Abstract
The X-linked inhibitor of apoptosis protein (XIAP) is a potent caspase inhibitor, best known for its anti-apoptotic function in cancer. During apoptosis, XIAP is antagonized by SMAC, which is released from the mitochondria upon caspase-mediated activation of BID. Recent studies suggest that XIAP is involved in immune signaling. Here, we explore XIAP as an important mediator of an immune response against the enteroinvasive bacterium Shigella flexneri, both in vitro and in vivo. Our data demonstrate for the first time that Shigella evades the XIAP-mediated immune response by inducing the BID-dependent release of SMAC from the mitochondria. Unlike apoptotic stimuli, Shigella activates the calpain-dependent cleavage of BID to trigger the release of SMAC, which antagonizes the inflammatory action of XIAP without inducing apoptosis. Our results demonstrate how the cellular death machinery can be subverted by an invasive pathogen to ensure bacterial colonization.
Keywords: inflammation, mitochondria, Shigella, SMAC, XIAP
See also: DN Bronner & MXD O'Riordan (October 2014)
Introduction
The X-linked inhibitor of apoptosis protein (XIAP) has been studied intensely in mammalian cancer on the basis of its anti-apoptotic function and its frequently elevated expression in malignant cells (Kashkar, 2010). The anti-apoptotic function of XIAP is regulated by several mitochondrial IAP binding motif (IBM)-containing proteins, including second mitochondria-derived activator of caspases (SMAC, also called DIABLO) and OMI (also called HtrA2), which are released from the mitochondrial intermembrane space (IMS) upon mitochondrial outer membrane permeabilization (MOMP) (Du et al, 2000; Martins et al, 2002). The critical role of XIAP and its antagonization during death receptor-induced apoptosis has recently been demonstrated in XIAP-deficient mice. Specifically, the Caspase-8-mediated proteolytic activation of BID (a BH3-only member of the BCL2 protein family) and the subsequent release of mitochondrial SMAC were shown to be crucial for XIAP antagonization and the execution of apoptosis upon FAS (also called APO-1 or CD95) ligation (Jost et al, 2009).
Although XIAP is typically thought of as an antagonist of cell death pathways, its role in innate immune processes is increasingly being recognized (reviewed in Gyrd-Hansen & Meier, 2010). In particular, XIAP has been identified as an essential factor in immune inflammatory signaling mediated by the nucleotide-binding domain leucine-rich repeat-containing (NLR) protein family members NOD1 and NOD2 (Krieg et al, 2009), which orchestrate the immune response to intracellular bacterial infection (Fritz et al, 2006; Elinav et al, 2011). Mechanistically, XIAP interacts with receptor interacting protein kinase 2 (RIPK2) via its baculoviral IAP repeat (BIR) 2 domain (Krieg et al, 2009; Damgaard et al, 2013), ubiquitylates RIPK2 and recruits the linear ubiquitin chain assembly complex (LUBAC), required for efficient activation of the transcription factor NF-κB, resulting in the production and secretion of pro-inflammatory cytokines (Damgaard et al, 2012). Importantly, XIAP-deficiency in patients causes X-linked lymphoproliferative syndrome (XLP type 2)—a primary immunodeficiency—that is associated with severe hemorrhagic colitis (Rigaud et al, 2006; Pachlopnik Schmid et al, 2011; Yang et al, 2012). Although the biochemical interaction of XIAP with the NOD signaling complex has recently been the subject of intense investigation (Krieg et al, 2009; Damgaard et al, 2012, 2013), the physiological role of XIAP and its antagonization during the course of intracellular bacterial infection remain elusive.
We employed Shigella flexneri, an invasive Gram-negative enteropathogenic bacterium, as an infection model to study the complex host–pathogen interactions that shape the immune response of intestinal epithelial cells in response to bacterial infection. The host response to intracellular Shigella involves NOD1 signaling followed by activation of RIPK2, leading to the activation of NF-κB and the production and secretion of pro-inflammatory cytokines such as IL-8 (Girardin et al, 2001). However, Shigella has evolved a number of strategies to actively down-regulate the host cell immune response to ensure bacterial survival and propagation within the human intestinal epithelium. Epithelial colonization by Shigella results in severe inflammatory colitis known as bacillary dysentery (or shigellosis) (Ashida et al, 2011; Marteyn et al, 2012). A better understanding of these mechanisms could open a novel therapeutic avenue in the fight against bacterial infections.
While we show that XIAP plays a pivotal role in orchestrating the immune response against bacterial infection, we also demonstrate how Shigella escapes this immune response by inducing the BID-dependent release of SMAC from the mitochondria, which in turn potently disrupts XIAP-mediated inflammation. Consequently, XIAP-deficiency in mice increased the susceptibility toward Shigella infection, whereas genomic ablation of SMAC or BID conferred resistance to Shigella, resulting in effective clearance of the bacterial infection. Our findings unravel a previously unknown strategy of an enteroinvasive bacterium that involves the BID-dependent release of SMAC to effectively neutralize XIAP-mediated inflammatory signaling. The targeting of host proteins that are co-opted by bacterial pathogens emerges as a potential alternative to antibiotics, avoiding the intense selection pressure, which has led many bacterial strains to become resistant (Baron, 2010).
Results
XIAP is required for Shigella-induced NF-κB activation
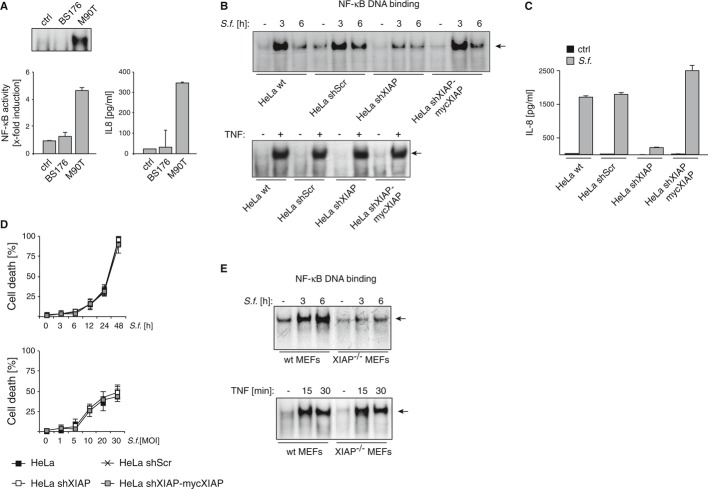
In line with previous reports (Philpott et al, 2000), only the invasive strain of Shigella flexneri M90T, but not the isogenic non-invasive Shigella flexneri strain BS176 is capable of inducing NF-κB activation and the production of the pro-inflammatory cytokine IL-8 in HeLa cells (Fig 1A). To address whether XIAP is involved in this process, HeLa cell lines stably expressing a XIAP-specific short hairpin RNA (HeLa-shXIAP) or a non-targeting control shRNA (HeLa-shScr) (Supplementary Fig S1A; Seeger et al, 2010) were infected with the invasive Shigella flexneri strain M90T (in the following referred to as Shigella). XIAP knockdown resulted in markedly reduced NF-κB activation and IL-8 production in response to Shigella infection (Fig 1B and C). Reintroduction of XIAP by stable overexpression of myc-tagged XIAP (lacking the 3′ UTR, that is targeted by the stably transduced shRNA) in XIAP knockdown HeLa cells (HeLa-shXIAP-mycXIAP) restored NF-κB activation and IL-8 production in response to Shigella infection (Fig 1B and C). These data identify XIAP as a potential mediator of the immune inflammatory signaling in response to intracellular Shigella infection. In contrast, TNF-induced NF-κB activation was not altered in XIAP-deficient HeLa cells (Fig 1B, lower panel), excluding a general defect in NF-κB signaling as a result of XIAP-deficiency. Importantly, the bacterial proliferation rate was not altered upon modification of XIAP expression as examined by a gentamycin protection assay (Supplementary Fig S1B), excluding the possibility that variations in the bacterial load between the cell lines could account for the observed difference in NF-κB activation. Furthermore, loss of XIAP expression neither affected cell viability (Fig 1D, Supplementary Fig S1C and D) nor impacted on MAPK activation (MEK1/2, JNK) (Supplementary Fig S1E) upon Shigella infection.
Figure 1. XIAP is required for Shigella-induced NF-κB activation.
- HeLa cells were left untreated (ctrl) or were infected with non-invasive Shigella strain BS176 or the invasive strain M90T (MOI 30). NF-κB (p65) DNA binding activity was analyzed by EMSA (upper panel) or ELISA (lower panel) 2 h post-infection (p.i.). IL-8 secretion was measured by ELISA in supernatants of cells 6 h p.i. (lower right panel). Data are presented as mean ± SEM (n = 3).
- HeLa wt, HeLa shScr, HeLa shXIAP and HeLa shXIAP-mycXIAP were left untreated (–) or were infected with Shigella M90T (MOI 30) (upper panel) or stimulated with TNF (10 ng/ml, 30 min) (lower panel). NF-κB (p65) DNA binding activity was analyzed by EMSA at the indicated time points p.i..
- HeLa wt, HeLa shScr, HeLa shXIAP and HeLa shXIAP-mycXIAP were left untreated (ctrl) or were infected with Shigella M90T (MOI 30). IL-8 secretion was monitored by ELISA in supernatants of cells 6 h p.i.. A representative experiment of three independent experiments is shown. Data are presented as mean ± SEM (n = 3).
- HeLa wt, HeLa shScr, HeLa shXIAP and HeLa shXIAP-mycXIAP were infected with Shigella M90T with MOI 30 (left panel) or the indicated MOI (right panel). Cell death was determined at the indicated time points p.i. by trypan blue exclusion. Data are presented as mean ± SD (n = 3).
- MEFs isolated from wt or XIAP−/− mice were infected with Shigella M90T (MOI 30) (upper panel) or were treated with TNF (10 ng/ml, 30 min) (lower panel). NF-κB (p65) DNA binding activity was analyzed by EMSA at the indicated time points p.i..
Corroborating the data obtained in HeLa cell lines, we show that in mouse embryonic fibroblasts (MEFs) derived from XIAP knockout mice (Supplementary Fig S1F) NF-κB activation was barely detectable following Shigella infection (Fig 1E, upper panel). In contrast, the lack of XIAP expression did not impair TNF-induced NF-κB activation in these XIAP knockout MEFs (Fig 1E, lower panel).
Together these data clearly establish XIAP as a crucial component of the pro-inflammatory signaling cascade in response to intracellular Shigella.
Shigella induces the release of mitochondrial SMAC and inhibits XIAP-mediated inflammatory signaling
It is well recognized that Shigella has evolved strategies to actively down-regulate the ‘immediate’ pro-inflammatory response in epithelial cells that is detrimental to the bacteria at early stages of infection (Ray et al, 2009; Ashida et al, 2011). Indeed, our data show that NF-κB activity declined significantly 3 h post-infection (p.i.) (Fig 2A, upper and middle panels). Furthermore, in line with previous data using synthetic NOD1 ligand (Krieg et al, 2009), our data show that Shigella infection induced complex formation of XIAP and RIPK2 within 3 h p.i. and this interaction was disrupted after 6 h p.i. (Supplementary Fig S1G). XIAP/RIPK2 dissociation was not due to the cytolytic activity of intracellular Shigella, as cell viability measurements clearly showed no increase in cell death up to 6 h p.i. (Fig 1D and Supplementary Fig S1C and D).
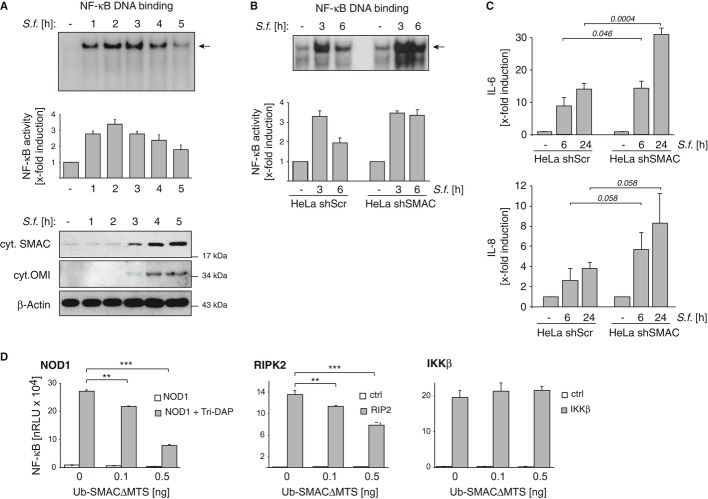
Figure 2. Shigella induces the release of mitochondrial SMAC and inhibits XIAP-mediated inflammatory signaling.
- HeLa wt cells were left untreated (−) or were infected with Shigella M90T (MOI 30). NF-κB DNA binding activity was analyzed by EMSA at the indicated time points p.i. (upper panel). Densitometric quantification of NF-κB DNA binding activity of five independent experiments (middle panel). Data are presented as mean ± SEM (n = 5). Cytosolic fractions from the same experiment were analyzed by Western blotting (lower panel).
- HeLa shScr or HeLa shSMAC cells were left untreated (−) or were infected with Shigella M90T (MOI 30). NF-κB DNA binding activity was analyzed by EMSA (upper panel) or by ELISA (lower panel) at the indicated time points p.i..
- Cells were treated as in (B). IL-8 and IL-6 secretion was measured by ELISA in supernatants of cells 6 h p.i.. Data are presented as mean ± SEM (n = 3).
- HEK293T cells were transiently transfected with expression vectors encoding NOD1, RIP2 or IKKβ alone or together with cytosolic SMAC (Ub-SMACΔMTS) with the indicated DNA amounts. NOD1-expressing cells were stimulated with Tri-DAP (10 μg/ml). NF-κB activity was analyzed by luciferase activity. A representative of three independent experiments is shown. Data are presented as mean ± SEM (n = 3). **P < 0.01; ***P < 0.001
Source data are available online for this figure.
Previous biochemical analyses concerning the role of XIAP in immune inflammatory signaling have shown that small molecule mimetics of SMAC (SMAC mimetics) can disrupt NOD-mediated NF-κB activation by antagonizing XIAP (Krieg et al, 2009; Bertrand et al, 2011; Damgaard et al, 2013). We therefore investigated whether Shigella employed the cellular XIAP antagonists, SMAC and OMI, to intercept XIAP-mediated NF-κB activation. Western blot analysis of cytosolic fractions of infected HeLa cells revealed the cytosolic appearance of SMAC and OMI in a time frame exactly corresponding to the decline in NF-κB activity upon infection with Shigella (Fig 2A, lower panel). Furthermore, cytosolic SMAC co-precipitated with XIAP (Supplementary Fig S2C) suggesting that the SMAC disrupts XIAP/RIPK2 interaction by direct binding to XIAP. Of note, SMAC and OMI were released only upon infection with the invasive Shigella strain M90T and not upon infection with the control strain BS176 (Supplementary Fig S2A). SMAC release was similarly observed in HCT116 cells following Shigella infection (Supplementary Fig S2B).
To further explore the role of SMAC in inflammatory signaling upon Shigella infection, we employed a HeLa cell line with stable knockdown of SMAC (HeLa-shSMAC, Seeger et al, 2010; Supplementary Fig S2D). As shown in Fig 2B and Supplementary Fig S2E, NF-κB activity increased within 3 h p.i. and persisted until 6 h p.i. in cells lacking SMAC but not in the HeLa control cell line (HeLa-shScr). Correspondingly, increased levels of inflammatory cytokines including IL-8 and IL-6 were measured in SMAC knockdown cells 6 h and 24 h p.i. (Fig 2C). Likewise, NF-κB activation was more persistent in SMAC/OMI double knockout MEFs than in wild-type MEFs (Supplementary Fig S2F). Importantly, no alteration of cell death could be induced by Shigella infection in SMAC knockdown cells (Supplementary Fig S2G).
To further demonstrate that the cytosolic appearance of SMAC was associated with the down-regulation of NOD1-induced NF-κB activity, we ectopically overexpressed cytosolic mature SMAC lacking the mitochondrial targeting sequence (MTS) by employing the ubiquitin (Ub) fusion system (Ub-SMACΔMTS) (Hunter et al, 2003; Kashkar et al, 2006). Ub-SMACΔMTS expression significantly attenuated Tri-DAP (NOD1)-mediated NF-κB activity as well as RIPK2-induced NF-κB activity in transiently transfected HEK293T cells (Fig 2D). Consistent with previous reports (Krieg et al, 2009; Damgaard et al, 2012), NF-κB activity induced by overexpression of IKKβ was not impaired by cytosolic SMAC demonstrating that XIAP is critical in regulating events upstream of IKKβ at the level of the RIPK2-containing signaling complex (Fig 2D, right panel).
Intracellular Shigella induces the release of mitochondrial SMAC without inducing mitochondrial damage
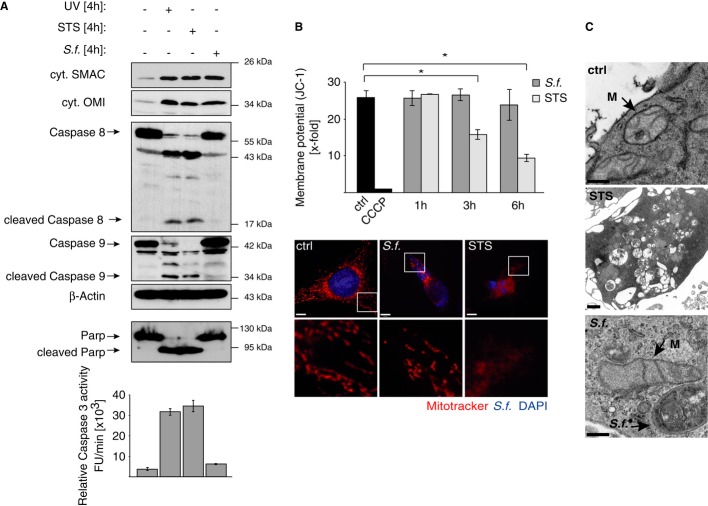
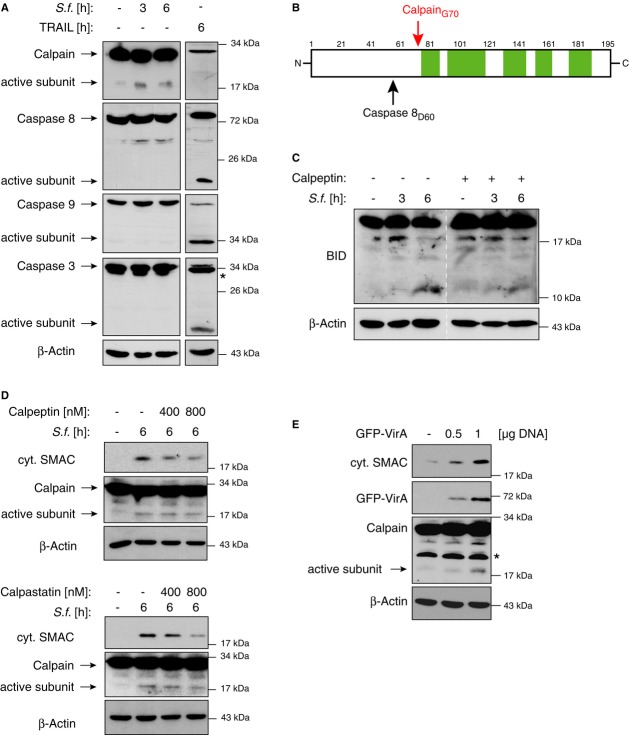
Many enteroinvasive bacterial pathogens interfere with the cellular death machinery in order to modulate host defense mechanisms and to ensure bacterial survival and propagation (Lamkanfi & Dixit, 2010). In particular, Shigella-infected epithelial cells have been shown to remain alive during early stages of infection (Mantis et al, 1996). In line with these observations, our data show that Shigella induced the mitochondrial release of SMAC and OMI without causing significant cytotoxicity for up to 12 h p.i. (Fig 1D and Supplementary Fig S1C and D). Although similar amounts of cytosolic SMAC were detected in cells infected with Shigella or after exposure to pro-apoptotic stimuli such as UV light or staurosporine (STS), no caspase activation was observed in Shigella-infected cells as demonstrated by the lack of Caspase-8 and Caspase-9 processing, Caspase-3 activity and PARP cleavage (Fig 3A and Supplementary Fig S3A and B). Furthermore, in contrast to apoptotic stimuli, Shigella infection was not associated with any profound structural alterations in the mitochondria as shown by the intact mitochondrial membrane potential (ΔΨm) (Fig 3B) and electron microscopic analyses of the mitochondrial ultrastructure (Fig 3C). These images show that the inner mitochondrial membrane is preserved after infection with Shigella, but not after treatment with STS, which led to rupture of the cell (Fig 3C).
Figure 3. Intracellular Shigella induces the release of mitochondrial SMAC without inducing mitochondrial damage.
- HeLa wt cells were left untreated (−), treated with UV light (10 mJ/m2), STS (0.5 μM) or were infected with Shigella M90T (MOI 30). After 4 h SMAC, OMI, Caspase-8 and Caspase-9 were analyzed in the cytosolic fractions by Western blotting. Actin served as a loading control. PARP cleavage was analyzed in nuclear fractions by Western blotting. Caspase-3 activity was determined fluorimetrically in cytosolic fractions using the substrate Ac-DEVD-AFC. Data are presented as mean ± SEM (n = 3).
- HeLa wt cells were left untreated (ctrl), infected with Shigella M90T (MOI 30) or treated with STS (0.5 μM). ΔΨm was analyzed after JC-1 or MitoTracker staining at the indicated time points using FACS (upper panel) or confocal microscopy (lower panel), respectively after 6 h. Control cells were treated with CCCP (50 mM). In confocal images, Shigella was stained blue by immunofluorescence. Data are presented as mean ± SEM (n = 3); *P < 0.05; (scale bar = 8 μm)
- HeLa wt cells were left untreated (ctrl), treated with STS (0.5 μM) or were infected with Shigella M90T (MOI 30). Mitochondrial cristae structure was analyzed 6 h p.i. by transmission electron microscopy. Arrows indicate mitochondria (M) or Shigella (scale bar: 300 nm (ctrl, S.f.); 1 μm (STS)).
Source data are available online for this figure.
Together, our data show that Shigella infection induced the release of SMAC and OMI in the absence of profound mitochondrial damage, to ensure the viability of the infected cells.
Calpain-cleaved BID induces the mitochondrial release of SMAC in Shigella-infected cells
Whereas intracellular Shigella induced the release of SMAC into the cytosol, activation of the NOD1 signaling cascade alone, either by overexpression of NOD1 or by stimulation with Tri-DAP or by overexpression of RIPK2, failed to induce SMAC release (Supplementary Fig S4A). Similarly, the specific knockdown of NOD1 had no impact on the Shigella-induced release of SMAC (Supplementary Fig S4B). This is in line with our observation, that modification of XIAP expression does not affect the release of SMAC (Supplementary Fig S4C), suggesting that Shigella engages more complex cellular mechanisms than NOD1 stimulation alone to induce the release of mitochondrial SMAC and OMI and to antagonize the XIAP-mediated inflammatory response.
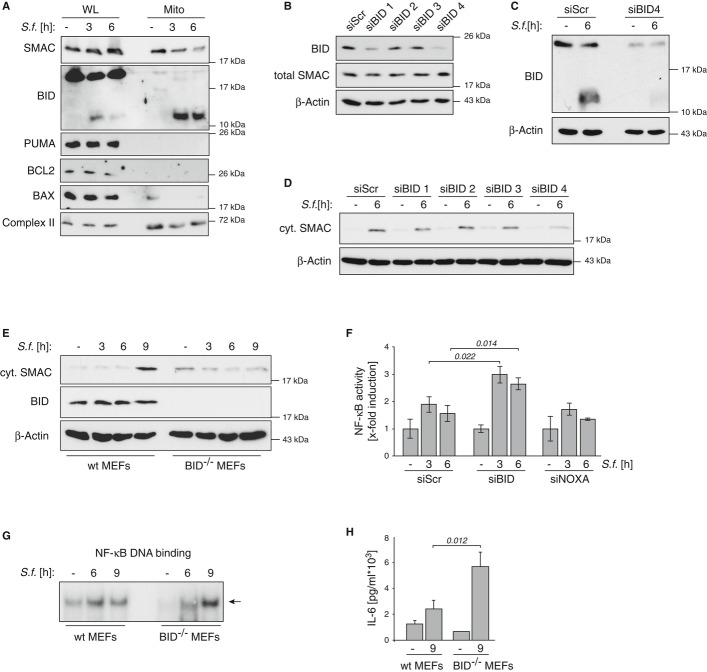
In apoptotic cells, the release of IMS proteins is regulated by members of the BCL-2 protein family, comprising three subgroups: pro- and anti-apoptotic multi BH-domain proteins, which are functionally controlled by a third divergent class of BH3-only proteins (Youle & Strasser, 2008). In order to investigate the regulation of Shigella-induced SMAC release, we analyzed the expression and the mitochondrial association of BCL-2 proteins in Shigella-infected cells. These experiments revealed that the BH3-only protein BID is processed in Shigella-infected cells and that the truncated C-terminal BID fragment accumulates in the mitochondrial fraction (Fig 4A). To explore if processed BID was required for Shigella-induced SMAC release, we employed specific siRNAs to knockdown BID expression (Fig 4B and C). Knockdown of BID, but not of any other member of the BH3-only protein family, including BAD, BIM or NOXA, prevented the release of SMAC upon Shigella infection (Fig 4D and Supplementary Fig S4D). Similarly, SMAC release upon Shigella infection was impaired in MEFs derived from BID knockout mice (Fig 4E). Together these data establish BID as an essential mediator of Shigella-induced SMAC release.
Figure 4. BID induces mitochondrial release of SMAC in Shigella-infected cells.
- HeLa wt cells were left untreated (−) or were infected with Shigella M90T (MOI 30). At the indicated time points whole-cell lysates (WL) and isolated mitochondria (Mito) were analyzed by Western blotting.
- HeLa wt cells were transiently transfected with four different siRNAs against BID (siBID1–4) or non-targeting ctrl (siScr). After 48 h cells were analyzed by Western blotting.
- HeLa wt cells were transiently transfected with BID-siRNA4. After 48 h cells were left untreated (−) or were infected with Shigella M90T (MOI 30). The mitochondrial fraction was analyzed 6 h p.i. by Western blotting.
- HeLa wt cells were transiently transfected with four different siRNAs against BID (siBID1–4) or non-targeting ctrl (siScr). After 48 h cells were left untreated (−) or were infected with Shigella M90T (MOI 30). Cytosolic extracts were analyzed 6 h p.i. by Western blotting.
- MEFs isolated from wt or BID−/− mice were left untreated (−) or infected with Shigella M90T (MOI 50). Cytosolic fractions were analyzed by Western blotting at the indicated time points p.i..
- HeLa wt cells were transiently transfected with specific siRNAs for BID, NOXA or non-targeting ctrl (siScr). After 48 h cells were infected with Shigella M90T (MOI 30). NF-κB DNA binding activity was analyzed by ELISA at the indicated time points p.i.. Data are presented as mean ± SD (n = 3).
- MEFs were treated as in (E). NF-κB DNA binding activity was analyzed by EMSA at the indicated time points p.i..
- MEFs were treated as in (E). IL-6 secretion was monitored by ELISA in supernatants of the cells 9 h p.i..
Source data are available online for this figure.
Consequently, specific knockdown of BID, but not of NOXA, was associated with increased NF-κB activity as shown by ELISA (Fig 4F). In line with these data, MEFs derived from BID knockout mice also showed increased NF-κB activity after Shigella infection (Fig 4G) leading to an increased production of IL-6 compared to wild-type MEFs (Fig 4H).
BID is known to be proteolytically processed upon death receptor ligation by active Caspase-8 (Li et al, 1998; Luo et al, 1998). The Caspase-8-cleaved BID fragment (BIDcasp8) then translocates to the mitochondria where it induces MOMP and the subsequent release of SMAC (Supplementary Fig S5A and B). However, Shigella infection, despite effectively inducing BID cleavage (Fig 4A and C), does not lead to caspase activation (Figs 3A and 5A), suggesting that the proteolytic activation of BID in response to Shigella infection occurs independently of caspase activity. Several other cellular proteases besides caspases are known to proteolytically activate BID including cathepsin, granzyme B and calpain (Billen et al, 2008). Detailed characterization of the truncated BID in the mitochondrial fractions of Shigella-infected cells by mass spectrometry indicated that the truncated C-terminal BID fragment contains the first amino acids corresponding to the calpain cleavage site, whereas the residues that would appear upon caspase cleavage were not detectable (Fig 5B and Supplementary Fig S5C and D). Intriguingly, calpain activation by the Shigella effector protein VirA has recently been described as a key step in the formation of the epithelial bacterial niche (Bergounioux et al, 2012). In line with these findings, we observed calpain activation upon Shigella infection but not after exposure to TNF-related apoptosis-inducing ligand (TRAIL), which potently induces caspase activation (Fig 5A). As reported for caspase-cleaved BID, calpain-cleaved BID also translocates to the mitochondria and induces the release of mitochondrial SMAC (Supplementary Fig S5A and B). To confirm that activated calpain was involved in BID processing, we inhibited the calpain activity with the calpain inhibitor calpeptin, leading to reduced proteolytic processing of BID upon Shigella infection (Fig 5C). Importantly, specific calpain inhibition, either by calpeptin or by a cell-permeable peptide corresponding to the calpain inhibitory sequence of calpastatin (the specific endogenous calpain inhibitor) potently inhibited the release of SMAC upon Shigella infection (Fig 5D). Our further analyses showed that ectopic expression of VirA induced calpain activation and SMAC release even in the absence of Shigella infection (Fig 5E). Unlike calpain inhibitors, the pan-caspase inhibitor Z-VAD or the Caspase-8 inhibitor z-IETD did not affect SMAC release following Shigella infection (Supplementary Fig S5E), although both potently inhibited Caspase-8 activation in response to stimulation with TRAIL (Supplementary Fig S5F).
Figure 5. BID is cleaved by Calpain after infection with Shigella.
- HeLa wt cells were left untreated (−), infected with Shigella M90T (MOI 30) or stimulated with TRAIL (50 ng/ml). At the indicated time points cytosolic extracts were analyzed by Western blotting (* marks unspecific bands).
- Schematic sequence coverage (green areas) of BID peptides identified by mass spectrometry. Caspase-8 and Calpain cleavage sites are indicated.
- HeLa wt cells were infected with Shigella M90T (MOI 30) and were exposed to the calpain inhibitor calpeptin (400 nM) after 30 min (time point zero, see Materials and Methods). Mitochondrial fractions were analyzed by Western blotting at the indicated time points p.i..
- HeLa wt cells were left untreated (−) or were infected with Shigella M90T (MOI 30) in combination with calpeptin (upper panel) or calpastatin (lower panel) at the indicated concentrations. Cytosolic extracts were analyzed by Western blotting 6 h p.i..
- HeLa wt cells were mock transfected (−) or transiently transfected with the indicated amounts of GFP-VirA. After 40 h cytosolic fractions were analyzed by Western blotting (* marks unspecific bands).
Source data are available online for this figure.
Together these data demonstrate that Shigella infection results in the activation of calpain, which induces the BID-mediated release of SMAC, which in turn dampens the XIAP-mediated inflammatory response toward bacterial infection.
XIAP confers immunity against Shigella infection in vivo
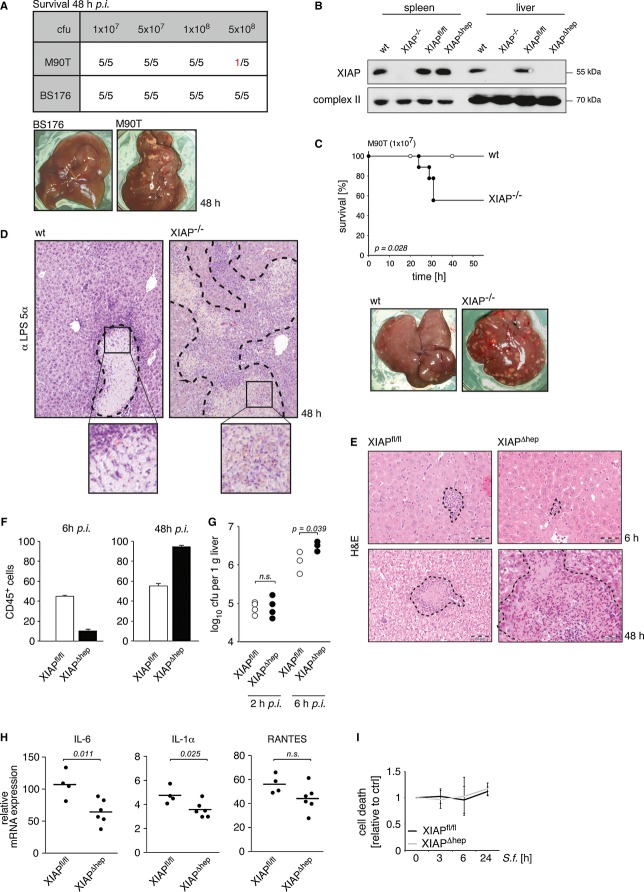
To investigate the physiological relevance of our findings concerning the role of XIAP in the anti-bacterial immune response toward Shigella, we infected mice intravenously (i.v.) with Shigella as previously described (Martino et al, 2005). As expected, only the invasive Shigella strain M90T and not the isogenic non-invasive control strain BS176 leads to the formation of liver abscesses and increased mortality at doses beyond 1 × 108 colony forming units (cfu) in wild-type mice (Fig 6A). To address the impact of XIAP on anti-bacterial immunity, we established a XIAP knockout mouse strain (XIAP−/−) (Fig 6B and Supplementary Fig S6A). Like the previously published XIAP-deficient mice (Harlin et al, 2001), our XIAP−/− mice do not appear to have obvious phenotypic defects. However, when compared to wild-type animals, XIAP knockout mice (XIAP−/−) were more susceptible to infection with the invasive Shigella strain M90T (Fig 6C) (but not to infection with the control strain BS176; Supplementary Fig S6B). Macroscopic as well as histological examinations of liver sections revealed large and coalescing necrotic areas in the livers of XIAP−/− mice (Fig 6C, lower panel, and D). These disseminated necrotic areas, containing a high bacterial burden (Fig 6D) are clear manifestations of the inefficient immune resolution of the bacterial infection in XIAP−/− mice. By contrast, in wild-type animals, liver necrosis was multifocal but well confined within a small peripheral margin of mixed inflammatory cells, barely containing bacteria (Fig 6D). These data demonstrate for the first time that XIAP−/− mice are profoundly impaired in their capacity to mount an efficient immune response toward Shigella, leading to pathological bacterial propagation and tissue damage.
Figure 6. XIAP confers immunity against Shigella infection in vivo.
- Wt mice were i.v. infected with invasive Shigella M90T or non-invasive Shigella BS176 with the indicated cfu. Survival was monitored for 48 h. Macroscopic inspection of representative livers (5 × 108 cfu, lower panel) (n = 5 in each group).
- Spleen and liver of wt, XIAP−/−, XIAPfl/fl and XIAPΔhep mice were analyzed for XIAP expression by Western blotting.
- Kaplan–Meier curve of wt and XIAP−/− mice infected with Shigella M90T (1 × 107 cfu). Mice were sacrificed 48 h p.i. and livers were inspected macroscopically (wt: n = 9; XIAP−/−: n = 9).
- Liver sections from mice treated as in (C) were analyzed microscopically after staining with hematoxylin & eosin and a Shigella-specific LPS 5α-antibody.
- XIAPfl/fl and XIAPΔhep mice were infected and treated as in (C). Liver sections were analyzed microscopically at the indicated time points as in (D) (scale bar = 50 μm).
- Quantification of leukocytes (CD45+ cells) was performed independently by two experienced pathologists. Immunostained slides for CD45 were evaluated and the number of CD45+ cells was counted per square millimeter in the hepatic parenchyma, with care taken to avoid areas of necrosis. Counts were expressed as numbers per square millimeter based on twenty 40× HP-field cells.
- XIAPfl/fl and XIAPΔhep mice were infected with Shigella M90T and sacrificed at the indicated time points p.i.. Bacterial load in the liver was determined. Each point represents log10 cfu per 1 g liver per individual animal.
- XIAPfl/fl and XIAPΔhep mice were infected with Shigella M90T and sacrificed 6 h p.i.. Relative mRNA expression was determined by RT-PCR and normalized to uninfected control mice.
- Primary hepatocytes isolated from XIAPfl/fl and XIAPΔhep mice were infected with Shigella M90T (MOI 50). Cell death was measured at the indicated time points by trypan blue exclusion. Data are presented as mean ± SD (n = 3).
Source data are available online for this figure.
Our data in 5Figures describe the role of XIAP in anti-bacterial immune signaling within epithelial cells. The systemic infection of mice with Shigella, however, does not only lead to the colonization of epithelial cells, but may also affect immune cell effector functions toward Shigella. To better understand how XIAP-deficiency specifically in the epithelial compartment affects the host's capacity to mount an immune response toward Shigella infection, we established a hepatocyte-specific XIAP knockout mouse strain (XIAPΔhep; Fig 6B and Supplementary Fig S6A). For this purpose, loxP-flanked XIAP (XIAPfl/fl) mice were crossbred with a hepatocyte-specific Cre transgenic strain (Kellendonk et al, 2000). XIAPΔhep mice and the corresponding control mice (XIAPfl/fl) were infected with Shigella and liver pathology and bacterial colonization pattern were analyzed at early and late stages of infection. At early stages of infection (6 h p.i.), the livers of XIAPΔhep mice showed significantly reduced leukocyte infiltration in comparison to wild-type mice as shown by histological staining for CD45-positive cells (Fig 6E and F, and Supplementary Fig S6C). In XIAPΔhep mice, the impaired ability to mount a cell-autonomous immune response in hepatocytes, results in a significantly increased bacterial burden in the liver compared with control mice as early as 6 h p.i. (Fig 6G and Supplementary Fig S6D). Reflecting the impaired immune response in the XIAPΔhep mice compared with control animals, the inflammatory cytokines IL-6 and IL-1α were significantly reduced 6 h p.i., measured by RT-PCR analysis (Fig 6H). Of note, the basal cytokine and NF-κB levels did not differ between control and XIAPΔhep mice (Supplementary Fig S6E and F). At later stages, the unrestrained bacterial proliferation in the livers of XIAPΔhep mice was associated with an increase in the size and frequency of necrotic liver areas (Fig 6E), elevated levels of CD45-positive cells compared to control mice (Fig 6F and Supplementary Fig S6C) and a high bacterial burden (Fig 6G and Supplementary Fig S6D). XIAPΔhep mice therefore recapitulate the disseminated liver damage observed in the whole-body XIAP knockout mice (XIAP−/−). The similar pathology observed in both, whole-body and hepatocyte-specific XIAP knockout mice, highlights the important function of XIAP in orchestrating an efficient cell-autonomous anti-bacterial immune defense in epithelial cells. However, this does not formally exclude additional effects associated with XIAP function in other effector cells including myeloid cells.
Genetic ablation of BID or SMAC restores immunity against Shigella infection
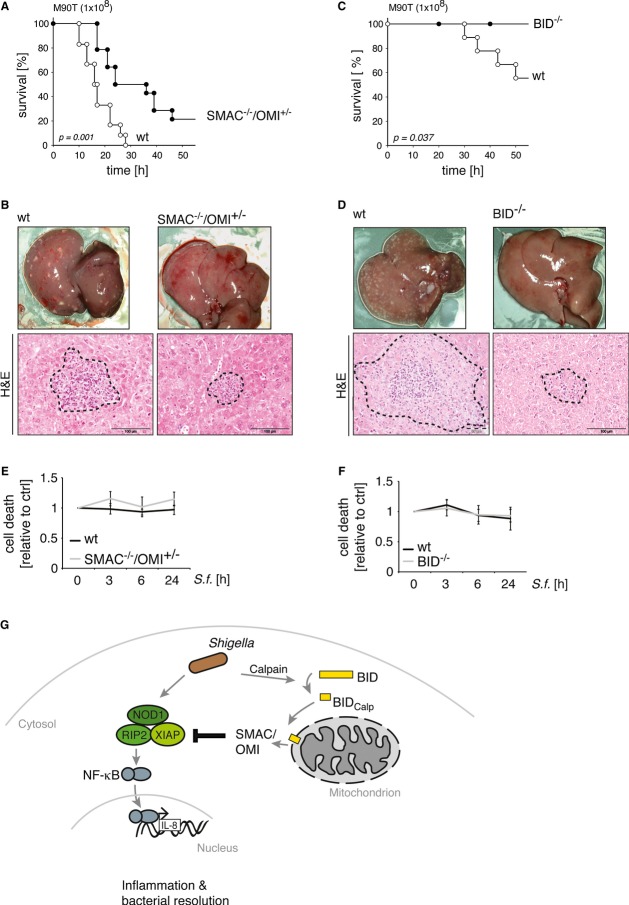
Our in vitro data demonstrate that XIAP-mediated immune signaling is efficiently antagonized through the BID-mediated release of SMAC (Figs 2–5). Accordingly, the lack of SMAC or BID should restore the XIAP-mediated immune response toward Shigella infection. Indeed, SMAC−/−/OMI+/− mice survived Shigella infection for significantly longer periods compared to wild-type animals (Fig 7A). Correspondingly, infected SMAC−/−/OMI+/− knockout mice showed significantly fewer liver abscesses on macroscopic or microscopic examination, consistent with an increased clearing efficiency of the anti-bacterial immune response in these animals (Fig 7B). Similarly, BID knockout mice (BID−/−) were highly resistant to Shigella infection (Fig 7C). SMAC−/−/OMI+/− mice, like BID−/− mice, survived bacterial infection and did not suffer from liver damage upon Shigella infection in sharp contrast to the corresponding wild-type mice (Fig 7D). While the basal NF-κB levels in SMAC−/−/OMI+/− and BID−/− mice compared to the corresponding wild-type animals were similar (Supplementary Fig S7B), NF-κB activity was increased in the livers of BID−/− mice compared to the corresponding wild-type animals 6 h p.i., consistent with a more efficient immune response toward Shigella (Supplementary Fig S7C). Importantly, the lack of XIAP, SMAC or BID expression had no effect on either Shigella-induced cell death in primary hepatocytes derived from the corresponding mice (Figs 6I, 7E and 7F) or on caspase activation in lysates of infected livers (Supplementary Figs S6G and S7A). In conclusion, these data highlight the central role played by XIAP antagonists in regulating the efficient immune inflammatory response toward invasive Shigella.
Figure 7. Genetic ablation of BID and SMAC restores immunity against Shigella infection.
- Kaplan–Meier curve of wt and SMAC−/−/OMI+/− mice infected with Shigella M90T (1 × 108 cfu) (upper panel) (wt: n = 12; SMAC−/−/OMI+/−: n = 14).
- Macroscopic and microscopic analysis of the liver from mice treated as in (A) (scale bar = 100 μm).
- Kaplan–Meier curve of wt and BID−/− mice infected with Shigella M90T (1 × 108 cfu) (wt: n = 10; BID−/−: n = 9).
- Macroscopic and microscopic analysis of the liver from mice treated as in (C).
- Primary hepatocytes isolated from wt and SMAC−/−/OMI+/− mice were infected with Shigella M90T (MOI 50). Cell death was measured at the indicated time points by trypan blue exclusion. Data are presented as mean ± SD (n = 3).
- Primary hepatocytes isolated from wt and BID−/− mice were infected with Shigella M90T (MOI 50). Cell death was measured at the indicated time points by trypan blue exclusion. Data are presented as mean ± SD (n = 3).
- Upon cytosolic appearance of Shigella, NOD1-signaling, involving RIPK2 and XIAP, is activated and promotes an inflammatory response (activation of NF-κB and IL-8 production) aiming at resolution of the bacteria. However, Shigella evolved a strategy to escape the innate immune defense by orchestrating a calpain-mediated BID-activation, which in turn potentiates the release of mitochondrial SMAC to neutralize XIAP-mediated anti-bacterial inflammatory signaling.
Discussion
Accumulating biochemical evidence has identified XIAP as a component of the NOD signaling cascade, implicating XIAP in immune inflammatory signaling (Krieg et al, 2009; Damgaard et al, 2012). However, the use of minimal peptidoglycan fragments to stimulate intracellular NOD signaling in these studies cannot fully account for the complexity of a bacterial infection. Our data now clearly implicate XIAP in NOD1-mediated NF-κB signaling upon infection with the Gram-negative enteropathogenic bacterium Shigella, both in vivo and in vitro. Notably, our preliminary analyses in cIAP1- or cIAP2-deficient cells showed no evidence for an involvement of other IAP members in Shigella-induced NF-κB activity (data not shown). Therefore, we here focused on the role of XIAP in the NOD-mediated response to Shigella infection and its antagonization by SMAC. However, since cytosolic SMAC indiscriminately targets cIAPs (Supplementary Fig S2H), we cannot formally exclude the possibility that cIAPs indirectly contribute to the observed phenotypes, for example, through their role in controlling the TNF superfamily signaling cascade (Vandenabeele & Bertrand, 2012).
That XIAP plays a pivotal role in the immune response to bacterial pathogens in mice was first demonstrated by the increased sensitivity of XIAP−/− mice to Listeria monocytogenes (Bauler et al, 2008) and Chlamydophila pneumonia infections (Prakash et al, 2010). These data indicated that XIAP plays a critical role in regulating the innate immune response provided by macrophages. In line with these observations, we here demonstrate that XIAP−/− mice infected with Shigella show a reduced survival rate compared with wild-type animals (Fig 6C). In view of the fact that XIAP−/− mice were not susceptible to the non-invasive Shigella strain BS176 (Supplementary Fig S6B), our data indicate the pivotal role of XIAP in orchestrating immunity against enteroinvasive pathogens. Notwithstanding the role of XIAP in myeloid cells, our findings in a newly established hepatocyte-specific XIAP knockout mouse model (XIAPΔhep) further demonstrate a specific function of XIAP in orchestrating the inflammatory response to bacterial infection in non-myeloid cells (Fig 6E–H).
The capacity of a host organism to mount an immediate early immune response toward infection is critical for its survival and the subversion of the cellular immune responses is one of the central strategies of enteroinvasive bacterial pathogens to ensure bacterial survival and colonization (Lamkanfi & Dixit, 2010). It is well established that Shigella has evolved various strategies to actively down-regulate intracellular pro-inflammatory responses (Ashida et al, 2011, 2013; Sanada et al, 2012; Kobayashi et al, 2013). Mitochondrial IBM-containing proteins including SMAC have been identified as potent antagonists of XIAP upon their release into the cytosol. Our data illustrate the critical role of XIAP antagonization by BID-mediated SMAC release (Figs 2–4), which is induced by intracellular bacteria to safeguard bacterial colonization.
Numerous studies have contributed to the current view that in response to systemic stress the release of mitochondrial proteins from the IMS is coincident with mitochondrial damage and cell death (Rehm et al, 2003; Munoz-Pinedo et al, 2006). We here shed light on this process by demonstrating that the release of SMAC upon Shigella infection is mechanistically controlled by BID and results in dampening of the XIAP-mediated immune response but not cell death, which in turn ensures bacterial propagation (Figs 4–7). Intriguingly, Shigella infection is known to activate calpain (Fig 5) which is recognized as an important step in ensuring bacterial propagation in epithelial cells (Bergounioux et al, 2012). The capacity of calpain to proteolytically process BID has already been demonstrated (Chen et al, 2001). Nevertheless, the physiological relevance of this activity and in particular its involvement in cell death remains controversial (Chen et al, 2001). Our data summarized in Fig 5 identify a novel physiological role for calpain-cleaved BID during the cell-autonomous immune response against intracellular bacterial infection.
In conclusion, our data highlight a previously unrecognized mode of intracellular host–pathogen crosstalk involving the mitochondria. We propose a model in which Shigella induces the release of SMAC to specifically inactivate intracellular immune signaling and thereby contributing to an immunosuppressive environment that supports effective bacterial propagation (Fig 7G). Our findings show how the non-apoptotic antagonization of XIAP by BID-mediated release of mitochondrial SMAC can control the delicate interaction between intracellular microbial pathogens and their hosts.
Materials and Methods
Antibodies and reagents
Primary antibodies
Polyclonal rabbit anti-Caspase-9, monoclonal mouse anti-Caspase-8 (clone 1C12), monoclonal rabbit anti-Caspase-3 (clone 8G10), polyclonal rabbit anti-Caspase-3, polyclonal mouse anti-SMAC, monoclonal mouse anti-BIM (C34C5), monoclonal rabbit anti-BAD (D24A9), polyclonal rabbit anti-BID antibody, polyclonal rabbit anti-phospho-JNK, polyclonal rabbit anti-JNK, polyclonal rabbit anti-phospho-MEK1/2 and polyclonal rabbit anti-MEK1/2 were obtained from Cell Signaling. Monoclonal mouse anti-β-Actin and anti-Flag (M2) were obtained from Sigma. Monoclonal mouse anti-BCL-2 (clone 7), monoclonal mouse anti-PARP (clone C2C10), polyclonal rabbit anti-BID, monoclonal rat anti-CD45 and monoclonal mouse anti-XIAP (clone 48) were obtained from BD Biosciences. Monoclonal mouse anti-Complex II (clone 2E3GC12FB2AE2) was obtained from Invitrogen. Polyclonal rabbit anti-OMI, monoclonal mouse anti-cIAP1 (clone 681732) and polyclonal goat anti-cIAP2 were obtained from R&D Systems. Monoclonal rabbit anti-SMAC (Y12) and monoclonal mouse anti-NOXA (114C307) were obtained from Millipore. Monoclonal rabbit anti-IκBα (clone C21) was obtained from Santa Cruz. Polyclonal rabbit anti-Shigella serotype 5α LPS was a kind gift of P. Sansonetti.
Secondary antibodies
HRP-conjugated anti-rabbit IgG and anti-mouse IgG were obtained from Cell Signaling. HRP-conjugated anti-goat IgG was obtained from Sigma.
Reagents
Staurosporine and z-VAD-FMK were obtained from Alexis Biochemicals, mTNF-α from Genentech. Carbonyl cyanide m-chlorophenylhydrazone (CCCP) was obtained from Merck. Tri-DAP was obtained from Invivogen. SuperKillerTRAIL™ and z-IETD-FMK were obtained from Enzo Life Sciences. Calpeptin was obtained from Calbiochem. Calpastatin was obtained from Merck Millipore. PD98059 (MEK1/2 inhibitor) was obtained from Cell Signaling.
DNA constructs
Ub-SMAC and SMACΔMTS were previously described (Kashkar et al, 2006). pUNO-IKKβ was from Invivogen (Toulouse, France). RIP2-VSV was a kind gift from Margot Thome (Lausanne, Switzerland). pcDNA3.1-NOD1 was a kind gift from Gabriel Nuñez (Ann Arbor, MI, USA). FLAG-NOD1 was previously described (Kufer et al, 2006). For construction of BID-GFP-fusion proteins, open reading frames (ORF) encoding human BID were amplified by PCR producing BIDcasp8 (f: 5′-ccccccaagcttatgggcaa ccgcagcagccactccc-3′) and BIDcalpain (f: 5′-ccccccaagcttatgagaatagaggcagattctgaaag-3′; r: 5′-ccccccggatcctcagtccatcccatttctggctaagc-3′). Both constructs were cloned into the pEGFP-C3 vector (Clontech). pKindling-Red mito was obtained from Evrogen. GFP-VirA was amplified by PCR from isolated Shigella DNA (f: 5′-gcggcgaagcttatgcagacatcaaacataactaacc-3′; r: 5′-ggccctcgagttaaacatcaggagatatgatggc-3′) and cloned into a pEGFP-C3 vector.
Bacterial infection
Bacterial infection of the indicated cell lines was performed using the Shigella flexneri strain M90T afaE as described previously (Philpott et al, 2000). The invasion plasmid-cured strain Shigella flexneri BS176 was used as non-invasive control. Prior to infection, cells were starved in DMEM without FCS for 1 h followed by incubation with Shigella (MOI = 30) for 15 min at RT and transferred to 37°C for 30 min (time point zero). Plates were then transferred into fresh DMEM containing 10% FCS and 50 μg/ml gentamycin to kill extracellular bacteria.
Gentamycin protection assay
Uptake of Shigella in different cell lines was analyzed by lysis of pellets of infected cells in 0.5% SDS/H2O. Serial dilutions of cell lysates were plated onto trypticase soy broth bacto agar plates without antibiotics and incubated at 37°C for 24 h. Colonies were counted and the recovery was determined as colony forming units (cfu).
NF-κB activity
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (Haubert et al, 2007) using the NF-κB-specific oligonucleotides (Applied Biosystems) end-labeled with γ-32P-ATP (Amersham Corp.).
ELISAs were performed after nuclear extraction using the TransAM® Nuclear Extract Kit and the TransAM® NF-κB p65 Kit (Active motif) according to the instructions of the manufacturer. Read-out was performed on an ELISA reader (Anthos HT2) at a wavelength of 450 nm/620 nm (Brinkmann et al, 2014).
Microscopy
Infected or treated HeLa cells were stained with MitoTracker (Invitrogen) and fixed with 3% paraformaldehyde at the indicated time points. Shigella was stained with anti-Shigella flexneri serotype 5α LPS antibody and nuclei were stained with DAPI. Imaging was performed on an inverted microscope (Olympus IX81 equipped with Cell^R Imaging Software; Tokyo, Japan) using a 60×/1.45 numerical aperture Plan Apo oil objective.
For transmission electron microscopy analyses, cells were washed in phosphate buffer, fixed in glutaraldehyde and collected as cell pellets after centrifugation. Cell pellets were then postfixed with osmium tetroxide and embedded in Epon (Fluka, Buchs, Switzerland). Ultrathin sections were cut using a Leica Ultracut EM UC6 and micrographs were obtained using a Philips CM 10 transmission electron microscopy.
For phase-contrast imaging, HeLa cells were seeded onto cover slips and infected with Shigella at MOI 30. After infection, cells were fixed with 3% paraformaldehyde and cover slips were mounted on microscope slides using Mowiol mounting medium. Imaging was performed on an Axioplan II fluorescence microscope (Zeiss, Jena, Germany).
Cell viability and cell death
HeLa cells were incubated in 96-well plates at 37°C in complete medium and infected with the indicated MOI of Shigella and further indicated incubation time. Cell viability was assessed by the Cell Proliferation Kit II (XTT) (Roche Applied Sciences, Mannheim, Germany) according to the instructions of the manufacturer. Cell death was assessed by trypan blue exclusion using an automated cell counter (CountessTM, Invitrogen, Karlsruhe, Germany) (Brinkmann et al, 2013).
LDH release was assessed with the Cytotoxicity Detection Kit (LDH) from Roche according to the instructions of the manufacturer.
Caspase-3 activity
Caspase-3 activity was performed as previously described (Kashkar et al, 2003).
Cell fractionation and Western blotting
Cellular and cytosolic extracts were prepared as previously described (Kashkar et al, 2003). Mitochondria were isolated with the Mitochondrial Isolation Kit from Miltenyi Biotec according to the instructions of the manufacturer. For Western blotting, equal amounts of protein were resolved by SDS-PAGE and transferred onto nitrocellulose membrane followed by incubation with the appropriate primary and secondary antibodies. Immunoprecipitation of myc-tagged XIAP was performed with cytosolic extracts by magnetic epitope labeling and separation using μMACS Epitope Tag Protein Isolation Kit from Miltenyi Biotec according to the instructions of the manufacturer.
RNAi interference
For siRNA-mediated knockdown, HeLa cells were transfected using Lipofectamine™ RNAiMAX (Invitrogen) with specific siRNAs for BID (siRNA1–3: BID Trilencer-27 human siRNA from Origene (SR300435); siRNA4: 5′-cuugcuccgugaugucuuutt-3), BAD (5′-guac uucccucaggccuau-3′), BIM (5′-ccucccuacagacagagcctt-3′) and NOXA (5′-ggugcacguuucaucaauu-3′). Cells were incubated for 48 h prior to infection with Shigella.
Mitochondrial membrane potential
For analysis of mitochondrial membrane potential, HeLa cells were stained with 10 μM JC-1 (Invitrogen) for 30 min at 37°C and analyzed by flow cytometry. For the analysis by confocal microscopy, HeLa cells were seeded on cover slips and stained with 25 nM MitoTracker® Red CMXRos (Invitrogen) in culture media at 37°C for 30 min (Coutelle et al, 2014).
Mass spectrometry
Tryptic in-gel digestion
Following electrophoresis the gel was washed thoroughly in water. The area of interest was cut out and minced using a scalpel. The gel pieces were destained with 50% 10 mM NH4HCO3/50% ACN at 55°C. After dehydration in 100%, ACN proteins were reduced with 10 mM DTT and carbamidomethylated in the dark using 50 mM iodoacetamide. Dehydrated gel pieces were equilibrated with 10 mM NH4HCO3 containing porcine trypsin (12.5 ng/μl; Promega) on ice for 2 h. Excess trypsin solution was removed and tryptic hydrolysis was performed overnight at 37°C in 10 mM NH4HCO3. The supernatant was collected and the peptides were further extracted. After acidification with 5%, TFA gel pieces were extracted twice with 1% TFA and then with 60% ACN/40% H2O/0.1% TFA followed by a subsequent two-step treatment using 100% ACN. The supernatant and the extractions were combined and concentrated using a SpeedVac concentrator (Christ). Prior to nano-LC-MS/MS analysis, the peptides were desalted using STAGE Tip C18 spin columns (Proxeon/Thermo Scientific) as described in (Rappsilber et al, 2007). Eluted peptides were concentrated in vacuo and then re-suspended in 0.5% acetic acid in water to a final volume of 10 μl.
Nano-LC ESI-MS/MS
Analyses using reversed-phase liquid chromatography coupled to nano-flow electrospray tandem mass spectrometry were carried out using an EASY nLC II nano-LC (Proxeon/Thermo Scientific) with a 150-mm C18 column (internal diameter 75 μm, Dr. Maisch GmbH) coupled to a LTQ Orbitrap mass spectrometer (LTQ Orbitrap Discovery, Thermo Scientific). Peptide separation using 8 μl of sample was performed at a flow rate of 250 nl/min over 120 min (10–40% acetonitrile, buffer A: 0.1% formic acid in H2O; buffer B: 0.1% formic acid in acetonitrile). Survey full scan MS spectra (m/z 350–2,000) of intact peptides were acquired in the Orbitrap at a resolution of 30,000 using m/z 445.12,003 as a lock mass. The mass spectrometer acquired spectra in ‘data dependent mode’ and automatically switched between MS and MS/MS acquisition. Signals with unknown charge state and + 1 were excluded from fragmentation. The ten most intense peaks were isolated and automatically fragmented in the linear ion trap using collision-induced dissociation (CID).
Mascot as implemented in the Proteome Discoverer 1.3 software (Thermo Scientific) was used for protein identification by searching the UniProt database of Homo sapiens using carbamidomethylation at cysteine residues as fixed modification. Oxidation at methionine residues was used as variable modification. Mass tolerance for intact peptide masses was 10 ppm for Orbitrap data and 0.8 Da for fragment ions detected the linear trap. Search results were filtered to contain only rank 1 peptides of high confidence (false discovery rate ≤ 1%) with a score of ≥ 20 and a mass accuracy of ≤ 5 ppm.
Mouse strains
XIAP-deficient mice (frozen embryos, Xiap/ICS/IR2785b/E114/2785c) were obtained from EUCOMM (The European Conditional Mouse Mutagenesis Program, ICS, Illkirch, France). For the generation of XIAP knockout mice (XIAP−/−, referring to both female and male KO mice), an L1L2-Bact-P cassette was inserted at position 39451542 of chromosome X upstream of exon 5 (corresponds to coding exon 3). The cassette is composed of an FRT site followed by a lacZ sequence and a loxP site. This first loxP site is followed by neomycin under the control of the human actin promoter, SV40 polyA, a second FRT site and a second loxP site. A third loxP site is inserted downstream of the targeted exon (exon 5) at position 39452298. Exon 5 is thus flanked by loxP sites. A ‘conditional ready’ (XIAPfl/fl) allele was created by crossbreeding with Flp deleter mice (Flp recombinase-expressing mice) (Dymecki, 1996). Subsequent crossbreeding with AFP-Cre mice (Kellendonk et al, 2000) resulted in a hepatocyte-specific knockout mouse (XIAPΔhep). Like the conventional XIAP knockout mouse (Harlin et al, 2001) our XIAP knockout (XIAP−/−) and hepatocyte-specific XIAP knockout mice do not have obvious phenotypic defects. SMAC/OMI knockout mice (C57BL/6J background; Martins et al, 2004) and BID knockout mice (C57BL/6J background; Kaufmann et al, 2007) were described previously.
Ethics statement
All animal procedures were performed in accordance with the German and Swiss animal protection legislations.
Infection of mice with Shigella
Mice received food and water ad libitum. Overnight cultures of Shigella strains M90T or BS176 were prepared as described and resuspended in sterile PBS to obtain the indicated concentrations. Mice were challenged by injection of the bacterial suspension into the lateral tail vein. Mice were inspected every 2 h and deaths were recorded for three consecutive days. For pathological analyses, the animals were sacrificed by cervical dislocation at the indicated time points. The liver was removed, photographed with a SC 100 digital color camera (Olympus) or prepared for immunohistochemical staining.
Bacterial counts
For determination of viable bacteria, the liver of infected mice was homogenized with the gentleMACS™ Dissociator (Miltenyi Biotech). The number of bacteria was determined by plating 10-fold serial dilutions of the homogenate on soy bean agar plates.
Tissue immunohistochemistry
Livers from infected mice were fixed in 4% paraformaldehyde over night. Whole livers were processed using an ASP300 S Tissue Processor (Leica). Embedded samples were cut using a sliding microtome HM 400 (Thermo Scientific, Fisher Scientific GmbH) to produce 5-μm-thick sections. Staining with hematoxylin/eosin (Thermo Scientific, Fisher Scientific GmbH), anti-Shigella flexneri serotype 5α LPS antibody, anti-Caspase-3 or anti-CD45 with the corresponding secondary antibody (DCS Super Vision2 Single Species 2-step-Polymersystem rabbit HRP) was performed.
Mouse cytokine antibody array proteome profiling
Mouse livers were homogenized with the gentleMACS Dissociator (Miltenyi Biotec). Cytokine expression was determined by the proteome profiler array (mouse cytokine array panel A, ARY006, R&D Systems). Signals were recorded on an electronic imaging system (LAS4000, Fujifilm). Signal intensity was determined with ImageJ software (NIH, Bethesda, MD). Cytokine levels are expressed as the average signal intensity of duplicate spots subtracted from signal background.
Gene expression analysis
Quantitative gene expression analysis was performed as previously described (Pal et al, 2013). Total cellular RNA from liver tissue of control and infected mice was isolated using a Qiagen RNeasy Kit (Qiagen, Germany). RNA was reversely transcribed with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and amplified by using TaqMan Gene Expression Master Mix (Applied Biosystems). Relative expression of target mRNAs IL-6 (Mm00446190_m1), IL-1α (Mm00439620_m1) and Rantes (Mm01302428_m1) was determined using standard curves based on cDNA derived from liver tissue, and samples were adjusted for total RNA content by Tbp (Mm00446971_m1) quantitative PCR. Calculations were performed by a comparative method (2CT). Assays were linear over 4 orders of magnitude. Quantitative PCR was performed on an ABI Prism 7900 sequence detector (Applied Biosystems).
Down-regulation of NOD1 was analyzed by RT-PCR as described in (Neerincx et al, 2012) with primer specific for NOD1 (f: TCCAAAGCCAAACAGAAACTC; r:CAGCATCCAGATGAACGTG).
Primary hepatocyte culture
Primary hepatocytes were isolated from control, XIAP−/−, SMAC−/−/OMI+/− and BID–/− mice as previously described (Wunderlich et al, 2012). Mouse livers were perfused via the vena carva with solution I (EBSS without Ca2+ and Mg2+ (Gibco), 0,5 mM EGTA) followed by perfusion with 50 ml of collagenase solution (EBSS with Ca2+ and Mg2+, 10 mM HEPES, 4,650 U collagenase type 4 (Worthington) and 2 mg Trypsin inhibitor (Sigma)). Hepatocytes were brushed through a 70-μm nylon mesh (BD Biosciences) and washed twice in high glucose DMEM (5% FCS) (Gibco). 2.5 × 105 cells were seeded on collagen-coated 6-well plates (BD BioCoat) and after 4 h the medium was renewed. The next day, hepatocytes were infected with the invasive Shigella strain M90T and analyzed for cell death and LDH release.
Statistical analysis
Data are presented as means of at least three independent experiments. Standard deviations were calculated as indicated in the figure legends. Two-tailed student's t-test was applied for statistical analyses. P-values for the Kaplan–Meier survival curves were calculated using chi-square statistics.
Acknowledgments
We thank P. Sansonetti (Paris, France) for the Shigella flexneri strains M90T and BS176, CS. Duckett (Ann Arbor, MI, USA), DL. Vaux (Victoria, Australia) and K. Rajalingam (Frankfurt, Germany) for XIAP−/− MEFs, H. Okada (Toronto, Canada) for SMAC−/− MEFs, J. Downward (London, UK) for OMI–/− MEFs and A. Strasser (Melbourne, Australia) for BID−/− MEFs and BID−/− mice. We especially thank Ute Sandaradura de Silva, Maureen Menning, Daniela Klubertz and Tanja Roth for excellent technical assistance. We also thank Esther Barth for preparing sections for electron microscopy, Irmgard Henke for help with IHC, Astrid Schauss and the CECAD Imaging Facility, the imaging facility of the Collaborative research center 670 (SFB670, Z2) and the Institute for Genetics (Prof. Jonathan Howard) as well as the animal facility of the Center for Molecular Medicine Cologne (CMMC). This work was supported by the Deutsche Forschungsgemeinschaft (DFG) SFB670.
Author contributions
MA performed the majority of the experiments; JMS, DWS were involved in generation of conditional XIAP KO mice; SS and KB performed the primary hepatocytes cell death analyses; MF contributed to XIAP pull down assay after infection; PB, AW and OC provided the independent experimental replicates of the central findings; KW performed EMSA analyses; CMW performed RT-PCR analyses and helped with isolation of primary hepatocytes; PM and EIR helped with electron microscopy; HB and TAK performed the NFκB luciferase assay; TL performed mass spectrometry; ASK performed the histopathologic analyses; OU carried out mouse infections; MK, AV, LMM and TK provided essential tools including cells or mouse strains; HK designed and conducted the study. All authors analyzed the data, discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Ashida H, Nakano H, Sasakawa C. Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to Inhibit PKC-NF-κB activity in invaded epithelial cells. PLoS Pathog. 2013;9:e1003409. doi: 10.1371/journal.ppat.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Ogawa M, Kim M, Suzuki S, Sanada T, Punginelli C, Mimuro H, Sasakawa C. Shigella deploy multiple countermeasures against host innate immune responses. Curr Opin Microbiol. 2011;14:16–23. doi: 10.1016/j.mib.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Baron C. Antivirulence drugs to target bacterial secretion systems. Curr Opin Microbiol. 2010;13:100–105. doi: 10.1016/j.mib.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Bauler LD, Duckett CS, O'Riordan MX. XIAP regulates cytosol-specific innate immunity to Listeria infection. PLoS Pathog. 2008;4:e1000142. doi: 10.1371/journal.ppat.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergounioux J, Elisee R, Prunier AL, Donnadieu F, Sperandio B, Sansonetti P, Arbibe L. Calpain activation by the shigella flexneri effector VirA regulates key steps in the formation and life of the bacterium's epithelial niche. Cell Host Microbe. 2012;11:240–252. doi: 10.1016/j.chom.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Lippens S, Staes A, Gilbert B, Roelandt R, De Medts J, Gevaert K, Declercq W, Vandenabeele P. cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1-4) PLoS ONE. 2011;6:e22356. doi: 10.1371/journal.pone.0022356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billen LP, Shamas-Din A, Andrews DW. Bid: a Bax-like BH3 protein. Oncogene. 2008;27:S93–S104. doi: 10.1038/onc.2009.47. [DOI] [PubMed] [Google Scholar]
- Brinkmann K, Hombach A, Seeger JM, Wagner-Stippich D, Klubertz D, Krönke M, Abken H, Kashkar H. Second mitochondria-derived activator of caspase (SMAC) mimetic potentiates tumor susceptibility toward natural killer cell-mediated killing. Leuk Lymphoma. 2014;55:645–651. doi: 10.3109/10428194.2013.807925. [DOI] [PubMed] [Google Scholar]
- Brinkmann K, Zigrino P, Witt A, Schell M, Ackermann L, Broxtermann P, Schüll S, Andree M, Coutelle O, Yazdanpanah B, Seeger JM, Klubertz D, Drebber U, Hacker UT, Krönke M, Mauch C, Hoppe T, Kashkar H. Ubiquitin C-Terminal Hydrolase-L1 potentiates cancer chemosensitivity by Stabilizing NOXA. Cell Rep. 2013;3:1–11. doi: 10.1016/j.celrep.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Chen M, He H, Zhan S, Krajewski S, Reed JC, Gottlieb RA. Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia/reperfusion. J Biol Chem. 2001;276:30724–30728. doi: 10.1074/jbc.M103701200. [DOI] [PubMed] [Google Scholar]
- Coutelle O, Hornig-Do H-T, Witt A, Andree M, Schiffmann LM, Piekarek M, Brinkmann K, Seeger JM, Liwschitz M, Miwa S, Hallek M, Krönke M, Trifunovic A, Eming SA, Wiesner RJ, Hacker UT, Kashkar H. Embelin inhibits endothelial mitochondrial respiration and impairs neoangiogenesis during tumor growth and wound healing. EMBO Mol Med. 2014;6:624–639. doi: 10.1002/emmm.201303016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Fiil BK, Speckmann C, Yabal M, Zur Stadt U, Bekker-Jensen S, Jost PJ, Ehl S, Mailand N, Gyrd-Hansen M. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol Med. 2013;5:1278–1295. doi: 10.1002/emmm.201303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, Rieser E, Rickard JA, Bankovacki A, Peschel C, Ruland J, Bekker-Jensen S, Mailand N, Kaufmann T, Strasser A, Walczak H, Silke J, Jost PJ, Gyrd-Hansen M. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell. 2012;46:746–758. doi: 10.1016/j.molcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Tournebize R, Mavris M, Page AL, Li X, Stark GR, Bertin J, DiStefano PS, Yaniv M, Sansonetti PJ. CARD4/Nod1 mediates NF-kappaB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- Harlin H, Reffey S, Duckett C, Lindsten T, Thompson C. Characterization of XIAP-Deficient Mice. Mol Cell Biol. 2001;21:3604–3608. doi: 10.1128/MCB.21.10.3604-3608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubert D, Gharib N, Rivero F, Wiegmann K, Hösel M, Krönke M, Kashkar H. PtdIns(4,5)P-restricted plasma membrane localization of FAN is involved in TNF-induced actin reorganization. EMBO J. 2007;26:3308–3321. doi: 10.1038/sj.emboj.7601778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AM, Kottachchi D, Lewis J, Duckett CS, Korneluk RG, Liston P. A novel ubiquitin fusion system bypasses the mitochondria and generates biologically active Smac/DIABLO. J Biol Chem. 2003;278:7494–7499. doi: 10.1074/jbc.C200695200. [DOI] [PubMed] [Google Scholar]
- Jost PJ, Grabow S, Gray D, McKenzie MD, Nachbur U, Huang DCS, Bouillet P, Thomas HE, Borner C, Silke J, Strasser A, Kaufmann T. XIAP discriminates between type I and type II FAS-induced apoptosis. Nature. 2009;460:1035–1039. doi: 10.1038/nature08229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkar H. X-linked inhibitor of apoptosis: a chemoresistance factor or a hollow promise. Clin Cancer Res. 2010;16:4496–4502. doi: 10.1158/1078-0432.CCR-10-1664. [DOI] [PubMed] [Google Scholar]
- Kashkar H, Haefs C, Shin H, Hamilton-Dutoit SJ, Salvesen GS, Kronke M, Jurgensmeier JM. XIAP-mediated caspase inhibition in Hodgkin's lymphoma-derived B cells. J Exp Med. 2003;198:341–347. doi: 10.1084/jem.20021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkar H, Seeger JM, Hombach A, Deggerich A, Yazdanpanah B, Utermohlen O, Heimlich G, Abken H, Kronke M. XIAP targeting sensitizes Hodgkin lymphoma cells for cytolytic T-cell attack. Blood. 2006;108:3434–3440. doi: 10.1182/blood-2006-05-021675. [DOI] [PubMed] [Google Scholar]
- Kaufmann T, Tai L, Ekert PG, Huang DCS, Norris F, Lindemann RK, Johnstone RW, Dixit VM, Strasser A. The BH3-only protein bid is dispensable for DNA damage- and replicative stress-induced apoptosis or cell-cycle arrest. Cell. 2007;129:423–433. doi: 10.1016/j.cell.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Kellendonk C, Opherk C, Anlag K, Schütz G, Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26:151–153. doi: 10.1002/(sici)1526-968x(200002)26:2<151::aid-gene17>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ogawa M, Sanada T, Mimuro H, Kim M, Ashida H, Akakura R, Yoshida M, Kawalec M, Reichhart J-M, Mizushima T, Sasakawa C. The Shigella OspC3 effector inhibits Caspase-4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host Microbe. 2013;13:570–583. doi: 10.1016/j.chom.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Krieg A, Correa RG, Garrison JB, Le Negrate G, Welsh K, Huang Z, Knoefel WT, Reed JC. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci USA. 2009;106:14524–14529. doi: 10.1073/pnas.0907131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Banks DJ, Philpott DJ. Innate immune sensing of microbes by Nod proteins. Ann N Y Acad Sci. 2006;1072:19–27. doi: 10.1196/annals.1326.020. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe. 2010;8:44–54. doi: 10.1016/j.chom.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Mantis N, Prevost MC, Sansonetti P. Analysis of epithelial cell stress response during infection by Shigella flexneri. Infect Immun. 1996;64:2474–2482. doi: 10.1128/iai.64.7.2474-2482.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteyn BS, Gazi AD, Sansonetti PJ. Shigella A model of virulence regulation in vivo. Gut Microbes. 2012;2:104–120. doi: 10.4161/gmic.19325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino MC, Rossi G, Tattoli I, Martini I, Chiavolini D, Cortese G, Pozzi G, Bernardini ML. Intravenous infection of virulent shigellae causes fulminant hepatitis in mice. Cell Microbiol. 2005;7:115–127. doi: 10.1111/j.1462-5822.2004.00441.x. [DOI] [PubMed] [Google Scholar]
- Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, Savopoulos J, Gray CW, Creasy CL, Dingwall C, Downward J. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem. 2002;277:439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, Creasy CL, Martin A, Hargreaves I, Heales SJ, Okada H, Brandner S, Schulz JB, Mak T, Downward J. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Pinedo C, Guio-Carrion A, Goldstein JC, Fitzgerald P, Newmeyer DD, Green DR. Different mitochondrial intermembrane space proteins are released during apoptosis in a manner that is coordinately initiated but can vary in duration. Proc Natl Acad Sci USA. 2006;103:11573–11578. doi: 10.1073/pnas.0603007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerincx A, Rodriguez GM, Steimle V, Kufer TA. NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J Immunol. 2012;188:4940–4950. doi: 10.4049/jimmunol.1103136. [DOI] [PubMed] [Google Scholar]
- Pachlopnik SJ, Canioni D, Moshous D, Touzot F, Mahlaoui N, Hauck F, Kanegane H, Lopez-Granados E, Mejstrikova E, Pellier I, Galicier L, Galambrun C, Barlogis V, Bordigoni P, Fourmaintraux A, Hamidou M, Dabadie A, Le Deist F, Haerynck F, Ouachee-Chardin M. Clinical similarities and differences of patients with X-linked lymphoproliferative syndrome type 1 (XLP-1/SAP deficiency) versus type 2 (XLP-2/XIAP deficiency) Blood. 2011;117:1522–1529. doi: 10.1182/blood-2010-07-298372. [DOI] [PubMed] [Google Scholar]
- Pal M, Wunderlich CM, Spohn G, Brönneke HS, Schmidt-Supprian M, Wunderlich FT. Alteration of JNK-1 signaling in skeletal muscle fails to affect glucose homeostasis and obesity-associated insulin resistance in mice. PLoS ONE. 2013;8:e54247. doi: 10.1371/journal.pone.0054247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott DJ, Yamaoka S, Israel A, Sansonetti PJ. Invasive Shigella flexneri activates NF-kappa B through a lipopolysaccharide-dependent innate intracellular response and leads to IL-8 expression in epithelial cells. J Immunol. 2000;165:903–914. doi: 10.4049/jimmunol.165.2.903. [DOI] [PubMed] [Google Scholar]
- Prakash H, Albrecht M, Becker D, Kuhlmann T, Rudel T. Deficiency of XIAP leads to sensitization for Chlamydophila pneumoniae pulmonary infection and dysregulation of innate immune response in mice. J Biol Chem. 2010;285:20291–20302. doi: 10.1074/jbc.M109.096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- Rehm M, Dussmann H, Prehn JH. Real-time single cell analysis of Smac/DIABLO release during apoptosis. J Cell Biol. 2003;162:1031–1043. doi: 10.1083/jcb.200303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, Fischer A, de Saint Basile G, Latour S. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, Oyama A, Ashida H, Kobayashi T, Koyama T, Nagai S, Shibata Y, Gohda J, Inoue J, Mizushima T, Sasakawa C. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- Seeger JM, Brinkmann K, Yazdanpanah B, Haubert D, Pongratz C, Coutelle O, Kronke M, Kashkar H. Elevated XIAP expression alone does not confer chemoresistance. Br J Cancer. 2010;102:1717–1723. doi: 10.1038/sj.bjc.6605704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Bertrand MJ. The role of the IAP E3 ubiquitin ligases in regulating pattern-recognition receptor signalling. Nat Rev Immunol. 2012;12:833–844. doi: 10.1038/nri3325. [DOI] [PubMed] [Google Scholar]
- Wunderlich CM, Delić D, Behnke K, Meryk A, Ströhle P, Chaurasia B, Al-Quraishy S, Wunderlich F, Brüning JC, Wunderlich FT. Cutting edge: inhibition of IL-6 trans-signaling protects from malaria-induced lethality in mice. J Immunol. 2012;188:4141–4144. doi: 10.4049/jimmunol.1102137. [DOI] [PubMed] [Google Scholar]
- Yang X, Miyawaki T, Kanegane H. SAP and XIAP deficiency in hemophagocytic lymphohistiocytosis. Pediatr Int. 2012;54:447–454. doi: 10.1111/j.1442-200X.2012.03683.x. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.