Abstract
The amino acid sequences of apolipoprotein E (apoE) from 63 different mammalian species have been downloaded from the protein database. The sequences were compared to human apoE4 to determine conserved and non-conserved sequences of amino acids. ApoE4 is the major risk factor for the development of late onset Alzheimer's disease while apoE3, which differs from apoE4 by a single amino acid change at position 112, poses little or no risk for the development of this disease. Thus, the two proteins appear to be structurally and functionally different. Seven highly conserved regions, representing approximately 47 amino acids (of 299) have been found. These regions are distributed throughout the protein and reflect ligand binding sites as well as regions proposed to be involved in the propagation of the cysteine–arginine change at position 112 to distant regions of the protein in the N- and C-terminal domains. Highly non-conserved regions are at the N- and C-terminal ends of the apoE protein.
Keywords: amino acid sequences, allosteric pathway, heparin binding, LDL receptor binding, lipoprotein binding, Aβ binding
Introduction
Humans, in contrast to other mammals, have three major alleles of the APOE gene designated as apoE2, apoE3, and apo4. The appearance of apoE3 and apoE2 appears to be a late event in evolution since other primates, such as chimpanzees and bonobos, have a single major protein characteristic of apoE4.1–3 Almost all apoE proteins are 299 amino acids in length. In humans, the alleles differ only by single amino acid changes4: ApoE2 has cysteines at positions 112 and 158, apoE3 has an arginine at position 158, and apoE4 has arginines at both 112 and 158.5 Yet, although the three isoforms have many similar properties, some properties appear to be quite different. It is now well established, for example, that apoE4 is the major risk factor for late onset Alzheimer's disease6 while apoE3 is benign in this regard and apoE2 might even be protective.7 The reasons for these differences are unknown.
Part of the difficulty in attempting to relate the cysteine–arginine change at position 112 (for apoE3 vs. apoE4) to functional differences is that, for many years after their discovery in the 1970s, no full-length apoE structure was available. Then, in 2011, Chen et al. published an NMR structure of full length apoE3, the first such structure to be determined for this important protein.8 The authors were able to accomplish this task by making 4–5 mutations in the C-terminal domain that prevented the protein from aggregating to larger species. While the N-terminal domain structure closely paralleled that determined for the isolated N-terminal domain,9 the C-terminal domain was totally different from earlier proposals.10 Thus, this domain was not exclusively helical as earlier proposed but importantly interacted with helix 4 (residues 131–164) of the N-terminal domain rather than helix 2 (residues 55–79)8 as had been suggested.10 An earlier publication discusses this issue.11 Based on experiments using hydrogen–deuterium exchange,12 Frieden and Garai identified regions of apoE that appeared to be structurally different between apoE isoforms.13 In particular, they concluded that these structurally different regions of the protein were distant from the residue change at position 112.13
In this article, I examine the role of amino acid residues of apoE that are conserved over a large database of known apoE sequences. It is shown that conserved residues not only identify ligand binding sites but also identifies the allosteric pathway by which the cysteine–arginine change at position 112 in the N-terminal domain could be propagated to the C-terminal domain.13
Results
An important contribution of structure/function relationships is that of conservation of amino acid residues throughout a series of homologous proteins. Similar to the active sites of enzymes, one can reasonably argue that conserved regions of protein that has no enzymatic activity, such as apoE, must have conserved regions related to function. Therefore, an approach to differences between apoE isoforms is to examine conservation (or non-conservation) of residues among the different mammalian species that have apoE. A search of the protein database reveals 63 sequences from different species that are over 60% similar to human apoE4. Supporting Information Table IS lists the NCBI protein sequence identifiers, the percent conserved relative to human apoE4, and the common name of the species. All 63 sequences were then aligned using the online alignment tool Muscle and compared to human apoE4. Figure 1(A) shows the percent of conserved amino acids for these 63 apoEs relative to human apoE4. Examining this figure it is immediately clear that there is both considerable variation among the sequences (see Supporting Information) as well as several regions of the protein where amino acids are highly conserved. These conserved regions were then examined to see whether they might provide information relevant to apoE function as well as answers to why apoE3 and apoE4 are functionally different. Among the 63 sequences, 15 amino acids were absolutely conserved in all sequences relative to apoE4. Because that appeared to be too small a database those amino acid residues were included that were identical in 62 of the 63 species as well, that is, those that were over 98% identical, adding another 32 amino acids. The total of 47 amino acid residues represents ∼16% of the 299 amino acid residues in apoE. Figure 1(B) shows those amino acids that are over 98% conserved over all species. I then examined whether there were conserved sequences of amino acids that would reflect conserved regions of apoE. Figure 1(B) shows that there is clustering of conserved amino acids. Given some latitude in selecting the larger regions, they are residues 34–48, 92–100, 142–151, 155–165, and 253–280. The amino acid residues in these regions are shown in Table I. In this table, the amino acids that are highly conserved are shown in bold. Discussed below, and as shown in Figure 2, these conserved regions can be grouped together based on their proximity to each other in the apoE structure as determined by Chen et al.8 These groupings are shown in red in Figure 2. Also noted in each apoE3 structure shown in Figure 2 is the location of cysteine 112. One small conserved region, residues 47–49, is not discussed because it does not appear to be related to other regions nor, at this time, is it known to be functionally important. As discussed later, Figure 1 also shows that there are residues that are poorly conserved (or conserved less than 30%) in all apoE sequences.
Table I.
Residues and Residue Sequence Number of Those that are Highly Conserved, Taken From Figure 1
| Residue | W | W | Y | L | W | Q | S | V | Q | E | E | L | L |
| Sequence number | 26 | 34 | 36 | 37 | 39 | 41 | 44 | 47 | 48 | 49 | 59 | 60 | 63 |
| Residue | T | E | R | K | E | Q | A | R | L | Y | R | G | R |
| Sequence number | 67 | 70 | 92 | 95 | 96 | 98 | 100 | 114 | 115 | 118 | 119 | 127 | 136 |
| Residue | R | K | K | R | R | D | L | R | A | Y | G | L | Q |
| Sequence number | 142 | 143 | 146 | 147 | 150 | 151 | 155 | 158 | 160 | 162 | 165 | 229 | 235 |
| Residue | K | Q | E | W | F | P | D | W | |||||
| Sequence number | 242 | 246 | 255 | 264 | 265 | 267 | 271 | 276 |
Those in bold and that are underlined reflect regions of amino acids in sequence as discussed in the text. The residues 47–49 represent a short sequence and are not discussed.
Figure 1.
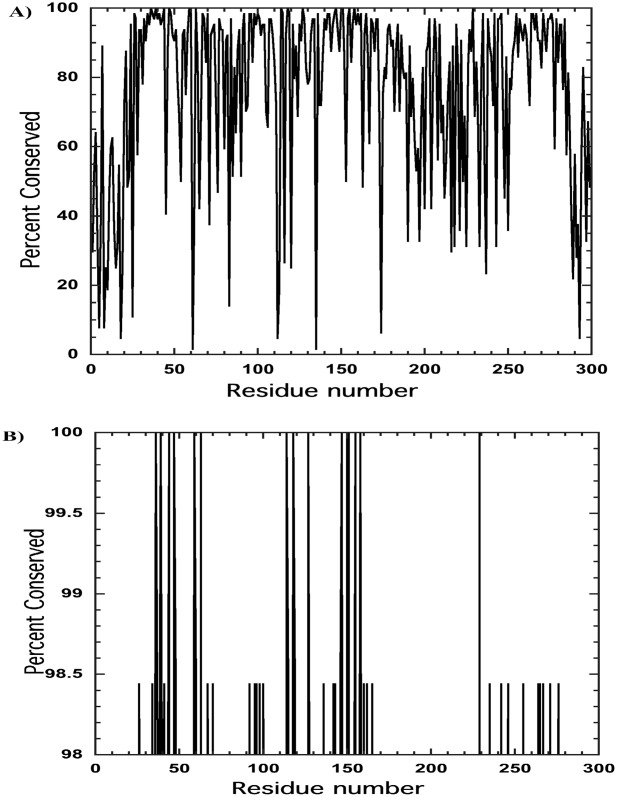
A: Percent conservation, relative to human apoE4, for 63 different species of apoE plotted as a function of residue number. A large number of apoE sequences were determined from analysis of the cellular DNA rather than direct amino acid sequencing. Determination of conservation was made using Muscle software. B: Data from Figure 1 plotted only for amino acid residues showing those over 98% conserved.
Figure 2.
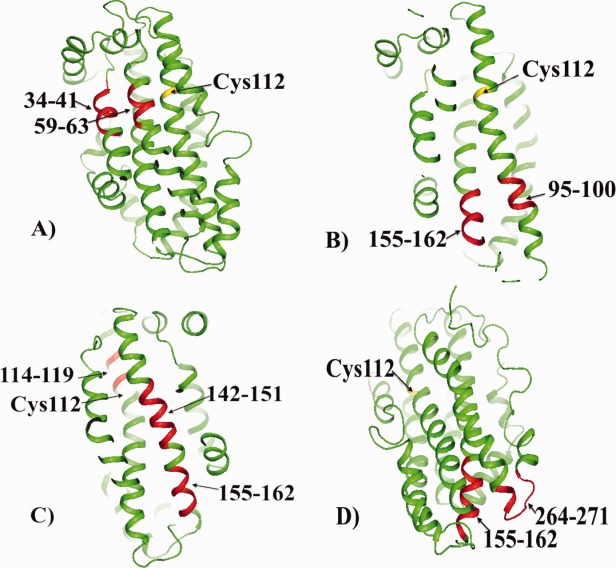
The highly conserved regions of apoE shown in red. A: The regions 34–41 and 59–63, (B) The regions 95–100 and 155–162; as described in the text, the region 155–162 is involved in the propagation of structural changes in the C-terminal domain. This region also contains Arg158 that is changed to Cys158 in apoE2 (see also Fig. 3). C: The regions 114–119, 142–151, and 155–162; as described in the text, the region 142–151 includes the heparin and LDL receptor binding sites. D: The regions 155–162 and 264–271. In all cases the cysteine residue at position 112 is indicated. Note that in (B,D) the cysteine residue at position 112 is distant from the highly conserved residues. The orientation of apoE differs in each figure to make easier viewing of the conserved regions.
The regions 34–41 and 59–63
These two regions of helix 1 (34–41) and helix 2 (59–63) are in close contact as shown by Figure 2(A). It is not clear at this time why these regions of apoE are conserved but, based on considerations discussed later, one could conclude that this region does have a functional role. In the hydrogen–deuterium exchange experiments12 and the chemical footprinting data previously published,14 the peptide 31–37 was not detected so it is unknown whether it is conformationally different between apoE isoforms or whether it could be involved in the oligomer formation of apoE. It is of interest that residues Glu59, Leu60, and Leu63 are highly conserved while, as has been previously noted,10 Arg61 is not conserved at all. There has been considerable discussion about the role of Arg61 because it was proposed, based partly on a Arg61Thr mutation, that this residue forms a salt bridge with Glu255 in the C-terminal domain of apoE4.15 As clearly shown by the NMR structure of apoE3, however, Arg61 is not close to Glu255.8 How the Arg61Thr mutation may affect the properties of apoE has been discussed in a previous publication.11 Briefly, however, it was proposed that the change from arginine to threonine at this position affected the behavior of apoE4 in the same way as the change of arginine to cysteine (i.e., the difference between apoE3 and apoE4) at position 112. As discussed below, other highly conserved regions appear to have important functional significance.
The region 95–100 and 155–162
As with regions 34–41 and 59–63, these two conserved regions, 95–100 and 155–162, are close as shown in Figure 2(B). Both sequences are distant from cysteine 112. Within the sequence 155–162 is Arg158. This residue is of special interest because the difference between apoE3 and apoE2 is an arginine to cysteine change at this position. In the hydrogen–deuterium exchange experiments, the peptide 94–104 showed slight differences between apoE2 compared to apoE3 and apoE4 (which showed no differences).12 This difference, which suggests greater stability in apoE2, could be a consequence of the effect to the arginine to cysteine change between apoE2 and the other isoforms. The conserved peptide 155–162 appears in three of the four conserved regions of interaction shown in Figure 2, the only peptide to do so, reflecting the important role this sequence plays in the behavior of apoE.
The regions 114–119, 142–151, and 155–162
The two regions, 142–151 and 155–162, make up a large percentage of helix 4 (residues 131–164) as shown in Figure 2(C). In a previous publication, we proposed that the cysteine–arginine change at position 112 was propagated through the apoE structure to a region of the C-terminal domain via helix 4.13 It was also suggested that the arginine–cysteine change at position 112 was transmitted to a region of helix 4, residues 140–158, by residues arginine114 and histidine140 that are in close proximity to Cys112. Thus, it is of interest that residue 114 is highly conserved. As shown by Chen et al.8 there are many interactions between helix 4 and the C-terminal domain.
But the region consisting of both conserved peptides, covering residues 142–162, may serve another function as well linking, as it does, regions from both the C- and N-terminal domains. It also appears to be involved in both low density lipoprotein (LDL) receptor and heparin binding as discussed later.
The regions 155–162 and 264–271
Residues 155–162 in helix 4 are adjacent to residues 264–271 of the C-terminal domain as shown in Figure 2(D). We had earlier proposed that region 155–162 (and the conserved region 142–151) were involved in the pathway leading to changes in the C-terminal domain as a consequence of the arginine–cysteine change at position 112.13 In particular, it was proposed that portions of the sequence 271–279 were conformationally different between apoE3 and apoE4. Figure 3 shows a larger region, residues 253–285, that includes the only turn in the C-terminal domain as well several charged residues with buried side chains. As discussed later, the region that appears conformationally different between apoE3 and apoE4 is not well conserved. Projecting into this region, and deeply buried, is arginine 158, a residue of great interest because, as noted earlier, the only difference between apoE3 and apoE2 is the change from arginine to cysteine at this position.
Figure 3.
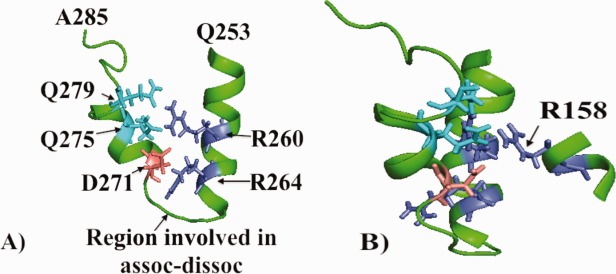
A: A representation of residues 253–285 encompassing a highly conserved region (residues 264–271) of apoE. These residues comprise the only turn in the C-terminal domain. Shown are charged residues as well as glutamine residues. B: The structure given in (A) but turned 90° to show how arginine 158 relates to residues shown in (A). The only amino acid change between apoE2 and apoE3 is a cysteine, rather than an arginine, at this position. Also noted is a region involved in the association of apoE to higher molecular weight forms.12
Non-conserved residues
Figure 1 also shows those residues that are not conserved (or conserved less than 30%) in all apoE sequences. In contrast to those highly conserved, there is no obvious pattern although the regions at the first 20 residues of the N-terminal domain and last 10 residues of the C-terminal appear to be clustered. Based on the hydrogen–deuterium exchange data there were two regions suggested to be structurally different between apoE3 and apoE4.12 One, as discussed above, includes residues 271–279 while a second region of comprising residues 5–30. This latter region in particular is not highly conserved while that of region 271–279 does include some conserved residues. Should these regions be conformationally altered by the arginine to cysteine changes at either position 112 or 158, it may not be surprising that they are not highly conserved. Below the relation of conserved residues to ligand binding are discussed.
ApoE receptors
The most important members of the low density lipoprotein receptor (LDLR) gene family, in terms of being primary receptors for apoE/lipoprotein, appear to be the LDLR and the low density lipoprotein receptor related protein (LRP1). In general, it appears that the region of apoE involved with binding to receptors encompasses residues 130–150. With regard to LDLR, Lalazar et al. made mutations in the region of 136–150 of apoE and found that all variants, and particularly the Lys143ALa mutant, displayed lower LDL receptor binding.16 With regard to LRP1, Croy et al. investigated the binding of two apoE peptides, 130–149 and a 141–155 dimer, to LDL-like domains of LRP117 and found that both bound with nM affinity to LRP1. These results certainly suggest that the conserved region 142–160, as shown in Figure 2(C), contains the residues essential for apoE binding to LDL receptors consistent with earlier studies.18,19 It is interesting to note that the arginine to cysteine change at position 158 in apoE2 results in poor binding of apoE2 to receptor.20
Lipoprotein particle/lipid binding
Nguyen et al.21 using surface plasmon resonance (SPR) methods analyzed the binding of two plasma lipoprotein particles, very low density lipoprotein (VLDL) and high density lipoprotein3 (HDL3) to apoE3 and apoE4. The data suggested two steps: an initial binding step and a slower step reflecting opening of the N-terminal domain. On measuring the binding constants, they found that both the isolated C-terminal domain and the isolated N-terminal domain could bind lipoprotein particles but that the C-terminal domain bound more tightly than either the N-terminal domain or the full-length protein. Their data were consistent with earlier reports, using truncated versions of apoE, that the region spanning residues 250–299 was important for lipid binding.22,23 These data indicate that, similar to binding of Aβ (below), multiple regions of the protein can bind lipids although not equally well. However, the region 253–285 is conserved and perhaps contains the binding site for the initial binding of the lipid. In their discussion of lipid binding, Chen et al. proposed the first step to be binding to the C-terminal domain but clearly the interaction with lipid is complex.8 Examination of the region 253–285 reveals at least 18 residues whose side chains are solvent exposed consistent with the idea that lipids bind to exposed hydrophobic regions.
Heparin binding
Heparin binding to apoE has been studied extensively.24–28 As early as 1986, Weisgraber et al. determined that there were at least two binding sites for heparin: the first located in the N-terminal domain in the vicinity of residues 142–147 and the second located in the C-terminal domain between residues 243 and 272.25 Both sites are highly conserved. More recently, Futumura et al.,28 using SPR, characterized binding as a two-step binding process. These authors determined large changes in the first step of binding in an apoE3 Lys146Glu mutant, a residue located in the 143–155 conserved region. This is the same region as discussed above with respect to binding of apoE to the LDL family of receptors.
Aβ binding
There is considerable evidence for the idea that the major mechanism by which apoE is correlated with the occurrence of late onset Alzheimer's disease is by its effect on the metabolism of Aβ, specifically Aβ clearance (i.e., Refs.29–34). Although some investigators have attempted to determine the apoE/Aβ binding site, the results have been controversial. Because Aβ, an intrinsically disordered protein, binds to several different proteins it seems unlikely that there would be a sequence of conserved residues common to such proteins. Indeed, strong evidence now exists that apoE does not bind to monomeric Aβ but rather to oligomers or clusters of Aβ aggregate35,36 suggesting non-specific binding to several regions of the apoE protein. Under these conditions, it may never be possible to accurately determine Aβ binding sites.
Association to higher molecular weight forms
ApoE is known to associate to higher molecular weight forms37 and Garai and Frieden characterized the association as an monomer–dimer–tetramer process when the apoE concentrations were below 10 μM.38 Huang et al. compared the monomeric forms with wild type protein and concluded that peptides 230–243 and 262–270 were involved12 consistent with the observation that the isolated C-terminal domain aggregates while the N-terminal domain is always monomeric. Interestingly, the region 264–271 is highly conserved [ Fig. 2(D)] in agreement with earlier reports that the region spanning residues 250–299 was important for lipid binding.22,23 On the other hand, the region of 230–243 is not conserved (Fig. 1) suggesting that apoEs from different species may show differences in their ability to form higher molecular weight forms. It remains puzzling as to whether the oligomerization of apoE has any physiological consequences.
The allosteric pathway
In a previous publication, we proposed an allosteric pathway to describe how a residue change at position 112 in the N-terminal domain could be transmitted to regions of the protein in the C-terminal domain.13 Here we find that there is a strong correlation between conserved residues and those involved in propagating the suggested structural difference between apoE3 and apoE4. Specifically, one such region, that of a portion of helix 4, is highly conserved.
Discussion
The critical question is whether highly conserved residues are linked to functional differences between apoE isoforms. In this article, I take advantage of the fact that there are 63 sequences to compare with human apoE4 and, more importantly, the determination of the domain–domain interaction as recently published by Chen et al.8 One can conclude that the highly conserved regions do indeed appear to be related to function. Another question that rises immediately is whether this approach, examining multiple apoE sequences for conserved regions, is valid for defining functional differences between apoE3 and apoE4. Typical database searches are directed toward conserved residues that may be involved, for example, in enzyme catalysis and substrate binding. In contrast, apoE proteins bind a number of different ligands. Thus, not only are we concerned with regions that may be involved in ligand binding but we wish to find those residues that might be involved in functional differences between apoE isoforms that occur as a consequence of the single amino acid change between apoE3 and apoE4. With regard to ligand binding, it was shown that critical properties of apoE, that of binding to lipoprotein particles, to members of the LDL receptor family and to heparin are highly conserved. Thus, ligand binding and conservation of residues appear to be linked.
The previous work suggested two regions, one from the N-terminal domain and one from the C-terminal domain that appeared to be structurally different between apoE isoforms.13 The allosteric pathway for both regions was the same. Surprisingly, although some residues within the regions in the C-terminal domain that we consider to be structurally different between apoE3 and apoE4 (residues 271–279)13 are conserved, several are not. Thus, it is not clear whether this region is conserved or not.
It should be noted, however, that there may be other regions that differ. The hydrogen–deuterium exchange experiments that have been discussed above did not cover the complete sequence.12 Missing from the data are results for residues 31–51, 61–78, 124–161, and 244–261. Two sequences where data are lacking include regions of conserved residues: 31–51 encompasses conserved region 34–45 and 124–161 encompasses conserved region 143–155. Thus, more regions that might show conformational differences may, and probably do, exist. Finally, it should be noted, perhaps as might be expected, that there is little conservation of amino acids at the ends (∼20 amino acids at the N-terminal end and 10 at the C-terminal end) of the protein. Nor is there any clear sequence of amino acids that are poorly conserved compared to those that are highly conserved.
Apo E is a highly plastic protein. Regions between structural elements, for example, are not tight turns but rather loops. The protein undergoes a large conformational change on binding to lipid in which the N-and C-terminal regions spread out along the surface of the lipid in a way described as a helical hairpin.39 Certainly, the relation of conserved sequences to functional differences is consistent with the structure determined by Chen et al.8 and inconsistent with earlier proposed structures that show no interaction between the C- and N-terminal domains in apoE3, a model that is clearly incorrect.
One can ask whether the differences between apoE3 and apoE4 are truly structural differences or rather reflect regions that exist in dynamic equilibrium between two or more conformations. The arginine to cysteine change at position 112 may then alter the equilibrium ratio of the different conformational forms, stabilizing one relative to the other(s). In this scenario, apoE would possess characteristics of both apoE3 and apoE4.
Acknowledgments
Author thanks Dr. Scott Wildman for obtaining and compiling the data shown in Figure 1. The author declares no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- 1.McIntosh AM, Bennett C, Dickson D, Anestis SF, Watts DP, Webster TH, Fontenot MB, Bradley BJ. The apolipoprotein E (APOE) gene appears functionally monomorphic in chimpanzees (Pan troglodytes) PLoS One. 2012;7:e47760. doi: 10.1371/journal.pone.0047760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finch CE. Evolution in health and medicine Sackler colloquium: evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finch CE, Sapolsky RM. The evolution of Alzheimer disease, the reproductive schedule, and apoE isoforms. Neurobiol Aging. 1999;20:407–428. doi: 10.1016/s0197-4580(99)00053-6. [DOI] [PubMed] [Google Scholar]
- 4.Weisgraber KH, Rall SC, Jr, Mahley RW. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem. 1981;256:9077–9083. [PubMed] [Google Scholar]
- 5.Rall SC, Jr, Weisgraber KH, Mahley RW. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982;257:4171–4178. [PubMed] [Google Scholar]
- 6.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 7.Rebeck GW, Kindy M, LaDu MJ. Apolipoprotein E and Alzheimer's disease: the protective effects of ApoE2 and E3. J Alzheimers Dis. 2002;4:145–154. doi: 10.3233/jad-2002-4304. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci USA. 2011;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 10.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 11.Frieden C, Garai K. Concerning the structure of apoE. Prot Sci. 2013;22:1820–1825. doi: 10.1002/pro.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang RYC, Garai K, Frieden C, Gross ML. Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemistry. 2011;50:9273–9282. doi: 10.1021/bi2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frieden C, Garai K. Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer's disease. Proc Natl Acad Sci USA. 2012;109:8913–8918. doi: 10.1073/pnas.1207022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gau B, Garai K, Frieden, Gross ML. Mass spectrometry-based protein footprinting characterizes the structures of oligomeric apolipoprotein E2, E3, and E4. Biochemistry. 2011;50:8117–8126. doi: 10.1021/bi200911c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 16.Lalazar A, Weisgraber KH, Rall SC, Jr, Giladi H, Innerarity TL, Levanon AZ, Boyles JK, Amit B, Gorecki M, Mahley RW, Vogel T. Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. J Biol Chem. 1988;263:3542–3545. [PubMed] [Google Scholar]
- 17.Croy JE, Brandon T, Komives EA. Two apolipoprotein E mimetic peptides, ApoE(130–149) and ApoE(141–155)2, bind to LRP1. Biochemistry. 2004;43:7328–7335. doi: 10.1021/bi036208p. [DOI] [PubMed] [Google Scholar]
- 18.Innerarity TL, Friedlander EJ, Rall SC, Jr, Weisgraber KH, Mahley RW. The receptor-binding domain of human apolipoprotein E. Binding of apolipoprotein E fragments. J Biol Chem. 1983;258:12341–12347. [PubMed] [Google Scholar]
- 19.Weisgraber KH, Innerarity TL, Harder KJ, Mahley RW, Milne RW, Marcel YL, Sparrow JT. The receptor-binding domain of human apolipoprotein E. Monoclonal antibody inhibition of binding. J Biol Chem. 1983;258:12348–12354. [PubMed] [Google Scholar]
- 20.Dong LM, Parkin S, Trakhanov SD, Rupp B, Simmons T, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol. 1996;3:718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen D, Dhanasekaran P, Phillips MC, Lund-Katz S. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry. 2009;48:3025–3032. doi: 10.1021/bi9000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 23.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J Biol Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 24.Cardin AD, Hirose N, Blankenship DT, Jackson RL, Harmony JA, Sparrow DA, Sparrow JT. Binding of a high reactive heparin to human apolipoprotein E: identification of two heparin-binding domains. Biochem Biophys Res Commun. 1986;134:783–789. doi: 10.1016/s0006-291x(86)80489-2. [DOI] [PubMed] [Google Scholar]
- 25.Weisgraber KH, Rall SC, Jr, Mahley RW, Milne RW, Marcel YL, Sparrow JT. Human apolipoprotein E. Determination of the heparin binding sites of apolipoprotein E3. J Biol Chem. 1986;261:2068–2076. [PubMed] [Google Scholar]
- 26.Dong J, Peters-Libeu CA, Weisgraber KH, Segelke BW, Rupp B, Capila I, Hernaiz MJ, LeBrun LA, Linhardt RJ. Interaction of the N-terminal domain of apolipoprotein E4 with heparin. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 27.Saito H, Dhanasekaran P, Nguyen D, Baldwin F, Weisgraber KH, Wehrli S, Phillips MC, Lund-Katz S. Characterization of the heparin binding sites in human apolipoprotein E. J Biol Chem. 2003;278:14782–14787. doi: 10.1074/jbc.M213207200. [DOI] [PubMed] [Google Scholar]
- 28.Futamura M, Dhanasekaran P, Handa T, Phillips MC, Lund-Katz S, Saito H. Two-step mechanism of binding of apolipoprotein E to heparin: implications for the kinetics of apolipoprotein E-heparan sulfate proteoglycan complex formation on cell surfaces. J Biol Chem. 2005;280:5414–5422. doi: 10.1074/jbc.M411719200. [DOI] [PubMed] [Google Scholar]
- 29.Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, Holtzman DM. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci USA. 2013;110:E1807–1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holtzman DM. Role of apoe/Abeta interactions in the pathogenesis of Alzheimer's disease and cerebral amyloid angiopathy. J Mol Neurosci. 2001;17:147–155. doi: 10.1385/JMN:17:2:147. [DOI] [PubMed] [Google Scholar]
- 31.Basak JM, Kim J, Pyatkivskyy Y, Wildsmith KR, Jiang H, Parsadanian M, Patterson BW, Bateman RJ, Holtzman DM. Measurement of apolipoprotein E and amyloid beta clearance rates in the mouse brain using bolus stable isotope labeling. Mol Neurodegener. 2012;7:14–27. doi: 10.1186/1750-1326-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wildsmith KR, Holley M, Savage JC, Skerrett R, Landreth GE. Evidence for impaired amyloid beta clearance in Alzheimer's disease. Alzheimers Res Ther. 2013;5:33–39. doi: 10.1186/alzrt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manelli AM, Stine WB, Van Eldik LJ, LaDu MJ. ApoE and Abeta1–42 interactions: effects of isoform and conformation on structure and function. J Mol Neurosci. 2004;23:235–246. doi: 10.1385/JMN:23:3:235. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Kanekiyo T, Shinohara M, Zhang Y, LaDu MJ, Xu H, Bu G. Differential regulation of amyloid-β endocytic trafficking and lysosomal degradation by apolipoprotein E isoforms. J Biol Chem. 2012;287:44593–44601. doi: 10.1074/jbc.M112.420224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ly S, Altman R, Petrlova J, Lin Y, Hilt S, Huser T, Laurence TA, Voss JC. Binding of apolipoprotein e inhibits the oligomer growth of amyloid-β peptide in solution as determined by fluorescence cross-correlation spectroscopy. J Biol Chem. 2013;288:11628–11635. doi: 10.1074/jbc.M112.411900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garai K, Verghese PB, Baban B, Holtzman DM, Frieden C. The binding of apolipoprotein E to oligomers and fibrils of amyloid-beta alters the kinetics of amyloid aggregation. Biochemistry. 2014;53:6323–6331. doi: 10.1021/bi5008172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perugini MA, Schuck P, Howlett GJ. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J Biol Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 38.Garai K, Frieden C. The association–dissociation behavior of the ApoE proteins: kinetic and equilibrium studies. Biochemistry. 2010;49:9533–9541. doi: 10.1021/bi101407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J Biol Chem. 2006;281:1073–1079. doi: 10.1074/jbc.M510851200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


