Abstract
Interaction between biomaterials and cells is a critical aspect for successful application of tissue engineering research. Technological advances within the past decade have enabled a number of studies to investigate how the spatial organization of cell-adhesive ligands impacts complex and rich cell behaviors ranging from adhesion to differentiation. Cells in their native environment are surrounded by chemical and physical factors spanning a range of length scales from nanometers to hundreds of microns. Furthermore, signals in the form of cell-adhesive ligands presented from this environment in different size scales and/or geometrical arrangements can change how a cell senses and responds to its surroundings. Biology can thus convey information not only in the concentration of a ligand but through its ability to change the spatial organization of these cues, raising questions both on the mechanisms by which it patterns such information and on the means by which a cell interprets it. This review discusses major findings associated with various systems developed to study cell-adhesive ligand presentation as well as an overview of the important material systems used in these studies. Promising material systems to further investigations in this field are also examined. Future directions will likely include determining how cells sense local and global ligand concentrations, understanding underlying mechanisms that regulate cell behaviors, and investigating the function of more complex cell types and diverse ligands.
Keywords: Biomaterials, Cell culture, Mammalian cells, Polymers
Abbreviations: —
E-beam, electron beam lithography; ECM, extracellular matrix; MSC, mesenchymal stem cell; PDMS, poly(dimethyl siloxane); PEG (PEO), polyethylene glycol (polyethylene oxide); PS, polystyrene; RGD, arginine-glycine-aspartic acid peptide sequence
1 Introduction
The engineering of biomaterial surfaces and cell-adhesive ligands is an active and growing field for both basic biological investigation and biotechnological application. A major challenge to the development of artificial environments that can control cell function is our currently incomplete mechanistic knowledge of how natural extracellular matrix (ECM), as well as other solid phase components within a microenvironment, regulate a cell. Thus, advances in basic knowledge of how, for example, the spatial organization of ECM cues modulate cell behavior will both advance cell biology and enable the design of synthetic, biomimetic versions of in vivo environments to aid biomedical applications including tissue engineering.
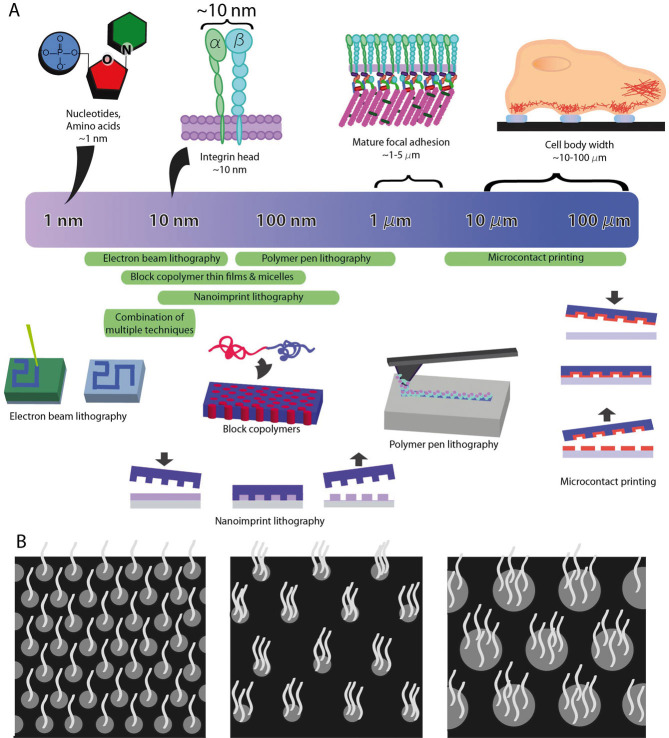
As depicted in Fig. 1A, biological processes on length scales ranging from nanometers to microns are important for understanding and engineering cell adhesion and downstream responses to ligand presentation. Integrins, a family of transmembrane heterodimeric adhesion receptors that interact with specific components of the ECM both biochemically and mechanically, connect the extracellular ECM with the intracellular cytoskeleton. The head diameter of an integrin is approximately 10 nm, which therefore sets the theoretical minimum scale at which these receptors can “resolve” adjacent ligands [1]. As receptors and ligands bind, cell-matrix interactions can be sorted into three additional, mechanistically defined size scales. At the smallest extreme, individual ECM proteins (e.g., laminin) and fiber-forming proteins (e.g., collagen, fibronectin, and elastin) have dimensions ranging from 50 to 300 nm [2, 3]. At the next larger size scale, mature focal adhesions, which concentrate both biomechanical linkages to the substrate and also regulatory information to the cell’s interior, are roughly 1–5 μm in size [4]. Finally, individual cells and their processes extend to approximately 10–100 μm in size, with features on this scale regulating processes such as motility and cellular orientation.
Figure 1.

(A) Length scales for cellular components and practical resolutions of micro/nanoscale patterning techniques. “Combinations of multiple techniques” refers to combination of electron beam lithography, nanoimprint lithography, metal evaporation and lift-off. (B) Schematic illustrating difference between global and local ligand concentrations. Global concentration refers to the average concentration of ligands presented across a surface: μm–mm in size (held constant in all three patterns). In contrast, local concentration refers to the concentration heterogeneity on size scales comparable to adhesive complex components: tens to hundreds of nanometers (different for the three patterns depicted), or an adhesive complex: ∼1 to 5 μm. In many studies discussed here, local concentration refers to the concentration of adhesive ligands within a cluster or isolated area on a patterned adhesive surface.
To date, a variety of different patterning techniques have been developed to exogenously present biomechanical cues on different biomaterial substrates and at a range of such size scales to investigate biological consequences, pushing the envelope on the minimum feature size down to the nanometer scale [5–7]. In this review, we discuss these technological developments and how they can be harnessed to gain further insight into the biomechanical regulation of cells. Additionally, we will also discuss how biologically diverse cells could be affected by presentation of a repertoire of cell-adhesive cues to aid our understanding cell-substrate interactions.
Numerous studies conducted with standard cell lines (such as fibroblasts, endothelial cells, and osteoblasts) [8, 9] have enabled a detailed understanding of focal adhesion structure and other downstream behaviors that result from cell regulation by the substrate [4, 10, 11]. These well-studied cells have very defined adhesive structures and are thus suitable for initial investigations of how cell processes and behaviors can be affected by the presentation of cell-adhesive ligands. However, much remains to be explored. For example, these approaches are increasingly being applied to other medically relevant cell types – such as stem cells and cancer cells – with a focus on how cell-specific processes are regulated by the geometry of ligand presentation. Additional stem cell types, such as pluripotent stem cells, are likely to be studied in the future. In addition, while some integrin–ligand interactions have been well investigated (e.g., the integrin–ligand Arg-Gly-Asp, RGD), future work is likely to encompass a broad range of cell-adhesive proteins and peptide sequences. Interfacing more complex cell types with more biomimetic surfaces may aid biotechnological applications such as tissue engineering.
2 Cell adhesion to ECM
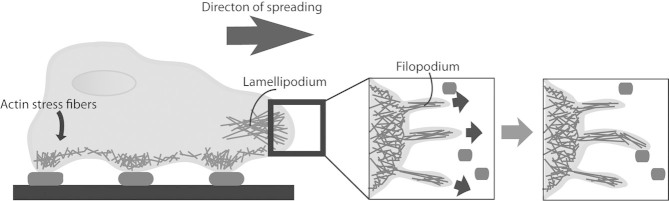
Before proceeding to a discussion of material fabrication technologies, it is important to discuss cell interactions with natural surfaces in greater depth. Cell adhesion is a critical process that mediates cell communication with its surrounding physical environment via many complex extracellular and intracellular interactions. An in-depth discussion of the actions occurring at cell–ECM focal adhesions can be found in many review articles and are only briefly mentioned here [4, 10, 11]. When a cell initially encounters cell-adhesive ligands, adhesion receptors attach to key peptide or glycan ligands. These cell-adhesive ligands are often short polypeptide sequences within ECM proteins that bind directly to integrins and initially form nascent adhesions for cell attachment [12]. Upon forming complex linkages with the cellular cytoskeleton, integrins mechanically pull on the ligands, and this cell-generated force from actomyosin contraction is balanced by ECM resistance and surrounding cells [11, 13]. Next, cell spreading occurs as actin-rich lamellipodia (sheet-like protrusions) extend from the cell body to enable a larger adhered area and in some cases subsequent cell migration. Lamellipodia extend approximately 2–4 μm away from main cell body in the direction of spread (Fig. 2) [4]. Additionally, filopodia are finger-like protrusions that – at a couple microns in length, a few hundred nanometers wide, and microns apart – extend even further from lamellipodium and search for cell-adhesive ligands in the environment [14].
Figure 2.
Cell spreading and searching for adhesive areas (dark ovals) using sheet-like lamellipodium and finger-like filopodia.
During these processes, focal adhesion maturation begins with integrin activation and clustering into larger complexes, which in turn recruit focal adhesion-associated proteins to connect the integrins to the cytoskeleton. Actin monomers are then recruited and polymerized to form actin microfilaments, which ultimately form the axial bundles termed stress fibers at the “bottom” of the cell, radially oriented fibers found near the leading edge, or networks found throughout the cytosol [15–17]. Nascent adhesions are less than 250 nm in size and contain only the focal adhesion-associated proteins paxillin and talin. However, upon increased mechanical tension from the actin cytoskeleton to the integrin–ECM connection, these adhesions mature into focal complexes approximately 500 nm in size that contain additional proteins including focal adhesion kinase (FAK) and α-actinin [4]. The complex either proceeds to mature focal adhesions that are 1–5 μm in size and contain vinculin and zyxin if the integrin–ECM connection can provide sufficient resistance to the cytoskeleton-generated tension, or alternatively the adhesion complex can dissociate at any step and allow the filopodia to search for another adhesive site [4, 18].
Focal adhesion information can be relayed to the rest of the cell to modulate cell function via biochemical or mechanical signals, and these two processes are highly integrated. The biochemical response from focal adhesions starts from recruited signaling proteins such as FAK and Src, which can initiate signaling cascades that propagate throughout the cell. Downstream signaling effectors and processes can include the PI3K/Akt pathway, RhoGTPases, and target gene expression, which collectively contribute to regulating cell motility, morphology, polarity, size, adhesion, division, and differentiation [19]. Mechanotransduction refers to the mechanisms by which cells sense mechanical stimuli (such as integrin–ECM resistance to cell-generated forces) and convert that information into biochemical signals that can alter cell function and cellular or extracellular organization. As with biochemical signaling, there are clear mechanisms by which mechanical information can be transduced from cell surface receptors through the cytoskeleton and to the nucleus. For instance, some speculate that the cytoskeleton may pull on the nucleus which could change its shape, allowing different components to transfer in or out [20]. In reality, as discussed in greater detail in other reviews, the processes of mechanotransduction and biochemical signal transduction are highly integrated and at many junctures are effectively indistinguishable [20–24]. Finally, advances in technologies to control nanoscale surface patterning can affect cell function though both signaling processes, which offers opportunities to gain basic biological knowledge and enhanced control over cell function for biotechnological application.
3 Types of presentation of cell-adhesive ligand
3.1 Micron-sized ligand-covered islands
3.1.1 Microcontact printing
As mentioned previously (Fig. 1), mature focal adhesions are roughly 1–5 μm in size, and cell-body widths are roughly 10–100 μm. A system that can pattern ligands onto substrates in this range is thus ideal for creating patterns that can probe focal adhesion size and investigate effects of limiting the area on which cells can attach and spread. This ∼5 to 100 μm range is possible with microcontact printing, a method that uses a soft polymeric stamp – typically poly(dimethyl siloxane) (PDMS) – replicated from a photolithography-made master mold to stamp self-assembled monolayers (SAMs) or biomolecules in specific patterns onto a substrate (see Fig. 1) [6]. The areas in between the SAMs or biomolecules deposited by a stamp can be passivated to avoid cell or protein adhesion to these regions, and hydrophilic polymers such as polyethylene glycol (PEG) and poly(hydroxyethylmethacrylate) (PHEMA) as well as proteins such as bovine serum albumin (BSA) and casein are often used for this blocking [25–27]. The aspect ratio (height/length of features) of the stamp and surface spreading of the substance being transferred create a lower limit on pattern size of roughly a few hundred nanometers [28, 29]. Possible ways to extend this technology exist to print patterns with much smaller length scales (nanoimprint lithography); however, this very elaborate process necessitates specialized coatings or rinse liquids to create a viable PDMS stamp without pattern collapse or other deformations [30, 31].
There has now been considerable cell biology work using microcontact printing, and multiple landmark studies will be mentioned here. Chen et al. [32] utilized microcontact printing to understand the effect of cell morphology and size on the proliferation and death of endothelial cells. Fibronectin was printed in various patch sizes, and as microscale adhesive island area increased, endothelial cell DNA synthesis increased, and apoptosis decreased [32]. In order to characterize whether this effect was due to ECM contact area or spread cell area, new patterns were created to allow cell spreading but offered much less actual ECM contact area within the patterns. Results showed that a larger spread area increased growth and decreased apoptosis, suggesting endothelial cells are more affected by spread area than actual ECM contact area [32].This discovery implies that cells do not require a continuous area of ECM in order to attach to a surface and instead need enough ECM contact area in order to resist cytoskeleton-generated forces and allow the cell to spread. A subsequent study found that fibronectin island size can also affect stem cell differentiation. Mesenchymal stem cells (MSCs) were cultured in medium that induces mixed lineage differentiation. Osteoblast (bone cell) differentiation increased, and adipocyte (fat cell) differentiation decreased as island size, therefore also cell spread area, increased [21]. Further analysis indicated that the GTPase RhoA and cytoskeletal tension underlay the effects of cell shape on lineage commitment [21]. These results showed that more biologically diverse cells can sense and respond to differences in ECM patterns, and therefore suggested that ECM presentation may impact many types of cellular behaviors in complex cells such as stem cells, cancer cells, etc.
Based on these studies and others using microcontact printing, cell spread area, ECM contact area, and distances between focal adhesion should all be taken into account in investigating substrate effects on cell behavior [33, 34].
3.2 Ligand clustering versus random presentation
At a smaller size scale, an interesting question is whether cells can sense differences in the spatial organization of adhesive ligands. For example, a cell may interact with its surroundings by clustering adhesive ligands in the formation of a nascent adhesion [16]. For this reason, clustered cell-adhesive ligands may allow the integrins to bind in an already clustered format, which could affect the dynamics or extent of a mature focal adhesion formation and affect both cell force generation and biochemical signaling.
3.2.1 PEO stars
In landmark studies investigating how cells respond to random versus clustered presentation of ligands, polyethylene oxide (PEO) star tethers were used [35, 36]. This system allowed for independent control over average surface ligand concentration (global concentration) and average number of ligands per cluster (ligand cluster size) with an inert PEO background that resisted cell adhesion and protein adsorption. Furthermore, the study used a minimal RGD-containing synthetic peptide to limit non-integrin receptor interactions with the substrate and avoid complexities such as unfolding of a substrate protein to reveal cryptic binding sites [36]. Cell speed increased as clus-ter size increased (from 1 to 9 RGDs/cluster) at similar global concentrations. Likewise, as global concentration increased so did the cell speed until saturation for each cluster size [36]. An analogous study showed that as global density increased, cell adhesion strength also increased at constant cluster size [35]. Additionally, increasing cluster size (from ∼1.7 to 5.4 RGDs/cluster) also increased the adhesion strength of the fibroblasts at constant global density [35]. These studies established that cells can sense and respond not only to overall, macroscopic surface ligand density but also local geometric features in the presentation of those ligands.
3.2.2 Alginate gels
Once ligand clusters were shown to have an effect on cell adhesion and motility, subsequent systems were developed to address additional questions about cell responses to ligand cluster density and global density. Alginate gels do not allow cell attachment in the absence of cell-adhesive ligands and are relatively biocompatible, and they therefore offer a tunable system in which cells attach only where cell-adhesive ligands are presented [37]. An alginate hydrogel system was designed to independently change overall ligand density (global concentration), number of ligands clustered in one location (local concentration), and idealized spacing between ligand clusters from roughly 40 to 170 nm. In this study, local concentration refers to the number of ligands clustered within the same area approximately tens of nanometers in size, spatially separated from other ligand clusters tens of nanometers away as depicted in Fig. 1B.
The global density of RGD peptide could be held constant while the number of peptides per cluster, and therefore also the spacing between clusters, was increased (increasing local density). Monte Carlo models were used in parallel to infer average numbers of peptides per cluster and distances between clusters [37]. Trends showed that at a fixed global ligand density, increasing the spacing between RGD clusters and therefore increasing the number of RGDs per cluster (local concentration) decreased the growth rate and increased cell spread area [37]. This result does not follow an anticipated trend that increased spreading is correlated with increased proliferation, potentially due to the nature of ligand presentation in clusters from the alginate.
3.3 Receptor-sized ligand patterns
Although microcontact printing allows for an in-depth analysis of cell responses to ligand patterns roughly the size of focal adhesions, an understanding of how cells respond to ligands presented at smaller length scales could elucidate more specific interactions of integrins with hierarchically structured proteins in the ECM. An integrin head diameter is roughly 10 nm, at least an order of magnitude smaller than the pattern size possible with microcontact printing, and therefore may interact with the surrounding ECM at a length scale of this order.
3.3.1 Block copolymer micelle lithography
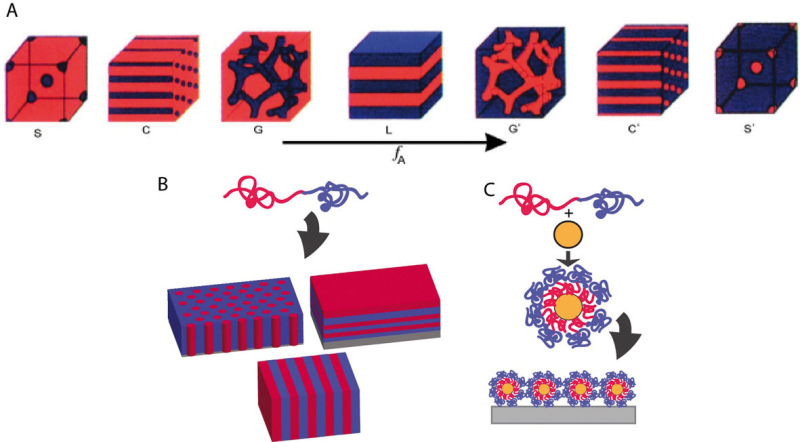
Block copolymers naturally assemble into micelles in solution on the appropriate length scale for investigating how ligand patterns on the order of integrin receptor sizes can affect cell adhesion. Block copolymers consist of two chemically dissimilar long molecules joined end-to-end, depicted as red and blue in Fig. 3. In the most well studied block copolymer micelle lithography system, gold nanoparticles are coated with the block copolymer in solution to form micelles as shown in Fig. 3A. Hard sphere repulsions between the micelles force them to generate roughly hexagonal networks when the solution is dip-coated on a substrate. The polymer can then be removed by heating, and the gold surfaces can be functionalized with adhesive peptides via thiol chemistry [38]. In studies using this approach, the gold nanoparticles have a diameter of roughly 10 nm so that only one integrin should be able to bind per RGD-coated gold particle, with the inter-particle spacing roughly controlled via the dip-coating process. As a result, the actual number of ligands attached to each gold particle is irrelevant, as each particle can presumably bind to only one integrin. This system is ideal for patterning cell-adhesive ligand on the same scale as integrin receptor sizes, though it does not allow for the average overall ligand concentration (global concentration) to be varied independently of the number of ligands per local, adhesive complex sized feature (local concentration).
Figure 3.
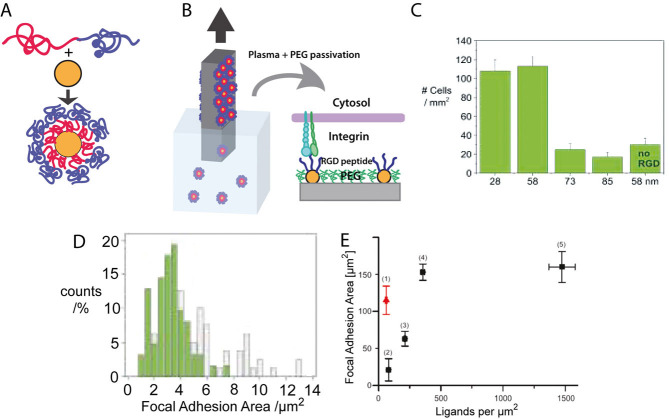
Block copolymer micelle lithography technique showing (A) Gold nanoparticle micelles forming from combination of block copolymer with gold nanoparticle. (B) Dip-coating and processing techniques to create gold nanoparticle patterns passivated with PEG containing nanoparticles similar in size to integrin head diameter. (C) Difference in adhesion between ligand spacings (D) comparison of focal adhesion characteristics between extensive (continuous) 58 nm spacing (white) and micropatterns of 58 nm spacing (green) [38]. (E) Focal adhesion area versus global ligand density of (1) micropatterns with 57 nm gold spacing, (2) extensive 120 nm gold spacing, (3) extensive 74 nm gold spacing, (4) extensive 57 nm gold spacing, and (5) extensive 28 nm gold spacing (previous data) [41]. Reprinted with permission from [38] and [41].
Spacings of individual ligands in a range roughly between 20 and 100 nm suggested that osteoblasts cannot adhere to ligands spaced on gold nanoparticles with global concentrations of 190 dots/μm2 or less (73 nm or more apart) as shown in Fig. 3C [38]. This system was also used to show that fibroblasts cannot form characteristic focal adhesions at the lower global concentrations and larger ligand spacings [39–41]. Processing by photolithography or electron beam lithography (E-beam) lithography was applied to the gold nanopattern with a global concentration of 280 dots/μm2 (58 nm spacing) to create micron-size patches (micropatches) of this nanopattern separated by micron-size unpatterned areas; these new “micro-nanopatterned surfaces” were created to determine if local ligand concentration within these micropatches was more important to cell attachment and spreading than ligand concentration across the entire surface (global concentration) [38, 41]. Higher values of focal adhesion area and detachment force needed to remove a cell were found when the cells grew on surfaces with micropatches than when cells grew on surfaces with extensive (continuous, unpatched) nanopatterns even at a lower global concentration (larger spacing). These results suggest that the micropatches improved cell adhesion despite the lower global concentration [41]. That said, such “micro-nanopatterns” have only been made with a local concentration of the micropatches around 300 dots/μm2, and therefore cell responses from micro-nanopatterns with higher and lower local concentrations have not yet been investigated.
Another study broaching the role of global and local concentrations, by comparing ordered and disordered ligand patterns at similar global concentrations, showed an increase in projected area and cell number with disordered patterns [42]. This increased adhesion was attributed to some ligands being arranged sufficiently close for integrins to cluster and form focal adhesions [42]. This system thus elucidated that there may be a minimum ligand concentration or maximum spacing between ligands to readily enable cell attachment. However, global and local concentration change in tandem when creating extensive nanopatterned surfaces, and pattern stability may be transient.
A modification of this system by the Ding group was recently used to investigate whether individual ligand spacing has an effect on MSC differentiation and morphology. The gold nanoparticle pattern was transferred to a PEG hydrogel by attaching an N, N-bis(acryloyl) cystamine linker to the gold nanoparticles for crosslinking to the hydrogel [43, 44]. Ligands on gold particles spaced 37–124 nm apart (global concentrations between ∼60 and 730 RGD/μm2) on the PEG hydrogels showed decreasing trends in cell number and cell spread area as RGD nanospacing increased and global concentration decreased. However, the dependence was continuous and did not exhibit a drastic drop in cell attachment below a ∼200 RGD/μm2 global concentration (above the ∼70 nm spacing) mark [44]. This effect could be due to differ-ences in mechanical properties between the hard plastic and the hydrogel surface or from a difference in focal adhesion formation and focal adhesion-associated proteins required between MSCs and non-stem cell lines. This system also showed that osteoblast differentiation increased and adipocyte differentiation decreased as global and local concentrations decreased in tandem (ligand spacing increased) when MSCs were cultured in medium that induced mixed differentiation (osteogenic and adipogenic) [44]. That said, work by other groups shows an opposite trend in which osteoblast differentiation increased with strong adhesion on large ECM-patterned areas and with stiffer surfaces [21, 45].
These block copolymer micelle lithography studies have been useful in elucidating a range of ligand spacings and global concentrations that affect cell adhesion and focal adhesion formation in common cell lines. Before these studies were performed, it was unknown whether cells could even sense ligand patterns at this small of length scale. The work with fibroblasts and MSCs will encourage additional studies with more complex ligand presentation and additional cell classes.
3.3.2 Block copolymer thin films
Block copolymer thin films provide the ability to independently change local and global concentration of ligand patterns with sizes and spacings on the order of ligand receptor dimensions (Fig. 1B). Block copolymers consist of two or more different types of polymers covalently attached to one another, and they can self-assemble into specific structures based on the competition between the free energy of mixing and entropy of the chains [46]. These specific morphologies include spheres, hexagonally close-packed cylinders, or lamellae depending on the volume fraction and molecular weights of each block (Fig. 4A) [47]. Varying the volume fraction and/or molecular weight of one or both of the blocks can change the size and spacing of the morphology (i.e., larger cylinders spaced further apart). The range of feature size (i.e., cylinder diameter or lamellae thickness) available using this technique is roughly 5–200 nm. Some of the issues with this method are the synthesis time to create the block copolymers, modifying substrate surface energy to create the desired morphology [7, 48], and for standard polymer chemistries subsequent challenges with selective ligand attachment to one block but not the other.
Figure 4.
(A) Block copolymer self-assembly structures with increasing volume fraction of the blue block [47]. Morphology changes from blue spheres (S) to blue cylinders (C) to blue gyroid (G) to lamellae (L) then back to red gyroid (G’) to red cylinders (C’) to red spheres (S’). (B) Self-assembly of block copolymer thin films. (C) Self-assembly of block copolymer micelles with gold nanoparticles. Reprinted with permission from [47].
One of the first block copolymer thin film methods to pattern adhesive ligands for cell growth used a polystyrene-block-poly(ethylene oxide) (PS-PEO) copolymer with a maleimide (Ma) group attached to the PEO chain end for subsequent RGD peptide attachment [49]. PS-PEO is commonly used to obtain well-ordered upright cylinders in thin films, and this block copolymer system was self-assembled into hexagonally close-packed cylinders that were perpendicular to the top surface such that PEO domains were visible in a PS matrix. PS homopolymer was blended with the block copolymer to increase the center-to-center PEO-Ma spacing from roughly 34 to 62 nm (∼860 to 260 RGD/μm2 if every PEO-Ma had only one RGD attached); however, the PEO domains become less uniform in size and spacing at around 50% block copolymer composition [49]. MSCs grown on these surfaces showed an increase in adipocyte differentiation and decrease in osteoblast differentiation as average PEO center-to-center spacing increased from 34 to 62 nm, though the effect of spacing may be convoluted by the uniformity of PEO domains [50]. One challenge with this chemistry, however, is PEO swells in water and may thus cover parts of the underlying pattern in aqueous conditions necessary for cell culture.
Another recent system used PS-PEO with perfluorooctanethiol (PFOT) as well as allyl glycidyl ether (AGE) groups attached within the PEO domain, the latter for RGD peptide conjugation [51]. PFOT was included to modify interfacial interactions and thus enhance phase segregation (due to the fluorine groups not mixing with hydrophilic domains). It also maintained the ordered morphology in aqueous conditions by not allowing the PEO domain to swell [51]. AGE groups connected the peptide to the block copolymer, and therefore increasing the number of such groups increases the amount of peptide present on the surface. This system allows one to change the size and spacing of adhesive areas but requires different syntheses for different peptide surface concentrations. To date, this system has been used with fibroblasts, which were able to attach and remain viable on the ligand-patterned surfaces [51]. A different type of block copolymer has been used to pattern biomolecules but has not yet been used with cells. This polystyrene-block-poly(2-hydroxyethyl methacrylate) PS-PHEMA system utilized Click chemistry to functionalize the PS domains with azide-tagged proteins in a protein-repellant PHEMA matrix [52].
These studies suggest a new ability to pattern ligand in an ordered manner that can allow independent changes in local and global concentration on the same length scales that were shown to modulate adhesion in studies using block copolymer micelle lithography (previous section). These studies may allow for more flexibility in the creation of patterns. Patterns that may define specifically where focal adhesions can start forming and limit how many integrins can bind could elucidate more about how cells respond to the specific size and locations of integrin–ECM contacts.
3.3.3 Electron beam lithography and nanoimprint lithography
Production of ligand patterns that can probe length scales on the order of integrin head size and smaller could allow more in-depth analyses of integrin interactions with the surroundings. E-beam provides a very precise and controlled means to pattern surfaces in the nanometer range; however, patterning using an E-beam entails serially creating one spot at a time. While E-beam technology has advanced significantly, this technique is thus more suited for complex patterning of small areas as opposed to large areas with imperfect periodicity created by self-assembly. That said, gold or other metals can be precisely deposited in nanometer-size patches with nanometer-sized distances between patches, allowing for exact production of patterns on a scale extremely difficult if not impossible to achieve with other techniques. Alkanethiol chemistry can then be used to attach cell-adhesive ligand to the patterns. Therefore this technique can probe length scales on the order of integrin head size and integrin clustering (see Fig. 1A). In an approach with related capabilities, nanoimprint lithography is a technique which uses a template created by lithography to imprint nanometer-sized features into a polymer resist [5].
E-beam lithography can be combined with nanoimprint lithography, along with some other nanofabrication techniques, to produce features that are sub-10 nm in size [53]. This combination has been used to explore whether fibroblast attachment requires a minimum number of RGD peptides per cluster (minimum local concentration) and whether spacing within and between clusters can also affect adhesion [53]. One result with this system showed that fibroblasts spread when a minimum number of peptides are presented per cluster [53]. The same percentage of spread cells (i.e. cells that switched from round to flat with filopodia and maintained this spread form) was observed independent of local concentration once local concentration exceeded a minimum threshold of 4 dots/cluster (and with a constant global concentration of 50 RGD/μm2) [53]. This suggests that fibroblasts can be more sensitive to local concentration of ligand than global concentration at this very low global density. This result also may suggest that fibroblasts need a minimum number of integrins clustered together to start forming a focal adhesion. Another aspect of this experiment noted that varying global ligand density between ∼50 and ∼200 RGD/μm2 showed the same percent of spread cells [53] even though previous work using block copolymer micelle lithography showed a difference in cell adhesion in this range of global concentrations when changing from extensive (continuous) nanopatterns to micropatches of nanopatterns [38]. This may suggest that the lower limit for global or local concentration necessary for cell adhesion is reduced upon clustering of the ligands. Performing similar studies using different global concentrations could provide a more detailed understanding of how cells respond to these types of ligand patterns.
3.4 Ligand-patch arrays
3.4.1 Polymer pen lithography
An array of ligand patterns or patch sizes/shapes could be used to screen cell responses to many variables simultaneously. Simple, continuous, and symmetric patterns can be easily fabricated via self-assembly techniques; however, they do not allow for multiple patterns on the same surface. Complex patterns can be created by microcontact printing or photolithography, yet require new masks or templates for any new patterns. In contrast to both of these options, dip pen nanolithography is a technique that can create arrays of different types of materials like alkanethiols or proteins in specific sizes and spacings with sub-50 nm resolution by taking advantage of the water meniscus formed when an atomic force microscopy (AFM) tip approaches a solid substrate, thereby depositing molecules in a controlled manner [54]. However, this technique does have similar limitations in throughput as E-beam lithography. To address this challenge, a variant technique called polymer pen lithography uses an array of soft polymer pens instead of an AFM tip and a scanning probe microscope to control the movement of the array [55]. Polymer pen lithography was recently used to create arrays of fibronectin patches of different sizes, based on the pressure and angle of the thousands of polymer pens depositing the fibronectin, to systematically screen MSC adhesion to different patch sizes [56]. This system showed that MSCs could differentiate to a greater extent into osteoblasts on the fibronectin patches with side lengths of 300 nm or 1 μm compared to nonpatterned surfaces, in the absence of differentiation-inducing media [56]. This type of system thus enables rapid screening of a wide range of conditions and sizes for numerous downstream applications.
3.5 Fluid surfaces with confinements
In addition to adhering to ligands presented from solid substrates, cells interface with the surfaces of neighboring cells. Ligands present in such fluid surfaces can undergo diffusion, and several synthetic systems have been developed to study how membrane fluidity can affect cell adhesion and function.
3.5.1 Lipid bilayers
Supported phospholipid bilayers, systems in which ligands are presented from a cell adhesion-resistant background surface that mimics the plasma membrane of a neighboring cell, have been used in many studies to mimic cell–cell interactions [57, 58]. For example, lipid bilayers can present ligands of many different kinds simply by attaching lipid tails to the ligands, which can thus easily be incorporated into the lipid bilayer. Surfaces can also be micropatterned with protein first, followed by lipid bilayer deposition around the protein [59, 60]. Lipid bilayers have recently been utilized to study stem cells, and in this study neural stem cells could adhere, proliferate, and differentiate on an RGD-presenting surface [29].
One interesting additional feature of these systems is that physical barriers microns in dimension can be added to restrict movement of ligands, for example to impede large scale receptor clustering and thereby gain insights into its role in cell function. Some methods to introduce barriers include manual scratching or blotting with a PDMS stamp, microcontact printing, or prefabricated barriers from photolithography [61]. One system included RGD peptide ligands in a lipid bilayer along with physical barriers to limit the movement of these integrins in order to understand the role of RGD peptide clustering in cell adhesion and spreading [62]. Results tracked the focal adhesions formed with these ligands and showed movement forward for active protrusion and then rearward for retraction [62]. Supported lipid bilayers allow for more biologically relevant responses such as clustering of ligands to occur because the presented ligands can freely diffuse.
4 Open questions
Over the past decade there have thus been many advances in the field of patterning cell-adhesive ligand to investigate how cell adhesion and other related behaviors are affected based on the presentation of ligand. We have learned that multiple responses – including proliferation, adhesion, viability, migration, and differentiation – of common or differentiated cell types are affected by spread cell area more than ECM contact area [21, 32]. This result suggests that as long as the focal adhesions can provide sufficient resistance to the cytoskeleton-generated tension, the actual area connecting to ECM may be less important for cell proliferation and viability. We have learned through several studies that ligand clustering improves adhesion and motility and that there may be a minimum number of ligands per cluster required for cell adhesion [35, 36, 53]. The minimum cluster size could result from a minimum number of integrins clustering together to recruit necessary focal adhesion-associated proteins and form a mature focal adhesion. Ligand clustering may also lower the minimum global concentration necessary for cell attachment and spreading. New surface engineering techniques have also shown that standard cell lines can detect differences in low global concentrations from ligands spaced apart on the order of receptor size [38]. Beforehand systems existed only to present ligands in a way that averaged out any differences in spacing between ligands. There may also be a difference between stem cell and fibroblast adhesion, as suggested with MSCs adhesion to surfaces at lower global concentrations than fibroblasts [44]. However, there are still many future questions that need to be explored.
4.1 Local concentration versus global concentration
Several studies have investigated whether local concentration or global concentration of cell-adhesive ligand is more important for cell adhesion and spreading. This question is difficult to address since both local (i.e., concentration heterogeneity on size scales comparable to an adhesive complex or adhesive complex components) and global (i.e., average concentration across a surface) concentration change simultaneously when ligand spacing changes. Several results indicate that local concentration may impact cell adhesion and spreading more than global concentration, including investigations of differences between confluent and micron-sized patches of nanopatterned ligand [38, 41], comparisons of ordered and disordered ligand presentation [42], and exploration of minimum numbers of ligands required per cluster for cell adhesion [53]. However, underlying molecular size scales and mechanisms remain to be investigated. Tightly controlled studies that hold as many variables as possible constant while only changing one or two in order to pinpoint exactly which surface property affects cell adhesion and other behaviors will enable even more precise statements about integrin and focal adhesion interactions with ligands (e.g., Fig. 1B). Many integrin ligands that exist in the ECM are multivalent and could therefore stimulate integrin clustering by themselves [1]. Local concentration patterning could thereby impact how many integrins can cluster together and how these clustered integrins recruit focal adhesion-associated proteins to start to form a mature focal adhesion. However, global concentration would affect the average number of adhesive bonds across the entire cell surface, also seemingly an important parameter. In addition, with the advent of fluid surfaces whose spatial patterns can be modulated, the roles of local and global adhesive patterning at cell–cell junctions can now be explored.
4.2 Cell sensing and responses to cell-adhesive ligand-patterned surfaces
In addition to trying to understand why local and/or global concentration may impact cell engagement with a surface, mechanisms by which ligand–receptor bonding regulates downstream cell behavior can be further explored. Considerable work has investigated how focal adhesion-associated proteins convey information from the ECM to the cytoskeleton [18]. In addition, increasing numbers of studies have established that these links can regulate complex downstream cellular responses to cell-adhesive ligand patterns on the micron scale, such as the work by McBeath et al. [21] showing that RhoGTPase activity and actin–myosin-generated tension affect MSC differentiation. However, an active area of research is how focal adhesions and the cytoskeleton convey mechanical information to the nucleus. Recently, Dupont et al. [63] have shown that transcriptional coactivators YAP and TAZ act as sensors and relay mechanical signals from ECM proteins to alter cell behaviors. Future work will likely establish causal connections between mechanical cues and such transcriptional regulators, as well as between these regulators and downstream cellular function.
4.3 Additional biological contexts
Most of the work with cell-adhesive ligands has been conducted with synthetic peptides containing RGD, a peptide sequence found in ECM proteins including fibronectin, laminin, and fibrinogen [12]. This canonical RGD sequence is clearly an important component of interactions between cell and ECM; however, the full range and richness of such interactions will extend beyond this motif. An increasing number of alternate cell-adhesive ligands have been used in other studies for their particular properties. For example, synthetic peptides were recently used to study the mechanisms of human embryonic stem cells (hESCs) binding to Matrigel surfaces as a step towards creating more defined culture substrates [64]. Others have used self-assembled monolayers as a platform to determine ideal peptide sequences for ESC growth and self-renewal [65]. In addition, bacterial peptide display was also used to screen many peptide sequences to determine which bind neural stem cells (NSCs) and support self-renewal and differentiation [66]. Using a different ligand could elicit recognition from other types of integrins or receptors, and different integrins may adopt different conformations upon connecting to alternate ECM motifs, which could change how cytoplasmic domains assemble and therefore affect how focal adhesion-associated proteins are recruited and how intracellular signals are relayed [1]. Therefore, different cell-adhesive ligands should increasingly be explored.
In addition to exploring more complex receptor–ligand interactions, additional cell types should be investigated. As mentioned previously, the majority of key research conducted with ligand-patterned surfaces uses model cell lines like fibroblasts, osteoblasts, endothelial cells, and others. These cell lines offer the advantages of being fairly straightforward to culture, having well-characterized adhesive structures and morphology, and exhibiting substrate-sensitive behavior such as adhesion, spreading, and migration shortly after engaging with a substrate. Translating such investigations to less characterized cell types that exhibit complex behaviors on longer time scales, such as stem cells, will both advance basic knowledge and take steps towards biomedical application. Several studies for example have investigated the responses of stem cells to RGD peptide sequences spaced at varying distances, primarily with MSCs [44, 50, 67]. This work has benefited from their similar adhesive behavior with fibroblasts, and investigating how more complex behaviors of MSCs and other stem cells – specifically their self-renewal and differentiation – will yield future advances and likely reveal further biological complexities. For example, based on the most recent work on individual RGD ligand spacing with MSCs, the ∼200 RGD/μm2 (∼70 nm spacing) limit found for osteoblasts, fibroblasts, and other common cell lines may not extend to other biologically diverse types of cells [44].
Additional complex behaviors may also be investigated in the future. Behaviors such as proliferation or apoptosis on ligand-patterned surfaces were investigated in foundational work by Chen et al. [32], and this work can be extended with different cells and patterns. Additionally, investigating cell motility, polarization, contractility, cell–cell signaling and organization, and other behaviors on ligand-patterned surfaces may provide useful information for designing biomaterials for implants.
5 Conclusions
Though the past decade has brought many advances in understanding how cells sense and respond to cell-adhesive ligand patterns, many important studies are still needed to elucidate many questions. A deeper understanding of how cells respond to the presentation of cell-adhesive ligands can aid in creating biomaterials that mimic characteristics of the in vivo environment more precisely. This will benefit basic biological studies of how biochemical and biophysical engagement with the ECM regulate cell function, as well as facilitate more controlled biological studies in general. Furthermore, this information could aid in vitro stem cell differentiation into specific lineages, which would benefit applications ranging from drug screening to scalable cell culture systems. Finally, regenerative medicine and tissue engineering efforts are likely to benefit from a deeper understanding of how cells respond to cell-adhesive cue presentation.
David Schaffer is a Professor of Chemical and Biomolecular Engineering, Bioengineering, and Neuroscience at the University of California, Berkeley, where he also directs the Berkeley Stem Cell Center. He received a B.S. from Stanford University in 1993 and Ph.D. from Massachusetts Institute of Technology in 1998, both in Chemical Engineering, then conducted a postdoctoral fellowship at the Salk Institute for Biological Studies. Since moving to Berkeley in 1999, Dr. Schaffer has applied engineering principles to enhance stem cell and gene therapy approaches for neuroregeneration, including the development of technologies to investigate and control stem cell fate decisions and to engineer novel viral gene delivery vehicles. He has received an NSF CAREER Award, Office of Naval Research Young Investigator Award, Whitaker Foundation Young Investigator Award, Technology Review Top 100 Innovator, Biomedical Engineering Society Rita Schaffer Young Investigator Award, and American Chemical Society BIOT Division Young Investigator Award.
Acknowledgments
This work was funded by NIH R01NS074831 and R01ES020903.
The authors declare no conflict of interest.
References
- 1.Springer TA, Wang J-H, Garcia KC. The three-dimensional structure of integrins and their ligands, andconformational regulation of cell adhesion. Adv. Protein Chem. 2004;68:29–63. doi: 10.1016/S0065-3233(04)68002-8. [DOI] [PubMed] [Google Scholar]
- 2.Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 1990;4:148–160. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 3.Structure and Function of Collagen Types , Academic Press, Inc., Orlando, FL 1987.
- 4.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Ann. Rev. Cell Dev. Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo LJ. Nanoimprint lithography: methods and material requirements. Adv. Mater. 2007;19:495–513. [Google Scholar]
- 6.Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat. Protocols. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 7.Segalman RA. Patterning with block copolymer thin films. Mater. Sci. Eng. R-Rep. 2005;48:191–226. [Google Scholar]
- 8.Lim JY, Donahue HJ. Cell sensing and response to micro- and nanostructured surfaces produced by chemical and topographic patterning. Tissue Eng. 2007;13:1879–1891. doi: 10.1089/ten.2006.0154. [DOI] [PubMed] [Google Scholar]
- 9.Altomare L, Farè S. Cells response to topographic and chemical micropatterns. J. Appl. Biomater. Biomech. 2008;6:132–143. [PubMed] [Google Scholar]
- 10.Legate KR, Wickström SA, Fässler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 11.Wehrle-Haller B. Assembly and disassembly of cell–matrix adhesions. Curr. Opin. Cell Biol. 2012;24:569–581. doi: 10.1016/j.ceb.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 13.Gallant ND, Michael KE, García AJ. Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol. Biol. Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogilner A, Rubinstein B. The physics of filopodial protrusion. Biophys. J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 16.Cluzel C, Saltel F, Lussi J, Paulhe F, et al. The mechanisms and dynamics of (alpha)v(beta)3 integrin clustering in living cells. J. Cell Biol. 2005;171:383–392. doi: 10.1083/jcb.200503017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodish, H., Berk, A., Zipursky, S., Cell Organization and Movement I: Microfilaments, in: Ahr, K. (Ed), Molecular Cell Biology,W. H. Freeman, New York 2008.
- 18.Roca-Cusachs P, Iskratsch T, Sheetz MP. Finding the weakest link – exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 2012;125:3025–3038. doi: 10.1242/jcs.095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar CC. Signaling by integrin receptors. Oncogene. 1998;17:1365–1373. doi: 10.1038/sj.onc.1202172. [DOI] [PubMed] [Google Scholar]
- 20.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 21.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 22.Mammoto A, Ingber DE. Cytoskeletal control of growth and cell fate switching. Curr. Opin. Cell Biol. 2009;21:864–870. doi: 10.1016/j.ceb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Allen WE, Jones GE, Pollard JW, Ridley AJ. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 1997;110:707–720. doi: 10.1242/jcs.110.6.707. [DOI] [PubMed] [Google Scholar]
- 24.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 2005;171:209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingshott P, Thissen H, Griesser HJ. Effects of cloud-point grafting, chain length, and density of PEG layers on competitive adsorption of ocular proteins. Biomaterials. 2002;23:2043–2056. doi: 10.1016/s0142-9612(01)00334-9. [DOI] [PubMed] [Google Scholar]
- 26.Reimhult K, Petersson K, Krozer A. QCM-D analysis of the performance of blocking agents on gold and polystyrene surfaces. Langmuir. 2008;24:8695–8700. doi: 10.1021/la800224s. [DOI] [PubMed] [Google Scholar]
- 27.Senaratne W, Andruzzi L, Ober CK. Self-Assembled monolayers and polymer brushes in biotechnology: current applications and future perspectives. Biomacromolecules. 2005;6:2427–2448. doi: 10.1021/bm050180a. [DOI] [PubMed] [Google Scholar]
- 28.Alom Ruiz S, Chen CS. Microcontact printing: a tool to pattern. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 29.Bernard A, Renault JP, Michel B, Bosshard HR, Delamarche E. Microcontact printing of proteins. Adv. Mater. 2000;12:1067–1070. [Google Scholar]
- 30.Jung G-Y, Li Z, Wu W, Chen Y, et al. Vapor-phase self-assembled monolayer for improved mold release in nanoimprint lithography. Langmuir. 2005;21:1158–1161. doi: 10.1021/la0476938. [DOI] [PubMed] [Google Scholar]
- 31.Beck M, Graczyk M, Maximov I, Sarwe EL, et al. Improving stamps for 10 nm level wafer scale nanoimprint lithography. Microelectron. Eng. 2002;61-2:441–448. [Google Scholar]
- 32.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 33.Lehnert D, Wehrle-Haller B, David C, Weiland U, et al. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J. Cell Sci. 2004;117:41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 34.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Micropatterned surfaces for control of cell shape, position, and function. Biotechnol. Progress. 1998;14:356–363. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 35.Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J. Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 36.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 37.Lee KY, Alsberg E, Hsiong S, Comisar W, et al. Nanoscale adhesion ligand organization regulates osteoblast proliferation and differentiation. Nano Lett. 2004;4:1501–1506. doi: 10.1021/nl0493592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold M, Cavalcanti-Adam EA, Glass R, Blümmel J, et al. Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 39.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, et al. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selhuber-Unkel C, Erdmann T, López-García M, Kessler H, et al. Cell adhesion strength is controlled by intermolecular spacing of adhesion receptors. Biophys. J. 2010;98:543–551. doi: 10.1016/j.bpj.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deeg JA, Louban I, Aydin D, Selhuber-Unkel C, et al. Impact of local versus global ligand density on cellular adhesion. Nano Lett. 2011;11:1469–1476. doi: 10.1021/nl104079r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Grater SV, Corbellini F, Rinck S, et al. Impact of order and disorder in rgd nanopatterns on cell adhesion. Nano Lett. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Ding J. Nanostructured interfaces with RGD arrays to control cell–matrix interaction. Soft Matter. 2010;6:3395–3401. [Google Scholar]
- 44.Wang X, Yan C, Ye K, He Y, et al. Effect of RGD nanospacing on differentiation of stem cells. Biomaterials. 2013;34:2865–2874. doi: 10.1016/j.biomaterials.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Bates FS, Fredrickson GH. Block copolymer thermodynamics – theory and experiment. Annu. Rev. Phys. Chem. 1990;41:525–557. doi: 10.1146/annurev.pc.41.100190.002521. [DOI] [PubMed] [Google Scholar]
- 47.Bates FS, Fredrickson GH. Block copolymers – designer soft materials. Phys. Today. 1999;52:32–38. [Google Scholar]
- 48.Albert JNL, Epps III TH. Self-assembly of block copolymer thin films. Mater. Today. 2010;13:24–33. [Google Scholar]
- 49.George PA, Doran MR, Croll TI, Munro TP, Cooper-White JJ. Nanoscale presentation of cell adhesive molecules via block copolymer self-assembly. Biomaterials. 2009;30:4732–4737. doi: 10.1016/j.biomaterials.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 50.Frith JE, Mills RJ, Cooper-White JJ. Lateral spacing of adhesion peptides influences human mesenchymal stem cell behaviour. J. Cell Sci. 2012;125:317–327. doi: 10.1242/jcs.087916. [DOI] [PubMed] [Google Scholar]
- 51.Killops KL, Gupta N, Dimitriou MD, Lynd NA, et al. Nanopatterning biomolecules by block copolymer self-assembly. ACS Macro Lett. 2012;1:758–763. doi: 10.1021/mz300153k. [DOI] [PubMed] [Google Scholar]
- 52.Shen L, Garland A, Wang Y, Li Z, et al. Two dimensional nanoarrays of individual protein molecules. Small. 2012;8:3169–3174. doi: 10.1002/smll.201200673. [DOI] [PubMed] [Google Scholar]
- 53.Schvartzman M, Palma M, Sable J, Abramson J, et al. Nanolithographic control of the spatial organization of cellular adhesion receptors at the single-molecule level. Nano Lett. 2011;11:1306–1312. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran, H., Killops, K. L., Campos, L. M., Advancements and challenges of patterning biomolecules with sub-50 nm features. Soft Matter2013.
- 55.Zheng Z, Daniel WL, Giam LR, Huo F, et al. Multiplexed protein arrays enabled by polymer pen lithography: addressing the inking challenge. Angew. Chem. Int. Ed. 2009;48:7626–7629. doi: 10.1002/anie.200902649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giam LR, Massich MD, Hao L, Shin Wong L, et al. Scanning probe-enabled nanocombinatorics define the relationship between fibronectin feature size and stem cell fate. Proc. Natl. Acad. Sci. 2012;109:4377–4382. doi: 10.1073/pnas.1201086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stroumpoulis D, Zhang H, Rubalcava L, Gliem J, Tirrell M. Cell adhesion and growth to peptide-patterned supported lipid membranes. Langmuir. 2007;23:3849–3856. doi: 10.1021/la062375p. [DOI] [PubMed] [Google Scholar]
- 58.Andersson A-S, Glasmästar K, Sutherland D, Lidberg U, Kasemo B. Cell adhesion on supported lipid bilayers. J. Biomed. Mater. Res. Part A. 2003;64A:622–629. doi: 10.1002/jbm.a.10442. [DOI] [PubMed] [Google Scholar]
- 59.Ochsenhirt SE, Kokkoli E, McCarthy JB, Tirrell M. Effect of RGD secondary structure and the synergy site PHSRN on cell adhesion, spreading and specific integrin engagement. Biomaterials. 2006;27:3863–3874. doi: 10.1016/j.biomaterials.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 60.Kam L, Boxer SG. Cell adhesion to protein-micropatterned-supported lipid bilayer membranes. J. Biomed. Mater. Res. 2001;55:487–495. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 61.Groves JT, Boxer SG. Micropattern formation in supported lipid membranes. Acc. Chem. Res. 2002;35:149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- 62.Yu, C.-H., Law, J. B. K., Suryana, M., Low, H. Y., Sheetz, M. P., Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc. Natl. Acad. Sci2011. [DOI] [PMC free article] [PubMed]
- 63.Dupont S, Morsut L, Aragona M, Enzo E, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 64.Meng Y, Eshghi S, Li YJ, Schmidt R, et al. Characterization of integrin engagement during defined human embryonic stem cell culture. FASEB J. 2010;24:1056–1065. doi: 10.1096/fj.08-126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Derda R, Li L, Orner BP, Lewis RL, et al. Defined substrates for human embryonic stem cell growth identified from surface arrays. ACS Chem. Biol. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- 66.Little LE, Dane KY, Daugherty PS, Healy KE, Schaffer DV. Exploiting bacterial peptide display technology to engineer biomaterials for neural stem cell culture. Biomaterials. 2011;32:1484–1494. doi: 10.1016/j.biomaterials.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsiong SX, Carampin P, Kong H-J, Lee K-Y, Mooney DJ. Differentiation stage alters matrix control of stem cells. J. Biomed. Mater. Res. Part A. 2008;85A:145–156. doi: 10.1002/jbm.a.31521. [DOI] [PubMed] [Google Scholar]