Abstract
Niphargus is a speciose amphipod genus found in groundwater habitats across Europe. Three Niphargus species living in the sulphidic Frasassi caves in Italy harbour sulphur-oxidizing Thiothrix bacterial ectosymbionts. These three species are distantly related, implying that the ability to form ectosymbioses with Thiothrix may be common among Niphargus. Therefore, Niphargus–Thiothrix associations may also be found in sulphidic aquifers other than Frasassi. In this study, we examined this possibility by analysing niphargids of the genera Niphargus and Pontoniphargus collected from the partly sulphidic aquifers of the Southern Dobrogea region of Romania, which are accessible through springs, wells and Movile Cave. Molecular and morphological analyses revealed seven niphargid species in this region. Five of these species occurred occasionally or exclusively in sulphidic locations, whereas the remaining two were restricted to nonsulphidic areas. Thiothrix were detected by PCR on all seven Dobrogean niphargid species and observed using microscopy to be predominantly attached to their hosts' appendages. 16S rRNA gene sequences of the Thiothrix epibionts fell into two main clades, one of which (herein named T4) occurred solely on niphargids collected in sulphidic locations. The other Thiothrix clade was present on niphargids from both sulphidic and nonsulphidic areas and indistinguishable from the T3 ectosymbiont clade previously identified on Frasassi-dwelling Niphargus. Although niphargids from Frasassi and Southern Dobrogea are not closely related, the patterns of their association with Thiothrix are remarkably alike. The finding of similar Niphargus–Thiothrix associations in aquifers located 1200 km apart suggests that they may be widespread in European groundwater ecosystems.
Keywords: amphipods, ecology, sulphide, symbiosis, systematics, taxonomy
Introduction
Since their discovery at hydrothermal vents in the late 1970s, myriad examples of symbioses between sulphur-oxidizing bacteria and invertebrates have been discovered worldwide in sulphidic marine environments (Dubilier et al. 2008). Dark, isolated and sulphide-rich habitats analogous to hydrothermal vents also exist in land-locked caves, such as Movile Cave in Romania and the Frasassi caves in Italy (Forti et al. 2002). Recently, ectosymbioses between sulphur-oxidizing Thiothrix bacteria and three species of the groundwater amphipod genus Niphargus were reported from Frasassi (Dattagupta et al. 2009; Bauermeister et al. 2012), extending the realm of such symbioses into nonmarine ecosystems.
The three ectosymbiotic Niphargus species in Frasassi harbour on their exoskeleton three distinct Thiothrix clades (T1–T3), which are predominantly attached to hairs (setae) and spines of their legs and antennae (Bauermeister et al. 2012). Clade T1 has so far only been found on Niphargus frasassianus, a species restricted to sulphidic locations, whereas clades T2 and T3 occur on Niphargus species in both sulphidic and nonsulphidic waters (T2 on Niphargus ictus and Niphargus montanarius, and T3 on all three Frasassi-dwelling species). The three ectosymbiont clades do not form a monophyletic group (Bauermeister et al. 2012), and neither do their host species (Flot et al. 2010a). The lack of congruence between the host and symbiont phylogenies suggests independent establishments of the symbioses and/or interspecies symbiont transfer (Bauermeister et al. 2012).
Although sulphide is a potent inhibitor of mitochondrial electron transfer (Bagarinao 1992) that is generally toxic to aquatic life (Theede et al. 1969; Oseid & Smith 1974; Sandberg-Kilpi et al. 1999), several niphargid species have been reported to thrive in sulphide-rich environments such as Frasassi (up to 415 μm sulphide; Flot et al. 2010a) and Acquapuzza (410 μm sulphide; Latella et al. 1999) in Italy as well as Movile Cave in Romania (up to 500 μm sulphide; Sarbu 2000). Other Niphargus species are found in the sulphidic cave of Melissotrypa in Greece (J.-F. Flot and S. Dattagupta, unpublished data) as well as in anchihaline caves in Croatia, where sulphide is present but has not yet been quantified (Sket 1996; Gottstein et al. 2007). This raises the question whether Niphargus–Thiothrix associations are restricted to Frasassi or also found in other sulphidic locations.
The Southern Dobrogea region (southwestern Romania) provides the ideal locality to start examining this question, as it has a sulphidic aquifer that can be accessed through artificial wells, springs and Movile Cave. Discovered in 1986, Movile Cave was the first terrestrial chemoautotrophic ecosystem described (Sarbu & Popa 1992; Sarbu et al. 1996; Sarbu 2000) and is one of the most thoroughly studied to date. It harbours two niphargid species (Sarbu et al. 1996), and amphipods are also known to occur in sulphidic and nonsulphidic wells and springs in the surrounding area. Our goals in this study were (i) to molecularly characterize the niphargids of the Southern Dobrogea region and compare them phylogenetically with Frasassi species and (ii) to examine them for Thiothrix epibionts using microscopy and molecular methods.
Materials and methods
Sampling and DNA extractions
Amphipods were collected between April 2011 and September 2012 in the Southern Dobrogea region of Romania (see Tables1, S1 and S2 for information on collection sites). Specimens were preserved in 70% ethanol for morphological examination and DNA sequencing, in RNAlater® (Sigma-Aldrich, Steinheim, Germany) for DNA sequencing and fluorescence in situ hybridization (FISH), and in 2.5% glutaraldehyde prepared in phosphate-buffered saline (PBS) for scanning electron microscopy (SEM). A microbial mat sample was collected in April 2011 from Airbell 2 of Movile Cave (see Sarbu et al. 1994 for a map) and kept frozen at −20 °C. This mat sample was later used for exploring the diversity of free-living Thiothrix found in Movile Cave.
Table 1.
Location details and groundwater geochemical characteristics of niphargid collection sites in this study
| Town | Measurement location | Latitude | Longitude | Measurement date | T (°C) | EC (μS/cm) | Eh (mV) | H2S (μm) | Niphargid species |
|---|---|---|---|---|---|---|---|---|---|
| Hagieni | Hagieni Spring | 43°48′08.90″N | 28°28′29.00″E | 09.2012 | 18.1 | 1080 | −245 | (172) | N. hrabei*, P. ruffoi |
| Mangalia | Movile Cave | 43°49′36.38″N | 28°33′43.48″E | 09.2011 | 21.2 | 1071 | −341 | 245 | N. cf. stygius, P. racovitzai |
| Mangalia | str. Matei Basarab 62 | 43°49′09.11″N | 28°34′16.10″E | 09.2012 | 19.8 | 1380 | −266 | (188) | N. cf. stygius, N. decui |
| Mangalia | str. Matei Basarab 74 | 43°49′10.61″N | 28°34′ 07.90″E | 09.2012 | 18.1 | 1460 | −174 | (126) | N. decui |
| Mangalia | str. Gheorge Netoi 1 | 43°49′10.87″N | 28°34′12.74″E | 09.2011 | 18.6 | 1078 | −263 | 133 | N. cf. stygius |
| Mangalia | str. Dumitru Ana 13 | 43°49′23.59″N | 28°34′01.45″E | 09.2011 | 19.1 | 1052 | −120 | 101 | N. cf. stygius |
| Mangalia | str. Ion Mecu 51 | 43°49′25.75″N | 28°34′29.40″E | 09.2011 | 19.9 | 1135 | −89 | 66 | N. cf. stygius |
| Mangalia | Aleea Cetăţii 1 | 43°48′53.21″N | 28°35′01.84″E | 05.2013 | 19.3 | 1650 | −64 | (48) | N. cf. stygius, P. racovitzai |
| Mangalia | str. Maior Giurescu 22 | 43°49′15.43″N | 28°34′46.19″E | 09.2012 | 18.4 | 2440 | 28 | (0) | N. gallicus |
| Mangalia | str. Horia Cloşca Crişan 13 | 43°49′18.67″N | 28°34′ 23.10″E | 09.2012 | 19.7 | 1450 | 40 | (0) | N. decui |
| Mangalia | str. Delfinului 16 | 43°48′34.73″N | 28°34′44.89″E | 09.2011 | 19.1 | 1473 | 66 | 0 | N. gallicus |
| Mangalia | str. Mihai Viteazu 20 | 43°48′49.30″N | 28°34′50.31″E | 09.2011 | 17.4 | 1193 | 68 | 0 | N. decui |
| Mangalia | str. Pictor Tonitza 1 | 43°49′09.05″N | 28°35′03.71″E | 09.2011 | 19.0 | 1242 | 104 | 0 | N. cf. stygius, N. decui, P. racovitzai |
| Mangalia | str. Vasile Pârvan 16 | 43°48′51.25″N | 28°35′07.32″E | 09.2012 | 15.0 | 2166 | 139 | (0) | N. gallicus |
| Mangalia | str. Delfinului 16 | 43°48′34.73″N | 28°34′44.89″E | 09.2011 | 19.1 | 1473 | 66 | 0 | N. gallicus |
| Albeşti | near road 393 | 43°47′47.50″N | 28°25′35.80″E | 09.2011 | 13.8 | 1445 | 72 | 0 | N. decui |
| Doi Mai | str. Mihail Kogălniceanu 393 | 43°47′25.72″N | 28°34′37.10″E | 09.2011 | 16.1 | 2235 | 56 | 0 | N. dobrogicus, N. decui |
| Dulceşti | near road 394 | 43°54′00.07″N | 28°32′39.10″E | 09.2011 | 15.4 | 1216 | 63 | 0 | N. gallicus, N. decui |
| Limanu | corner str. Mărului/str. Traian Vuia | 43°48′10.01″N | 28°31′24.83″E | 09.2011 | 16.2 | 1092 | 69 | 0 | N. dobrogicus, N. decui |
| Vama Veche | str. Mihail Kogălniceanu 23 | 43°45′ 07.07″N | 28°34′18.59″E | 05.2013 | 14.3 | 1690 | 55 | (0) | N. dobrogicus |
| Vama Veche | str. Plajei 100 | 43°45′09.10″N | 28°34′38.40″E | 05.2013 | 14.0 | 1760 | 73 | (0) | N. decui |
H2S values in parentheses were not measured directly but inferred from measured redox potentials (taking advantage of the quasi-linear relationship observed between these two parameters).
This species was collected at a time when the spring was almost dry and no smell of sulphide was perceived.
DNA extractions for niphargid sequencing and PCR screenings (see below) were performed as described in Flot et al. (2010a) using Qiagen DNeasy kits, whereas DNA extractions for 16S rRNA gene clone library constructions followed Bauermeister et al. (2012). DNA from the microbial mat sample was extracted using the method of Neufeld et al. (2007). All sequencing was performed on an ABI 3730xl DNA Analyser.
Niphargid sequence analysis
A total of 71 amphipod specimens were analysed molecularly (Table S1, Supporting information), comprising 69 niphargids and two outgroups (Synurella sp. and Gammarus sp.). PCR amplifications and sequencing of a fragment of the mitochondrial cytochrome c oxidase (COI) gene, of the complete nuclear internal transcribed spacer (ITS) and of a fragment of the nuclear 28S rRNA gene were performed as reported previously (Flot et al. 2010a). The sequences of length-variant heterozygotes (Flot et al. 2006) were unravelled using the online program Champuru (Flot 2007; http://www.mnhn.fr/jfflot/champuru), and the haplotypes of other heterozygotes were determined using Clark's method (Clark 1990).
Sequences were aligned by eye in MEGA5 (Tamura et al. 2011) for COI or using MAFFT's E-INS-i option (Katoh et al. 2002) for the 28S rRNA gene and for ITS. Only the 1st and 2nd codon positions were taken into account for COI. Maximum-likelihood (ML) phylogenetic analyses were performed in MEGA5 under the GTR+G+I model (using all sites) and with 1000 bootstrap replicates (Felsenstein 1985); additional bootstrap analyses were conducted using neighbour-joining (under the K2P model, pairwise deletion) and parsimony approaches. The ITS phylogenetic tree was turned into a haploweb by adding connections between haplotypes found co-occurring in heterozygous individuals (Flot et al. 2010b, 2011).
Thiothrix epibiont detection using SEM and FISH
Ten specimens from Movile Cave and from surrounding sulphidic and nonsulphidic wells in the town of Mangalia were examined for filamentous Thiothrix epibionts using SEM (Table S2, Supporting information). Sample preparation and analysis were done as described previously (Bauermeister et al. 2012).
Three other individuals (JFF_12.19, JFF_12.32 and JFF_12.39) were investigated using FISH. The Thiothrix-specific oligonucleotide probe G123T (5′-CCT TCC GAT CTC TAT GCA-3′) and its competitor probe G123T-C (5′-CCT TCC GAT CTC TAC GCA-3′; Kanagawa et al. 2000) were synthesized at Eurofins MWG Operon (Ebersberg, Germany), with G123T being 5′-fluorescently labelled with cyanine-3. Both probes were mixed in equimolar amounts to enhance binding specificity to Thiothrix as recommended (Kanagawa et al. 2000). Niphargid samples were fixed in 4% paraformaldehyde for 3 h at 4 °C and transferred to a 1:1 ethanol–PBS solution. Several legs and antennae of each specimen were dissected and transferred into individual tubes. FISH experiments were carried out as described by Amann (1995) using the 1:1 G123T/G123T-C probe mix. Formamide concentration was 40% (following Kanagawa et al. 2000) and hybridization time was set to 2 h. The niphargid appendages were subsequently transferred onto glass slides and mounted with Vectashield (Vector Laboratories, Burlingame, CA, USA). Confocal epifluorescence micrographs of attached Thiothrix filaments were collected on a Zeiss 510 Meta scanning microscope equipped with argon and helium–neon lasers (wavelengths 488 and 543 nm, respectively).
Design and optimization of Thiothrix-specific PCR primers
The Thiothrix-specific forward primer THIO714F (5′-ATG CAT AGA GAT CGG AAG G-3′; Bauermeister et al. 2012) and the newly designed reverse primer THIO1492R (5′-GGC TAC CTT GTT ACG ACT T-3′) were used for constructing partial 16S rRNA gene clone libraries and for direct PCR screening of niphargids. Using PRIMROSE (Ashelford et al. 2002), THIO1492R was designed to match nearly all publicly available Thiothrix sequences. Gradient PCRs were performed to determine the optimal annealing temperature for the primer pair. THIO714F worked well for PCRs but not for sequencing. Thus, another Thiothrix-specific forward primer (THIO718Fseq; 5′-ATA GAG ATC GGA AGG AAC A-3′) was designed as described above and used in combination with THIO1492R for direct sequencing of PCR products.
Thiothrix clone library construction
Partial 16S rRNA gene clone libraries were constructed from DNA extracts of the Movile microbial mat sample and of two niphargid specimens (AH_10.4 and SS_10.1). PCR mixtures (50 μL) contained 1× ammonium buffer (Bioline, Luckenwalde, Germany), 2 mm MgCl2, 0.3 mm dNTP mix (Bioline), 25–30 ng of DNA (quantified using a ND-1000 Nanodrop, PEQLAB Biotechnology, Erlangen, Germany), 1.25 units of BioTaq DNA polymerase (Bioline) and 25 pm each of the primers THIO714F and THIO1492R. PCRs were performed in a Labcycler (SensoQuest, Göttingen, Germany), with an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 56 °C for 25 s, 72 °C for 2.5 min and a final extension at 72 °C for 5 min. PCR products were checked on a 1% agarose gel. Bands of the expected size (∼800 bp) were excised and extracted using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). PCR products were cloned and sequenced as described previously (Bauermeister et al. 2012). Sequences were manually checked using CodonCode Aligner version 3.7.1.2 (CodonCode Corporation, Dedham, MA, USA) and screened for chimeras using Pintail version 1.0 (Ashelford et al. 2005). Putative chimeras were excluded from subsequent analyses. Using MOTHUR (Schloss et al. 2009), a rarefaction curve was created from all sequences obtained from the Movile mat clone library in order to evaluate the number of clones to be sequenced to cover the most abundant Thiothrix species.
PCR detection of Thiothrix epibionts
DNA extracts obtained from the 71 amphipod specimens (Table S1, Supporting information) were examined for Thiothrix DNA by direct PCR screenings. PCR mixtures (10 μL) contained the same ingredients as described for Thiothrix clone library construction. Cycling conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 50 cycles of 94 °C for 45 s, 56 °C for 30 s and 72 °C for 1.5 min. PCR products were checked on a 1% agarose gel, and samples revealing bands of the expected size (∼800 bp) were sequenced directly using the primers THIO718Fseq and THIO1492R. In cases where mixtures of two overlapping Thiothrix sequences were obtained from the same PCR product, the individual sequences were resolved by comparison with Thiothrix epibiont sequences from the clone libraries. All sequences were assembled and screened for chimeras as described above.
Phylogenetic analysis of Thiothrix sequences
Sequences obtained from 16S rRNA gene clone libraries and direct PCR screenings were compared with sequences in the public GenBank database using nucleotide BLAST (Altschul et al. 1990). 84 sequences (60 of 68 obtained from the clone libraries and 24 of 35 obtained by direct PCR) turned out to be closely related to sequences of cultivated Thiothrix species and to sequences previously obtained from Niphargus species and microbial mats in Frasassi. 81 of these 84 sequences (leaving out three redundant Thiothrix sequences from samples AH_10.4 and SS_10.1 that were obtained in both PCR screenings and clone libraries) were used for phylogenetic analyses, together with 65 closely related Thiothrix sequences downloaded from GenBank. All sequences were aligned using the MAFFT version 6 multiple sequence alignment tool (Katoh & Toh 2010) with the Q-INS-i strategy for consideration of RNA secondary structure (Katoh & Toh 2008). The alignment was manually refined, and a 50% consensus filter was applied in MOTHUR (Schloss et al. 2009), resulting in 743 nucleotide positions used for phylogenetic analysis. jModelTest version 0.1.1 (Posada 2008) was used to determine the best-suited nucleotide model among 88 possible models following the Bayesian Information Criterion. The selected model (TIM3+I+G) was used to build a ML phylogenetic tree (1000 bootstrap replicates) using PhyML 3.0 (Guindon & Gascuel 2003). The tree was rooted with an epibiont clone sequence from the hydrothermal vent galatheid crab Shinkaia crosnieri (GenBank accession number AB476284; Watsuji et al. 2010). In addition, neighbour-joining bootstrap values for all nodes were calculated based on the same alignment using the BioNJ algorithm (Kimura 2-parameter model; 1000 bootstrap replicates) implemented in SeaView version 4 (Gouy et al. 2010).
Results
Seven niphargid species were identified in Southern Dobrogea
All 71 amphipod specimens yielded 28S rRNA gene sequences (Table S1, Supporting information), whereas sequencing of the ITS marker failed for one specimen and the COI sequences of four of them were of bacterial origin. Both the nuclear ITS and the mitochondrial COI markers were congruent in delineating seven species among our samples (Fig.1); each of these species was monophyletic and supported by bootstrap values >90% using all three methods (maximum likelihood, distance and parsimony). Although most species were also distinguishable using the 28S rRNA gene, two of them had identical sequences and could not be resolved using this marker (Fig.2).
Fig 1.
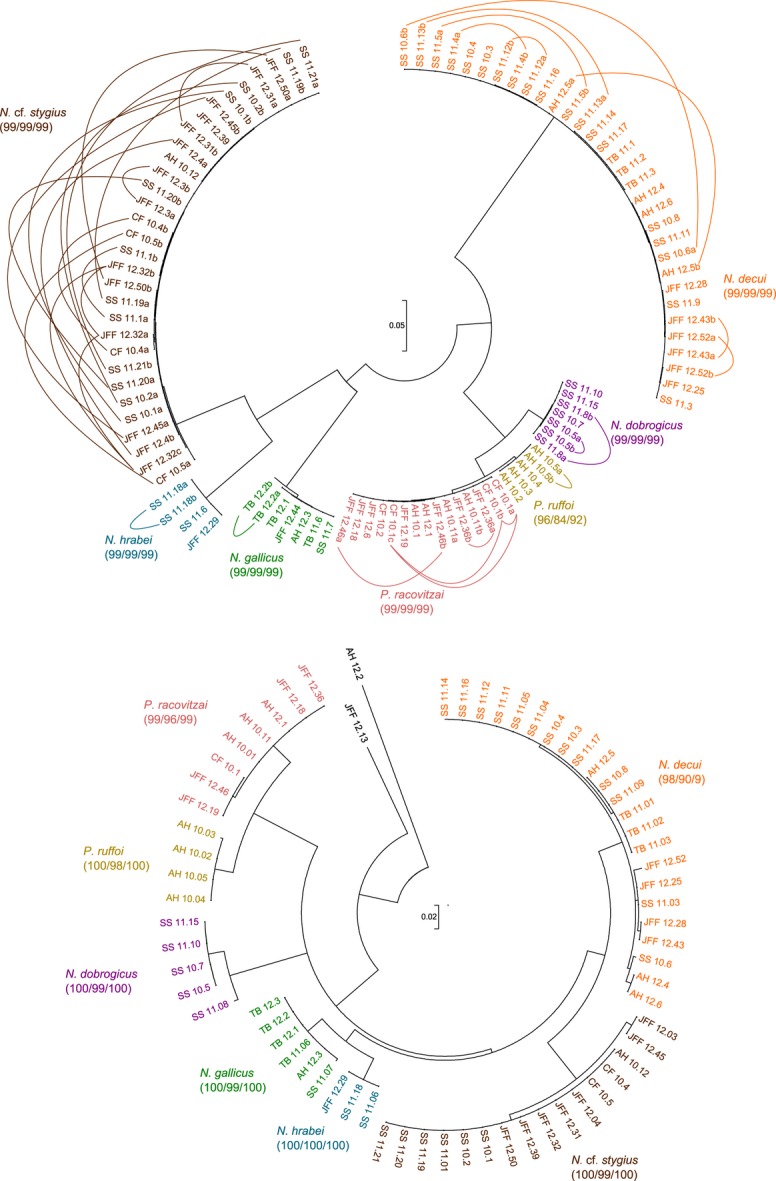
Top: Haploweb of ITS sequences of the 68 niphargid samples successfully sequenced for this marker. The two outgroups AH_12.2 (Synurella sp.) and JFF_12.13 (Gammarus sp.) were not included in the alignment, as their sequences were too divergent. The underlying phylogeny was obtained using a maximum-likelihood approach (model: GTR+G+I), following which connections were added between the sequences found co-occurring in heterozygous individuals. Bottom: Haplotree of COI sequences of the 67 amphipods samples successfully sequenced for this marker. The underlying phylogeny was obtained using a maximum-likelihood approach (model: GTR+G+I). Both approaches delineated seven species (represented by different colours); bootstrap values obtained from maximum-likelihood/parsimony/neighbour-joining are shown next to the name of each species.
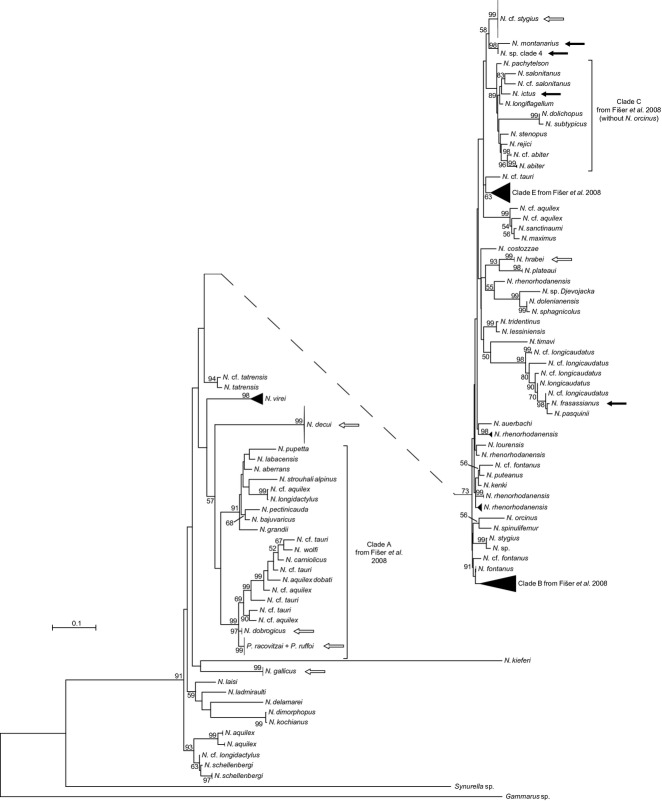
Fig 2.
Maximum-likelihood 28S rRNA gene phylogeny of the amphipods collected in the present study. This phylogeny includes sequences from Lefébure et al. (2006, 2007), Fišer et al. (2008), Trontelj et al. (2009), Flot (2010), Flot et al. (2010a) and Hartke et al. (2011). Filled arrows point at Niphargus sequences from the Frasassi caves in Italy, whereas empty arrows point at niphargid sequences from the present study.
The seven niphargid species delineated molecularly among our samples were further identified morphologically. Six of them were already known from Southern Dobrogea: Niphargus cf. stygius (Schiödte 1847), Niphargus decui Karaman & Sarbu 1995, Niphargus dobrogicus Dancău 1964, Niphargus gallicus Schellenberg 1935, Pontoniphargus racovitzai Dancău 1970 and Pontoniphargus ruffoi Karaman & Sarbu 1993. The seventh species, Niphargus hrabei Karaman 1932, had never been reported from southeastern Romania (Brad 1999). To confirm our morphological identification of N. hrabei, we compared its COI, ITS and 28S rRNA gene sequences with those of one individual collected from a side arm of the Danube River near Budapest, about 50 km away from its type locality. The sequences obtained were very close (for COI) or identical (for ITS and the 28S rRNA gene) to the ones from Dobrogea, despite a geographical distance of over 1000 km. We were not able to obtain material from the type locality of N. gallicus in France to verify whether it is really the same species (as hypothesized by Dancău 1963).
Comparison of the 28S rRNA gene sequences obtained from Southern Dobrogean niphargids with previously published ones (Fig.2) revealed that the sequences of N. cf. stygius were markedly different from those of the actual N. stygius from Slovenia, confirming that these are distinct species. The 28S rRNA gene phylogeny did not support the putative sister-genus relationship between Niphargus and Pontoniphargus; instead, P. racovitzai and P. ruffoi were nested within the genus Niphargus and closely related to N. dobrogicus.
Thiothrix epibionts were detected on all niphargid species from Southern Dobrogea
SEM revealed accumulations of filamentous bacteria on two P. ruffoi specimens from Hagieni, on one N. cf. stygius individual from Movile Cave and on two N. cf. stygius specimens from sulphidic wells in Mangalia (Table S2, Supporting information). The bacteria were found attached predominantly to hairs and spines on legs and antennae of the niphargids (Fig.3). The Thiothrix-specific oligonucleotide probe G123T bound to filamentous bacteria on appendages of all three niphargid individuals investigated using FISH (one P. ruffoi and two N. cf. stygius).
Fig 3.
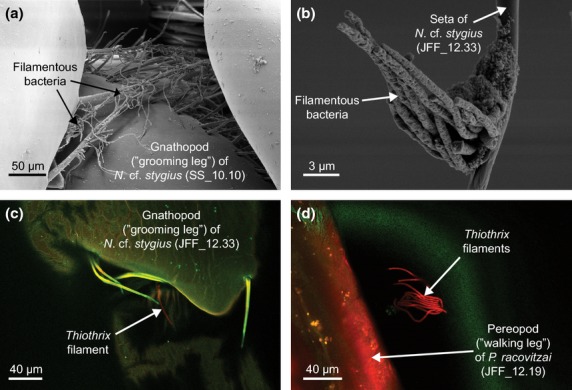
Thiothrix epibionts on niphargids from Southern Dobrogea. Panels a & b: Scanning electron micrographs (SEM) of filamentous bacteria attached to hairs on the legs of two N. cf. stygius individuals. The filaments closely resemble Thiothrix bacteria previously identified on Niphargus species from the Frasassi caves in Italy (Bauermeister et al. 2012). Panels c & d: Confocal epifluorescence micrographs showing a Thiothrix-specific FISH probe (red) bound to bacterial filaments on legs of N. cf. stygius and P. racovitzai.
Thiothrix-related partial 16S rRNA gene sequences were obtained from 21 of the 71 amphipod DNA extracts using PCR screenings with Thiothrix-specific primers (Table S1, Supporting information). From three of these 21 DNA extracts (samples SS_10.1, SS_10.2 and JFF_12.39), mixtures of two overlapping Thiothrix sequences were obtained, which were resolved by comparison with Thiothrix epibiont sequences in clone libraries obtained from P. ruffoi (AH_10.4) and N. cf. stygius (SS_10.1). From 11 of the remaining 50 amphipod samples, bands of the expected size (∼800 bp) were obtained on the agarose gel, but the top BLAST hits of the corresponding sequences belonged to bacteria other than Thiothrix (Table S1, Supporting information). The clone library constructed from the Movile microbial mat DNA yielded 52 Thiothrix sequences, which were used for comparison with Thiothrix sequences obtained from the niphargids.
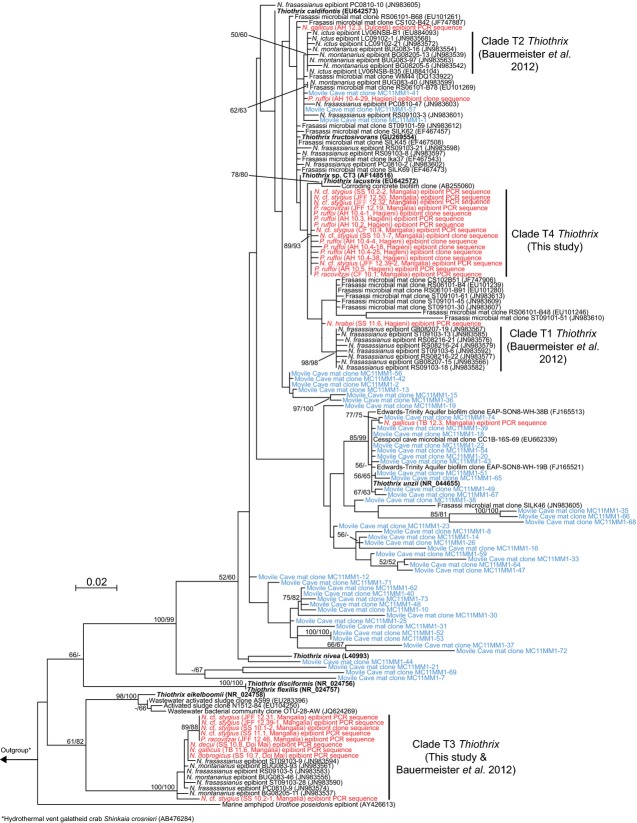
The majority (86%) of Thiothrix epibiont sequences obtained in this study fell into two clades (Fig.4). One of these clades (100% ML bootstrap support) contained Thiothrix sequences from P. racovitzai, N. cf. stygius, N. decui, N. gallicus and N. dobrogicus from both sulphidic and nonsulphidic areas, and this clade was indistinguishable from the T3 ectosymbiont clade of Frasassi-dwelling Niphargus species. The other clade, hereafter named T4, was supported by an 89% ML bootstrap value and exclusively contained sequences from individuals of P. ruffoi, P. racovitzai and N. cf. stygius collected in sulphidic locations (Table1). Four remaining Thiothrix epibiont sequences obtained from samples of N. gallicus, P. ruffoi and N. hrabei fell neither within clade T3 nor T4, but either formed individual branches in the Thiothrix tree or clustered with Thiothrix sequences from the Movile mat sample (this study) and from Frasassi microbial mats (Macalady et al. 2006, 2008).
Fig 4.
Maximum-likelihood 16S rRNA gene phylogeny of Thiothrix. Sequences obtained from Southern Dobrogean niphargid samples are in red, those contained in the Movile mat clone library in blue. Cultivated Thiothrix strains are in bold. Accession numbers of sequences downloaded from GenBank are given in parentheses. Maximum-likelihood/neighbour-joining bootstrap values greater than 50 are displayed next to the respective nodes.
Discussion
Invertebrates harbouring ectosymbionts are common in sulphidic marine habitats (Dubilier et al. 2008; Goffredi 2010). Well-known examples include different deep-sea alvinocaridid shrimp (Tokuda et al. 2008; Petersen et al. 2010), hydrothermal vent crabs of the genus Kiwa (Goffredi et al. 2008; Thurber et al. 2011) and stilbonematid nematodes dwelling in marine coastal sediments (Polz et al. 1992; Bayer et al. 2009). The discovery of filamentous, sulphur-oxidizing Thiothrix bacteria on Niphargus amphipods living in the land-locked Frasassi caves showed that ectosymbioses might also be prevalent in sulphidic freshwater environments (Dattagupta et al. 2009). Here, we demonstrate that Niphargus–Thiothrix associations are not restricted to Frasassi but are also found in the partly sulphidic aquifers of the Southern Dobrogea region of Romania. The regular presence of Thiothrix bacteria on several geographically and phylogenetically distant members of the genus Niphargus makes these relationships particularly suitable for evolutionary studies. Moreover, as the genus Niphargus contains over 300 species distributed across Europe (Fišer et al. 2008), it is possible that Niphargus–Thiothrix associations are even more diverse than we have uncovered so far.
In this study, we used molecular and morphological analyses to delineate the niphargid species inhabiting sulphidic and nonsulphidic aquifers in Southern Dobrogea. The combination of ITS and COI sequence markers proved sufficient to resolve five Niphargus and two Pontoniphargus species with a high level of confidence (Fig.1) and to find their position in a phylogenetic tree of niphargid amphipods. The less variable 28S rRNA gene adjacent to ITS in ribosomal DNA was easier to amplify and sequence consistently and could be used to construct a rooted phylogeny of Niphargus (Fig.2). However, it was not variable enough to distinguish the two closely related Pontoniphargus species. Contrary to previous morphological hypotheses (Dancău 1970; Karaman 1989; Karaman & Sarbu 1993), both Pontoniphargus species turned out to be firmly nested within Niphargus clade A (Fišer et al. 2008) and closely related to N. dobrogicus. No new species was discovered, which was surprising considering previous reports of rampant cryptic species in amphipods in general and in the genus Niphargus in particular (Lefébure et al. 2006, 2007; Trontelj et al. 2009).
Direct PCR screenings revealed that individuals of all seven niphargid species harboured Thiothrix epibionts, most of which belonged to two distinct clades named T3 and T4 (Fig.4 and Table S1, Supporting information). While T3 had previously been reported as an ectosymbiont clade of three Niphargus species from the Frasassi caves in central Italy (Bauermeister et al. 2012), T4 is a new discovery. Both T3 and T4 were found each on multiple individuals of different niphargid species, and they were distinct from Thiothrix bacteria identified in a Movile microbial mat sample (Fig.4). Therefore, T3 and T4 may be regarded as putative ectosymbionts of Southern Dobrogean niphargids pending future studies. Four Thiothrix epibiont sequences clustered with Movile and Frasassi microbial mat sequences instead of with sequences of T3 or T4 (Fig.4). Further, sequences belonging to bacteria other than Thiothrix were obtained from 11 of the examined niphargid samples (Table S1, Supporting information). On the basis of the present data, it is not possible to say whether these additional bacteria represent yet unknown symbionts or free-living bacteria that were loosely attached to the niphargid individuals at the time of collection.
There are many similarities between the Niphargus–Thiothrix epibioses found in Southern Dobrogea and Frasassi. First, the Thiothrix filaments are attached to the base of hairs on the amphipod appendages in both cases (Fig.3; cf. Dattagupta et al. 2009; Bauermeister et al. 2012). Second, more than one Thiothrix clade occurs on some Niphargus individuals (Table S1, Supporting information). Third, clade T3 is present in both sulphidic and nonsulphidic waters, whereas the other clades (T4 in the case of Southern Dobrogea and T1-T2 in the case of Frasassi) are only abundant in sulphidic locations (Bauermeister et al. 2012). Given that the niphargids of Southern Dobrogea are not closely related to the three species described from Frasassi (Fig.2), the parallels between the Niphargus–Thiothrix associations found in these aquifers that are more than 1200 kilometres apart from each other are particularly striking.
T3 Thiothrix were found to associate with Niphargus in nonsulphidic locations in both Southern Dobrogea and Frasassi. Although Thiothrix are commonly considered sulphur-oxidizing bacteria, several heterotrophic strains have been shown to grow in the absence of reduced sulphur (Howarth et al. 1999; Aruga et al. 2002; Chernousova et al. 2009). If T3 is capable of growth without sulphide, it may be widely distributed on niphargids throughout Europe. Our results provide grounds for looking further into the distribution and evolution of the Niphargus–T3 association.
Epibiotic Thiothrix filaments were previously identified on the marine amphipod Urothoe poseidonis living in coastal sediments (Gillan & Dubilier 2004). Just as in the case of Niphargus from Frasassi and Southern Dobrogea, Thiothrix were found attached predominantly to hairs and spines of the legs of U. poseidonis. Thus, it is possible that Thiothrix bacteria have a tendency to associate with both freshwater and marine amphipods and a preference to attach to their appendages. Whether this attachment location is advantageous for either Thiothrix or their amphipod hosts is not yet known. A previous study showed that the Thiothrix ectosymbionts of Frasassi-dwelling Niphargus probably do not play a role in sulphide detoxification for their hosts (Bauermeister et al. 2013). In the present study, Thiothrix sequences were not amplified from DNA of 48 of 71 amphipods screened with PCR (Table S1, Supporting information), and Thiothrix filaments were missing on 5 of 10 niphargid specimens examined by SEM (Table S2, Supporting information). It seems unlikely that all these apparently Thiothrix-lacking individuals had lost their epibionts due to moulting. Instead, our results suggest that the Thiothrix ectosymbionts may not be obligate for their hosts. Whether the Thiothrix bacteria benefit from associating with amphipods remains to be investigated.
Conclusion
Six of the seven niphargid species identified in the Southern Dobrogea region of Romania harbour putative Thiothrix ectosymbionts of clades T3 and/or T4, which are attached to their hosts in a manner similar to that previously observed in the Frasassi caves in Italy. Thus, Niphargus–Thiothrix associations are not restricted to Frasassi, where they were first discovered, but may be widespread in European sulphidic and nonsulphidic aquifers.
Acknowledgments
Many thanks to Melanie Heinemann for logistic assistance in the laboratory and to Cene Fišer, Radu Popa, Boris Sket and Fabio Stoch for useful comments. We gratefully acknowledge Mihai Baciu, Andreea Cohn, Vlad Voiculescu, Sandra Iepure and Marjeta Konec for their help during fieldwork, the Grupul de Explorări Subacvatice şi Speologice (GESS) for logistic support in Mangalia, Colin Murrell and Jason Stephenson for sending us a DNA extract from the Movile microbial mat they collected, and Péter Borza for sending us specimens of Niphargus hrabei from Hungary. We also thank three anonymous reviewers for their valuable suggestions. This article is contribution number 122 from the Courant Research Center Geobiology funded by the German Initiative of Excellence.
Sample collections and geochemical measurements: J.F.F., T.B., A.H.V., S.M.S., S.D.; DNA extractions and molecular analyses: J.F.F., J.B.; morphological identifications: J.F.F.; SEM and FISH: J.B.; manuscript drafting: J.F.F., J.B. and S.D.; manuscript revision: T.B., A.H.V. and S.M.S.
Data accessibility
All DNA sequences obtained in the present study were deposited in GenBank (accession numbers KF290023-KF290376 and KF362044-KF362049). Alignments were deposited in Dryad (doi:10.5061/dryad.qm636).
Supporting Information
Additional supporting information may be found in the online version of this article.
Niphargid samples analysed molecularly in this study. Samples are colour-coded according to niphargid species.
Niphargid samples analysed by scanning electron microscopy (SEM). Samples are colour-coded according to niphargid species.
References
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann RI. In situ identification of microorganisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkerman ADL, van Elsas DJ, de Bruijn FJ, editors. Molecular Microbial Ecology Manual. Dortrecht: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- Aruga S, Kamagata Y, Kohno T, Hanada S, Nakamura K, Kanagawa T. Characterization of filamentous Eikelboom type 021N bacteria and description of Thiothrix disciformis sp. nov. and Thiothrix flexilis sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2002;52:1309–1316. doi: 10.1099/00207713-52-4-1309. [DOI] [PubMed] [Google Scholar]
- Ashelford KE, Weightman AJ, Fry JC. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Research. 2002;30:3481–3489. doi: 10.1093/nar/gkf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Applied and Environmental Microbiology. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagarinao T. Sulfide as an environmental factor and toxicant: tolerance and adaptations in aquatic organisms. Aquatic Toxicology. 1992;24:21–62. [Google Scholar]
- Bauermeister J, Ramette A, Dattagupta S. Repeatedly evolved host-specific ectosymbioses between sulfur-oxidizing bacteria and amphipods living in a cave ecosystem. PLoS ONE. 2012;7:e50254. doi: 10.1371/journal.pone.0050254. doi: 10.1371/journal.pone.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauermeister J, Assig K, Dattagupta S. Exploring the sulfide tolerance of ectosymbiotic Niphargus amphipods from the Frasassi caves, central Italy. International Journal of Speleology. 2013;42:141–145. [Google Scholar]
- Bayer C, Heindl HR, Rinke C, Lücker S, Ott JA, Bulgheresi S. Molecular characterization of the symbionts associated with marine nematodes of the genus Robbea. Environmental Microbiology Reports. 2009;1:136–144. doi: 10.1111/j.1758-2229.2009.00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brad T. The present stage of our knowledge concerning the spreading of subterranean amphipods in Romania. Studii Şi Cercetări (Biologie) Bistriţa. 1999;5:157–164. [Google Scholar]
- Chernousova E, Gridneva E, Grabovich M, et al. Thiothrix caldifontis sp. nov. and Thiothrix lacustris sp. nov., gammaproteobacteria isolated from sulfide springs. International Journal of Systematic and Evolutionary Microbiology. 2009;59:3128–3135. doi: 10.1099/ijs.0.009456-0. [DOI] [PubMed] [Google Scholar]
- Clark A. Inference of haplotypes from PCR-amplified samples of diploid populations. Molecular Biology and Evolution. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- Dancău D. Niphargus gallicus Schell., amfipod subteran nou pentru fauna R.P.R. Comunicările Academiei Republicii Populare Române. 1963;13:123–129. [Google Scholar]
- Dancău D. Noi contribuţii la studiul amfipodelor subterane Niphargus dobrogicus n. sp. Lucrările Institutului de speologie “Emil Racoviţă”. 1964;3:397–403. [Google Scholar]
- Dancău D. Sur un nouvel amphipode souterrain de Roumanie, Pontoniphargus racovitzai, n. g., n. sp. In: Orghidan T, Dumitresco M, editors. Livre du centenaire. Emile G. Racovitza 1868–1968. Bucarest: Académie de la République socialiste de Roumanie; 1970. pp. 275–285. [Google Scholar]
- Dattagupta S, Schaperdoth I, Montanari A, et al. A novel symbiosis between chemoautotrophic bacteria and a freshwater cave amphipod. The ISME Journal. 2009;3:935–943. doi: 10.1038/ismej.2009.34. [DOI] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nature Reviews Microbiology. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fišer C, Sket B, Trontelj P. A phylogenetic perspective on 160 years of troubled taxonomy of Niphargus (Crustacea: Amphipoda) Zoologica Scripta. 2008;37:665–680. [Google Scholar]
- Flot J-F. Champuru 1.0: a computer software for unraveling mixtures of two DNA sequences of unequal lengths. Molecular Ecology Notes. 2007;7:974–977. [Google Scholar]
- Flot J-F. Vers une taxonomie moléculaire des amphipodes du genre Niphargus: exemples d'utilisation de séquences d'ADN pour l'identification des espèces. Bulletin de la Société des Sciences Naturelles de l'Ouest de la France. 2010;32:62–68. [Google Scholar]
- Flot J-F, Tillier A, Samadi S, Tillier S. Phase determination from direct sequencing of length-variable DNA regions. Molecular Ecology Notes. 2006;6:627–630. [Google Scholar]
- Flot J-F, Wörheide G, Dattagupta S. Unsuspected diversity of Niphargus amphipods in the chemoautotrophic cave ecosystem of Frasassi, central Italy. BMC Evolutionary Biology. 2010a;10:171. doi: 10.1186/1471-2148-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot J-F, Couloux A, Tillier S. Haplowebs as a graphical tool for delimiting species: a revival of Doyle's “field for recombination” approach and its application to the coral genus Pocillopora in Clipperton. BMC Evolutionary Biology. 2010b;10:372. doi: 10.1186/1471-2148-10-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot J-F, Blanchot J, Charpy L, et al. Incongruence between morphotypes and genetically delimited species in the coral genus Stylophora: phenotypic plasticity, morphological convergence, morphological stasis or interspecific hybridization? BMC Ecology. 2011;11:22. doi: 10.1186/1472-6785-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti P, Galdenzi S, Sarbu S. The hypogenic caves: a powerful tool for the study of seeps and their environmental effects. Continental Shelf Research. 2002;22:2373–2386. [Google Scholar]
- Gillan DC, Dubilier N. Novel epibiotic Thiothrix bacterium on a marine amphipod. Applied and Environmental Microbiology. 2004;70:3772–3775. doi: 10.1128/AEM.70.6.3772-3775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffredi SK. Indigenous ectosymbiotic bacteria associated with diverse hydrothermal vent invertebrates. Environmental Microbiology Reports. 2010;2:479–488. doi: 10.1111/j.1758-2229.2010.00136.x. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Jones WJ, Erhlich H, Springer A, Vrijenhoek RC. Epibiotic bacteria associated with the recently discovered Yeti crab, Kiwa hirsuta. Environmental Microbiology. 2008;10:2623–2634. doi: 10.1111/j.1462-2920.2008.01684.x. [DOI] [PubMed] [Google Scholar]
- Gottstein S, Ivković M, Ternjej I, Jalžić B, Kerovec M. Environmental features and crustacean community of anchihaline hypogean waters on the Kornati islands, Croatia. Marine Ecology. 2007;28:24–30. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hartke TR, Fišer C, Hohagen J, Kleber S, Hartmann R, Koenemann S. Morphological and molecular analyses of closely related species in the stygobiontic genus Niphargus (Amphipoda) Journal of Crustacean Biology. 2011;31:701–709. [Google Scholar]
- Howarth R, Unz RF, Seviour EM, et al. Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater-treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. International Journal of Systematic Bacteriology. 1999;49:1817–1827. doi: 10.1099/00207713-49-4-1817. [DOI] [PubMed] [Google Scholar]
- Kanagawa T, Kamagata Y, Aruga S, Kohno T, Horn M, Wagner M. Phylogenetic analysis of and oligonucleotide probe development for Eikelboom Type 021N filamentous bacteria isolated from bulking activated sludge. Applied and Environmental Microbiology. 2000;66:5043–5052. doi: 10.1128/aem.66.11.5043-5052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman S. Beitrag zur Kenntnis der Süsswasser-Amphipoden. Prirodoslovne Razprave. 1932;2:179–232. [Google Scholar]
- Karaman GS. New data on genus Pontoniphargus Dancău, 1970 (fam. Niphargidae) from Romania (Contribution to the knowledge of the Amphipoda 1999) Glasnik Republičkog Zavoda za Zaštitu Prirode i Prirodnjačkog Muzeja u Titogradu. 1989;22:79–94. [Google Scholar]
- Karaman GS, Sarbu S. A new species of the genus Pontoniphargus Dancău, 1970 (Amphipoda Gammaridea, family Niphargidae) from Romania, P. ruffoi n. sp. Bollettino del Museo Civico di Storia Naturale di Verona. 1993;20:569–582. [Google Scholar]
- Karaman GS, Sarbu SM. Niphargus decui n. sp. (Amphipoda, Gammaridea, Niphargidae), a new species from Romania. Travaux de l'Institut de Spéologie Emile Racovitza. 1995;34:77–87. [Google Scholar]
- Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212. doi: 10.1186/1471-2105-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella L, Di Russo C, de Pasquale L, Dell'Anna L, Nardi G, Rampini M. Preliminary investigations on a new sulfurous cave in central Italy. Mémoires de Biospéologie. 1999;26:131–1335. [Google Scholar]
- Lefébure T, Douady CJ, Gouy M, Trontelj P, Briolay J, Gibert J. Phylogeography of a subterranean amphipod reveals cryptic diversity and dynamic evolution in extreme environments. Molecular Ecology. 2006;15:1797–1806. doi: 10.1111/j.1365-294X.2006.02888.x. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Douady CJ, Malard F, Gibert J. Testing dispersal and cryptic diversity in a widely distributed groundwater amphipod (Niphargus rhenorhodanensis. Molecular Phylogenetics and Evolution. 2007;42:676–686. doi: 10.1016/j.ympev.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Lyon EH, Koffman B, et al. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Applied and Environmental Microbiology. 2006;72:5596–5609. doi: 10.1128/AEM.00715-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalady JL, Dattagupta S, Schaperdoth I, Jones DS, Druschel G, Eastman DK. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. The ISME Journal. 2008;2:590–601. doi: 10.1038/ismej.2008.25. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Schafer H, Cox MJ, Boden R, McDonald IR, Murrell JC. Stable-isotope probing implicates Methylophaga spp and novel Gammaproteobacteria in marine methanol and methylamine metabolism. The ISME Journal. 2007;1:480–491. doi: 10.1038/ismej.2007.65. [DOI] [PubMed] [Google Scholar]
- Oseid DM, Smith LL., Jr Chronic toxicity of hydrogen sulfide to Gammarus pseudolimnaeus. Transactions of the American Fisheries Society. 1974;103:819–822. [Google Scholar]
- Petersen JM, Ramette A, Lott C, Cambon-Bonavita M-A, Zbinden M, Dubilier N. Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environmental Microbiology. 2010;12:2204–2218. doi: 10.1111/j.1462-2920.2009.02129.x. [DOI] [PubMed] [Google Scholar]
- Polz MF, Felbeck H, Novak R, Nebelsick M, Ott JA. Chemoautotrophic, sulfur-oxidizing symbiotic bacteria on marine nematodes: morphological and biochemical characterization. Microbial Ecology. 1992;24:313–329. doi: 10.1007/BF00167789. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Sandberg-Kilpi E, Vismann B, Hagerman L. Tolerance of the Baltic amphipod Monoporeia affinis to hypoxia, anoxia and hydrogen sulfide. Ophelia. 1999;50:61–68. [Google Scholar]
- Sarbu SM. Movile Cave: a chemoautotrophically based groundwater ecosystem. In: Humphreys WF, editor; Wilkens H, Culver DC, editors. Ecosystems of the World. Subterranean Ecosystems. Amsterdam: Elsevier; 2000. pp. 319–343. [Google Scholar]
- Sarbu SM, Popa R. A unique chemoautotrophically based cave ecosystem. In: Camacho AI, editor. The Natural History of Biospeleology. Madrid: Museo Nacional de Ciencias Naturales; 1992. pp. 637–666. [Google Scholar]
- Sarbu SM, Kinkle BK, Vlasceanu L, Kane TC, Popa R. Microbiological characterisation of a sulfide-rich groundwater ecosystem. Geomicrobiology Journal. 1994;12:175–182. [Google Scholar]
- Sarbu SM, Kane TC, Kinkle BK. A chemoautotrophically based cave ecosystem. Science. 1996;272:1953–1955. doi: 10.1126/science.272.5270.1953. [DOI] [PubMed] [Google Scholar]
- Schellenberg A. Schlüssel des Amphipodengattung Niphargus mit Fundortangaben und mehreren neuen Formen. Zoologischer Anzeiger. 1935;111:204–211. [Google Scholar]
- Schiödte JC. Undersögelser over den underjordiske Fauna i Hulerne i Krain og Istrien. Oversigt over det Kongelige danske Videnskabernes Selskabs Forhandlinger og dets Medlemmers Arbeider i Aaret. 1847;1847:75–82. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sket B. The ecology of anchihaline caves. Trends in Ecology & Evolution. 1996;11:221–225. doi: 10.1016/0169-5347(96)20031-x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theede H, Ponat A, Hiroki K, Schlieper C. Studies on the resistance of marine bottom invertebrates to oxygen-deficiency and hydrogen sulphide. Marine Biology. 1969;2:325–337. [Google Scholar]
- Thurber AR, Jones WJ, Schnabel K. Dancing for food in the deep sea: bacterial farming by a new species of Yeti crab. PLoS ONE. 2011;6:e26243. doi: 10.1371/journal.pone.0026243. doi: 10.1371/journal.pone.0026243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda G, Yamada A, Nakano K, Arita NO, Yamasaki H. Colonization of Sulfurovum sp. on the gill surfaces of Alvinocaris longirostris, a deep-sea hydrothermal vent shrimp. Marine Ecology. 2008;29:106–114. [Google Scholar]
- Trontelj P, Douady CJ, Fišer C, et al. A molecular test for cryptic diversity in ground water: how large are the ranges of macro-stygobionts? Freshwater Biology. 2009;54:727–744. [Google Scholar]
- Watsuji T, Nakagawa S, Tsuchida S, et al. Diversity and function of epibiotic microbial communities on the galatheid crab, Shinkaia crosnieri. Microbes and Environments. 2010;25:288–294. doi: 10.1264/jsme2.me10135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Niphargid samples analysed molecularly in this study. Samples are colour-coded according to niphargid species.
Niphargid samples analysed by scanning electron microscopy (SEM). Samples are colour-coded according to niphargid species.
Data Availability Statement
All DNA sequences obtained in the present study were deposited in GenBank (accession numbers KF290023-KF290376 and KF362044-KF362049). Alignments were deposited in Dryad (doi:10.5061/dryad.qm636).