Abstract
Inositol phospholipids are critical regulators of membrane biology throughout eukaryotes. The general principle by which they perform these roles is conserved across species and involves binding of differentially phosphorylated inositol head groups to specific protein domains. This interaction serves to both recruit and regulate the activity of several different classes of protein which act on membrane surfaces. In mammalian cells, these phosphorylated inositol head groups are predominantly borne by a C38:4 diacylglycerol backbone. We show here that the inositol phospholipids of Dictyostelium are different, being highly enriched in an unusual C34:1e lipid backbone, 1-hexadecyl-2-(11Z-octadecenoyl)-sn-glycero-3-phospho-(1'-myo-inositol), in which the sn-1 position contains an ether-linked C16:0 chain; they are thus plasmanylinositols. These plasmanylinositols respond acutely to stimulation of cells with chemoattractants, and their levels are regulated by PIPKs, PI3Ks and PTEN. In mammals and now in Dictyostelium, the hydrocarbon chains of inositol phospholipids are a highly selected subset of those available to other phospholipids, suggesting that different molecular selectors are at play in these organisms but serve a common, evolutionarily conserved purpose.
Keywords: Dictyostelium, ether lipids, phosphoinositides, PI3K, plasmanylinositol
See also: GRV Hammond & T Balla (October 2014)
Introduction
Inositol phospholipids are believed to be ubiquitous amongst the eukaryotes, where they play crucial roles in organising a wide variety of cellular functions, particularly vesicular trafficking and signal transduction (Balla, 2013; Di Paolo & De Camilli, 2006; Michell, 2008). Eight of these lipids are commonly described, the most abundant of which is phosphatidylinositol (PtdIns). PtdIns is distributed throughout the intracellular membrane systems of eukaryotes and usually represents approximately 10% of total cellular phospholipids. The other inositol phospholipids, where present, are much less abundant and carry one or more phosphate groups on their inositol ring, namely PtdIns3P, PtdIns4P, PtdIns5P, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2 and PtdIns(3,4,5)P3. These more highly phosphorylated inositol lipids are metabolically interconverted by a series of reactions catalysed by specific kinases and phosphatases, and characteristically exhibit a much more restricted cellular distribution relating to their function.
Phylogenetic surveys indicate that genes encoding type-III PI3K and PI4K inositol lipid kinases (making PtdIns(3)P and PtdIns(4)P, respectively) and PI(4)P5K (making PtdIns(4,5)P2) are likely to be present across the eukaryotes. In contrast, Class I PI3Ks, producing PtdIns(3,4,5)P3 at the plasma membrane, are absent in plants and fungi, but are found in the Amoebozoa, such as Dictyostelium discoideum (Brown & Auger, 2011; Engelman et al, 2006; Zhou et al, 1995).
In mammalian cells, PtdIns(3)P and PtdIns(3,5)P2 play important roles in endosomal/lysosommal trafficking and the induction of autophagy (Burman & Ktistakis, 2010; Raiborg et al, 2013). PtdIns(4)P and PtdIns(4,5)P2 play roles in plasma membrane identity, cytoskeletal organisation and secretion (Di Paolo & De Camilli, 2006; Saarikangas et al, 2010). PtdIns(4,5)P2 also plays a major role as the substrate for two major signal transduction pathways; PLC-catalysed conversion of PtdIns(4,5)P2 to the messenger molecules diacylglycerol and inositol 1,4,5-trisphosphate (Kadamur & Ross, 2013) and Class I PI3K-catalysed conversion of PtdIns(4,5)P2 to the messenger PtdIns(3,4,5)P3 (Hawkins et al, 2006).
To a greater or lesser extent, all of the phosphorylated inositol lipids signal via the binding of their specific inositol phosphate head groups to several defined protein domains, the best studied of which are the binding of PtdIns(3,4,5)P3 to a subfamily of PH-domain containing proteins at the inner leaflet of the plasma membrane and the binding of PtdIns(3)P to FYVE or PX domains at the cytoplasmic face of endosomes and autophagosomes (Lemmon, 2008). These lipids are thus envisaged to act as regulatable membrane scaffolds dictating the localisation and function of proteins at the surface in which they reside.
Thus far, the composition of the hydrocarbon chains in inositol phospholipids has received very little attention. In part, this is because they are envisaged to merely anchor the inositol phospholipid in the bilayer, with minimal influence on the functional interaction of the head group with specific protein domains. However, it is also because most of the widely used techniques to measure these lipids in cell extracts do not yield the composition of the hydrocarbon chains and, indeed, often rely on anion-exchange chromatography of their deacylated derivatives (Guillou et al, 2007). Mass spectrometry-based approaches have the potential to define the full structures of lipids, but they have generally lacked sufficient sensitivity to accurately measure the more highly phosphorylated inositol lipids (Pettitt et al, 2006). We have recently solved some of these problems through chemical derivatisation of the phosphate groups on the inositol ring, allowing sensitive detection of PI, PIP, PIP2 and PIP3 species (the abbreviation PI(P)n is used here to refer to inositol phospholipids without implying the nature of the hydrocarbon linkage to the glycerol unit; PtdIns(P)n is used where the linkages are known to be esters, that is defining a ‘phosphatidyl’ unit) (Clark et al, 2011; Kielkowska et al, 2014). However, these methods do not distinguish between regioisomers of these lipids, that is, PI3P/PI4P/PI5P and PI(3,4)P2/PI(3,5)P2/PI(4,5)P2; PI(3,4,5,)P3 is the only known isomer of PIP3 found in cells. The mass spectrometric analysis of the fatty acyl/alkyl composition of highly phosphorylated inositol lipids is still in its infancy and has focused thus far on lipid extracts of mammalian origin, confirming that these lipids possess the characteristic diacylglycerol backbone of their precursor, PtdIns (Holub, 1986). The acyl groups that make up this backbone are highly selected: typically stearoyl (C18:0; 18 carbons: 0 double bonds) at the sn-1 position and arachidonoyl (C20:4) at the sn-2 position (C38:4 in total), particularly in primary tissue samples, with less abundant species containing palmitoyl (C16:0), oleoyl (C18:1) and linoloyl (C18:2) groups (Anderson et al, 2013; Lee et al, 2012; Milne et al, 2005; Rouzer et al, 2006). This raises the questions of whether the same C38:4 backbone is used in other organisms and whether there is the same strong selection for this (or another) backbone in the inositol lipids compared to the heterogeneity of backbone available in the other phospholipids.
The social amoeba Dictyostelium discoideum has genetic and cell biological features commending it as a model organism for investigating biological organisation across species (King & Insall, 2009; Muller-Taubenberger et al, 2013). Dictyostelium cells possess 7 recognisable PI3K genes, 5 of which have Ras-binding domains indicative of regulation through Ras (Hoeller & Kay, 2007; Zhou et al, 1995), a clear PTEN homologue (Iijima & Devreotes, 2002), and a number of PIP3-binding effector proteins, including a homologue of the protein kinase PKB/AKT (Meili et al, 2000; Zhang et al, 2010). Genetic studies show that PI3K signalling is required for efficient macropinocytosis (Buczynski et al, 1997; Hoeller et al, 2013; Zhou et al, 1998) and for the relay of cyclic-AMP signals during aggregation (Insall et al, 1994; Loovers et al, 2006). Its role in chemotaxis to cyclic-AMP and folic acid is much more controversial (Kay et al, 2008). Although PIP3 is made in response to both chemoattractants (see later) and can be polarised towards the leading edge of chemotaxing cells (Parent et al, 1998), genetic elimination of PIP3 signalling does not prevent efficient chemotaxis to either chemical and indeed improves chemotaxis to folic acid (Hoeller et al, 2013; Hoeller & Kay, 2007; Takeda et al, 2007; Veltman et al, 2014). There is also evidence that PIP3 is involved in bleb-driven cell movement (Zatulovskiy et al, 2014), and as PIP3 is produced at phagosomes (Dormann et al, 2004), it is likely to be involved in phagocytosis. Despite this intense interest in phosphoinositide signalling, very few measurements have been published quantifying changes in the inositol lipids themselves in this organism.
We applied our recent mass spectrometry techniques to analyse inositol phospholipids from Dictyostelium. To our surprise, we discovered that the vast majority of the inositol phospholipids in this organism are ether lipids, with a novel plasmanyl-C34:1e structure (the ‘e’ is used here to indicate that one of the hydrocarbon chains is linked to the glycerol via an ether linkage). This C34:1e species is much more enriched in the inositol phospholipids than the other major phospholipid classes, suggesting it has been selected to convey specific properties to this pool. We also show through the use of appropriate mutants that these plasmanylinositols are used by the PI3K signalling pathway and define the kinetics of C34:1e PIP3 production in response to chemoattractants.
Results
Inositol phospholipids in Dictyostelium are C34:1e plasmanylinositols
We recently described a new HPLC-ESI mass spectrometry method to analyse phosphorylated inositol lipids in cellular lipid extracts (Kielkowska et al, 2014). This method uses methylation of acidic phosphate groups with TMS-diazomethane, reverse phase chromatography on a C4 column, fragmentation of the lipids at the phosphodiester phosphate and measuring the charged molecular species derived from the neutral loss of a specific methylated head group. We applied this methodology to measure inositol phospholipids in lipid extracts from Dictyostelium discoideum grown in axenic medium. Neutral loss scans corresponding to the loss of the inositide head groups (inositol, methylated inositol phosphate or methylated inositol bisphosphate) indicated that the most abundant inositide species had unexpected and unusual masses (m/z 837.6, 945.6 and 1053.6 respectively; Fig 1). The corresponding glycerol fragment had an m/z of 563.6, which suggested either it was derived from a diacylglycerol containing a fatty acyl group with an odd number of carbons or, that one chain was attached to the glycerol via an ether linkage. Both of these possibilities would appear to have the same mass at the resolution of the mass spectrometer used, although fatty acids with an odd carbon chain length are rarely found in eukaryotes.
Figure 1. The major molecular species of inositol phospholipids present in lipid extracts of Dictyostelium discoideum possess a C34:1e backbone.
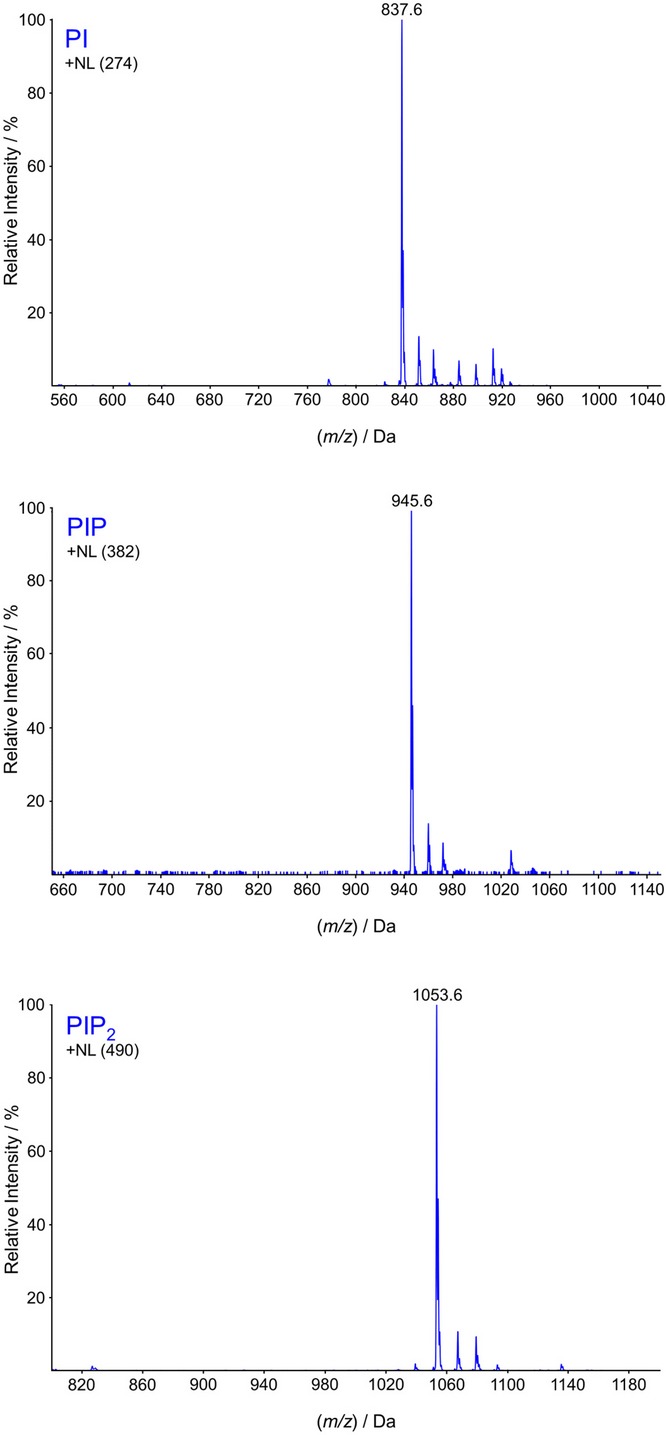
Lipid extracts were prepared from D. discoideum grown in axenic medium, then methylated with TMS-diazomethane and analysed by HPLC-ESI mass spectrometry. Neutral loss scans are shown which describe the major species of PI, PIP and PIP2 present (the mass of the individual neutral fragments corresponding to the mass of methylated inositol phosphate ‘head groups’ are listed in parentheses and differ by multiples of 108, the mass of a methylated phosphate). The most abundant species detected in each case corresponded to the generation of a glycerol fragment with an m/z of 563.6, suggesting the presence of either one ether-linked hydrocarbon chain plus one acyl chain (C34:1e) or two acyl chains with an odd number of total carbon atoms (C33:1). Further high-resolution mass analysis (Supplementary Fig S1A) and fragmentation (Supplementary Fig S1B) confirmed the presence of C16:0 alkyl and C18:1 acyl chains. Similar results were obtained when lipid extracts were prepared from D. discoideum grown on bacteria (Supplementary Fig S1C) or in a fully defined medium containing no added fatty acids (Supplementary Fig S1D).
To resolve this ambiguity, a sample of methylated PIP2 was isolated by HPLC and an accurate mass obtained on an Orbitrap mass spectrometer capable of working at a higher mass resolution. A value of m/z for MH+ of 1053.5809 was obtained (Supplementary Fig S1A). The theoretical m/z for a C34:1e ether/acyl PIP2 is 1053.5804, whereas the theoretical m/z for the alternative C33:1 diacyl compound would be 1053.5440 (which was not seen). Further fragmentation studies of the PIP2 species revealed an ion with a m/z of 265.4 (Supplementary Fig S1B), suggestive of a C18:1 acyl cation. If this was indeed a C18:1 acyl cation, then the other chain would most likely be a C16:0 ether-linked chain, which was consistent with the other ions observed (see Supplementary Fig S1B). Previous work with other organisms has shown that ether-containing phospholipids are synthesised by a metabolic pathway that first exchanges an acyl chain in the sn-1 position of the glycerol for an alcohol, in a reaction catalysed by alkyl-DHAP synthase (Nenci et al, 2012). We therefore attempted to confirm the structure and pathway for synthesis of a putative C34:1e lipid by feeding Dictyostelium cells a C16:0 alcohol (hexadecan-1-ol or palmitol) which contained two deuterium nuclei at the C1 position. Both deuteriums were efficiently incorporated into the C34:1e structure, indicating that the palmityl ether was present (Fig 2). The high proportion of deuterium incorporation (80% of PIP2 molecules had incorporated two deuteriums by 490 min; Supplementary Fig S2), and the lack of any significant incorporation of a single deuterium nucleus, ruled out significant metabolism at the C1 of D2-palmitol and hence more indirect routes of deuterium incorporation. Importantly, the lack of incorporation of a single deuterium also ruled out the presence of a vinyl ether linkage, a characteristic feature of a group of ether lipids called plasmalogens (Brites et al, 2004).
Figure 2. Determination of the alkyl chain structure in Dictyostelium discoideum inositol lipids.
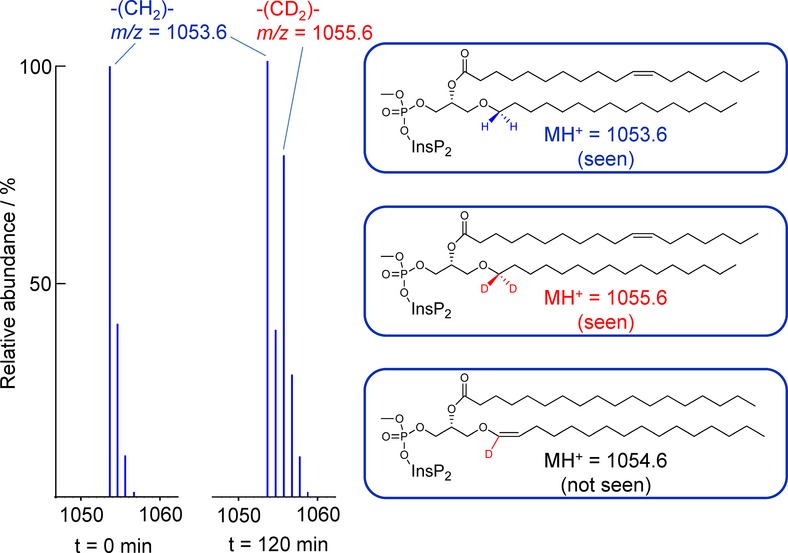
Methylated lipid extracts were prepared from D. discoideum grown in axenic medium in the absence (left panel) or presence (right panel) of D2-hexadecan-1-ol for 120 min and then analysed by HPLC-ESI mass spectrometry. The mass data are shown with a centroid presentation to allow differences of one mass unit to be more easily discerned. Both the unlabelled and labelled signals show the typical pattern obtained in mass spectra of compounds predominantly made up of carbon, hydrogen and oxygen atoms, which is the result of the natural abundance of 13C. D2-hexadecan-1-ol was synthesised with both deuterium nuclei in the C1 position, and the mass data indicate both deuteriums were efficiently incorporated into PIP2; that is, m/z peaks were shifted by precisely two mass units. There was no detectable increase in the m/z 1054.6 signal, indicating no significant incorporation of a single deuterium nucleus, and hence the absence of a vinyl ether linkage to the C16 (palmityl) chain (illustrated by the coloured structures). A more detailed description of the rate and extent of D2-hexadecan-1-ol incorporation is shown in Supplementary Fig S2.
Dictyostelium C34:1e lipids were sensitive to hydrolysis by phospholipase A2 (Scott et al, 1990), indicating that the C18:1 acyl chain is in the sn-2 position (Fig 3A). Finally, the position of the C=C double bond in the C18:1 chain was identified by ozonolysis to be delta-11, identifying the acyl chain as the 11-ocatadecenoyl group (Fig 3B). It is most likely that the double bond is the cis isomer because this is the isomer which is most commonly found in biologically important fatty acids and because Z11-ocatadecenoic acid has recently been shown to be an abundant fatty acid in Dictyostelium discoideum (Blacklock et al, 2008). Thus, the structures of the most abundant species of inositol lipids in Dictyostelium discoideum are defined as 1-hexadecyl-2-(11Z-octadecenoyl)-sn-glycero-3-phospho-(1’-myo-inositol) and phosphates thereof.
Figure 3. Determination of the acyl chain structure in Dictyostelium discoideum inositol lipids.
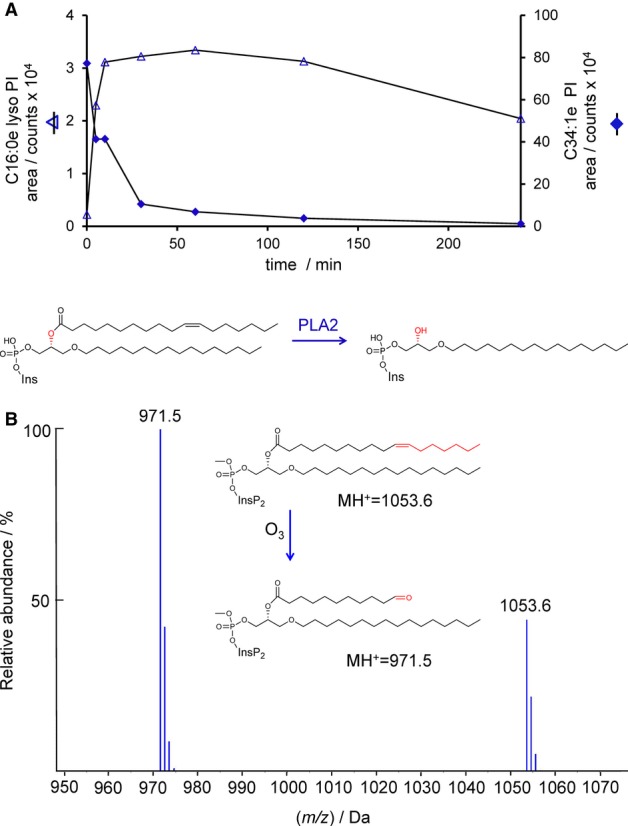
- The PI in D. discoideum is susceptible to hydrolysis by PLA2. Lipid extracts prepared from D. discoideum were mixed with phospholipase A2 (PLA2; from bee venom), and the levels of PI and lyso-PI measured over time by HPLC-ESI mass spectrometry. The quantitative conversion of PI to lyso-PI demonstrates the presence of an acyl chain in the sn-2 position.
- The PIP2 in D. discoideum contains an 11-octadecenoyl acyl chain. Methylated lipid extracts prepared from D. discoideum were subjected to ozonolysis. The mass of the fragment generated indicates the presence of an 11-octadecenoyl chain.
We also analysed the major species of inositol phospholipids in Dictyostelium discoideum grown on bacteria or in a fully defined medium with no added fatty acids, SIH medium (Han et al, 2004) and, in each case, found a similarly high proportion of the C34:1e species (Supplementary Fig S1C and D), indicating that this structure is independent of the availability of particular fatty acyl chains in the medium. Further, we additionally examined three other species of social amoebae—Dictyostelium purpureum, Dictyostelium mucoroides and Polysphondylium violaceum—and in each case found that the major species of PIP2 is the same mass as that from D. discoideum (m/z 1053.6), suggesting that the use of plasmanylinositols is common amongst these amoebae (unpublished observations).
The C34:1e plasmanyl motif is enriched in inositol lipids compared to other phospholipid classes
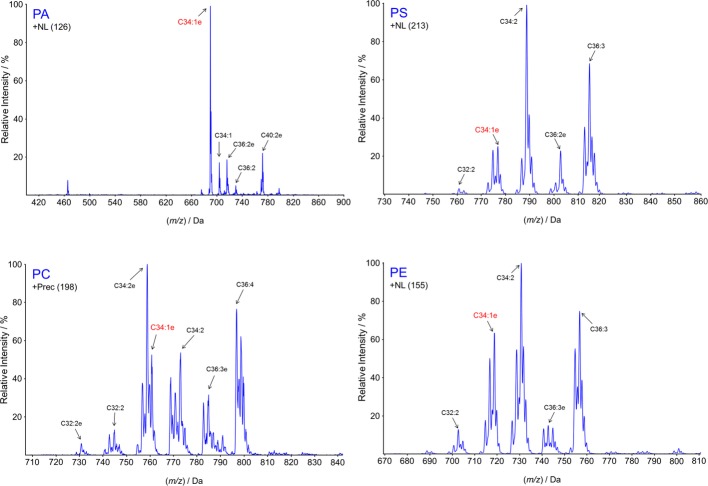
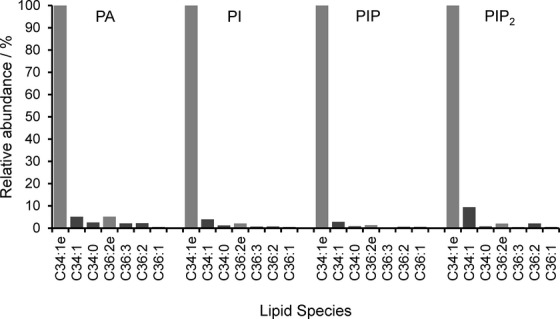
The striking selection for a particular combination of acyl tails in mammalian phosphoinositides raises the speculation that this selectivity could be functionally important; but equally with only this single example, it could be happenstance. We therefore asked whether a similar molecular selectivity exists in the Dictyostelium phosphoinositides. The relative abundances of molecular species of PC, PS, PE and PA in Dictyostelium were assessed by analogous neutral loss scans to those described above for the inositol phospholipids (Fig 4). It is immediately apparent that the C34:1e species is the most abundant form of PA, but is a smaller component of the other major phospholipid pools, which have the greater proportion comprised of a mixture of diacyl- and acyl/ether-species (relative comparison of these molecular species is given in Supplementary Fig S3).
Figure 4. The molecular species of the major phospholipid classes in Dictyostelium discoideum are highly heterogeneous.
Neutral loss (PA, PS, PE) or precursor ion (PC) scans are shown describing the major molecular species of abundant phospholipids present in methylated lipid extracts of D. discoideum grown under axenic conditions (see Materials and Methods for the MRM transitions monitored). The C34:1e species highlighted in red is analogous to the major species of inositol lipids found in D. discoideum. Relative quantification of some of these species is given in Supplementary Fig S3.
The most abundant species of PI, PIP, PIP2 and PA were targeted for more careful comparison by multiple reaction monitoring (MRM), which indicated that the vast majority of each of these lipid pools was comprised of the C34:1e species under both axenic (Fig 5) and bacterially fed conditions (unpublished observations). The accepted pathway for de novo synthesis of PI in eukaryotes is via the formation of CDP-DG from PA and CTP, catalysed by CDP-DG synthase (Saito et al, 1997), followed by the formation of PI from CDP-DG and inositol, catalysed by PI synthase (Paulus & Kennedy, 1960). Thus, the enrichment of C34:1e species of PI/PIP/PIP2 is naturally explained by the relative abundance of C34:1e PA. However, the other major phospholipids are derived from diacylglycerol by base activation (the Kennedy Pathway; Vance & Vance, 2004). Analogous neutral loss scans of DG were not possible, so we quantified selected DG species based on the major species detected for PI and PC, described above. The results indicated that the DG pool appears to be comprised mostly of diacyl-species, with only small quantities of C34:1e (Supplementary Fig S4). This suggests that the major pools of PA and DG are maintained with very different molecular compositions, consistent with their differential roles as the sources of PI and other phospholipids, respectively.
Figure 5. Measurement of the relative abundance of the major molecular species of inositol phospholipids and PA in Dictyostelium discoideum.
Methylated lipid extracts prepared from D. discoideum were grown under axenic conditions and analysed by HPLC-ESI mass spectrometry. MRM traces were integrated to provide relative abundances of the major species of PA, PI, PIP and PIP2 present.
C34:1e plasmanylinositols are used in Dictyostelium signalling pathways
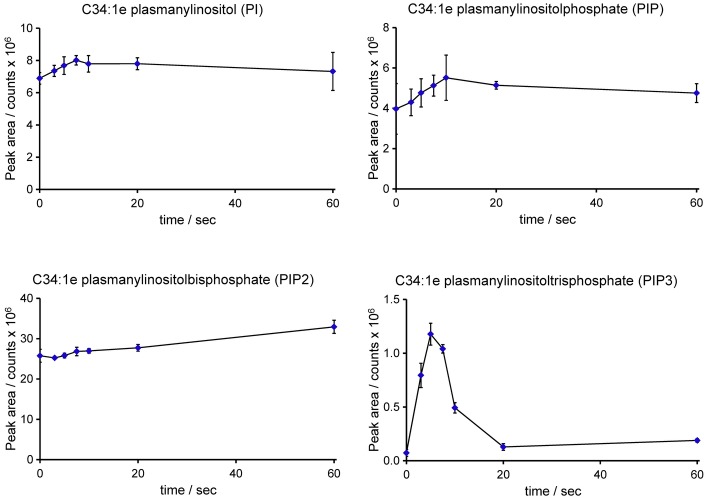
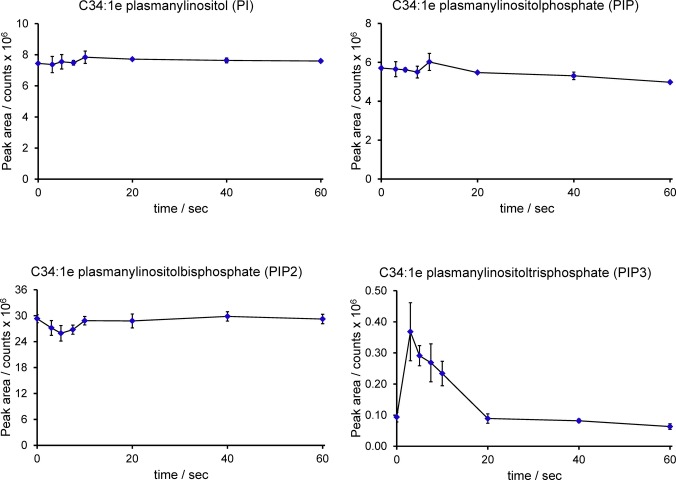
Previous work has characterised important roles for the PI3K signalling pathway in the response of Dictyostelium amoebae to chemoattractants. Thus far, however, measurements of the inositol phospholipids themselves in this organism have rarely been attempted, with most work using in vivo reporters instead. Although the levels of PIP3 were too low to obtain good neutral loss spectra (see above), specifically targeting the C34:1e species produced a very good signal/noise ratio and accurate measurement was readily achievable (see Supplementary Fig S5). We first measured changes in the levels of C34:1e-PI, -PIP, -PIP2 and -PIP3 in response to added cyclic-AMP using an adenylyl cyclase mutant to reduce background levels of cyclic-AMP and thus sharpen the response (acaA−; Fig 6). Addition of cyclic-AMP produced a very fast and transient rise in PIP3, with a minimal associated drop in PIP2. The level of PIP3 peaked at approximately 5 s. A very similar pattern of changes was seen in the response of the Ax2 strain to a different chemoattractant, folic acid (Fig 7).
Figure 6. Changes in inositol phospholipids in response to cAMP in Dictyostelium discoideum (acaA−).
The acaA− strain of D. discoideum was starved and rendered competent to respond to cAMP. Individual samples of cells were then stimulated with 10 μM cAMP for the times shown. Methylated lipid extracts were prepared and analysed by HPLC-ESI mass spectrometry. Integrated MRM values (mean ± SD, n = 3 individual cell incubations) for the C34:1e species of PI, PIP, PIP2 and PIP3 are shown; for example, traces from which the integrations were performed are shown in Supplementary Fig S5. This experiment has been repeated three times with qualitatively very similar results. These data are uncorrected for differences in extraction and ionisation of different lipid classes, and so, no significance can be placed on differences between the signal intensity of PI, PIP, PIP2 or PIP3.
Figure 7. Changes in inositol phospholipids in response to folic acid in Dictyostelium discoideum (Ax2).
The Ax2 strain of D. discoideum was grown on bacteria and stimulated with 100 μM folic acid for the times shown. Methylated lipid extracts were prepared and analysed by HPLC-ESI mass spectrometry. Integrated MRM values (mean ± SD, n = 3 individual cell incubations) for the C34:1e species of PI, PIP, PIP2 and PIP3 are shown. This experiment has been repeated three times with qualitatively very similar results. These data are uncorrected for differences in extraction and ionisation of different lipid classes, and so, no significance can be placed on differences between the signal intensity of PI, PIP, PIP2 or PIP3.
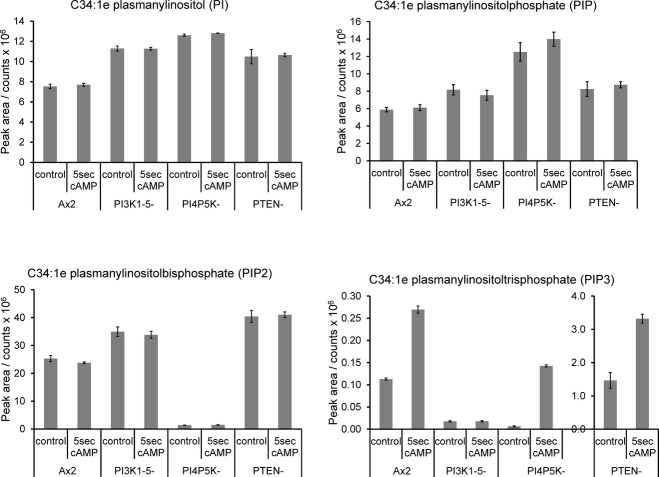
To examine the genetic dependence of the response to cyclic-AMP, we used mutants lacking key enzymes in inositol phospholipid signalling pathways. C34:1e inositol lipids were measured in these mutants before and after a 5-s stimulation with cyclic-AMP (Fig 8; note that because these mutants were created in the Ax2 strain, endogenous production of cyclic-AMP likely led to a somewhat de-sensitised response compared to the acaA− strain described above). Loss of all of the five PI3Ks with a Ras-binding domain (Hoeller & Kay, 2007) prevented any PIP3 response to cyclic-AMP, though a basal level of PIP3 was still detectable; this indicates that these PI3Ks are the cyclic-AMP-sensitive enzymes responsible for PIP3 synthesis, but another minor pathway of PIP3 synthesis must also exist in this organism. Loss of the PIP3 phosphatase PTEN (Iijima & Devreotes, 2002) resulted in a hugely elevated basal and stimulated level of PIP3, indicating it is a major PIP3 phosphatase in this organism. Loss of PI4P5K dramatically reduced the levels of PIP2 as previously reported (Fets et al, 2014), but remarkably, this diminished level could still support substantial cyclic-AMP-stimulated PIP3 synthesis, conflicting with previous measurements made using an ELISA.
Figure 8. Changes in inositol phospholipids in response to cAMP in mutants of Dictyostelium discoideum.
The parental Ax2 strain of D. discoideum, or the indicated mutant strains derived from it (PI3K1-5-, pikA−, pikB−, pikC−, pikF−, pikG−; PI4P5K-, pikI−; PTEN-, ptenA−) were grown on bacteria and then rendered competent to respond to cAMP. Individual samples of cells were then stimulated with 10 μM cAMP or vehicle for 5 s. Methylated lipid extracts were prepared and analysed by HPLC-ESI mass spectrometry. Integrated MRM values (mean ± SD, n = 3 individual cell incubations) for the C34:1e species of PI, PIP, PIP2 and PIP3 are shown. This experiment has been repeated three times with qualitatively very similar results. These data are uncorrected for differences in extraction and ionisation of different lipid classes and so no significance can be placed in differences between the signal intensity of PI, PIP, PIP2 or PIP3.
Discussion
Most work on phosphoinositide signalling in Dictyostelium hitherto has relied on in vivo reporters, such as the PIP3-binding PH-domain of CRAC fused to GFP (Parent et al, 1998), leaving the nature of the acyl/alkyl-glycerol backbone unknown. We have used a combination of mass spectrometry and further analytical techniques to define the major molecular species of inositol phospholipids: over 95% of the PI, PIP and PIP2 pools contain the C34:1e structure, with an ether-linked C16:0 chain (palmityl) in the sn-1 position and an ester-linked C18:1 chain (11-ocatadecenoyl) in the sn-2 position. The correct prefix for phospholipids with a saturated alkyl chain in the sn-1 position is plasmanyl, to distinguish them from plasmenyl-lipids (which contain a vinyl ether linkage in the sn-1 position, often referred to as plasmalogens) and phosphatidyl-lipids (which contain ester linkages in both sn-1 and 2 positions). The correct abbreviation for phosphatidylinositols is PtdIns(P)n, but often the more shorthand PI(P)n is used in the literature; we suggest the PI(P)n nomenclature is retained to include both phosphatidyl- and plasmanyl-inositols.
Ether lipids have been widely described in other organisms, for example in humans plasmalogens make up approximately 20% of total phospholipid mass, with plasmenyl ethanolamine and plasmenyl choline being particularly abundant in mammalian brain and heart, respectively (Brites et al, 2004). Myeloid cells such as macrophages and neutrophils are also known to contain high levels of plasmanyl choline, which is the synthetic precursor of platelet activating factor (which has a 1-O-ether linkage at sn-1 and an acetyl-chain at sn-2) (Montrucchio et al, 2000). However, ether-linked inositol phospholipids have only been reported previously in trace quantities (Ivanova et al, 2010) or, as components of glycophosphatidylinositol (GPI) anchors (Kanzawa et al, 2009), and this is the first example where they represent the major species of inositol phospholipids present in any organism or tissue. The standard methods of inositol lipid analysis rely on base induced deacylation followed by anion-exchange chromatography of glycerophosphoesters (Guillou et al, 2007). This approach is not appropriate for Dictyostelium phosphoinositide analysis because the ether linkage is resistant to these conditions, and this may explain past failures to measure significant quantities of these lipids.
A large body of literature implicates PI3K signalling pathways in the regulation of macropinocytosis and cell movement in Dictyostelium. Our results indicate that the previously identified ‘Class 1' PI3Ks in this organism are responsible for chemoattractant stimulated formation of PIP3 and that the phosphatase PTEN is a major regulator of PIP3 levels. Further, our results define the extremely rapid time course for the formation of PIP3 in response to chemoattractants and imply that C34:1e PIP3 is responsible for PH-domain-mediated translocation of downstream effectors such as CRAC or PhdA (Funamoto et al, 2001; Parent et al, 1998). The relatively small and transient drop in the level of PIP2 in response to chemoattractants suggests pathways for resynthesis of this lipid are well matched to PLC and PI3K induced pathways of consumption, which is consistent with many analogous contexts in mammalian cells, although not easily reconciled with recent theories suggesting large drops in PIP2 enhance PI3K signalling via loss of membrane-bound PTEN (Kortholt et al, 2007). The ability to accurately measure the levels of inositol phospholipids in Dictyostelium should now allow future studies to gain a much better understanding of these signalling pathways in this organism.
The composition of the inositol phospholipids in Dictyostelium is strikingly biased, with the C34:1e configuration comprising about 95% of the total, whereas diacyl or other alkyl/acyl configurations are preferred in the other phospholipid classes. In principle, the molecular selector that causes this bias could act at various levels in the biosynthesis or turnover of these lipids, but our data suggest it acts prior to the synthesis of PA. Phosphoinositides arise from PA, which in most organisms is synthesised de novo and can be dephosphorylated to produce DGs. These DGs are then used for the synthesis of the other phospholipid classes (Kent, 1995). However, the PA and DG pools in Dictyostelium comprise very different fatty alkyl/acyl compositions. PA is mostly the C34:1e species that is highly represented in the PI pool, but the DGs contain higher proportions of the other species which predominate in other phospholipids. Ether-linked PA is synthesised via DHAP in a pathway that originates in peroxisomes (Wanders & Waterham, 2006). The key step is the formation of 1-alkyl-DHAP by alkyl-DHAP synthase from acyl-DHAP and a long chain alcohol (a reaction in which the original acyl chain is essentially swapped for the alcohol) (Nenci et al, 2012). Our data suggest that the predominant alcohol used for this reaction in Dictyostelium is palmitol. Alkyl-DHAP is then thought to leave the peroxisome, after which it is reduced to 1-alkyl-glycerol 3-phosphate by an acyl/alkyl-DHAP reductase and then acylated at the sn-2 position by a fatty acyl-CoA acyl transferase located in the ER (Wanders & Waterham, 2006). Given the enrichment of delta11-ocatadecenoyl in Dictyostelium PA, this would predict the presence of an acyl transferase with specificity for 1-palmityl-glycerol-3-phosphate and delta11-ocatadecenoyl-CoA. In this regard, an active and unusual fatty acyl-CoA elongase has recently been described in Dictyostelium, which converts delta9-palmitoyl-CoA to delta11-ocatadecenoyl-CoA, providing the likely source of substrate for this reaction (Blacklock et al, 2008). In contrast, diacyl PA is thought to be synthesised by successive acylation of glycerol 3-phosphate directly in the ER (Shindou et al, 2009). It seems unlikely that there are novel and active pathways in Dictyostelium which re-model C34:1e-PA into diacyl DGs, and therefore, it seems most probable that there is a pool of diacyl PA which is synthesised de novo but does not accumulate to significant levels, perhaps because it is rapidly channelled into diacylglycerols. However, further work will be needed to establish the synthetic route of C34:1e PA in Dictyostelium and delineate how the diverse pools of PA and DGs are created.
A key issue raised by our results is why evolution has driven the creation of such an unusual structure for inositol phospholipids in Dictyostelium and why the inositol phospholipids are relatively molecularly homogeneous and distinct from the much more heterogeneous species of the other major classes of phospholipid. Inositol phospholipids are crucial signalling molecules designed to interact with extrinsic membrane proteins via their phosphorylated inositol lipid head groups. During macropinosome formation and in other situations, PIP3 forms a dense signalling patch on the membrane, and it is possible that the 1-O-ether linkage conveys molecular properties that are particularly suited to this signalling role. The lack of a carbonyl in the sn-1 position may allow the lipid to be more tightly integrated into the membrane, alter its packing with other lipids or subtly alter the orientation of the head group. Previous studies with plasmalogens suggest ether-linked lipids are more resistant to oxidants, but this property is thought to be due primarily to the vinyl ether bond (Brites et al, 2004; Zoeller et al, 1999), which is absent in plasmanyl lipids. The lack of an ester linkage at sn-1 would also make these lipids insensitive to hydrolysis by a PLA1 activity. It seems unlikely however that any such physical or chemical properties of ether-linked lipids would be more pertinent to the roles of inositol phospholipids in Dictyostelium than in other organisms.
There are still very few careful studies of the molecular composition of inositol phospholipids across species. Most of the work has been done in mammalian cells, where phosphatidylinositols are highly enriched in C38:4 species (C18:0, C20:4) compared to most other lipid classes (Holub, 1984; MacDonald et al, 1975). The mechanisms creating this enrichment are unclear, but are probably derived from the substrate specificity of key enzymes in the de novo synthesis pathway for PtdIns and remodelling via specific acylCoA-PtdIns transferases (Anderson et al, 2013; D'Souza & Epand, 2013; Lee et al, 2012). The specific properties conveyed by this enrichment in C38:4 species are also unclear. It has been suggested that these lipids may act as a source of PLA2-liberated C20:4 for lipoxygenases and cycloxygenases (Gijon et al, 2008), or it represents a mechanism to segregate PA and DG derived from inositol lipids from PA and DG involved in the metabolism of triacylglcerol or other phospholipids. Some evidence in favour of this notion is the specificity of some PKC isoforms for C38:4 DG derived from PLC-catalysed hydrolysis of PtdInsP2 (Madani et al, 2001). Of relevance here, neither eicosanoid biosynthesis, nor DG-regulated PKCs, have been convincingly described in Dictyostelium.
Dictyostelium, though having distinct phosphoinositides from mammals, also displays lipid tails chemically distinct from other phospholipids, which suggests that the evolutionary selective processes that gave rise to this distinction may have a common functional significance. Thus, it is plausible that the enrichment of C34:1e species of inositol phospholipids in Dictyostelium represents a molecular signature for these lipids that allows common intermediates to be distinguished by their lipid origin; thus, enabling intermediates such as PA or DG to be used as specific signals and/or selectively reused for re-synthesis of inositol phospholipids. Whatever the answers, these are clearly important questions to resolve since they impact on our understanding of how inositol phospholipid signalling pathways are organised and how they may interact with other metabolic pathways regulating lipid synthesis and nutrition.
Materials and Methods
Dictyostelium methods
Ax2 (Kay laboratory strain of Dictyostellium discoideum) was used as wild-type, with the following mutant strains derived from it: HM1366 (acaA−; adenylyl cyclase null, isolated by Oliver Hoeller); HM1200 (pikA−, pikB−, pikC−, pikF−, pikG−; PI3K quintuple null) (Hoeller & Kay, 2007); HM1289 (ptenA−; PTEN null, isolated by Oliver Hoeller) and HM1513 (pikI−; PIP5K null) (Fets et al, 2014). Dictyostelium cells were grown at 22°C either in shaken suspension in HL5 axenic medium + glucose (ForMedium) at 180 rpm, or on SM plates in association with Klebsiella aerogenes bacteria (Kay, 1987). Axenic growth was used as standard, but growth on bacteria was used for folate stimulation, the phosphoinositide signalling mutants and for Dictyostelium purpureum, Dictyostelium mucoroides and Polysphondylium violaceum. In some experiments, Dictyostellium discoideum was also grown in fully defined SIH medium (Han et al, 2004).
Cells were labelled with D2-palmitol by first preparing an emulsion of 25 mM D2-palmitol (from 1 M stock in chloroform) in 25 mg/ml BSA by sonicating in a bath-sonicator at 100 watts for 40 s. This emulsion was diluted to 0.4 mM D2-palmitol, 0.4 mg/ml BSA into either HL5 growth medium or KK2 (16.5 mM KH2PO4, 3.8 mM K2HPO4, 2 mM MgSO4, 0.1 mM CaCl2, pH 6.2); labelling was rapid with around 20% deuterated PIP2 seen after 10 min.
For lipid measurements, cells were washed free of nutrients by repeated centrifugation (200 g × 2 min) and resuspension in KK2, then starved at 107/ml in KK2 with shaking at 180 rpm. Folate stimulation was performed after 30 min of starvation, whereas cyclic-AMP stimulation was performed after 5.5 h, during which time the cells were pulsed with 100 nM cyclic-AMP every 6 min after the first hour, to bring them to a responsive state, then pelleted, washed once and resuspended to the original concentration in KK2. Samples were prepared in 2-ml Safelock tubes by adding 750 μl acidified chloroform–methanol (chloroform/methanol/1M HCl, 1:2:0.097) to 170 μl of cell suspension and stored on dry ice for further processing. In stimulation experiments, 160 μl cells were added to 10 μl of 170 μM cyclic-AMP or 1.7 mM folic acid (to give 10 μM and 100 μM respectively), then the response was terminated as above. All samples were prepared in triplicate. Protein was determined after solubilisation of pellets in 10 mM NaOH using the Bradford dye-binding assay, with BSA as a standard.
Lipid extraction and methylation
Incubations of Dictyostelium discoideum (170 μl) terminated as above, and forming a single phase was extracted using an acidified Folch phase partition, as described previously (Folch et al, 1957; Kielkowska et al, 2014). The final lower phase from the lipid extraction (approx. 1 ml) was mixed with 50 μl TMS-diazomethane (hazardous) for 10 min at RT; excess TMS-diazomethane was then quenched with 6 μl acetic acid and lipids re-extracted through a neutral pH Folch phase partition, as described previously (Clark et al, 2011; Kielkowska et al, 2014). The final lower phases were dried in vacuo or under a stream of N2 and then lipids re-dissolved by bath sonication in 100 μl of methanol:H2O (80:20 v/v).
HPLC-ESI mass spectrometry
Methylated lipid extracts (typically 40 μl) were separated by HPLC on an in-line C4 column before infusion into a triple quadrapole mass spectrometer in positive ion mode, as described previously (Clark et al, 2011; Kielkowska et al, 2014).
Mass spectrometer parameters used were the same as described in Clark et al ( 2011) unless specified otherwise. Fragmentations were carried out with a range of collision energies using the same settings for the other machine parameters, as described previously (Clark et al, 2011; Kielkowska et al, 2014). MRM transitions monitored and neutral loss masses are shown in the Table 1. Note that the PC data were acquired as parents of the 198.09.
Table 1.
The parent masses, neutral loss masses and glycerol fragment ions for the different mass spectrometry experiments used are shown in the table.
| Neutral loss mass | 213.03 | 155.03 | 198.09 | 126 | 274 | 382.04 | 490.05 | 598 | |
|---|---|---|---|---|---|---|---|---|---|
| Lipid chain | Glycerol MRM fragment | PS | PE | PC | PA | PI | PIP | PIP2 | PIP3 |
| C34:1 ae | 563.503 | 776.53 | 718.53 | 761.59 | 689.5 | 837.503 | 945.543 | 1053.6 | 1161.5 |
| C34:1 aa | 577.535 | 790.57 | 732.57 | 775.63 | 703.54 | 851.535 | 959.575 | 1067.6 | 1175.54 |
| C36:3 ae | 579.53 | 792.56 | 734.56 | 777.62 | 705.53 | 853.53 | 961.57 | 1069.6 | 1177.53 |
| C36:2 ae | 589.52 | 802.55 | 744.55 | 787.61 | 715.52 | 863.52 | 971.56 | 1079.6 | 1187.52 |
| C36:1 ae | 591.55 | 804.58 | 746.58 | 789.64 | 717.55 | 865.55 | 973.59 | 1081.6 | 1189.55 |
| C36:3 aa | 601.519 | 814.55 | 756.55 | 799.61 | 727.52 | 875.519 | 983.559 | 1091.6 | 1199.52 |
| C36:2 aa | 603.535 | 816.57 | 758.57 | 801.63 | 729.54 | 877.535 | 985.575 | 1093.6 | 1201.54 |
| C36:1 aa | 605.55 | 818.58 | 760.58 | 803.64 | 731.55 | 879.55 | 987.59 | 1095.6 | 1203.55 |
| Parent mass for each species | |||||||||
Synthesis of d2-palmitol
Methyl palmitate (2.5 g, 9.24 mmol) was dissolved in 20 ml dry tetrahydrofuran and cooled in an ice bath. 1 M lithium aluminium deuteride (20 ml, 20 mmol) was added in tetrahydrofuran under an inert atmosphere. The mixture was stirred for 30 min, then removed from the ice bath, allowed to warm up and stirred further at room temperature for 1 h. The mixture was then poured into 100 ml ice and 50 ml 2 M hydrochloric acid added, extracted into ethyl acetate, dried over magnesium sulphate and concentrated under vacuum. Yield was 2.1 g, 93%. GC-MS data showed the compound to be correct, 226.27 (M-18)+. Fragmentation data were as expected by comparison with the literature. Purity by GC and TLC was greater than 90%.
PLA2 hydrolysis
Preparation of the liposomes: lipids from 4 × 107 cells grown under axenic conditions were extracted (acid Folch), dried under nitrogen and then dried further under vacuum for 2 h. 1.8 ml of HEPES buffer (20 mM, pH 7.2) and 200 μl of 1 M CaCl2 were added, and then, the solution was vortexed and bath sonnicated for a total time of 15 min. The sample was then subjected to nine cycles of freezing in CO2(s)/acetone and thawing, which yielded a cloudy solution of liposomes; the liposomes were stored at −20°C until use.
A stock solution of phospholipase A2 was prepared (Sigma-Aldrich; 1 μg/μl in H2O). 10 μl of PLA2 solution was mixed with 50 μl of liposomes and samples incubated at room temperature for 5, 10, 30, 60, 120 or 240 min (a sample without PLA2 was prepared as a control). Enzyme activity was quenched with 170 μl 2 M HCl, and then, each sample was vortexed and prepared for HPLC-ESI mass spectrometry, as described above.
Ozonolysis
Ozone was generated at the anode of an electrolytic cell made from two platinum electrodes separated by a glass walled tube in 0.4 M sulphuric acid. The ozone was carried into the samples using a flow of nitrogen across the top of the glass tube containing the anode. A standard laboratory power pack was used set at 30 V, and a current of between 500 and 700 mA was measured.
Samples were extracted and methylated as usual and suspended in 200 μl methanol. This was then cooled to −78°C, and the ozone/nitrogen mix bubbled into the samples for 45 min. 2 μl dimethyl sulphide was then added and the sample allowed to warm up to room temperature, when 40 μl water was added and the sample loaded onto the mass spectrometer.
Acknowledgments
We are grateful to Oliver Hoeller for creating mutant strains and both the Medical Research Council (MRC file reference number U105115237) and the Biotechnology and Biological Sciences Research Council (ISPG BB/J004456/1) for core support. We would also like to thank RH Michell for helpful discussions.
Author contributions
JC, RRK, PTH and LRS designed the study, performed experiments and wrote the paper. AK designed and performed experiments and contributed to writing the paper. IN synthesised critical reagents. LF provided a valuable mutant. DO performed high-resolution mass spectroscopy.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Anderson KE, Kielkowska A, Durrant TN, Juvin V, Clark J, Stephens LR, Hawkins PT. Lysophosphatidylinositol-acyltransferase-1 (LPIAT1) is required to maintain physiological levels of PtdIns and PtdInsP(2) in the mouse. PLoS One. 2013;8:e58425. doi: 10.1371/journal.pone.0058425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklock BJ, Kelley D, Patel S. A fatty acid elongase ELO with novel activity from Dictyostelium discoideum. Biochem Biophysl Res Comm. 2008;374:226–230. doi: 10.1016/j.bbrc.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Brites P, Waterham HR, Wanders RJ. Functions and biosynthesis of plasmalogens in health and disease. Biochim Biophys Acta. 2004;1636:219–231. doi: 10.1016/j.bbalip.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Brown JR, Auger KR. Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol Biol. 2011;11:4. doi: 10.1186/1471-2148-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczynski G, Grove B, Nomura A, Kleve M, Bush J, Firtel RA, Cardelli J. Inactivation of two Dictyostelium discoideum genes, DdPIK1 and DdPIK2, encoding proteins related to mammalian phosphatidylinositide 3-kinases, results in defects in endocytosis, lysosome to postlysosome transport, and actin cytoskeleton organization. J Cell Biol. 1997;136:1271–1286. doi: 10.1083/jcb.136.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584:1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, Wakelam MJ, Stephens LR, Hawkins PT. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat Meth. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Dormann D, Weijer G, Dowler S, Weijer CJ. In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J Cell Sci. 2004;117:6497–6509. doi: 10.1242/jcs.01579. [DOI] [PubMed] [Google Scholar]
- D'Souza K, Epand RM. Enrichment of phosphatidylinositols with specific acyl chains. Biochim Biophys Acta. 2013;1838:1501–1508. doi: 10.1016/j.bbamem.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Fets L, Nichols JM, Kay RR. A PIP5 kinase essential for efficient chemotactic signaling. Curr Biol. 2014;24:415–421. doi: 10.1016/j.cub.2013.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Funamoto S, Milan K, Meili R, Firtel RA. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J Cell Biol. 2001;153:795–810. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijon MA, Riekhof WR, Zarini S, Murphy RC, Voelker DR. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283:30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou H, Stephens LR, Hawkins PT. Quantitative measurement of phosphatidylinositol 3,4,5-trisphosphate. Meth Enzymol. 2007;434:117–130. doi: 10.1016/S0076-6879(07)34007-X. [DOI] [PubMed] [Google Scholar]
- Han SI, Friehs K, Flaschel E. Improvement of a synthetic medium for Dictyostelium discoideum. Process Biochem. 2004;39:925–930. [Google Scholar]
- Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Hoeller O, Bolourani P, Clark J, Stephens LR, Hawkins PT, Weiner OD, Weeks G, Kay RR. Two distinct functions for PI3-kinases in macropinocytosis. J Cell Sci. 2013;126:4296–4307. doi: 10.1242/jcs.134015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub BJ. 1982 Borden Award lecture. Nutritional, biochemical, and clinical aspects of inositol and phosphatidylinositol metabolism. Can J Physiol Pharmacol. 1984;62:1–8. doi: 10.1139/y84-001. [DOI] [PubMed] [Google Scholar]
- Holub BJ. Metabolism and function of myo-inositol and inositol phospholipids. Ann Rev Nutr. 1986;6:563–597. doi: 10.1146/annurev.nu.06.070186.003023. [DOI] [PubMed] [Google Scholar]
- Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Insall R, Kuspa A, Lilly PJ, Shaulsky G, Levin LR, Loomis WF, Devreotes P. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1994;126:1537–1545. doi: 10.1083/jcb.126.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova PT, Milne SB, Brown HA. Identification of atypical ether-linked glycerophospholipid species in macrophages by mass spectrometry. J Lipid Res. 2010;51:1581–1590. doi: 10.1194/jlr.D003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadamur G, Ross EM. Mammalian phospholipase C. Ann Rev Physiol. 2013;75:127–154. doi: 10.1146/annurev-physiol-030212-183750. [DOI] [PubMed] [Google Scholar]
- Kanzawa N, Maeda Y, Ogiso H, Murakami Y, Taguchi R, Kinoshita T. Peroxisome dependency of alkyl-containing GPI-anchor biosynthesis in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:17711–17716. doi: 10.1073/pnas.0904762106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RR. Cell differentiation in monolayers and the investigation of slime mold morphogens. Meth Cell Biol. 1987;28:433–448. doi: 10.1016/s0091-679x(08)61661-1. [DOI] [PubMed] [Google Scholar]
- Kay RR, Langridge P, Traynor D, Hoeller O. Changing directions in the study of chemotaxis. Nat Rev Mol Cell Biol. 2008;9:455–463. doi: 10.1038/nrm2419. [DOI] [PubMed] [Google Scholar]
- Kent C. Eukaryotic phospholipid biosynthesis. Annu Rev Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- Kielkowska A, Niewczas I, Anderson KE, Durrant TN, Clark J, Stephens LR, Hawkins PT. A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv Biol Reg. 2014;54:131–141. doi: 10.1016/j.jbior.2013.09.001. [DOI] [PubMed] [Google Scholar]
- King JS, Insall RH. Chemotaxis: finding the way forward with Dictyostelium. Trend Cell Biol. 2009;19:523–530. doi: 10.1016/j.tcb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Kortholt A, King JS, Keizer-Gunnink I, Harwood AJ, Van Haastert PJ. Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol Biol Cell. 2007;18:4772–4779. doi: 10.1091/mbc.E07-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Inoue T, Sasaki J, Kubo T, Matsuda S, Nakasaki Y, Hattori M, Tanaka F, Udagawa O, Kono N, Itoh T, Ogiso H, Taguchi R, Arita M, Sasaki T, Arai H. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Mol Biol Cell. 2012;23:4689–4700. doi: 10.1091/mbc.E12-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Loovers HM, Postma M, Keizer-Gunnink I, Huang YE, Devreotes PN, van Haastert PJ. Distinct roles of PI(3,4,5)P3 during chemoattractant signaling in Dictyostelium: a quantitative in vivo analysis by inhibition of PI3-kinase. Mol Biol Cel. 2006;17:1503–1513. doi: 10.1091/mbc.E05-09-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald G, Baker RR, Thompson W. Selective synthesis of molecular classes of phosphatidic acid, diacylglycerol and phosphatidylinositol in rat brain. J Neurochem. 1975;24:655–661. [PubMed] [Google Scholar]
- Madani S, Hichami A, Legrand A, Belleville J, Khan NA. Implication of acyl chain of diacylglycerols in activation of different isoforms of protein kinase C. FASEB J. 2001;15:2595–2601. doi: 10.1096/fj.01-0753int. [DOI] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Firtel RA. A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr Biol: CB. 2000;10:708–717. doi: 10.1016/s0960-9822(00)00536-4. [DOI] [PubMed] [Google Scholar]
- Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- Milne SB, Ivanova PT, DeCamp D, Hsueh RC, Brown HA. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J Lipid Res. 2005;46:1796–1802. doi: 10.1194/jlr.D500010-JLR200. [DOI] [PubMed] [Google Scholar]
- Montrucchio G, Alloatti G, Camussi G. Role of platelet-activating factor in cardiovascular pathophysiology. Physiol Rev. 2000;80:1669–1699. doi: 10.1152/physrev.2000.80.4.1669. [DOI] [PubMed] [Google Scholar]
- Muller-Taubenberger A, Kortholt A, Eichinger L. Simple system–substantial share: the use of Dictyostelium in cell biology and molecular medicine. Eur J Cell Biol. 2013;92:45–53. doi: 10.1016/j.ejcb.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Nenci S, Piano V, Rosati S, Aliverti A, Pandini V, Fraaije MW, Heck AJ, Edmondson DE, Mattevi A. Precursor of ether phospholipids is synthesized by a flavoenzyme through covalent catalysis. Proc Natl Acad Sci USA. 2012;109:18791–18796. doi: 10.1073/pnas.1215128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froelich WM, Murphy DB, Devreotes PN. G Protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Paulus H, Kennedy EP. The enzymatic synthesis of inositol monophosphatide. J Biol Chem. 1960;235:1303–1311. [PubMed] [Google Scholar]
- Pettitt TR, Dove SK, Lubben A, Calaminus SD, Wakelam MJ. Analysis of intact phosphoinositides in biological samples. J Lipid Res. 2006;47:1588–1596. doi: 10.1194/jlr.D600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Schink KO, Stenmark H. Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J. 2013;280:2730–2742. doi: 10.1111/febs.12116. [DOI] [PubMed] [Google Scholar]
- Rouzer CA, Ivanova PT, Byrne MO, Milne SB, Marnett LJ, Brown HA. Lipid profiling reveals arachidonate deficiency in RAW264.7 cells: structural and functional implications. Biochem. 2006;45:14795–14808. doi: 10.1021/bi061723j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- Saito S, Goto K, Tonosaki A, Kondo H. Gene cloning and characterization of CDP-diacylglycerol synthase from rat brain. J Biol Chem. 1997;272:9503–9509. doi: 10.1074/jbc.272.14.9503. [DOI] [PubMed] [Google Scholar]
- Scott DL, Otwinowski Z, Gelb MH, Sigler PB. Crystal structure of bee-venom phospholipase A2 in a complex with a transition-state analogue. Science. 1990;250:1563–1566. doi: 10.1126/science.2274788. [DOI] [PubMed] [Google Scholar]
- Shindou H, Hishikawa D, Harayama T, Yuki K, Shimizu T. Recent progress on acyl CoA: lysophospholipid acyltransferase research. J Lipid Res. 2009;50(Suppl):S46–S51. doi: 10.1194/jlr.R800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Sasaki AT, Ha H, Seung HA, Firtel RA. Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium. J Biol Chem. 2007;282:11874–11884. doi: 10.1074/jbc.M610984200. [DOI] [PubMed] [Google Scholar]
- Vance JE, Vance DE. Phospholipid biosynthesis in mammalian cells. Biochem Cell Biol. 2004;82:113–128. doi: 10.1139/o03-073. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Lemieux MG, Knecht DA, Insall RH. PIP3-dependent macropinocytosis is incompatible with chemotaxis. J Cell Biol. 2014;204:497–505. doi: 10.1083/jcb.201309081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Ann Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- Zatulovskiy E, Tyson R, Bretschneider T, Kay RR. Bleb-driven chemotaxis of Dictyostelium cells. J Cell Biol. 2014;204:1027–1044. doi: 10.1083/jcb.201306147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci USA. 2010;107:11829–11834. doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KM, Takegawa K, Emr SD, Firtel RA. A phosphatidylinositol (PI) kinase gene family in Dictyostelium discoideum: biological roles of putative mammalian p110 and yeast Vps34p PI 3-kinase homologs during growth and development. Mol Cell Biol. 1995;15:5645–5656. doi: 10.1128/mcb.15.10.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Pandol S, Bokoch G, Traynor KA. Disruption of Dictyostelium PI3K genes reduces [32P]phosphatidylinositol 3,4 bisphosphate and [32P]phosphatidylinositol trisphosphate levels, alters F-actin distribution and impairs pinocytosis. J Cell Sci. 1998;111:283–294. doi: 10.1242/jcs.111.2.283. [DOI] [PubMed] [Google Scholar]
- Zoeller RA, Lake AC, Nagan N, Gaposchkin DP, Legner MA, Lieberthal W. Plasmalogens as endogenous antioxidants: somatic cell mutants reveal the importance of the vinyl ether. Biochem J. 1999;338:769–776. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.