3+ and XiEZH2−/XiH3K27me3+ patterns, respectively.
(H) Quantitation of the immunostaining experiment in (G), giving the proportion of NANOG+ cells with XiEZH2+ or XiH3K27me3+ at indicated time points.
(I) Summary of Xi and global dynamics of PRC2 and H3K27me3 during reprogramming, relative to CHD1 and NANOG expression. Female-specific features are shown in orange/red, and those occurring in both female and male reprogramming are shown in blue. The width of the boxes represents the level of the epigenetic mark considered.
(J) Experimental design for the isolation and characterization of CDH1+/− or SSEA1+/− reprogramming subpopulations.
(K) Number of NANOG+ colonies with or without XiEZH2+ in CDH1+ and CDH1− sorted cell populations isolated as shown in (J), at indicated days after replating.
(L) Proportion of NANOG+ colonies with or without XiEZH2+ in SSEA1+ and SSEA1− sorted cell populations isolated as shown in (J), at indicated days after replating. n = 6 for each SSEA1+ time point, and n = 1 for the SSEA1− count at +d11. Colonies appearing in SSEA1+ replated cells become larger throughout this time course as shown in (M).
(M) Visualization of XiEZH2+ changes in replated SSEA1+ reprogramming intermediates over time from the experiment shown in (J) and (L). Replated cells were immunostained for EZH2 (green in merge) and NANOG (red) at the indicated days. Note the increase in colony size and disappearance of XiEZH2+ (yellow arrowhead) with time in culture.
See also Figure S1.
We first analyzed the dynamics of PRC2 on the Xi. EZH2 did not accumulate on the Xi within the first 6 days of reprogramming (Figures 1B and 1C, i). However, after CDH1 (E-cadherin) became expressed, which marks the MET (Li et al., 2010), and before the pluripotency factor NANOG was detectable, a strong nuclear EZH2 staining focus characteristic of Xi accumulation (XiEZH2+) arose in a small fraction of the cells (Figures 1B, 1C, ii, and 1D). The same result was obtained for SUZ12, another PRC2 subunit, and the PRC2-recruitment factor JARID2 (da Rocha et al., 2014) (Figure S1A available online). The XiEZH2+ was restricted to CDH1+ cells (Figure 1E) and only occurred in a subset of CDH1+ cells, around 50% at day 10 of reprogramming. These findings show that XiEZH2+ arises after an epithelial cell character is established during reprogramming, indicative of the existence of a reprogramming stage immediately downstream to MET that is more restrictive than CDH1 expression. In agreement with this, XiEZH2+ was also present in known late reprogramming intermediates such as pre-iPSCs (Figures S1B–S1D) and was only detectable in reprogramming cultures co-transduced with viruses encoding Oct4, Sox2, and Klf4 with or without cMyc, but not when fewer reprogramming factors were employed (Figure S1E), demonstrating that the PRC2 composition of the Xi only changes when reprogramming factor combinations able to induce pluripotency are used.
Notably, we also observed that the level of nuclear EZH2 (i.e., on autosomes) gradually increased during reprogramming in both male and female cells, which was initiated specifically in a subset of CDH1+ cells before XiEZH2+ was induced (Figure 1C, progression from i to iv). However, ectopic EZH2 expression in female MEFs did not induce XiEZH2+ (Figure S1F), indicating that the global EZH2 increase during reprogramming is not sufficient for XiEZH2+. These results reveal that, downstream of MET, PRC2 is gradually upregulated at the global level, irrespective of sex chromosome content, and additionally relocalizes to the Xi in female cells, providing a direct readout of reprogramming progression.
To uncover whether CDH1-positive (CDH1+)/XiEZH2+ cells are intermediates on the path to the NANOG-positive (NANOG+) reprogramming stage, we determined the presence of XiEZH2+ in the first cells that express the NANOG protein during reprogramming. We found that NANOG activation initiated within a subset of XiEZH2+ colonies, with nearly all NANOG+ cells that first appeared in reprogramming cultures carrying the XiEZH2+ (Figures 1C, iii, and 1F). Later in reprogramming, and usually in large NANOG+ colonies, almost all NANOG+ cells lacked XiEZH2+ (Figures 1C, iv, and 1F), and the absolute number of XiEZH2+ cells and colonies decreased accordingly (data not shown). Moreover, NANOG+ cells were initially surrounded by NANOG-negative (NANOG−)/XiEZH2+ cells (Figure S1G), which is consistent with the induction of NANOG occurring in a subset of CDH1+/XiEZH2+ cells followed by removal of XiEZH2+ within NANOG+ colonies.
H3K27me3, the downstream mark of PRC2, enriched on the Xi in NANOG− cells, was lost from the Xi exclusively within NANOG+ cells with kinetics slightly delayed compared to the loss of XiEZH2+, such that NANOG+ cells with XiH3K27me3+ but without XiEZH2+ could be briefly detected (Figures 1G and 1H). These data suggest that the loss of XiH3K27me3+ is a consequence of the removal of EZH2 from the Xi. Thus, NANOG expression precedes both loss of XiEZH2+ and XiH3K27me3+.
Taken together, our findings suggest that cells go through defined epigenetic steps as they progress toward pluripotency. Specifically, we reveal four steps by simply following PRC2, at the Xi-specific and global level, relative to CDH1 and NANOG. Downstream of MET, PRC2 proteins increase in overall levels and accumulate on the Xi in a subset of CDH1+ cells. Then, a subset of CDH1+/XiEZH2+ cells reactivates NANOG, which precedes EZH2 and H3K27me3 removal from the Xi specifically in these NANOG+ cells (Figure 1I). Importantly, the reacquisition of XiEZH2+ represents the inversed sequence of events of developmental XCI, where PRC2 accumulates on the Xi immediately after Xist RNA initially coats the X and disappears from the Xi later in differentiation (Plath et al., 2003; Silva et al., 2003), suggesting that XiEZH2+ reflects the extent of reversal of the differentiated state established during reprogramming.
The histone variant macroH2A1 associates with the Xi late during developmental XCI and has been shown to act as a barrier to reprogramming (Pasque et al., 2012). We found that the XimacroH2A1+ of MEFs was maintained during reprogramming until XiEZH2+ was lost (Figures S1H–S1K). Unexpectedly, the global level of macroH2A1 first increased from the somatic level before dropping again to the lower level of pluripotent cells in both female and male cells (Figure S1I; data not shown), indicating that an epigenetic mark associated with resistance to reprogramming is transiently induced during the reprogramming process (Figure S1O). Altogether, these results strengthen the conclusion that the Xi-specific and global epigenetic states of cells define multiple stages of reprogramming. Unlike XiEZH2+, the kinetics of XimacroH2A1+ loss during reprogramming does not represent the reversed sequence of developmental XimacroH2A1+ dynamics, suggesting that distinct mechanisms regulate the temporal Xi accumulation of different epigenetic marks during reprogramming.
Subpopulations with Increased Reprogramming Capacity Recapitulate Xi Events
To test whether the steps identified based on fixed cultures represent dynamics of cells that would, if not fixed, continue along the path to pluripotency, we considered the use of pluripotency reporters such as Oct4-GFP or Nanog-GFP. However, we found that their activation occurred well after the endogenous NANOG protein was detectable, at a time when EZH2 is already removed from the Xi (Figures S1L–S1N), precluding their use for monitoring reprogramming events that occur when NANOG becomes initially expressed.
Instead, we asked whether NANOG+/XiEZH2+ cells arise from CDH1+ cells by sorting CDH1+ and CDH1− cells at day 7 of reprogramming and assessing their ability to give rise to NANOG+/XiEZH2+ cells after replating an equal number of both sorted cell populations (Figures 1J and S1P). We found that CDH1+-sorted cells preferentially gave rise to NANOG+/XiEZH2+ colonies compared to NANOG+/XiEZH2− colonies (Figure 1K), supporting the conclusion that NANOG+/XiEZH2+ cells originate from CDH1+ cells. Furthermore, in the time frame considered, replated CDH1−-sorted cells also proceeded to the NANOG+/XiEZH2+ state but with delayed kinetics and reduced efficiency (Figure 1K), which is in agreement with the notion that cells reprogram with variable latencies (Hanna et al., 2009). We also performed sorting experiments employing SSEA1, a marker of a reprogramming intermediate arising within CDH1+ cells (Polo et al., 2012) (Figures 1J and S1Q). As expected, shortly after replating, NANOG+ colonies were detected specifically from the SSEA1+ population, and cells within these colonies were initially exclusively XiEZH2+ (Figures 1L and 1M). Remarkably, as these NANOG+ colonies grew bigger over time, they completely lost XiEZH2+ (Figures 1L and 1M). SSEA1− cells gave rise to NANOG+ cells later, and these were all first XiEZH2+ (Figure 1L). Therefore, we conclude that the reprogramming steps defined based on our fixed time courses correctly capture the trajectory of cells moving toward pluripotency.
XCR Occurs after Loss of Xist RNA in NANOG+ Cells
To determine the dynamics of Xist RNA, the key regulator of developmental XCI, during reprogramming, we combined immunostainings with RNA FISH for Xist. Early in reprogramming, virtually all cells showed Xist RNA coating, detectable as a large “cloud” of RNA FISH signal (Figure 2A, i). At late reprogramming time points, Xist RNA was specifically absent from the Xi within NANOG+ colonies, whereas NANOG− cells still exhibited Xist RNA coating (Figure 2A, iii). However, the first NANOG+ cells to appear in culture were always XiXist+ (Figures 2A, ii, and 2B), indicating that Xist repression follows NANOG activation. Furthermore, we found that XiEZH2+ in NANOG+ cells highly correlated with the presence of Xist RNA and that their loss occurred with similar dynamics (Figures S2A and S2B), which is consistent with Xist-dependent recruitment of PRC2 to the Xi (Plath et al., 2003).
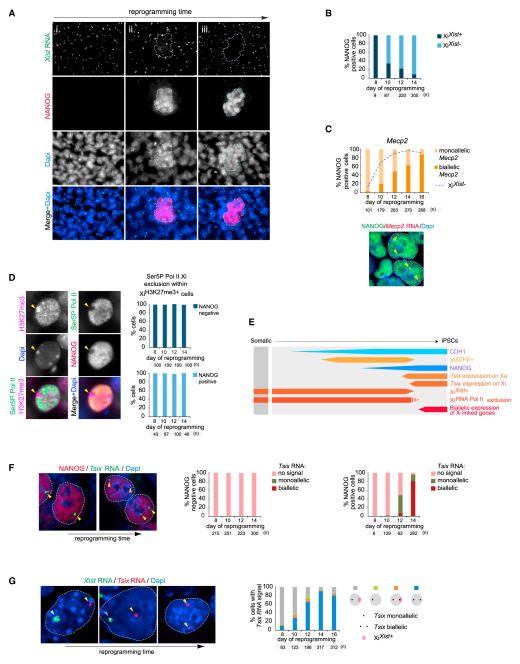
Figure 2. Kinetics of Xist and Tsix RNA and XCR during Reprogramming.
(A) Representative images of Xist RNA (green in merge), NANOG (red), and Dapi (blue) from immunoFISH stainings at different time points of reprogramming reflecting different states of XiXist+ and NANOG expression as determined in (B). Dotted lines indicate the position of NANOG+ colonies across different channels. (i) day 8, (ii) day 10, and (iii) day 14. Each image represents a series of ten Z-sections merged onto a single plane.
(B) Proportion of NANOG+ cells with or without XiXist+ at different time points.
(C) ImmunoFISH analysis of NANOG expression and nascent transcripts of the X-linked gene Mecp2 (seen as a strong pinpoint) during reprogramming. In the image, the biallelic Mecp2 expression pattern is indicated (two arrowheads), and the proportion of NANOG+ cells with mono- or biallelic Mecp2 expression is given below. The dotted line indicates the proportion of XiXist− cells from the same time course.
(D) Images depict an immunostaining for H3K27me3, Ser5P polymerase II (Ser5P Pol II), and NANOG at day 12 of reprogramming. Quantification gives the proportion of Ser5P Pol II Xi-exclusion cells in NANOG−(top) or NANOG+ (bottom) cells that also display XiH3K27me3+ at indicated time points.
(E) Summary of reprogramming stages related to this figure, displayed as described in Figure 1I. Dashed lines indicate the window of time we narrowed down for the feature to occur or disappear.
(F) ImmunoFISH analysis as in (C), except for NANOG (red) and Tsix RNA (green). Proportion of NANOG− (left) and NANOG+ (right) cells, respectively, displaying monoallelic, biallelic, or no Tsix RNA FISH signal.
(G) RNA FISH analysis of the relationship between Xist and Tsix RNA in reprogramming. In the images, yellow arrowheads represent Tsix expression without Xist RNA present on the same X chromosome, and the white and gray arrowheads represent an Xist RNA cloud that does not or does overlap a Tsix signal, respectively. The quantification of cells with Tsix expression showing mono- or biallelic Tsix expression with and without XiXist+ is shown.
See also Figure S2.
Next, we used RNA FISH to examine when genes on the Xi reactivate during reprogramming (Figure S2C) and found that cells mostly displayed monoallelic expression of the X-linked genes Mecp2, Atrx, Gpc4, and Rlim when NANOG+ cells first appeared, indicative of maintenance of XCI in these cells (Figures 2C and S2D–S2F). Later in reprogramming, NANOG+ cells exhibited biallelic expression of these genes, a sign of XCR (Figures 2C and S2D–S2F). For all tested genes, reactivation occurred with delay relative to the loss of Xist RNA. We conclude that XCR is a very late event of reprogramming that occurs in a coordinated fashion along the chromosome after Xist RNA coating has disappeared. Our results also suggest that XCR takes place independently in multiple cells of a given NANOG+ colony because all cells in NANOG+ colonies initially are XiXist+/XiEZH2+, whereas, at later reprogramming time points, the cells in larger NANOG+ colonies are not (Figure S2A).
To confirm that XCR occurs only in NANOG+ cells, we performed two additional assays. First, we observed that the exclusion of RNA polymerase II from the Xi domain was maintained in all NANOG+ or NANOG− cells that carried XiH3K27me3+ (Figure 2D), which is consistent with the maintenance of silencing at these reprogramming stages. Second, we examined the expression of an Xi-linked GFP reporter (Maherali et al., 2007) and did not detect GFP reporter reactivation in NANOG− cells, whereas NANOG+ cells consistently expressed GFP at late reprogramming time points (Figure S2G). We conclude that, late in reprogramming, NANOG expression precedes the loss of Xist RNA, which coincides with loss of XiEZH2+ and occurs before XCR (Figure 2E).
Reprogramming Reverses the Developmental Sequence of Tsix Expression
To establish the dynamics of activation of Tsix (transcribed anti-sense to Xist) RNA, a critical regulator of Xist during initiation of XCI (Lee and Bartolomei, 2013), we used strand-specific RNA FISH and found that Tsix was not expressed during the early stages of reprogramming (i.e., in NANOG− cells) or in the first NANOG+ cells that appear (Figure 2F). Within maturing NANOG+ cells, however, Tsix became first monoallelically expressed in cells still carrying XiXist+. The monoallelic Tsix signal occurred specifically from the active X chromosome (Xa), as it never overlapped with XiXist+ (Figure 2G). Tsix activation on the Xi took place later, at the very tail end of XiXist loss (Figures 2G and S2H). Together, these results show that reprogramming to pluripotency recapitulates the expression of Tsix in the reverse order from that of developmental XCI, where Tsix is first downregulated on the future Xi and then becomes repressed on the Xa (Lee and Lu, 1999) (Figure 2E).
Kinetics of XCR in Relation to Pluripotency Gene Activation
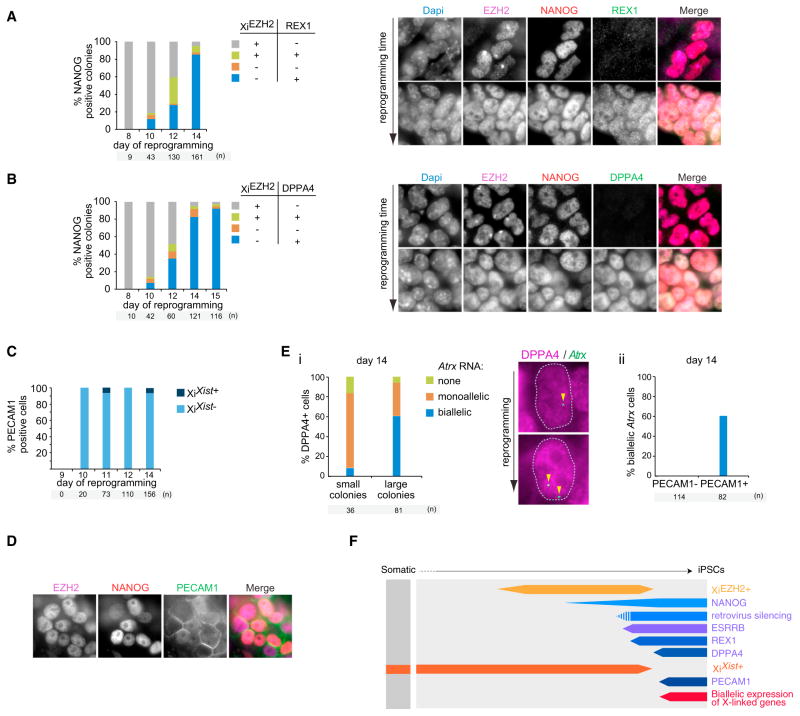
Given the stepwise changes of Xi hallmarks late in reprogramming, we aimed to determine the dynamics of these features in relation to the activation of pluripotency-associated factors. In agreement with the reported hierarchical activation of pluripotency factors late in reprogramming based on single-cell transcript analysis (Buganim et al., 2012), we observed the sequential induction of the pluripotency factors ESRRB, REX1, DPPA4, and PECAM1 at the single-cell level using multicolor immunostaining, which only occurred in NANOG+ cells (Figures S3A–S3F). In addition, silencing of the reprogramming factor-expressing retroviruses can be placed early in this hierarchy at around the time of ESRRB/REX1 activation (Figures S3G–S3I), which is consistent with the shift to endogenous pluripotency factor activation. XiEZH2+ was lost after REX1 expression and just after DPPA4 activation (Figures 3A and 3B). PECAM1 expression, which is very late in the pluripotency factor hierarchy, marked cells that are devoid of XiXist+ and XiEZH2+ (Figures 3C and 3D). Consistent with a delay of XCR relative to Xist RNA loss, XCR took place after DPPA4 activation, as small DPPA4+ colonies expressed the X-linked gene Atrx monoallelically and large colonies expressed the X-linked gene mostly biallelically (Figure 3E). Together, these results demonstrate the molecular timeline of Xi changes relative to hierarchical pluripotency gene activation (summarized in Figure 3F).
Figure 3. Xi Features in Relation to the Sequential Expression of Pluripotency Factors during Reprogramming.
(A) Quantitation of an immunostaining analysis of NANOG, XiEZH2+, and REX1, presenting the proportion of NANOG+ colonies with XiEZH2+ and/or REX1 expression at indicated time points. Right, representative immunostaining images for EZH2 (magenta in the merge), NANOG (red), and REX1 (green).
(B) As (A) for NANOG, XiEZH2+, and DPPA4.
(C) Quantitation of an immunoFISH analysis for PECAM1 and Xist RNA displaying the proportion of PECAM1+ cells with XiXist+.
(D) Representative immunostaining image for EZH2 (magenta in the merge), NANOG (red), and PECAM1 (green), demonstrating the absence of XiEZH2+ in PE-CAM1+ cells.
(E) (i) Quantitation of mono- and biallelic expression of the X-linked gene Atrx within cells of small (<12 cells) and large (>20 cells) DPPA4+ colonies and representative images from the DPPA4 (magenta) and Atrx (green) immunoFISH staining. Arrowheads indicate Atrx nascent transcription signals. (ii) Quantification of biallelic Atrx expression in PECAM1+/− cells.
(F) Summary of reprogramming stages identified in this figure as in Figure 2E.
See also Figure S3.
Sequential Xi States Are Conserved across Different Reprogramming Systems
To establish whether the sequential changes on the Xi during reprogramming are specific to the reprogramming system used, we reprogrammed MEFs carrying a single dox-inducible, poly-cistronic reprogramming cassette encoding Oct4, Sox2, Klf4, and cMyc in a defined locus instead of retroviral infection. In addition, we reprogrammed another starting cell type, mouse embryonic endoderm cells, and also used different culture conditions. The dynamics of XiEZH2+ and sequential pluripotency gene activation were reproduced in each case (Figures S3J–S3N). We conclude that the epigenetic states identified represent fundamental changes inherent to reprogramming, applicable to multiple starting cell types and reprogramming systems.
CDH1 and NANOG Are Required, but Not Sufficient, for the Efficient Induction of Reprogramming Steps Leading to XCR
Our data indicate that CDH1 expression marks the cells that subsequently induce XiEZH2+ and NANOG and that only those cells that activate NANOG are fated to induce Xist loss, Tsix activation on the Xi and Xa, pluripotency-associated factor activation, and XCR, suggesting that both CDH1 and NANOG are critical for this hierarchy of events. To address the role of CDH1 and NANOG for epigenetic changes taking place downstream of their expression, we performed both knockdown and overexpression experiments. We found that knockdown of Cdh1 during reprogramming with shRNAs decreased the number of XiEZH2+, NANOG+, and DPPA4+ colonies (Figures 4A and S4A). In contrast, Cdh1 overexpression did not promote any of the epigenetic events that normally take place downstream of CDH1 induction (Figures S4B–S4E). Depletion of Nanog transcripts during reprogramming using an inducible shRNA (Figures S4F and S4G) did not prevent CDH1 activation, global upregulation of EZH2, and XiEZH2+ (Figures S4H and S4I), in agreement with its induction later during reprogramming (Silva et al., 2009). By contrast, the activation of Tsix on the Xa and Xi, as well as of pluripotency-associated transcription factors, was strongly reduced by Nanog depletion (Figures S4J–S4M). The lack of biallelic Tsix expression in the absence of Nanog also suggests that XCR was impaired without Nanog. Thus, NANOG orchestrates the efficient transition through the later molecular events, including XCR, although the requirement for Nanog can be bypassed (Carter et al., 2014; Schwarz et al., 2014). Overexpression of Nanog late in reprogramming promoted steps toward XCR as judged by the increased number of DPPA4+/XiEZH2− colonies, but not those before NANOG is normally induced (i.e., XiEZH2+) (Figures 4B, S4N, and S4O). However, most NANOG-overexpressing cells did not induce the subsequent reprogramming steps. Together, these results demonstrate that both Cdh1 and Nanog are required, but not sufficient, for the induction of the epigenetic events leading to XCR.
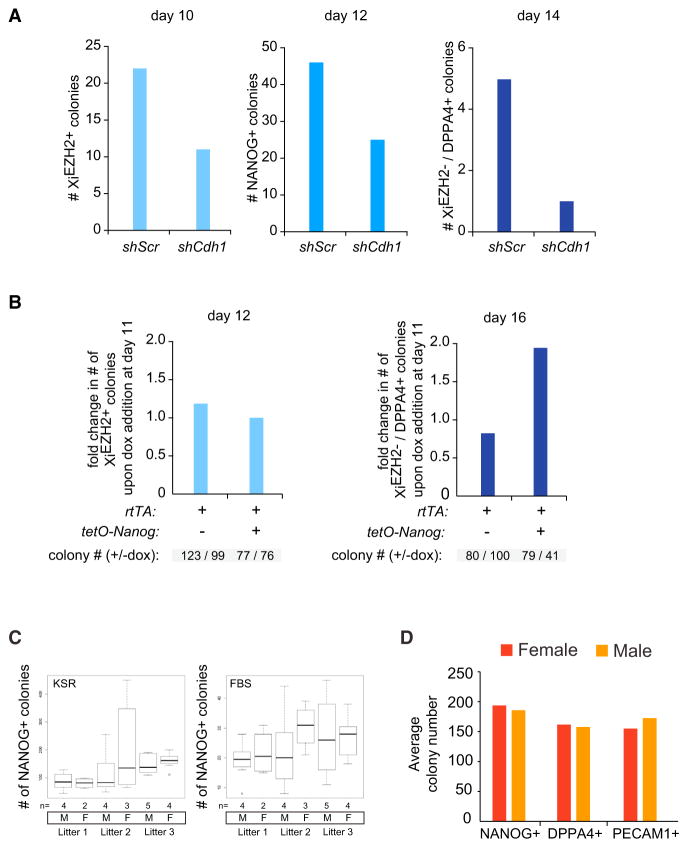
Figure 4. Cdh1 and Nanog Modulate the Efficiency of Reprogramming and Dynamics of Xi Hallmarks, whereas XCR Does Not Represent a Reprogramming Barrier.
(A) Number of XiEZH2+, NANOG+, and DPPA4+/XiEZH2− colonies obtained when Cdh1 is knocked down by shRNAs (shCdh1) throughout reprogramming, compared to scrambled shRNA (shScr) reprogramming experiments.
(B) Reprogramming experiments with female MEFs carrying rtTA only or rtTA and the tetO-Nanog allele, and with and without dox addition at day 11. The number of XiEZH2+ and DPPA4+/XiEZH2− colonies was determined at day 12 and 16 of reprogramming, respectively, and was plotted as fold change between dox relative to no dox treatment per cell line. Note that XiEZH2+ counts are similar at day 12, whereas DPPA4+/XiEZH2− counts differ at day 14 when Nanog is overexpressed.
(C) Comparison of reprogramming efficiency between male (M) and female (F) MEFs. Box plots depict the number of NANOG+ colonies for reprogramming experiments in KSR and FBS media, respectively, with male and female MEFs, at day 14 and day 25 of reprogramming, respectively. MEFs are grouped by litter and the number of male and female MEF populations per litter is given (n). Whiskers demarcate the minimum and maximum of the data.
(D) Quantitation of different late reprogramming stages for MEFs isolated from seven female and seven male embryos, as judged by the number of NANOG+, DPPA4+, or PECAM1+ colonies at day 14.
See also Figure S4.
The above finding raised the question of whether XCR represents a barrier to reprogramming. To test this, we obtained a large number of female and male MEFs preparations from four independent litters and measured the efficiency with which NANOG+, DPPA4+, or PECAM1+ colonies formed, without prior knowledge of the sex. This experiment revealed no difference in the reprogramming efficiency between male and female MEFs in KSR or FBS culture media and in the transition to different reprogramming stages (Figures 4C and 4D). Thus, even though the Xi represents the most extreme form of facultative heterochromatin, XCR does not limit reprogramming to induced pluripotency.
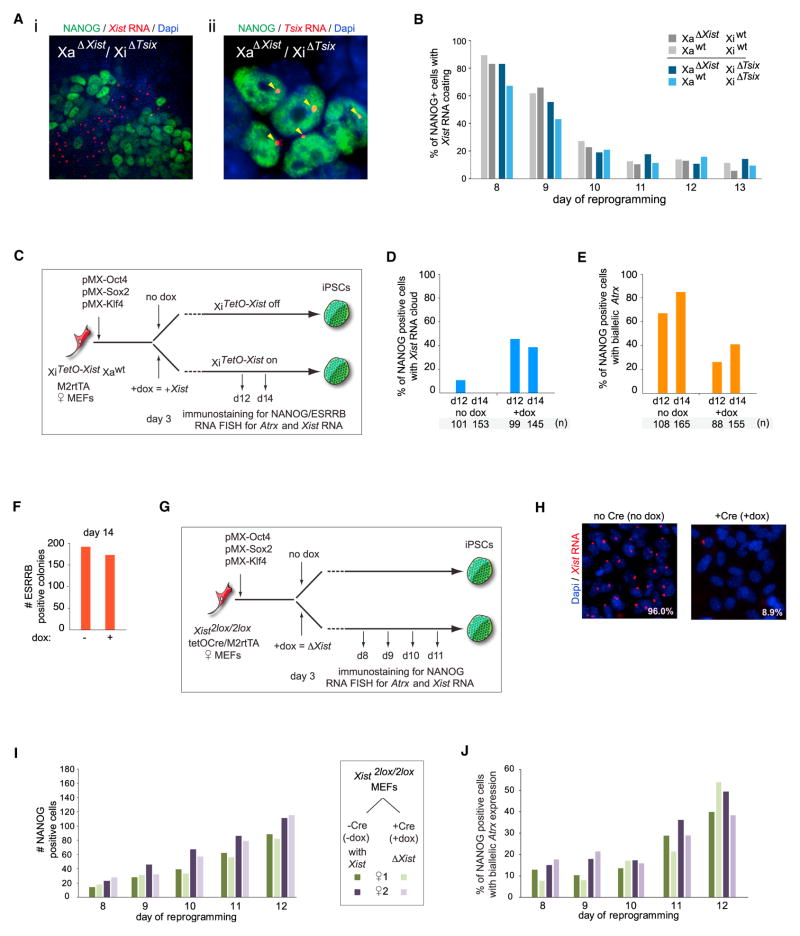
Requirement of Xist Silencing, but Not Tsix Expression, for XCR
To examine the molecular mechanism of XCR during reprogramming, we focused on the requirement for Xist and for Tsix, its negative regulator in pluripotent cells (Lee and Bartolomei, 2013). Despite Tsix becoming expressed on the Xi as Xist RNA disappears (Figure 2), deletion of Tsix did not alter the kinetics of Xist repression in NANOG+ cells (Figures 5A and 5B), indicating that Tsix does not negatively regulate Xist at the end of reprogramming. Conversely, to test whether repression of Xist RNA is required for XCR, we ectopically expressed Xist from the Xi during reprogramming (Figure 5C). Constitutive Xist expression did not alter the efficiency by which ESRRB+ colonies appeared but resulted in a decrease in XCR within NANOG+ cells, as measured by the extent of biallelic Atrx expression (Figures 5D–5F). Thus, Xist silencing at the end of reprogramming is necessary for XCR.
Figure 5. Xist Silencing Is Necessary, but Not Sufficient, for XCR.
(A) (i) Representative image of an immunoFISH analysis for NANOG (green) and Xist RNA (red) at day 14 of reprogramming with MEFs carrying a deletion of Tsix on the Xi and of Xist on the Xa, illustrating that NANOG+ cells lose Xist RNA accumulation on the Xi even in the absence of Tsix on the Xi. (ii) iPSCs derived from the experiment in (i) were stained for NANOG (green) and Tsix RNA (red), confirming monoallelic expression of Tsix due to deletion on one X chromosome (arrowheads).
(B) Kinetics of Xist RNA loss in the absence of Tsix on the Xi. Proportion of NANOG+ cells with XiXist+ in reprogramming time courses performed with MEFs with (gray bars) and without (blue bars) Tsix on the Xi. Just like Tsix deletion, the additional deletion of Xist on the Xa (dark versus lighter bars) does not affect the kinetics of Xist RNA loss in NANOG+ cells.
(C) Diagram of Xist overexpression reprogramming experiments using MEFs in which the promoter of Xist on the Xi is replaced with a tet-inducible promoter.
(D) Proportion of NANOG+ cells with XiXist+ in reprogramming cultures described in (C) with and without ectopic Xist induction conditions (+/−dox), based on immunoFISH analysis.
(E) As (D), but for NANOG+ cells with biallelic Atrx expression.
(F) As (D), but for the number of ESRRB+ colonies.
(G) Diagram of the Xist deletion reprogramming experiments with female conditional Xist MEFs.
(H) Xist RNA FISH for MEFs described in (G) under control (−dox) and +dox conditions, the latter leading to Xist RNA loss in the majority of cells.
(I) Number of NANOG+ colonies at various time points of reprogramming for the experiment described in (G) under control (no dox/−Cre) and the Xist deletion (+dox/+Cre) conditions.
(J) As in (I), but quantitation of NANOG+ cells with biallelic Atrx expression based on immunoFISH analysis.
To determine whether XCR depends solely on Xist repression, we asked whether Xist deletion leads to precocious activation of the Xi. Specifically, we deleted Xist early in the reprogramming process using female MEFs homozygous for a conditional (2lox) Xist allele (Csankovszki et al., 2001), which also carried a dox-inducible Cre recombinase (Figures 5G and 5H). Xist ablation had no effect on the efficiency with which NANOG+ colonies were generated (Figure 5I) and, surprisingly, did not alter XCR kinetics (Figure 5J). Therefore, Xist repression is necessary, but not sufficient, for XCR to occur, indicating the existence of other mechanisms that can maintain Xi silencing throughout reprogramming and even initially in NANOG+ cells.
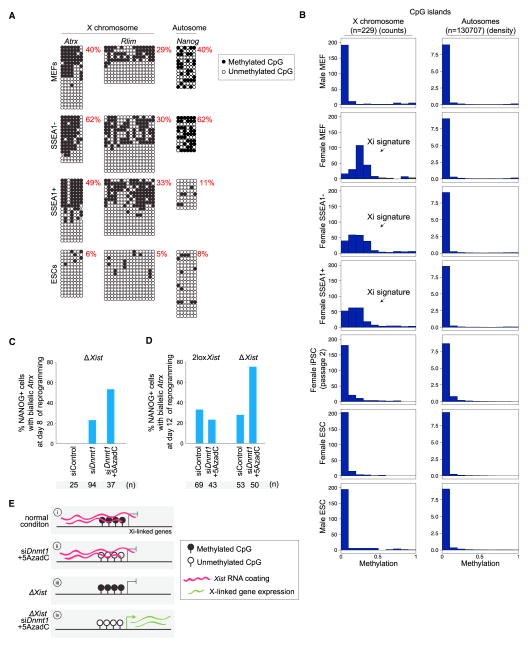
High Persistence of Xi DNA Methylation during Reprogramming
We considered the possibility that DNA methylation could maintain the silent state of the Xi during reprogramming in the absence of Xist. DNA methylation at CpG islands of the Xi arises late in the sequence of epigenetic changes on the Xi during development (Gendrel et al., 2012). We examined the DNA methylation pattern of X-linked genes in SSEA1− and SSEA1+ subpopulations, isolated from day 9 reprogramming cultures, representing cell populations with different reprogramming capabilities (Figures 1L and 1M; Polo et al., 2012). Traditional bisulfite sequencing at promoters of the X-linked genes Atrx and Rlim demonstrated the presence of the hypermethylated Xi in female MEFs, as well as in SSEA1− and SSEA1+ subpopulations, but not in female ESCs (Figure 6A). In contrast, the Nanog promoter region, methylated at an intermediate level in MEFs and SSEA1− cells, displayed demethylation characteristic of pluripotent cells already in SSEA1+ cells. These findings suggested a differential persistence of the methylation mark between Nanog and Xi-linked genes.
Figure 6. Analysis of DNA Methylation on the X Chromosome during Reprogramming.
(A) Bisulfite PCR analysis of the promoter regions of the X-linked genes Atrx and Rlim and of Nanog in female MEFs, ESCs, and day 9 SSEA1−/+ reprogramming intermediates. Black circles indicate methylated CpGs, and open circles indicate unmethylated CpGs. The proportion of methylated CpGs is given. For MEFs and SSEA1+/− cells, hemimethylation represents Xi methylation.
(B) Histograms showing the distribution of methylation levels across CpG islands on the X chromosome and autosomes in indicated cell types based on RRBS data (n, number of CpG islands). The arrow indicates the Xi-specific DNA methylation signature.
(C) Proportion of NANOG+ cells with biallelic Atrx expression based on immunoFISH analysis at day 8 of reprogramming with 2lox/2lox Xist MEFs in which Xist was deleted by activation of the dox-inducible Cre-recombinase, and siControl, siDnmt1, and siDnmt1 plus 5AzadC, respectively, the latter were added at day 5. All NANOG+ cells present in the culture were counted (n).
(D) Similar to (C), except that Xist deletion was performed only in half of the reprogramming culture, and siControl or siDnmt1+5AzadC were applied on day 5 and day 8. At day 12, all NANOG+ cells present in the culture were assessed for biallelic Atrx expression.
(E) Summary of the role of Xist RNA and DNA methylation in the control of gene silencing on the Xi during reprogramming.
See also Figure S5.
To determine the DNA methylation status along the entire X chromosome, we employed reduced representative bisulfite sequencing (Meissner et al., 2008), which provides genome-scale single-base-resolution maps of DNA methylation. For this analysis, we additionally included early passage female iPSCs, as well as male MEFs and male ESCs for comparison. CpG islands on the Xa in male cells were hypomethylated to the same degree as those on autosomes in male or female cells (Figure 6B). By contrast, in female MEFs, CpG islands across the X chromosome showed an average of 20%–50% methylation, which is consistent with an Xi-specific methylation signature. This pattern was present in both SSEA1− and SSEA1+ subpopulations but absent in early-passage female iPSCs and female ESCs (Figure 6B). A similar result was obtained for CpG-island shores, high and low CpG-containing promoters (Figure S5A). These results indicate that DNA methylation established on the Xi late during differentiation (Gendrel et al., 2012) is preserved on the Xi until very late in reprogramming. The persistence of Xi-DNA methylation in reprogramming is Xist independent (Figures S5B and S5C), supporting the hypothesis that this Xi mark could maintain the silent state of the Xi until late in reprogramming, even when Xist is experimentally deleted. Because Xi-DNA methylation is not yet reversed when Nanog (Figure 6A) and many other ESC-specific enhancer elements have already become demethylated (V.P., R.K., C. Chronis, A.M., and K.P., unpublished data), we conclude that this Xi mark has a remarkable stability during reprogramming.
Xist RNA and DNA Methylation Both Maintain Xi Silencing Throughout Reprogramming
We determined whether XCR is mechanistically linked to the loss of both Xist and DNA methylation by deleting Xist and inhibiting Dnmt1, the maintenance DNA methyltransferase, during the late phase of reprogramming. The block of Dnmt1 activity and loss of DNA methylation within Xi-linked CpG islands was confirmed (Figures S5D and S5E). In reprogramming cultures in which Xist on the Xi was experimentally deleted, 23% of NANOG+ cells displayed biallelic expression of the X-linked gene Atrx upon brief Dnmt1 depletion, and this proportion was more than doubled when Dnmt1 knockdown was combined with 5AzadC treatment to enhance DNA demethylation (Figures 6C and S5E). XCR was not detected at this time point in NANOG+ cells in control reprogramming cultures (Figure 6C). Importantly, we found that inhibition of DNA methylation only enhances XCR in NANOG+ cells in the absence of Xist, but not in its presence (Figure 6D). This finding also excludes the possibility that the acceleration of XCR upon inhibition of DNA methylation is simply due to faster overall reprogramming. We conclude that Xist RNA is able to maintain the Xi when DNA methylation is reduced and that DNA methylation is sufficient for Xi maintenance in the absence of Xist. Therefore, both DNA demethylation and Xist silencing are required for XCR late in reprogramming and occur downstream to the reactivation and demethylation of Nanog (Figure 6E).
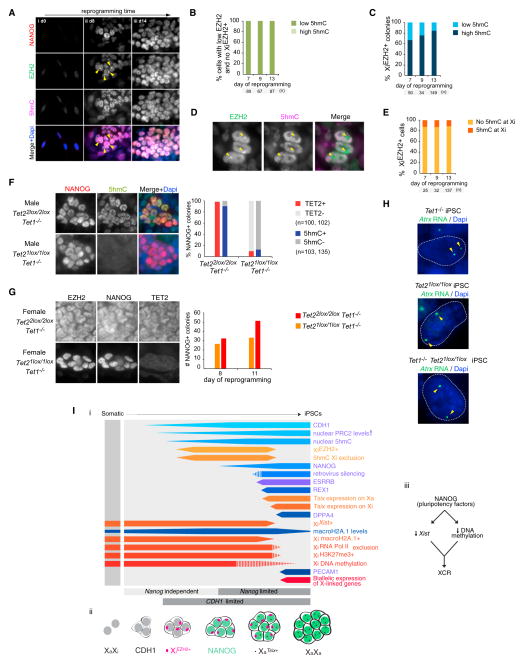
Tet1, Tet2, and High Global 5hmC Levels Are Dispensable for XCR
Given the implication of conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in DNA demethylation processes (Wu and Zhang, 2014), we defined the Xi-specific and global dynamics of 5hmC during reprogramming. We found a striking increase in the global 5hmC level, specifically in those cells that globally upregulate EZH2 and gain XiEZH2+ (Figures 7A–7C). Global upregulation of 5hmC also took place in male reprogramming cultures and in the absence of Vitamin C (Figure S6A), indicating that this epigenetic remodeling event is intrinsic to reprogramming across different culture conditions and sex chromosome content. Despite overall elevated 5hmC levels, this mark was depleted on the Xi in XiEZH2+ cells (Figures 7D and 7E). Thus, during reprogramming, cells start off with low levels of 5hmC and EZH2 and then increase 5hmC and EZH2 downstream of MET, with PRC2 accumulating on the Xi and 5hmC remaining excluded from the Xi, all of which precedes the reactivation of pluripotency genes and transition to a pluripotent state with XCR devoid of XiPRC2+ and 5hmC Xi exclusion.
Figure 7. Tet1 and Tet2 and Global 5hmC Are Dispensable for XCR.
(A) Representative immunostaining images for different patterns of NANOG (red in merge), EZH2 (green), and 5hmC (magenta) arising at indicated days of reprogramming. Arrowheads indicate XiEZH2+.
(B) Proportion of cells with low nuclear EZH2 levels and no XiEZH2+ that display either low or high nuclear levels of 5hmC at indicated time points.
(C) Proportion of XiEZH2+ colonies that display either low or high 5hmC at indicated time points.
(D) Representative immunostaining image for EZH2 (green in merge) and 5hmC (magenta) in the XiEZH2+ reprogramming intermediate. 5hmC Xi exclusion is indicated by arrowheads.
(E) Proportion of XiEZH2+ cells that display 5hmC Xi exclusion (Xi5hmC−) at indicated time points.
(F) Representative immunostaining images of male Tet22lox/2loxTet1−/− reprogramming cultures infected with Ad5 (top) or AdCre (bottom) adenoviruses, stained for NANOG (red in merge) and 5hmC (green) at day 14 of reprogramming. AdCre induces Tet2 deletion (Tet21lox/1loxTet1−/−). The graph gives the proportion of NANOG+ colonies positive for 5hmC and TET2, respectively, at day 14 based on immunostaining. The absence of the TET2 signal in NANOG+ cells confirms effective Tet2 deletion. Loss of both Tet1 and Tet2 leads to loss of the 5hmC immunostaining signal (loss of global 5hmC).
(G) As in (F), except for female Tet22lox/2loxTet1−/− and Tet21lox/1loxTet1−/− reprogramming cultures, immunostained for EZH2, NANOG, and TET2. The number of NANOG+ colonies at indicated time points in these cultures is given in the graph.
(H) RNA FISH for Atrx nascent transcription on female Tet1−/−, Tet21lox/1lox and Tet21lox/1loxTet1−/− iPSCs. Arrowheads indicate the biallelic Atrx signal.
(I) Stages of XCR and somatic cell reprogramming to induced pluripotency. Our view of the stages leading to XCR and the induction of pluripotency, shown as described in Figures 1I and 2E. Female-specific events are shown in orange/red, and those occurring in both female and male cells are shown in blue. With the exception of retroviral silencing in male reprogramming, all results presented are based on experimental evidence in both female and male reprogramming.
See also Figures S6 and S7.
Given the dynamics of 5hmC, we tested the requirement of Tet1 and Tet2 for XCR using female MEFs carrying Tet1 knockout (Tet1−/−) and Tet2 conditional (Tet22lox/2lox) alleles, in which genetic deletion of Tet2 could be induced by addition of Cre-expressing adenoviruses (AdCre) (Figure S6B). Strikingly, genetic ablation of both Tet1 and Tet2, but not that of either Tet1 or Tet2 individually, prevented the global induction of 5hmC during reprogramming (Figures 7F, S6C, and S6D). Importantly, Tet1/Tet2 double knockout and absence of global 5hmC did not affect the upregulation of nuclear EZH2 and occurrence of XiEZH2+, nor the efficiency with which NANOG+ colonies were obtained, nor the activation of the late pluripotency marker PE-CAM1 and XCR (Figures 7G, 7H, and S6E–S6G). Reprogramming experiments with ablation of either Tet1 or Tet2 resulted in similar results, and the resulting iPSCs contributed to chimeras and were effectively demethylated at cis-regulatory regions of the Pou5f1 (Oct4) gene and Xi-linked promoters (Figures S6H–S6K and S7A–S7D). Additional shRNA-mediated depletion of Tet3 transcripts in pre-iPSCs also carrying the Tet1 and Tet2 genetic deletion still enabled XCR (Figures S7E–S7K). We conclude that Tet1 and Tet2 and the global increase in 5hmC nuclear levels are dispensable for XCR and the transition through the reprogramming hierarchy that we have established.
DISCUSSION
A dramatic reorganization of the epigenome occurs during the reprogramming of somatic cells to iPSCs. Our findings demonstrate that changes in global and Xi-specific chromatin states, noncoding RNA expression, and pluripotency-associated factor expression are highly reproducible and reveal the existence of a multitude of epigenetic steps that occur in a defined sequence throughout the reprogramming process (Figure 7I, i). For instance, focusing only on XiEZH2+ dynamics relative to CDH1 and NANOG expression, transition through four steps can be defined: (1) CDH1+/XiEZH2−/NANOG−; (2) CDH1+/XiEZH2+/NANOG−; (3) CDH1+/XiEZH2+/NANOG+; and (4) CDH1+/XiEZH2−/NANOG+ (Figure 7I, ii). These stages are likely going to be generally applicable to female cells and not cell-type specific as the Xi enrichment of PRC2 is also expected to occur in epithelial cells at an intermediate step of reprogramming. The relocalization of EZH2 (PRC2) and its cofactor JARID2 to the Xi, along with global increases in PRC2, macroH2A1, and 5hmC downstream of MET and upstream of NANOG expression, indicate that major changes in chromatin structure take place in cells undergoing reprogramming, before pluripotency is reached.
Compared to the establishment of Xi features during differentiation, we find that these have different propensities for reversal during reprogramming. Whereas XiEZH2+ and the activation of Tsix from the Xa and Xi take place in an apparent reverse order of the developmental XCI program, macroH2A1 and DNA methylation, both associated with the differentiated state and resistance to reprogramming (Pasque et al., 2012; Mikkelsen et al., 2008), are reversed on the Xi only very late in reprogramming, despite the fact that they are established on the Xi late in differentiation. Similarly, the activation of Xi-linked genes during reprogramming only occurs after Xist RNA loss, even though Xist RNA coating precedes silencing of the X chromosome during differentiation. Thus, based on a subset of Xi hallmarks, reprogramming proceeds in a manner that would be expected for developmental reversal, indicating progressive dedifferentiation. However, based on another set of marks, cells undergoing reprogramming remain epigenetically distinct from those traversing differentiation. Thus, during reprogramming, certain epigenetic features follow the differentiation state of the cell, whereas others are uncoupled from this regulation.
During differentiation, Xist is required to initiate XCI, and its experimental silencing in the first days after the establishment of the Xi leads to immediate reactivation of the X chromosome (Wutz and Jaenisch, 2000). However, later in differentiation, Xist can be deleted from the Xi without dramatically affecting the stability of the silent chromosome, which, at this point, is thought to be maintained through the action of multiple repressive chromatin pathways (Csankovszki et al., 2001). In contrast to a recent study that used a different system to reduce Xist expression (Chen et al., 2014), we made the surprising observation that Xist ablation on the Xi does not alter the kinetics of XCR during reprogramming. Our result indicates that the extended, several-day-long window of Xist dependency of silencing seen during the initiation of XCI in differentiation (Wutz and Jaenisch, 2000) is not re-established during reprogramming. Silencing of the Xi in the absence of Xist remains stable until the very end of reprogramming because it is functionally maintained by DNA methylation, which has an extraordinarily high persistence on the Xi during reprogramming and is only erased after the pluripotency factor Nanog is already demethylated. Notably, the experimental interference with DNA methylation alone does not lead to precocious XCR, indicating that Xist RNA also actively contributes to the silencing of the Xi late in reprogramming. In agreement with this, we also discovered that forced Xist expression prevents XCR during reprogramming. Therefore, XCR requires loss of both Xist RNA and DNA methylation at the end of the reprogramming process. Because both events take place only late during hierarchical pluripotency-associated gene activation, these ensure that XCR only occurs in cells that establish faithful pluripotency (Figure 7I, iii). Accordingly, a block early in the pluripotency hierarchy blocks XCR (Figure S4). Notably, the pluripotency factor PRDM14 has been reported to be required for XCR during reprogramming (Payer et al., 2013), but whether Prdm14 deletion blocks the reprogramming process at a stage prior to XCR needs to be resolved to understand its specific role in XCR.
The generation of 5hmC by Tet proteins has been suggested to play important roles during reprogramming to iPSCs and potentially mediates DNA demethylation through active and passive mechanisms (Hu et al., 2014; Wu and Zhang, 2014). Our findings reveal that Tet1, Tet2, and global 5hmC are dispensable for XCR. This raises the question of which DNA demethylation pathway, either active or passive, leads to XCR. We posit that loss of DNA methylation on the Xi during reprogramming likely occurs in a synchronous manner across the entire chromosome, requiring a mechanism that can act across a large number of CpG islands in a relatively short time frame. We expect that the characterization of the Xi DNA demethylation event will yield critical insights into mechanisms that control the final stages of reprogramming.
Notably, during reprogramming by SCNT, developmental defects are caused by misregulation of the XCI system, particularly due to ectopic Xist expression from the Xa (Inoue et al., 2010). By contrast, our data indicate that, during reprogramming to iPSCs, the reactivation of the Xi (Figure 4) or ectopic XCI on the Xa (Figures 5B and S7L) does not seem to act as barriers to reprogramming, pointing to mechanistic differences between transcription factor- and oocyte-induced somatic cell reprogramming. Furthermore, in contrast to our findings in iPSC reprogramming, the activation of X-linked genes during XCR in preimplantation development occurs in the presence of Xist (Williams et al., 2011).
Importantly, our study defines many sequential reprogramming steps, extending previous reports based on gene expression studies that identified a limited number of reprogramming stages (Parchem et al., 2014; O’Malley et al., 2013; Buganim et al., 2012; Polo et al., 2012). We propose that the global epigenetic state of cells as they reprogram to iPSCs, and that of the Xi, is less variable than transcriptional states. However, our data do not exclude stochastic gene expression differences in cells with the same epigenetic state. One advantage of our analyses is that the stage of any cell in a reprogramming culture can be easily assessed, taking into account criteria such as colony growth and positional information of cells, as well as protein levels and sub-cellular localization. Notably, although most of our analyses focused on the female-specific XCR process, our work led to the identification of many reprogramming stages that are also applicable to male reprogramming (Figure 7I, i). For example, the global increase in EZH2 and 5hmC levels that occurs in both female and male cells was uncovered during our analysis of the localization of these marks on the Xi in female cells.
Our study provides an easily applicable platform for assaying the effects of interference with intrinsic and extrinsic factors on the stages of reprogramming and on the transitions between them. Additionally, we anticipate that the analysis of the transcriptome and other epigenetic features such as DNA methylation in the multiple reprogramming intermediates that we have identified will reveal insights into reprogramming. Another task ahead remains the continuous imaging of the transitions between the reprogramming steps identified here to quantitatively model the reprogramming process.
In conclusion, our comprehensive study yields insights into XCR and provides unprecedented details on the epigenetic dynamics of somatic cell reprogramming to induced pluripotency, establishing a valuable foundation exploitable for many applications, including staging of reprogramming cultures, isolation of intermediates, and to uncover mechanistically how cells transition toward pluripotency.
EXPERIMENTAL PROCEDURES
Reprogramming Experiments and Time Courses
Reprogramming was carried out using cells derived from reprogrammable mice or directly infected with retroviruses encoding Oct4, Sox2, and Klf4, as described in detail in the Extended Experimental Procedures. For time course analyses, reprogramming cultures on 22 × 22 mm gelatinized glass coverslips were fixed every other day, usually from day 6 to day 14, before carrying out immunostaining and RNA FISH analyses.
Flow Cytometry
Flow cytometry for SSEA1 and CDH1 was done starting from large reprogramming cultures using methods previously reported (Stadtfeld et al., 2008) with modifications described in the Extended Experimental Procedures.
Immunostaining and RNA FISH
Immunostainings and RNA FISH were carried out on 22 × 22 coverslips obtained from reprogramming cultures and as described previously (Maherali et al., 2007). Details are given in the Extended Experimental Procedures.
Bisulfite Analysis
Bisulfite-converted DNA was subjected to RRBS or analyzed by PCR as detailed in Table S1. Details are given in the Extended Experimental Procedures.
Data Analyses
See the Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to Drs. Nakamura, Xu, Eng, Berk, Silva, Pei, and Besser for providing reagents; Dr. Chronis for help with bioinformatics; Dr. Ch’ng and A. Sahakyan for help with microscopy and western blot analysis; F. Codrea and J. Scholes at the Broad Stem Cell Center FACS Core for help; and Dr. Lowry and members of the Plath lab for advice and critical reading of the manuscript. V.P., S.P., and A.S.D. are supported by CIRM Training Grants TG2-01169 and TB1-01183; J.T. is supported by a fellowship of the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research; A.M. is a NYSCF Robertson Investigator and is supported by P01GM099117; K.P. is supported by the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research and NIH P01 GM099134 and CIRM (RN1-00564); D.C. is supported by the NIH Ruth L Kirschstein National Research Service Award (GM007185); G.B. is supported by the Whitcome Pre-doctoral Training Program and a UCLA Dissertation Year Fellowship; M.U. is supported by R25GM055052; R.H. is supported by an NIH Training Grant (5T32AI060567-07) and the UCLA Graduate Division Dissertation Year Fellowship; and R.S. is supported by the UCLA Graduate Division Dissertation Year Fellowship.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the RRBS data reported in this paper is GSE58109.
AUTHOR CONTRIBUTIONS
V.P., J.T., and K.P. designed experiments; V.P., J.T., R.K., M.U., A.S.D., D.C., B.P., S.P., R.M., and K.P. performed experiments; V.P., J.T., R.K., M.U., A.S.D., D.C., B.P., G.B., S.P., R.M., A.M., and K.P. analyzed data; R.H. helped with experiments; R.S., T.S., and T.T. generated reagents; and V.P. and K.P. wrote the manuscript with edits from J.T., R.K., B.P., and A.M.
Supplemental Information includes Extended Experimental Procedures, seven figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.11.040.
References
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Gribnau J. X chromosome inactivation and embryonic stem cells. Adv Exp Med Biol. 2010;695:132–154. doi: 10.1007/978-1-4419-7037-4_10. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AC, Davis-Dusenbery BN, Koszka K, Ichida JK, Eggan K. Nanog-independent reprogramming to iPSCs with canonical factors. Stem Cell Reports. 2014;2:119–126. doi: 10.1016/j.stemcr.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Gao S, He W, Kou X, Zhao Y, Wang H, Gao S. Xist repression shows time-dependent effects on the reprogramming of female somatic cells to induced pluripotent stem cells. Stem Cells. 2014;32:2642–2656. doi: 10.1002/stem.1775. [DOI] [PubMed] [Google Scholar]
- Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Hochedlinger K, Rideout W, 3rd, Yanagimachi R, Jaenisch R. X-Chromosome inactivation in cloned mouse embryos. Science. 2000;290:1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, Tang YA, Huntley D, Montana G, Taylor S, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell. 2012;23:265–279. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, Tomlinson SR, Linnarsson S, Kaji K. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature. 2013;499:88–91. doi: 10.1038/nature12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem RJ, Ye J, Judson RL, LaRussa MF, Krishnakumar R, Blelloch A, Oldham MC, Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V, Radzisheuskaya A, Gillich A, Halley-Stott RP, Panamarova M, Zernicka-Goetz M, Surani MA, Silva JCR. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J Cell Sci. 2012;125:6094–6104. doi: 10.1242/jcs.113019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Rosenberg M, Yamaji M, Yabuta Y, Koyanagi-Aoi M, Hayashi K, Yamanaka S, Saitou M, Lee JT. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol Cell. 2013;52:805–818. doi: 10.1016/j.molcel.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Schwarz BA, Bar-Nur O, Silva JCR, Hochedlinger K. Nanog is dispensable for the generation of induced pluripotent stem cells. Curr Biol. 2014;24:347–350. doi: 10.1016/j.cub.2013.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AHFM, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Williams LH, Kalantry S, Starmer J, Magnuson T. Transcription precedes loss of Xist coating and depletion of H3K27me3 during X-chromosome reprogramming in the mouse inner cell mass. Development. 2011;138:2049–2057. doi: 10.1242/dev.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.