Abstract
Aims
To check the usefulness of a standardized protocol of PTEN FISH in 31 endometrial carcinomas (ECs) in comparison with SNP array (SNPA), multiplex ligation-dependent probe amplification (MLPA), and immunohistochemistry.
Methods and results
Fluorescence in-situ hybridization analysis showed two PTEN copies in 17 cases, three copies in nine cases, hemizygous deletion in two cases, and diverse cell populations with different PTEN copy number in three cases. A good correlation was seen between FISH and SNPA, particularly in cases with three copies. FISH identified two cases with entire deletion of chromosome 10, but did not identify a focal deletion of PTEN. Five cases with PTEN deletion and duplication of the second allele by SNPA were interpreted as normal by FISH. Concordance between FISH and MLPA was seen in 15 cases with two copies, and in two cases with PTEN deletion. Six cases were interpreted as amplified by MLPA, but showed polyploidy by FISH. FISH was superior to SNPA and MLPA in assessing the tumours with diverse cell populations with different PTEN copies.
Conclusions
The results show good concordance between FISH, SNPA and MLPA. SNPA was superior in tumours with deletion of one copy and duplication of the second allele. FISH was superior in assessing tumour heterogeneity.
Keywords: deletions, fluorescence in-situ hybridization, polysomy, preanalytical variables, protocol, PTEN
Introduction
PTEN (phosphatase and tensin homologue deleted on chromosome 10) has been identified as a tumour suppressor gene,1 and is mutated in a large number of cancers.2–10 It is rather than cancers, it is located at 10q23.31. PTEN is a phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase containing a tensin-like domain as well as a catalytic domain similar to that of the dual-specificity protein tyrosine phosphatases.11 Unlike most of the protein tyrosine phosphatases, this protein preferentially dephosphorylates phosphoinositide substrates.12 It negatively regulates the PI3K–Akt signalling pathway.13
Somatic alterations of PTEN, including point mutations, insertions, and deletions, occur throughout the whole gene in a significant proportion of sporadic cancers. In some cases, PTEN deletion occurs in the setting of large chromosomal losses involving the whole of chromosome 10.14 In other instances, deletions are restricted to the entire PTEN gene, or to a few exons.15 Loss of heterozygosity (LOH) at 10q23 occurs in 40% of endometrial carcinomas (ECs).16 Deletions may be hemizygous or homozygous. PTEN being a tumour suppressor gene, two hits are common for PTEN loss of function, and deletion of PTEN is commonly the second hit. However, more recently, it has become clear that one alteration may be sufficient for PTEN inactivation.17,18 Interestingly, when a tumour has chromosomal instability, three copies of PTEN can be seen, as a result of polysomy of chromosome 10.19 The biological significance of chromosome 10 polysomy, and the consequences for PTEN levels, have not been fully elucidated.
In some types of cancer, PTEN alteration is an early event, being involved in the initial steps of the process of neoplastic development.20 In these cases, alterations of PTEN are seen to be homogeneously distributed in the tumours. However, in other cases, PTEN abnormalities occur in the process of tumour progression,8,21 and PTEN alterations can be detected in specific regions of the tumour. The heterogeneous involvement of PTEN in a tumour may be demonstrated by immunohistochemistry.22 However, decreased PTEN expression does not necessarily reflect the presence of LOH at 10q23, as it may result from PTEN point mutations, and also from promoter hypermethylation, transcriptional repression, microRNA regulation, disruption of competitive endogenous RNA networks, or post-translational modification.19,23
Fluorescence in-situ hybridization (FISH) may be used to detect cytogenetic aberrations,24 such as deletions,25 duplications,26 amplifications27 and structural abnormalities of specific genes, loci, or chromosomal sequences.28 The regions assessed with FISH are typically larger than those studied with polymerase chain reaction (PCR), but smaller than those visualized microscopically with standard cytogenetics. Studies have demonstrated that FISH analysis of PTEN can identify groups of tumours with different prognoses among prostatic carcinomas28 and rectal carcinomas.29
In the present study, we developed a standardized protocol for PTEN FISH to detect PTEN copy number alterations (see Supporting Information) in a series of ECs in comparison with results obtained by single nucleotide polymorphism array (SNPA) and multiplex ligation-dependent probe amplification (MLPA). We used the fast IQ hybridization buffer technology (IQFISH; Dako, Glostrup, Denmark).30 We assessed the role of FISH in evaluating the heterogeneous presence of PTEN copy number alterations in the tumours. Finally, we compared the molecular results with the levels of PTEN protein determined by immunohistochemistry, in an attempt to correlate PTEN copy number alterations with PTEN loss.
Materials and methods
Case Selection
A series of 31 ECs were studied. Formalin-fixed paraffin-embedded (FFPE) blocks were available for FISH and immunohistochemistry. Matched frozen tissue samples were selected for SNPA and MLPA. Haematoxylin and eosin (H&E) sections were obtained to confirm that frozen samples corresponded to the same areas as the FFPE material. The tumours were obtained from the files of Hospital Universitari Arnau de Vilanova, Lleida, Spain (HUAV). The local Ethics Committee approved the study. Informed consent was obtained from each patient. Pathological features of the tumours (histological type, grade, and stage) are summarized in Table1.
Table 1.
Summary of pathological features of the tumours and analysis by FISH
| No | PTEN/CEN10 ratio | % | FISH result | Histological type | Grade | Stage |
|---|---|---|---|---|---|---|
| 1 | 0.97 | 72 | Polysomy (three copies) | SC | III | IA |
| 2 | 1.02 | 76 | Normal (two copies) | EEC | II | IB |
| 3 | 1.13 | 81 | Normal (two copies) | EEC | II | IB |
| 4 | 0.84 | 77 | Normal (two copies) | EEC | I | IA |
| 5 | 1.1 | 86 | Normal (two copies) | EEC | I | IA |
| 6 | 1.13 | 73 | Polysomy (three copies) | EEC | II | IB |
| 7 | 1.05 | 79 | Normal (two copies) | EEC | II | IA |
| 8 | 0.96 | 71 | Polysomy (three copies) | SC | III | IB |
| 9 | 0.91 | 83 | Normal (two copies) | EEC | III | IB |
| 10 | Clone 1: 0.87 Clone 2: 0.98 |
44 56 |
Heterogeneous (C1, polysomy; C2, normal) |
SC | III | IA |
| 11 | 0.93 | 74 | Normal (two copies) | EEC | II | IB |
| 12 | 1.02 | 85 | Polysomy (three copies) | EEC | II | IB |
| 13 | 1.05 | 76 | Normal (two copies) | EEC | II | IA |
| 14 | 0.98 | 78 | Normal (two copies) | EEC | II | IA |
| 15 | Clone 1: 0.66 Clone 2: 1.10 |
42 58 |
Heterogeneous (C1, hemizygous deletion; C2, normal) |
EEC | I | IA |
| 16 | 1.1 | 74 | Polysomy (three copies) | EEC | II | IA |
| 17 | 1.12 | 79 | Normal (two copies) | EEC | III | IB |
| 18 | 0.95 | 71 | Normal (two copies) | EEC | III | IB |
| 19 | 0.63 | 81 | Hemizygous deletion (one copy) | EEC | III | IA |
| 20 | 1.08 | 73 | Polysomy (three copies) | EEC | II | IB |
| 21 | 0.95 | 77 | Normal (two copies) | EEC | III | IB |
| 22 | Clone 1: 1.06 Clone 2: 0.89 |
40 60 |
Heterogeneous (C1, polysomy; C2, normal) |
EEC | II | IA |
| 23 | 0.94 | 83 | Normal (two copies) | SC | III | IB |
| 24 | 0.98 | 81 | Normal (two copies) | EEC | I | IA |
| 25 | 1.12 | 74 | Polysomy (three copies) | EEC | II | IB |
| 26 | 0.56 | 77 | Hemizygous deletion (one copy) | EEC | III | II |
| 27 | 1.04 | 78 | Normal (two copies) | EEC | III | II |
| 28 | 0.92 | 75 | Normal (two copies) | EEC | II | IA |
| 29 | 1.13 | 72 | Polysomy (three copies) | EEC | III | IB |
| 30 | 1.1 | 74 | Normal (two copies) | EEC | III | IB |
| 31 | 0.97 | 82 | Polysomy (three copies) | EEC | I | IB |
EEC, Endometrioid carcinoma; SC, serous carcinoma.
For evaluation of preanalytical variables, two additional surgical specimens were used, obtained from the surgical pathology files of HUAV. They corresponded to ECs removed by hysterectomy and bilateral salpingoophorectomy. Surgical specimens were obtained from the operating room, immediately after resection. Cases were recruited because the size of the tumours allowed the selection of different small fragments to test the high number of variables. Tissue subjected to preanalytical factor evaluation of PTEN FISH was obtained from the excess material after selection of tissue samples for appropriate diagnosis.
Fluorescence In-Situ Hybridization
Formalin-fixed paraffin-embedded blocks were sectioned at a thickness of 3 μm, dried for 1 h at 65°C, deparaffinized, and rehydrated. A pretreatment was performed in a microwave oven with a sensing capability for 10 min at 95°C, and the sections were then left for 15 min at room temperature (20–25°C). Pepsin digestion was performed using pepsin RTU (ready to use) at 37°C. The PTEN IQFISH probe used for hybridization was made as a mixture of a DNA-based Texas Red (TxR) probe specific for PTEN and a PNA-based, fluorescein isothiocyanate (FITC)-labelled reference probe specific for the centromeric region of chromosome 10 (CEN10); the probe was a gift from Dako. We used the fast IQ hybridization buffer technology (IQFISH), which utilizes ethylene carbonate rather than formamide as the DNA-destabilizing component. Consequently, the IQFISH buffer is non-toxic, and also reduces the time needed for hybridization to genomic targets. The total turn-around time for FISH staining with an IQFISH-based probe is therefore <4 h. The IQFISH probe was incubated in Dako Hybridizer in two steps: denaturation at 66°C for 10 min, and hybridization at 45°C for a maximum of 2 h. After hybridization, a stringent wash was performed for 10 min at 63°C with a stringent wash buffer. Finally, samples were dehydrated, and nuclei were stained with DAPI. All reagents (wash buffer, pretreatment buffer, stringent buffer, and pepsin RTU) were obtained from the Histology FISH Accessory Kit (K5799; Dako). Signals were counted under a microscope equipped with a ×100 objective, with a tetramethylrhodamine isothiocyanate (TRITC) philtre, an FITC philtre, and a TxR/FITC double philtre.
PTEN Fish Scoring
For evaluation of the results of FISH, several factors were taken into account to select the optimal tumour sections for PTEN FISH scoring, as rigorous quality criteria for suitability for FISH analysis. They included: preservation of tissue architecture, fixation artefacts, tumour cellularity within the section, and the absence of truncated nuclei and irregular glandular distortions. Areas selected for FISH evaluation were marked on an H&E-stained section that was directly adjacent to the section being used for FISH. Tissue areas were considered to be appropriate for FISH analysis after meeting rigorous criteria: (i) at least 85% of all nuclei in the target area were easily enumerable; (ii) the FISH signal was consistently greater than background intensity; (iii) areas where the borders of individual nuclei were not clearly identifiable were avoided; (iv) areas with excessive nuclear overlap were disregarded; (v) similar signal staining in the nuclei; and (vi) uniform DAPI staining. Staining of adjacent normal tissue was checked as reference. We required ∼100 tumour cell nuclei to be analysed for each sample, depending upon the density of cancer cells. In complex tissue where there was more than one type of aberration, each of them was scored individually (scoring 100 cells for each).
We established five different statuses for PTEN FISH. First, normal PTEN copy number was characterized by the presence of two green (CEN10) and two red (PTEN) signals in a minimum of 70% of nuclei, and a PTEN/CEN10 ratio (number of PTEN signals/number of CEN10 signals) between 0.8 and 1.2. Second, PTEN hemizygous deletion was characterized by the presence of two green (CEN10) signals and one red (PTEN) signal in a minimum of 70% of nuclei, and a PTEN/CEN10 ratio of <0.8. Third, PTEN gene homozygous deletion was characterized by the presence of two green (CEN10) signals and no red (PTEN) signal in a minimum of 70% of nuclei, and a PTEN/CEN10 ratio between 0 and 0.2. Fourth, whole chromosome 10 deletion was characterized by the presence of one green (CEN10) and one red (PTEN) signal in a minimum of 70% of nuclei, and a PTEN/CEN10 ratio between 0.8 and 1.2. Fifth, increased PTEN copy number (polysomy) was characterized by the presence of three green (CEN10) and red (PTEN) signals in a minimum of 70% of nuclei, and a PTEN/CEN10 ratio between 0.8 and 1.2.31 The criteria for scoring PTEN by FISH were obtained after taking into account the previous experience of the research team and data from the literature.32 The scoring system was validated with 25 cases of normal endometrial tissue. In all cases, the PTEN score fitted within the ranges of normal PTEN copy number.
SNP Array
The Human CytoSNP array (Illumina; San Diego, CA, USA) was used to detect 240 000 SNPs evenly distributed across the genome, according to the manufacturer's protocol. ASCAT (allele-specific copy number analysis of tumours) was used to dissect the allele-specific copy numbers of solid tumours. On the basis of the observed B-allele frequency and logR ratio of each SNP, chromosomal amplifications, deletions and copy-neutral loss of heterozygosity were detected at the genome-wide level.
Multiplex Ligation-Dependent Probe Amplification
We used SALSA MLPA probe-mix P225-C1 PTEN (MRC-Holland; Amsterdam, The Netherlands). This probe-mix contains at least two probes for each of the nine exons; the total number of PTEN probes included is 20. It also contains: 13 probes located elsewhere on chromosome 10 that can help to distinguish focal PTEN deletions in tumour-derived DNA samples from loss of the complete 10q arm or a chromosome 10 aneuploidy; two probes for copy number detection of the pseudogene PTENP1 (at 9p13.3); and 15 reference probes. A DNA extraction was used (DNeasy Blood and Tissue Kit; Qiagen, Hilden, Germany). The MLPA assay was performed according to the manufacturer's protocol (MLPA DNA protocol version MDP-v002). After fragment separation by capillary electrophoresis (Applied Biosystems; Foster City, CA, USA), an evaluation of the raw data was performed using GeneMapper. The results were always normalized and compared with strict external controls such as blood samples and non-tumour samples.
Immunohistochemistry
Formalin-fixed paraffin-embedded sections were evaluated by applying our optimized PTEN staining immunohistochemistry assay.33 Sections were obtained at a thickness of 3 μm, dried for 1 h at 65°C, deparaffinized, rehydrated, and subjected to target retrieval in the pretreatment module PT Link (Dako) at 95°C for 20 min in Target Retrieval Solution (pH 9) (Dako). Endogenous peroxidase was blocked using peroxidase-blocking reagent (Dako), and this was followed by incubation with primary PTEN antibody (clone 6H2.1; Dako). The incubation time for the primary antibody was 40 min. EnVision FLEX+/HRP (Dako) (20 + 15 min) was used to amplify signal, and detection was done using 3,3′-diaminobenzidine tetrahydrochloride (DAB) as chromogen (Dako). Slides were counterstained with haematoxylin, dehydrated, and mounted. PTEN expression was nuclear and cytoplasmic. Appropriate negative controls were also tested: these involved omission of the primary antibody, and the inclusion of tissue samples known to show PTEN silencing. We used a histological staining score (Hscore) that takes into consideration the intensity of the staining, and the percentage of positive cells, by applying the formula: Hscore = (1 × % light staining) + (2 × % moderate staining) + (3 × % strong staining). This gives a score ranging from 0 (no immune reaction) to 300 (maximal immunoreactivity).
Statistical Analysis
Fisher's exact test was used to correlate PTEN expression as detected by immunohistochemistry with PTEN copies as detected by SNPA, MLPA, and FISH.
Results
Fish Scoring
Results from the analysis of the 31 ECs are shown in Table1. Seventeen cases showed a normal PTEN copy number, nine showed three copies (polysomy 3), and two cases had hemizygous deletion of PTEN (one copy). The presence of more than one tumour cell population with differences regarding PTEN copy number was seen in three cases. In one case (case 10), 44% of tumour cells showed three copies of PTEN (polysomy), and 56% showed a normal copy number. In the second case (case 15), 42% of tumour cells showed hemizygous deletion of PTEN, and 58% showed a normal copy number. In the third case (case 22), 40% of tumour cells showed three copies of PTEN (polysomy), and 60% showed a normal copy number. Representative pictures are shown in Figures 1–7.
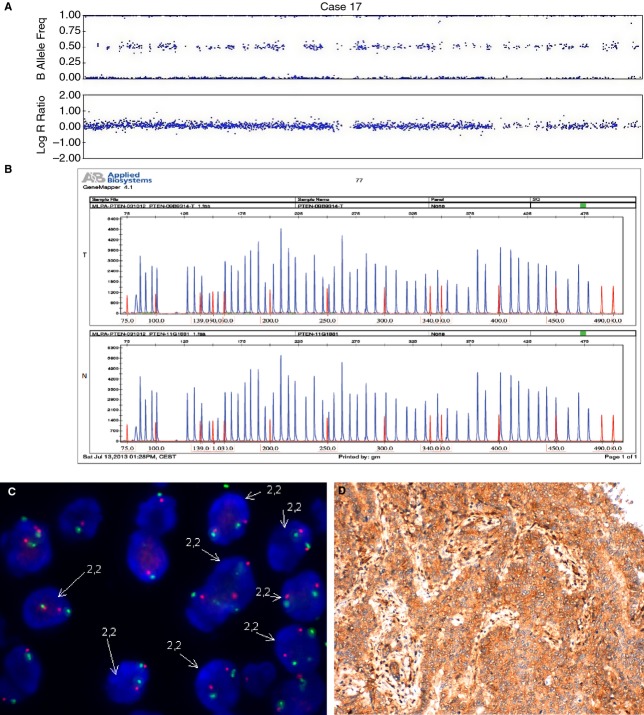
Figure 1.
An endometrial carcinoma with normal PTEN status (case 17). A, SNPA image showing normal PTEN. B, MLPA with normal PTEN in tumour tissue (T) as compared with normal tissue control (N). C, FISH image showing cells with two copies of PTEN. D, Positive immunostaining for PTEN.
Figure 7.
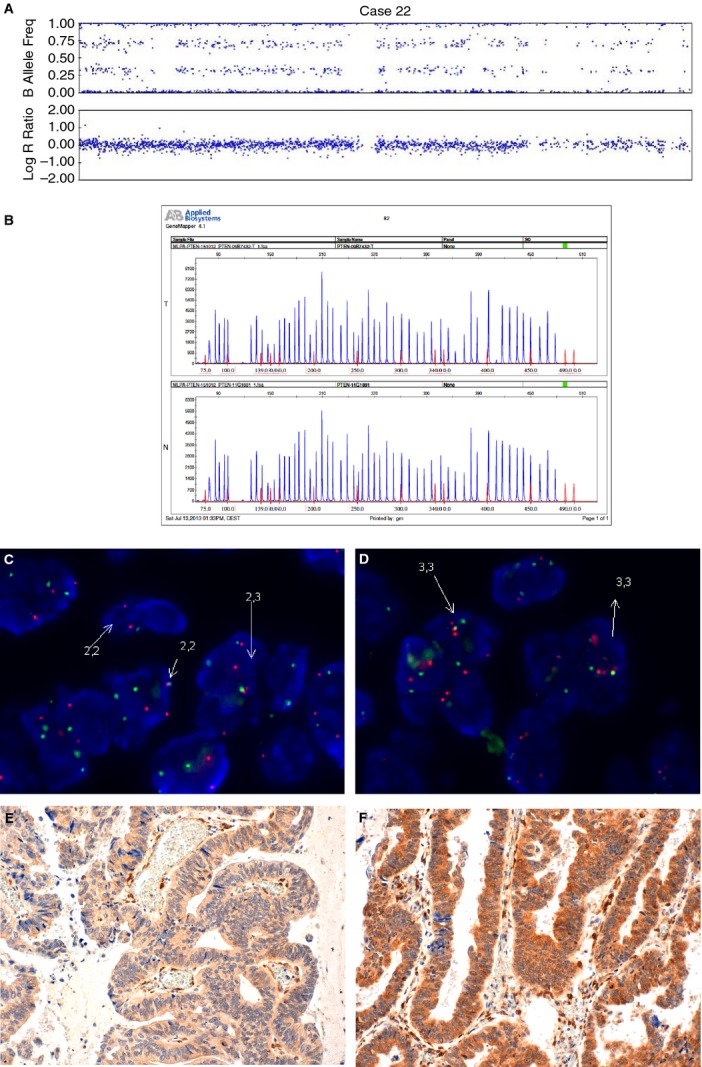
An example of an endometrial carcinoma with heterogeneous distribution of PTEN copy number (case 22). A, SNPA showing one allele of PTEN deleted and the second duplicated. B, MLPA with normal PTEN in both tumour tissue (T) and normal tissue control (N). C, D, FISH image showing a group of cells with two copies of PTEN (C) and a group of cells with three copies of PTEN (D). E, F, Immunohistochemistry shows cells with negative staining (E) and positive staining (F) for PTEN.
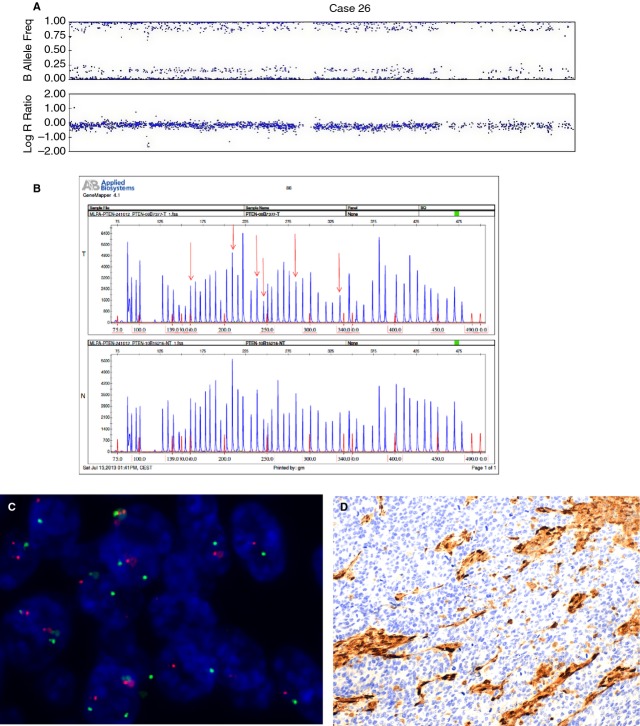
Figure 2.
An endometrial carcinoma with a PTEN deletion (case 26). A, SNPA image showing deletion of chromosome 10. B, MLPA with deleted PTEN; arrows indicate deleted areas in tumour tissue (T) as compared with normal tissue control (N). C, FISH image showing one copy of chromosome 10. D, Negative immunostaining for PTEN.
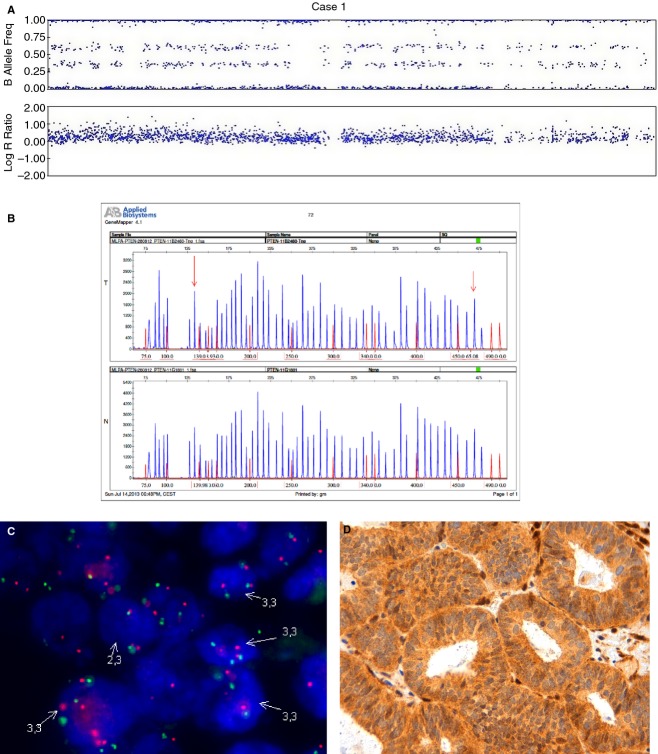
Figure 3.
An endometrial carcinoma with polysomy of chromosome 10 (case 1). A, SNPA image showing three copies of PTEN. B, MLPA with amplification of exons 1 and 9 in tumour tissue (T) as compared with normal tissue control (N). C, FISH image showing cells with three copies of chromosome 10. D, Positive immunostaining for PTEN.
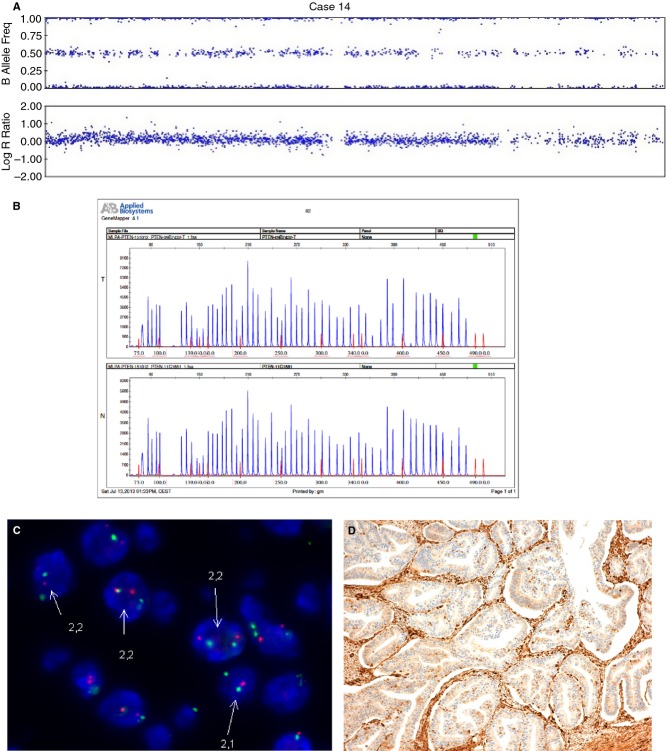
Figure 4.
An endometrial carcinoma with deletion of PTEN and duplication of the second allele (case 14). A, SNPA image showing one PTEN allele deleted and the second allele duplicated. B, MLPA with normal PTEN status in tumour tissue (T) as compared with normal tissue control (N). C, FISH image showing cells with two copies of PTEN. D, Negative immunostaining for PTEN.
Figure 6.
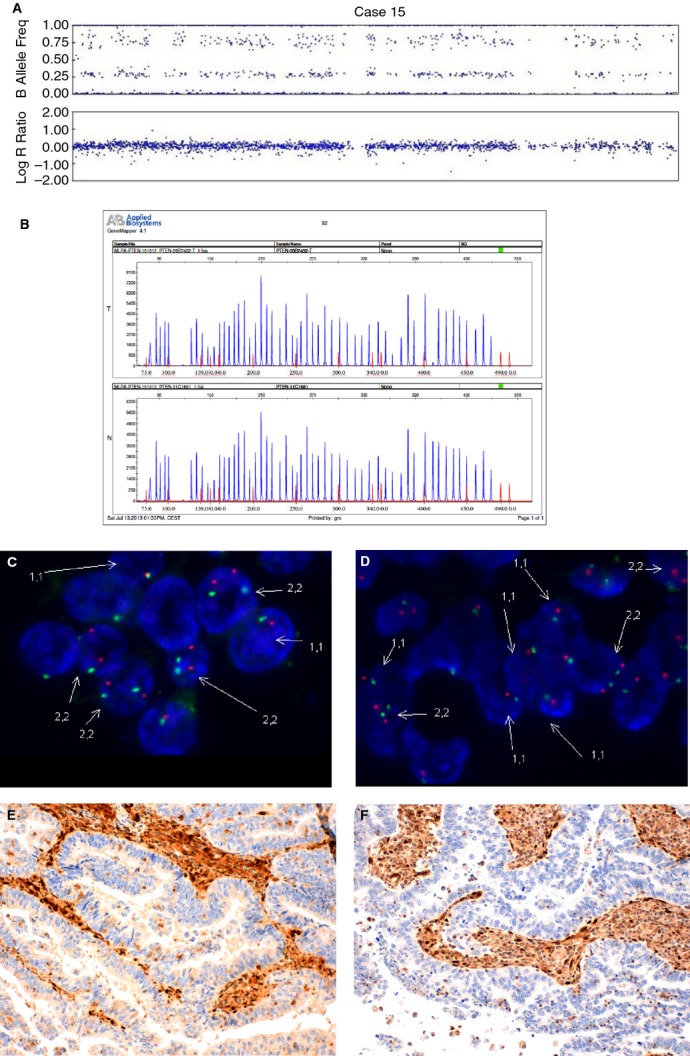
An example of an endometrial carcinoma with heterogeneous distribution of PTEN copy number (case 15). A, SNPA showing one allele of PTEN deleted and the second allele duplicated. B, MLPA with normal PTEN in both tumour tissue (T) and normal tissue control (N). C, D, FISH image showing a group of cells with two copies of PTEN (C) and a group of cells with complete deletion of chromosome 10 (D). E, F, Negative PTEN immunostaining in the two areas.
SNP Array
Table2 shows the correlation between FISH and SNPA after exclusion of the three cases that showed heterogeneous populations by FISH, which will be presented separately. A normal PTEN copy number was seen in 11 of 31 cases. Three copies of PTEN were identified in nine cases. An absolute correlation between FISH and SNPA was achieved in cases showing three copies of PTEN. Hemizygous deletion of the whole of chromosome 10 (one copy of chromosome 10) was seen in two cases. A hemizygous deletion restricted to PTEN was seen in one case. In five cases, SNP analysis showed hemizygous deletion of one PTEN allele with duplication of the second allele. Overall, deletion of PTEN was seen in eight of 31 cases (26%), although, in five of them, there was also duplication of the second allele. Fluorescence in-situ hybridization identified the two cases with deletion of the whole of chromosome 10. However, FISH did not identify the only case with focal deletion of PTEN. The five cases of PTEN deletion and duplication of the second allele were interpreted as normal by FISH.
Table 2.
Correlation between FISH and SNP array in 28 cases; three cases with heterogeneous alterations of PTEN are excluded
| SNP array | FISH | ||
|---|---|---|---|
| Normal (two copies) | Polysomy (three copies) | Deletion (one copy) | |
| Two copies | 11 | ||
| Three copies | 9 | ||
| One copy | 2 | ||
| Focal deletion | 1 | ||
| PTEN deletion plus duplication of the second allele | 5 | ||
Multiplex Ligation-Dependent Probe Amplification
Table3 shows the correlation between MLPA results and those obtained by FISH; and Table4 the correlation between MLPA results and those obtained by SNPA. In both Tables the three cases that showed heterogeneous populations by FISH have been excluded. A normal PTEN copy number was seen in 17 cases, four cases showed one copy of PTEN, six cases showed PTEN amplification, and one case showed a complex PTEN status (amplification and deletion). Fifteen cases had a concordant normal diploid pattern by FISH and MLPA, and two had two copies by MLPA and three copies by FISH. Two cases had a concordant pattern of PTEN deletion by FISH and MLPA, whereas results were discordant in two additional cases. Six cases were interpreted as amplified by MLPA, but showed polyploidy by FISH. One case with a complex MLPA pattern (amplification and deletion) was interpreted as normal by FISH.
Table 3.
Correlation between multiplex ligation-dependent probe amplification (MLPA) and FISH in 28 cases; three cases with heterogeneous alteration of PTEN by FISH are excluded
| MLPA | FISH | ||
|---|---|---|---|
| Normal (two copies) | Polysomy (three copies) | Deletion (one copy) | |
| Normal | 15 | 2 | |
| Deletion | 1 | 1 | 2 |
| Amplification | 6 | ||
| Amplification plus deletion | 1 | ||
Table 4.
Correlation between SNP array and multiplex ligation-dependent probe amplification (MLPA) in 28 cases; three cases with heterogeneous alteration of PTEN by FISH are excluded
| SNP array | MLPA | |||
|---|---|---|---|---|
| Normal | Deletion | Amplification | Amplification plus deletion | |
| Two copies | 11 | |||
| Three copies | 2 | 1 | 6 | |
| One copy | 2 | |||
| Focal deletion | 1 | |||
| Deletion plus duplication of the second allele | 4 | 1 | ||
A concordant normal pattern (two copies) was seen in 11 cases with SNPA and MLPA. Six cases showed amplification by MLPA and polysomy by SNPA, but discordant results were obtained in three additional cases, which were interpreted as polyploid by SNPA. Five cases showed deletion and duplication of the second allele by SNPA, and four of them were interpreted as normal by MLPA and one as having a PTEN deletion.
Immunohistochemistry
Tables 5, 6 and S1 show the results of analysis of the relationship between immunohistochemistry and the results obtained with the three molecular techniques (FISH, SNPA, and MLPA), after exclusion of the three cases that showed heterogeneous populations by FISH. Interestingly, tumours showing polysomy by FISH and SNPA (interpreted as amplified by MLPA) showed similar PTEN expression as tumours with a normal PTEN copy number (P = 0.68 and P = 1, respectively). In contrast, cases showing deletion of PTEN by FISH, SNPA or MLPA showed decreased expression of PTEN, even in the presence of duplication of the second allele, although statistical significance was not reached.
Table 5.
Correlation between FISH and immunohistochemistry (IHC) in 28 cases; three cases with heterogeneous alteration of PTEN are excluded
| IHC Hscore | FISH | ||
|---|---|---|---|
| Normal (two copies) | Polysomy (three copies) | Deletion (one copy) | |
| 0–40 | 9 | 4 | 2 |
| 41–100 | 4 | 4 | |
| 101–200 | 2 | ||
| 201–300 | 2 | 1 | |
Table 6.
Correlation between SNP array and immunohistochemistry (IHC) in 28 cases; three cases with heterogeneous alteration of PTEN are excluded
| IHC Hscore | SNP array | ||||
|---|---|---|---|---|---|
| Two copies | Three copies | One copy | Focal deletion | Deletion plus duplication of the second allele | |
| 0–40 | 6 | 4 | 2 | 3 | |
| 41–100 | 2 | 4 | 2 | ||
| 101–200 | 2 | ||||
| 201–300 | 1 | 1 | 1 | ||
Tumours with Heterogeneous PTEN Alterations
The three cases that showed a heterogeneous distribution of PTEN by FISH were interpreted separately (Table7; Figures 5–7). The first case (case 10) contained one cell population with three copies of chromosome 10 (44%) and a second population with a normal copy number (two copies) (56%). The case was interpreted as containing three copies by SNPA and containing two copies by MLPA. Interestingly, immunohistochemical expression of PTEN was heterogeneous in the tumour. Although there was not an absolute correlation, the areas with polysomy showed increased PTEN immunostaining in comparison with tumour areas with a normal PTEN copy number. The second case (case 15) contained a tumour cell population with a PTEN deletion (42%), and a population with a normal copy number (58%). The tumour was interpreted as having one allele deleted and duplication of a second allele by SNPA, and as having a normal copy number by MLPA. Immunohistochemical expression of PTEN was also homogeneously negative in the two components. The third case (case 22) contained one cell population with three copies of chromosome 10 (40%) and a second population with a normal copy number (two copies) (60%). The tumour was interpreted as having one allele deleted and duplication of a second allele by SNPA, and as having a normal copy number by MLPA. Immunohistochemical expression of PTEN was also heterogeneous.
Table 7.
Correlation between FISH, SNP array and multiplex ligation-dependent probe amplification (MLPA) in three cases with heterogeneous PTEN FISH patterns
| SNP array | FISH | MLPA | IHC staining | |
|---|---|---|---|---|
| Case 10 | Polysomy (three copies) | Polysomy (44%) Normal (56%) |
Normal | Heterogeneous |
| Case 15 | One allele deleted plus duplication of the second allele | Deleted (42%) Normal (58%) |
Normal | Negative |
| Case 22 | One allele deleted plus duplication of the second allele | Polysomy (40%) Normal (60%) |
Normal | Heterogeneous |
Figure 5.
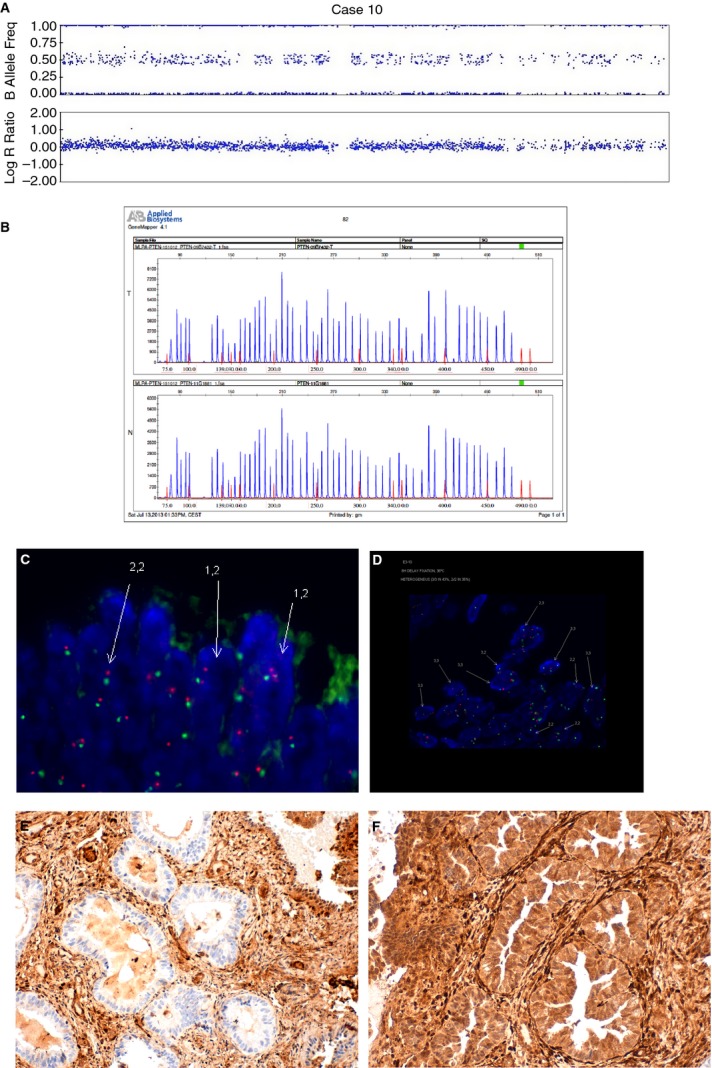
An example of an endometrial carcinoma with heterogeneous distribution of PTEN copy number (case 10). A, SNPA showing three copies of PTEN. B, MLPA with normal PTEN in both tumour tissue (T) and normal tissue control (N). C, D, FISH image showing a group of cells with two copies of PTEN (C) and a group of cells with three copies of PTEN (D). E, F, Immunohistochemistry shows negative staining for PTEN in some areas (E), and positive immunostaining in others (F).
Discussion
Tumour suppressor genes (TSGs) such as PTEN are important in tumorigenesis. Although inactivation of TSGs may take place at the transcriptional and protein levels (enhanced protein degradation, or decreased expression because of promoter hypermethylation), it usually results from partial loss of the gene, or even loss of the entire gene. According to Knudson,34 the inactivation of a TSG takes place in two steps. The first is usually a point mutation, or a small deletion, whereas the second typically consists of a large deletion of part of a chromosome that involves the gene. Although it has recently been demonstrated that monoallelic inactivation may be sufficient,35 demonstration of deletions is still quite informative regarding assessment of the role of a specific TSG in a particular type of tumour. Searching for specific areas of the genome where gene deletions (LOH) commonly occur in a determinate type of tumour is important, because it may lead to the recognition of TSGs that are involved in the development or progression of such a tumour. PTEN is among the most frequently lost or mutated TSGs, with a frequency of monoallelic mutations estimated at 50–80% in EC, glioblastoma, and prostate cancer, and at 30–50% in breast, colon and lung tumours.14 Complete loss of PTEN is observed at the highest frequencies in EC and glioblastoma.7 Detection of gene deletions not only helps in understanding the molecular mechanisms underlying the development of cancer, but also provides important information for disease diagnosis and prognosis.20,25 Identification of deletions (LOH) may be carried out by different techniques, including karyotyping,36 microsatellite polymerase chain reaction (PCR),37 comparative genomic hybridization,38 FISH,39 MLPA,40 and SNPA.41
Fluorescence in-situ hybridization is a good method for identifying deletions in tumour cells.25 It usually requires the identification of the deleted target gene with a locus-specific probe, in combination with preservation of the corresponding chromosomal centromere, with the appropriate centromeric probe. It has limited value in detecting small deletions.42 FISH analysis has some advantages over molecular analysis, particularly regarding correlation with morphology and assessment of tumour heterogeneity.43 However, one of the major limitations of detecting deletions in tissue sections by FISH is that part of the cell can be lost during the sectioning process, leading to artificially induced deletions in a significant percentage of nuclei.42 To decrease the false-positive rate, the literature recommends using centromeric probes as controls, as well as evaluation of a substantial number of cells.32 The usefulness of FISH in the evaluation of PTEN has been evaluated in carcinomas of the prostate,25 rectum,44 and stomach.45. In prostate cancer, PTEN alterations, as analysed by FISH, have been shown to have prognostic importance.25
In the MLPA technique, genomic DNA is hybridized in solution to probe sets, each of which consists of two halves. One half consists of a target-specific sequence (20–30 bp),40 and the other half also has a target-specific sequence at one end (25–43 bp) and a universal primer sequence at the other, but has a variable-length random fragment in between (19 and 370 bp) to generate the size differences in the probes that are necessary to allow electrophoretic resolution. The amounts of ligated probe produced will be proportional to the target copy number, and, after PCR amplification, the relative peak heights indicate deletion or duplication of the target sequence. MLPA has proven to be a highly sensitive and accurate tool for detecting copy number changes.
Single nucleotide polymorphism array enables simultaneous detection of a large number of DNA polymorphic loci in a simple way. Single nucleotide polymorphism array provides a high-throughput, high-resolution approach to genotypic analysis that includes assessment of LOH of chromosomal regions based on paired normal and tumour samples from the same patients.41 The application of informatics tools to this technique may allow the identification of LOH in many loci, and also the hierarchical clustering of cancers on the basis of shared LOH patterns. Resolution varies according to the type of array, but LOH may be resolved at least to approximately 3 Mb, or 100 SNPs, in 100K arrays. Further technical developments make SNPA capable of analysing both signal intensity variations and changes in allelic composition in parallel. Single nucleotide polymorphism array can also detect both copy number changes and copy-neutral LOH events. The Human CytoSNP array (Illumina) was used in this study, and is able to detect 240 000 SNPs evenly distributed across the genome.
ECs can be divided into two subtypes with different clinicopathological and molecular characteristics.46 Endometrioid carcinomas (EECs) (85%) are usually low-grade, oestrogen-dependent tumours.47 Serous carcinomas (SCs), which represent the prototype of non-endometrioid carcinomas (15%), are more aggressive, are independent of oestrogen, and develop in postmenopausal women. The molecular alterations of EEC and SC are different; abnormalities in PTEN occur in 40% of EECs. LOH at 10q23 frequently coexists with somatic PTEN mutations, which are also found in 37–61% of EECs.48 Assessment of PTEN by immunohistochemistry or FISH may help in classifying difficult cases.
In the present study, we developed a standardized protocol for evaluation of PTEN by FISH, and we also assessed PTEN status by FISH in a series of 31 ECs. The same cases were subjected to SNPA, MLPA, and immunohistochemistry. The results show good concordance between FISH and SNPA or MLPA for ECs with a normal PTEN copy number (all 11 cases with two copies of PTEN by SNPA had a normal copy number by FISH, and a concordant diploid pattern between FISH and MLPA was seen in 15 of 17 cases). Moreover, there was good correlation between FISH and SNPA in assessment of the presence of large deletions (two cases), if we exclude the tumours in which a deletion of PTEN coexisted with duplication of the second allele (five cases). In these cases, SNPA was superior to FISH and MLPA, because FISH could not identify one of the two alleles as a duplicated one. Finally, FISH was superior to SNPA and MLPA in detecting the presence of gene copy number variation, in the context of morphology. This was particularly obvious for the three tumours with a heterogeneous distribution of tumour cells with different pattern of PTEN copy number variation. In a previous study,33 a heterogeneous distribution of PTEN staining was seen in 27% of cases, and correlated with different expression levels of putative targets of AKT (BAD, p27, and NFκB; data not shown).
Interestingly, nine ECs had three copies of PTEN (polysomy), and in this group there was complete agreement between FISH and SNPA. Polysomy was more frequent among SCs (three of four cases, one of them heterogeneous) than among EECs (nine of 27 cases, one of them heterogeneous), probably reflecting the frequent presence of chromosomal instability in SCs. Six of these nine tumours were interpreted as showing PTEN amplification by MLPA. The results suggest that FISH and SNPA are superior to MLPA in assessing PTEN triploidy. Interestingly, the PTEN levels in the nine tumours with an extra copy of PTEN, as measured by immunohistochemistry, were similar to the levels in tumours with a normal PTEN copy number (P = 0.68). This suggests that the presence of an additional copy of PTEN does not have an impact on protein production. However, it is worth mentioning that the area that contained three copies of PTEN had a greater correspondence with the area showing increased PTEN immunostaining than the area with two copies in two of the three heterogeneous tumours. The biological role of polysomy in altering the function of oncogenes or TSGs is under debate. Polyploidy at chromosome 17, where HER2 is located, occurs in one-third of breast carcinomas. It has been shown that polysomy 17 is mechanistically distinct from HER2 amplification. Polysomy at chromosome 17, without HER2 amplification, does not seem to significantly increase HER2 mRNA or HER2 protein levels in many cases, although there is a low to moderate increase in HER2 level in some cases. Breast cancers with extra copies of chromosome 17 resemble HER2-negative tumours according to standard pathological criteria, including tumour grade, and hormonal status. Even more perplexing is the presence of extra copies of TSGs, such as PTEN, which are typically inactivated by loss of function molecular events. In ECs, polysomy at chromosome 10 has been described previously. It may occur separately, or may coexist with polysomy in other chromosomes, such as chromosome 1 and chromosome 8. Several studies have shown that polysomy at chromosome 10 is more frequent in EC than in endometrial hyperplasia, and that it is more frequent in tumours with higher histological grade.48 Whether polysomy at chromosome 10 is related to disturbance of PTEN expression, or simply reflects the presence of chromosomal instability, is still unknown. However, the results of our study show that, as for HER2 in breast carcinoma, polysomy at the PTEN loci does not result in increased PTEN protein levels.19,36 Interestingly, polysomy at chromosomal areas involving well-known TSGs or genes involved in DNA repair (RB1, p53, APC, BRCA1, BRCA2, and NF1) has been reported.49
It is worth mentioning that five tumours showed deletion of one copy of PTEN and duplication of the second allele. Although not statistically significant, perhaps because of the small number of cases, these tumours showed a trend for decreased protein expression levels (eight of nine had Hscores of <100), suggesting that the biological significance of the deleted copy is more important than that of duplication of the second allele. Interestingly, one of the heterogeneous tumours showed a PTEN deletion in one area and a normal PTEN number in another area by FISH (a PTEN deletion with duplication of the second allele by SNPA), and PTEN immunostaining was negative in both. We may speculate about the molecular basis of this interesting subgroup of tumours. Polysomy is usually a consequence of mis-segregation during mitosis, which happens in tumours with chromosomal instability, and there are many findings showing that ECs with PTEN alterations have genomic instability. We may hypothesize that, in the presence of PTEN alteration, chromosomal instability can lead to the development of polysomy at some chromosomal loci, chromosome 10 among them. In this regard, it is worth noting that deletion of PTEN was seen in eight of 31 cases (26%), and in five of these eight cases there was duplication of the second allele. In other words, the polysomy at chromosome 10 was seen in more than half of the tumours showing deletion of PTEN.
In summary, a combination of these techniques is helpful for assessment of PTEN copy number changes in ECs. The combination of FISH and SNPA seems to be particularly helpful. Fluorescence in-situ hybridization allows much better assessment of tumour heterogeneity, whereas SNPA is helpful in identifying tumours with deletion of PTEN and duplication of the second allele, which appear as normal diploid by FISH. The combination of all of these techniques, together with immunohistochemistry, is helpful in interpreting the biological significance of polysomy at the PTEN locus in chromosome 10. Discrepancies between the results obtained with these techniques and those obtained with immunohistochemistry may be explained by the presence of PTEN mutations and alterations in PTEN translation. Our results suggest that a combination of FISH and immunohistochemistry may be of some help in assessing PTEN status in routine practice.
Acknowledgments
The study was supported by a research agreement with Dako, Denmark. The research team was also supported by grants FIS PI100922, Fundación Mutua Madrileña AP75732010, 2009SGR794, RD12/0036/0013, Fundación Asociación Española contra el Cancer, Programa de Intensificación de la Investigación, Instituto Carlos III, Verelst Baarmoederkankerfonds, Leuven, and ENITEC (European Network for Individualized Treatment of Endometrial Carcinoma). F. Amant is senior researcher for the research fund Flandersb (FWO). Tumour samples were obtained with the support of Xarxa Catalana de Bancs de Tumours, and Plataforma de Biobancos ISCIII (PT13/0010/0014).
Author contributions
X. Matias-Guiu and F. Amant were responsible for the study design. X. Matias-Guiu, O. Maiques, D. Cuevas and A. Velasco performed and interpreted FISH assays. O. Maiques, S. Müller and H. C. Pedersen worked on optimization of the protocol. O. Maiques, D. Cuevas, A. Velasco and M. Romero performed and interpreted MLPA assays. X. Matias-Guiu, O. Maiques, M. Santacana and S. Gatius performed and interpreted immunohistochemical staining. D. A. G. Dios, L. Coenegrachts, D. Lambrechts and F. Amant performed and interpreted SNPA assays. X Matias-Guiu, O. Maiques and S. Gatius reviewed the pathological data. X. Matias-Guiu and O. Maiques were involved in drafting the manuscript. All authors reviewed the Results and Discussion sections.
Conflicts of interest
The study was performed in a research collaboration with Dako Denmark A/S. The sponsor did not participate in the interpretation of results. Two authors (S. Müller and H. C. Pedersen) work for Dako, and were involved in optimization of the protocol. The other authors do not have any conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article
Preanalytical variables.
Optimization of PTEN FISH
Preanalytical variables did not have a significant influence on PTEN FISH staining.
Correlation of MLPA with IHC in 28 cases.
References
- 1.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumor suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 2.Cairns P, Okami K, Halachmi S, et al. Frequent inactivation of PTENMMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 3.Xie CC, Lu L, Sun J, et al. Germ-line sequence variants of PTEN do not have an important role in hereditary and non-hereditary prostate cancer susceptibility. J. Hum. Genet. 2011;56:496–502. doi: 10.1038/jhg.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shugart YY, Cour C, Renard H, et al. Linkage analysis of 56 multiplex families excludes the Cowden disease gene PTEN as a major contributor to familial breast cancer. J. Med. Genet. 1999;36:720–721. [PMC free article] [PubMed] [Google Scholar]
- 5.Kurose K, Zhou XP, Araki T, Eng C. Biallelic inactivating mutations and an occult germline mutations of PTEN in primary cervical carcinomas. Genes Chromosom. Cancer. 2000;29:166–172. [PubMed] [Google Scholar]
- 6.Kwabi-Addo B, Giri D, Schmidt K, et al. Haploinsufficiency of the PTEN tumor suppressor gene promotes prostate cancer progression. Proc. Natl Acad. Sci. USA. 2001;98:11563–11568. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li DM, Sun H. PTENMMAC1TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc. Natl Acad. Sci. USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birck A, Ahrenkiel V, Zeuthen J, Hou-Jensen K, Guldberg P. Mutation and allelic loss of the PTENMMAC1 gene in primary and metastatic melanoma biopsies. J. Invest. Dermatol. 2000;114:277–280. doi: 10.1046/j.1523-1747.2000.00877.x. [DOI] [PubMed] [Google Scholar]
- 9.Poetsch M, Lorenz G, Kleist B. Detection of new PTENMMAC1 mutations in head and neck squamous cell carcinomas with loss of chromosome 10. Cancer Genet. Cytogenet. 2002;132:20–24. doi: 10.1016/s0165-4608(01)00509-x. [DOI] [PubMed] [Google Scholar]
- 10.De Vivo I, Gertig DM, Nagase S, et al. Novel germline mutations in the PTEN tumor suppressor gene found in women with multiple cancers. J. Med. Genet. 2000;37:336–341. doi: 10.1136/jmg.37.5.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JO, Yang H, Georgescu MM, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 12.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhou XP, Marsh DJ, Morrison CD, et al. Germline inactivation of PTEN and dysregulation of the phosphoinositol-3-kinase/Akt pathway cause human Lhermitte-Duclos disease in adults. Am. J. Hum. Genet. 2003;73:1191–1198. doi: 10.1086/379382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonneau D, Longy M. Mutations of the human PTEN gene. Hum. Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Nagase S, Sato S, Tezuka F, Wada Y, Yajima A, Horii A. Deletion mapping on chromosome 10q25–q26 in human endometrial cancer. Br. J. Cancer. 1996;74:1979–1983. doi: 10.1038/bjc.1996.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong D, Suzuki A, Zou TT, et al. PTEN1 is frequently mutated in primary endometrial carcinomas. Nat. Genet. 1997;17:143–144. doi: 10.1038/ng1097-143. [DOI] [PubMed] [Google Scholar]
- 17.Muresu R, Cossu A, Scarpa AM, et al. Numerical abnormalities of chromosomes 1 and 10 in endometrial adenocarcinoma: fluorescence in situ hybridization analysis of 23 archival paraffin-embedded samples. Cancer Genet. Cytogenet. 1998;107:37–42. doi: 10.1016/s0165-4608(98)00059-4. [DOI] [PubMed] [Google Scholar]
- 18.Qian J, Weber D, Cochran R, Hossain D, Bostwick DG. Detection of chromosomal anomalies in endometrial atypical hyperplasia and carcinoma by using fluorescence in situ hybridization. Cancer Cytopathol. 2010;118:97–104. doi: 10.1002/cncy.20072. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg CL. Polysomy 17 and HER-2 amplification: true, true and unrelated. J. Clin. Oncol. 2008;26:4856–4858. doi: 10.1200/JCO.2008.17.2684. [DOI] [PubMed] [Google Scholar]
- 20.Mutter GL, Lin HC, Fitzgerald JT, et al. Altered PTEN expression as a diagnostic marker for the earliest endometrial precancers. J. Natl Cancer Inst. 2000;92:924–930. doi: 10.1093/jnci/92.11.924. [DOI] [PubMed] [Google Scholar]
- 21.Wen S, Stolarov J, Myers MP, et al. PTEN controls tumor-induced angiogenesis. Proc. Natl Acad. Sci. USA. 2001;98:4622–4627. doi: 10.1073/pnas.081063798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanamori Y, Kigawa J, Itamochi H, et al. Correlation between loss of PTEN expression and Akt phosphorylation in endometrial carcinoma. Clin. Cancer Res. 2001;7:892–895. [PubMed] [Google Scholar]
- 23.Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability–high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 24.Pinkel D, Landegent J, Collins C, et al. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc. Natl Acad. Sci. USA. 1998;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br. J. Cancer. 2007;97:678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson SA, Cheng Z, Wang ML, et al. Comparative fluorescence in situ hybridization mapping of a 431-kb Arabidopsis thaliana bacterial artificial chromosome contig reveals the role of chromosomal duplications in the expansion of the Brassica rapa genome. Genetics. 2000;156:833–838. doi: 10.1093/genetics/156.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kallioniemi OP, Kallioniemi A, Kurisu W, et al. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA. 1992;89:5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bubendorf L, Kononen J, Koivisto P, et al. Survey of gene amplifications during prostate cancer progression by high-throughput fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999;59:803–806. [PubMed] [Google Scholar]
- 29.Bohn BA, Mina S, Khron A, et al. Altered PTEN function caused by deletion or gene disruption is associated with poor prognosis in rectal but not in colon cancer. Hum. Pathol. 2013;44:1524–1533. doi: 10.1016/j.humpath.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Matthiesen SH, Hansen CM. Fast and non-toxic in situ hybridization without blocking of repetitive sequences. PLoS ONE. 2012;7:e40675. doi: 10.1371/journal.pone.0040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaffer LG, Tommerup N. An international system for human cytogenetic nomenclature. Basel: S. Karger in Collaboration with Cytogenetics and Genome Research; 2005. [Google Scholar]
- 32.Ventura RA, Martin-Subero JI, Jones M, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J. Mol. Diagn. 2006;8:141–151. doi: 10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiques O, Santacana M, Valls J, et al. Optimal protocol for PTEN immunostaining; role of analytical and preanalytical variables in PTEN staining in normal and neoplastic endometrial, breast, and prostatic tissues. Hum. Pathol. 2014;45:522–532. doi: 10.1016/j.humpath.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 34.Knudson AG., Jr Hereditary cancer, oncogenes, and antioncogenes. Cancer Res. 1985;45:1437–1443. [PubMed] [Google Scholar]
- 35.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veldman T, Vignon C, Schroeck E, Rowley JD, Ried T. Hidden chromosome abnormalities in haematological malignancies detected by multicolour spectral karyotyping. Nat. Genet. 1997;15:406–410. doi: 10.1038/ng0497-406. [DOI] [PubMed] [Google Scholar]
- 37.Edwards MC, Gibbs RA. Multiplex PCR: advantages, development, and applications. Genome Res. 1994;3:S65–S75. doi: 10.1101/gr.3.4.s65. [DOI] [PubMed] [Google Scholar]
- 38.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 39.Lowery MC, Morris CA, Ewart A, et al. Strong correlation of elastin deletions, detected by FISH, with Williams syndrome: evaluation of 235 patients. Am. J. Hum. Genet. 1995;57:49–53. [PMC free article] [PubMed] [Google Scholar]
- 40.Sellner LN, Taylor GR. MLPA and MAPH: new techniques for detection of gene deletions. Hum. Mutat. 2004;23:413–419. doi: 10.1002/humu.20035. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X, Li C, Paez JG, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 42.Perry A, Nobori T, Ru N, et al. Detection of p16 gene deletions in gliomas: a comparison of fluorescence in situ hybridization (FISH) versus quantitative PCR. J. Neuropathol. Exp. Neurol. 1997;56:999–1008. doi: 10.1097/00005072-199709000-00005. [DOI] [PubMed] [Google Scholar]
- 43.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Perrone F, Lampis A, Orsenigo M, et al. PI3KCAPTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann. Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 45.Mina S, Bohn BA, Simon R, et al. PTEN deletion is rare but often homogeneous in gastric cancer. J. Clin. Pathol. 2012;65:693–698. doi: 10.1136/jclinpath-2011-200525. [DOI] [PubMed] [Google Scholar]
- 46.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 47.Carcangiu ML, Chambers JT, Voynick IM, Pirro M, Schwartz PE. Immunohistochemical evaluation of estrogen and progesterone receptor content in 183 patients with endometrial carcinoma. Part I: clinical and histologic correlations. Am. J. Clin. Pathol. 1990;94:247–254. doi: 10.1093/ajcp/94.3.247. [DOI] [PubMed] [Google Scholar]
- 48.Tashiro H, Blazes MS, Wu R, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 49.Zhang W, Yu Y. The important molecular markers on chromosome 17 and their clinical impact in breast cancer. Int. J. Mol. Sci. 2011;12:5672–5683. doi: 10.3390/ijms12095672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Preanalytical variables.
Optimization of PTEN FISH
Preanalytical variables did not have a significant influence on PTEN FISH staining.
Correlation of MLPA with IHC in 28 cases.