Abstract
Neurons employ a set of homeostatic plasticity mechanisms to counterbalance altered levels of network activity. The molecular mechanisms underlying homeostatic plasticity in response to increased network excitability are still poorly understood. Here, we describe a sequential homeostatic synaptic depression mechanism in primary hippocampal neurons involving miRNA-dependent translational regulation. This mechanism consists of an initial phase of synapse elimination followed by a reinforcing phase of synaptic downscaling. The activity-regulated microRNA miR-134 is necessary for both synapse elimination and the structural rearrangements leading to synaptic downscaling. Results from miR-134 inhibition further uncover a differential requirement for GluA1/2 subunits for the functional expression of homeostatic synaptic depression. Downregulation of the miR-134 target Pumilio-2 in response to chronic activity, which selectively occurs in the synapto-dendritic compartment, is required for miR-134-mediated homeostatic synaptic depression. We further identified polo-like kinase 2 (Plk2) as a novel target of Pumilio-2 involved in the control of GluA2 surface expression. In summary, we have described a novel pathway of homeostatic plasticity that stabilizes neuronal circuits in response to increased network activity.
Keywords: homeostatic plasticity, microRNA, miR-134, neuronal activity, Pum2
Introduction
It is well established that neurons employ a variety of mechanism to homeostatically stabilize firing rates in response to bidirectional perturbations of network activity that would deviate firing rates from a defined set point. These mechanisms include regulation of intrinsic excitability (Marder & Goaillard, 2006), shift in the inhibition excitation balance (Maffei et al, 2004; Gonzalez-Islas & Wenner, 2006), compensatory changes in excitatory synaptic strength (Turrigiano & Nelson, 2004), and modulation of synapse number (Kirov et al, 1999; Wierenga et al, 2006). Which form of homeostatic response is used depends on the developmental stage of the neurons, the form and time of stimulation and can involve both cell-wide and local adaptations (Turrigiano, 2008; Yu & Goda, 2009). A large body of work has begun to elucidate the molecular mechanisms of adaption to a chronic decrease in network activity, as, for example, induced by the application of tetrodotoxin (Turrigiano, 2012). However, little is known about the signaling pathways underlying homeostatic compensation of increased network activity, a mechanism which we refer to as homeostatic synaptic depression (HSD). A transcriptional program that requires the CAMKK/CAMKIV pathway is necessary for HSD in organotypic hippocampal slices (Goold & Nicoll, 2010). During synaptic downscaling, activation of the polo-like kinase 2 (Plk2) is necessary for the reduction of synaptic AMPA receptors (Seeburg et al, 2008). In particular, it is not understood how different mechanisms contributing to homeostatic synaptic depression, such as synaptic downscaling and synapse elimination, are coordinated in time and space. A detailed understanding of homeostatic plasticity mechanisms is particularly important since evidence accumulates that it plays an essential role in vivo during the refinement of neuronal circuits, such as during ocular dominance plasticity in the mammalian visual cortex (Hengen et al, 2013; Keck et al, 2013). Furthermore, the failure of neuronal homeostasis results in common neuropsychiatric phenotypes, including mental retardation, autism, and schizophrenia (Ramocki & Zoghbi, 2008). Neuronal microRNAs (miRNAs), small non-coding RNAs that regulate translation of mRNAs by binding to target sequences in the 3′UTR, are attractive candidates for molecules that could regulate homeostatic plasticity mechanisms. First, miRNAs are capable of fine-tuning the expression of hundreds of target mRNAs, thereby globally buffering cellular protein production (Bartel, 2004). Second, they have been shown to regulate key functional and morphological parameters of neuronal physiology that are hallmarks of homeostatic synaptic plasticity, including dendritic arborization and dendritic spine morphogenesis. Lastly, several neuronal miRNAs are regulated by synaptic activity as part of homeostatic feedback control mechanisms at the transcriptional and post-transcriptional level (Schratt, 2009). One of the best studied examples is the dendritically enriched miR-134 which negatively regulates dendritic spine size (Schratt et al, 2006) and is required for activity-dependent dendritogenesis in primary hippocampal neurons (Fiore et al, 2009). MiR-134 activity and expression are regulated by multiple activity-dependent mechanisms at the transcriptional and post-transcriptional level (Schratt et al, 2006; Fiore et al, 2009; Bicker et al, 2013). We therefore decided to study a potential contribution of miR-134-dependent target gene regulation to homeostatic adaptations in response to chronic elevation of network activity.
In this study, we show that miR-134 regulates key aspects of HSD induced by a chronic increase in network activity. We provide first evidence that HSD is a sequential process, consisting of an initial phase of synapse elimination followed by a downscaling of the unitary strength of remaining synapses at a later stage. MiR-134 is specifically required for early synapse elimination and late structural rearrangements of excitatory synapses, but not for functional downscaling. We identify the RNA-binding protein Pumilio-2 (Pum2), which was previously implicated in the regulation of neuronal homeostasis, as a key target for miR-134-dependent HSD. Pum2 downregulation is limited to the synapto-dendritic compartment, suggesting an important contribution of miR-134-dependent local translation of Pum2 to HSD. Furthermore, we show that the Plk2 mRNA is bound by Pum2 and that the Pum2/Plk2 interaction is functionally involved in the downscaling of GluA2 surface receptors in response to chronic activity. Taken together, our results uncover a new translational control mechanism which is essential for the expression of homeostatic plasticity in response to increased neuronal activity.
Results
A two-stage mechanism of homeostatic synaptic depression
HSD in response to sustained activation of neural networks requires functional and structural modification of excitatory synapses, including both a uniform downscaling of synaptic strength and a complete elimination of synapses (Seeburg et al, 2008; Goold & Nicoll, 2010). The precise timing of these events however is unresolved. In order to characterize the time course of homeostatic rearrangements at synapses of primary hippocampal pyramidal neurons, we induced chronic network activity by treating mature hippocampal cultures (days in vitro DIV18) with the GABA receptor antagonist picrotoxin (Ptx, 100 μM) over a period of 48 h.
The impact of increased excitability on excitatory synapses was evaluated by assessing morphological parameters for synaptic strength (dendritic spine volume, size of GluA2-containing AMPA receptor surface clusters) and synapse density (PSD-95/synapsin co-clusters) in vehicle versus Ptx-treated, GFP-transfected pyramidal neurons at 8 and 48 h after Ptx treatment (Fig1A, C and E). As expected, a 48-h Ptx treatment induced a significant reduction of all morphological parameters examined compared to control conditions (Fig1B, D, F and G), indicating that neurons respond homeostatically to increased excitability by reducing both the strength of individual synapses and the number of excitatory synapses that form onto these neurons. These results are in agreement with previous results (Goold & Nicoll, 2010). Surprisingly, whereas homeostatic downscaling of synaptic strength was not observed after 8-h Ptx treatment (Fig1A–D), this treatment already led to a robust elimination of synaptic co-clusters. Forty-eight hours of Ptx treatment had no adverse effect on cell viability, as assessed by counts of total cells and fragmented nuclei, suggesting that the observed synapse elimination upon Ptx was not due to toxic effects of the drug (Supplementary Fig S1A). Together, these results indicate the presence of a two-stage mechanism of HSD. First, a subset of excitatory synapses is eliminated to reduce excitability. Second, remaining synapses are downscaled, presumably to further strengthen the homeostatic response. This so-called synaptic downscaling occurs later and was first observed 24 h after Ptx treatment (Supplementary Fig S1B).
Figure 1. Homeostatic synaptic depression involves synapse elimination followed by synaptic downscaling.
- Effect of different times of Ptx treatment on spine volume. DIV18 GFP-transfected hippocampal neurons were incubated for the indicated times with either 100 μM Ptx or solvent, fixed and analyzed by confocal laser scanning microscopy. Pictures of representative dendrites are shown for each condition and time point. Scale bar: 20 μm.
- Quantification of relative spine volume in Ptx-treated neurons as shown in (A). The spine volume from 8 cells for each condition was quantified for each experiment. The average of 3 (8 h Ptx) to 4 (48 h Ptx) independent experiments ± SD is shown. P = 0.0471 (8 h); P = 0.0054 (48 h).
- Effect of different times of Ptx treatment on the size of GluA2-containing AMPA receptor surface clusters. The surface GluA2 subunit (red) was detected by immunofluorescence in GFP-transfected (green) rat hippocampal neurons treated for the indicated time with Ptx. Pictures of representative dendrites are shown for each condition and time point.
- Quantification of the size of GluA2-containing surface AMPA receptor clusters as shown in (C). The size of the GluA2 clusters from 10 cells for each condition was quantified for each experiment. The average of 3 (8 h Ptx) to 4 (48 h Ptx) independent experiments ± SD is shown. P = 0.0074.
- Effect of Ptx on the density of synaptic co-clusters. DIV18 GFP-transfected (green) hippocampal neurons were incubated for the indicated times with either 100 μM Ptx or solvent, fixed, processed for immunofluorescence using antibodies against PSD-95 (red) and synapsin (blue), and analyzed by confocal laser scanning microscopy.
- Quantification of synaptic co-cluster density in Ptx-treated neurons as shown in (E). Twelve cells per condition were quantified for each experiment. The average of 3 (8 h Ptx) to 5 (48 h Ptx) independent experiments ± SD is shown. P = 0.0477 (8 h) P = 0.0196 (48 h).
- Summary of the effect of different times of Ptx treatment on dendritic spine volume, GluA2 cluster size, and synapse density. Data are presented as the average ± SD of the ratio for the indicated parameter between Ptx- and vehicle-treated cells.
MiR-134 is necessary for homeostatic synapse elimination
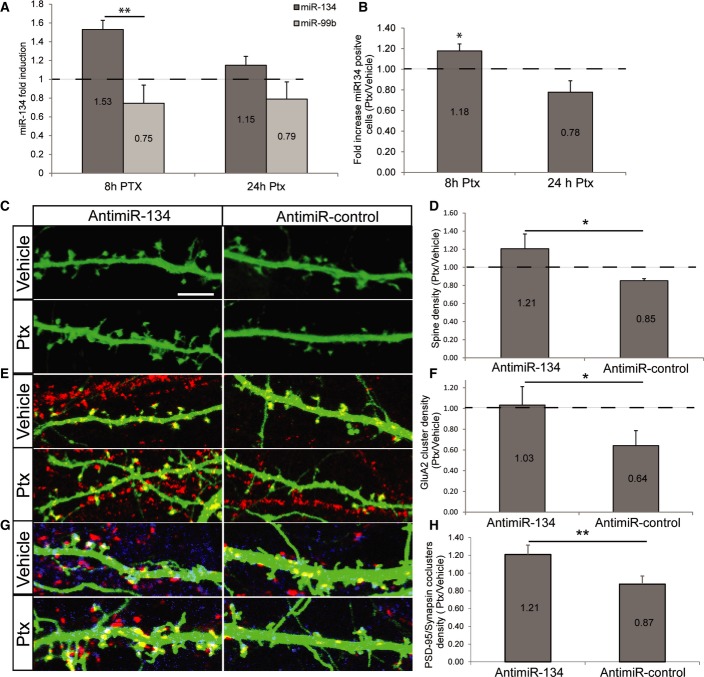
Little is known regarding the molecular mechanisms underlying HSD, in particular with regard to homeostatic synapse elimination that we showed is the initial process in HSD. We considered an involvement of miRNAs, since miRNAs were reported to control cellular homeostasis in multiple systems (Herranz & Cohen, 2010). The synaptic miRNA miR-134 appeared to be particularly promising, since it is regulated by neuronal activity and it controls neuronal morphogenesis (Schratt et al, 2006; Fiore et al, 2009). We first assessed the effect of Ptx on the level and activity of miR-134 in hippocampal cultures. Using qPCR, we found a small but reproducible induction of miR-134 expression after 8 h, but not 24 h of Ptx treatment (Fig2A). The observed increase in mature miR-134 levels translated into increased miR-134 activity, since 8 h, but not 24 h of Ptx treatment resulted in an increased cleavage of a miR-134-dependent fluorescent sensor construct (Fiore et al, 2009) in hippocampal neurons (Fig2B). As expected, Ptx bath application induces a prolonged increase in polo-like Kinase 2 (Plk2) mRNA levels (Supplementary Fig S2A), a kinase known to be induced by sustained network activity. Together, our results are consistent with an involvement of miR-134 in the regulation of early events during HSD, in particular synapse elimination.
Figure 2. MiR-134 is necessary for homeostatic synapse elimination.
- Ptx treatment increases mature miR-134 expression. Quantitative real-time PCR analysis of RNA extracted from DIV18 hippocampal neurons treated with either Ptx or vehicle for the indicated times. The average fold induction relative to mock-treated neurons from 3 independent experiments ± SD is shown.
- Ptx treatment increases miR-134 activity. A miRNA sensor assay was performed in hippocampal neurons that were transfected with a miR-134 sensor construct and treated with Ptx for the indicated times. After Ptx treatment, cells were fixed and scored as miR-134 positive if the miR-134-dependent fluorescent signal was absent. The average from three independent experiments ± SD is shown.
- Representative pictures of neurons co-transfected with GFP and the indicated anti-miRs (10 nM) and stimulated with Ptx or vehicle at DIV18 for 48 h. Scale bar: 20 μm.
- Quantification of the effect of miR-134 inhibition on spine density, the effect of Ptx is expressed as the ratio between the spine density in Ptx-treated neurons divided by the spine density of mock-treated neurons. The average ± SD of four independent experiments (eight cells per condition) is shown. P = 0.0206.
- Neurons transfected with GFP and the indicated anti-miRs were processed as in Fig1C to detect GluA2 surface clusters.
- Quantification of the effect of miR-134 inhibition on GluA2 cluster density. The average ± SD of four independent experiments (10 cells per condition) is shown. P = 0.0411.
- Neurons transfected with GFP and the indicated anti-miRs were processed as in Fig1E to detect PSD-95/synapsin I co-clusters.
- Quantification of the effect of miR-134 inhibition on PSD-95/synapsin I co-cluster density. The average ± SD of five experiments (12 cells per condition) is shown. P = 0.0028.
The peak of miR-134 induction during chronic increase in network activity coincides with the onset of synaptic elimination. We thus tested whether perturbation of miR-134 function affected the ability of hippocampal neurons to homeostatically adapt to increased network activity, using three different parameters for synapse density: dendritic spine, GluA2 surface cluster, and PSD-95/synapsin co-cluster densities. We transfected primary rat hippocampal neurons (DIV13) with a specific miR-134 inhibitory oligonucleotide (anti-miR-134) or a corresponding control molecule of unrelated sequence (anti-miR-control) and then measured the density of dendritic spines, GluA2-containing surface clusters, and synaptic co-clusters. Since we observed that the effects on HSD parameters (synapse downscaling and elimination) were most robust 48 h after the induction of chronic activity, we used 48-h Ptx treatment (DIV18–20) for all following experiments. MiR-134 inhibition completely abolished the reductions in the density of dendritic spines, GluA2-containing AMPA receptor surface clusters, and synaptic co-clusters induced by Ptx in transfected neurons (Fig2C–H). Thus, multiple lines of evidence suggest that miR-134 is required for homeostatic synapse elimination in response to chronic Ptx treatment. This function of miR-134 is restricted to excitatory synapses since neither Ptx treatment nor miR-134 inhibition affected the density of inhibitory synapses in primary neurons (Supplementary Fig S1C). MiR-134 is required for homeostatic synapse elimination already after 8 h of Ptx treatment, suggesting that miR-134-dependent mechanisms are engaged in the early phase of HSD (Supplementary Fig S3A).
MiR-134 is necessary for structural but not functional homeostatic synaptic downscaling
Given the prominent function of miR-134 in synapse elimination, we wanted to further determine the consequences of prolonged miR-134 inhibition on the later phase of HSD that is mainly characterized by a unitary downscaling of synaptic strength of the remaining excitatory synapses. The analysis of dendritic spine volume and GluA2-containing AMPA receptor surface cluster size in neurons transfected with anti-miR-134 revealed that miR-134 was also necessary for the structural rearrangements associated with synaptic downscaling. Accordingly, the decrease in both spine volume and GluA2-containing clusters size was completely abolished upon miR-134 inhibition (Fig3A and B). Inhibition of miR-134 blocked the reduction in spine volume induced by 48-h incubation with Ptx in DIV20 neurons without affecting this parameter in vehicle-treated neurons. As expected, anti-miR-134 had no effect on spines and GluA2 clusters upon 8 h of Ptx treatment, a time when no significant morphological downscaling of spine volume and GluA2 cluster size was observed (Supplementary Fig S3B and C). To investigate the functional consequences of these miR-134-dependent alterations in postsynaptic morphological parameters, we measured miniature excitatory postsynaptic currents (mEPSCs) at DIV20 (Fig3C–E). The observed reductions in synapse number should translate into reduced mEPSC frequency, whereas synaptic downscaling is expected to result in lower mEPSC amplitudes (Seeburg et al, 2008). In agreement, we observed in control-transfected neurons a trend toward reduced mean mEPSC frequencies (Fig3D and Supplementary Fig S5) and a significant reduction in mean mEPSC amplitudes (Fig3E and see KS test in Supplementary Fig S6) upon 48-h Ptx treatment. Importantly, in Ptx-treated neurons, mEPSC frequencies were significantly higher in anti-miR-134 compared to control-transfected neurons (Fig3D and Supplementary Fig S4), suggesting that functional synapse elimination in chronically activated neurons is compromised when miR-134 activity is blocked. Surprisingly, the Ptx-mediated reduction in mEPSC amplitudes was still observed in anti-miR-134-transfected neurons (Fig3E and Supplementary Fig S5). Accordingly, in contrast to mEPSC frequencies, mEPSC amplitudes in anti-miR-134-transfected neurons did not significantly differ between Ptx- and vehicle-treated cultures (Fig3E). Two lines of evidence strongly suggest that miR-134 is regulating mEPSC frequency postsynaptically. First, we previously found using a very sensitive fluorescent in situ hybridization protocol that miR-134 is enriched within the postsynaptic dendritic compartment of hippocampal neurons (Schratt et al, 2006; Fiore et al, 2009). Second, due to the low transfection efficiency, neurons in which miR-134 activity was blocked and which we recorded from were only connected with the presynaptic terminals of non-transfected neurons. Thus, together with our morphological results, our data argue that miR-134 is required in the postsynaptic cell during both phases of HSD, being first necessary to reduce synapse density and later on to trigger the structural rearrangements associated with synaptic downscaling. The reduction in unitary synaptic strength upon chronic activity, however, does not require miR-134 activity. Therefore, manipulating miR-134 activity allowed us to uncouple mechanisms responsible for morphological and functional synaptic downscaling.
Figure 3. MiR-134 is necessary for structural but not functional synaptic downscaling.
- Quantification of the effect of miR-134 inhibition on spine volume. The average ± SD of four independent experiments (eight cells per condition) is shown. P = 0.0232.
- Quantification of the effect of miR-134 inhibition on surface GluA2 cluster size. The average ± SD from four independent experiments (10 cells per condition) is shown. P = 0.0070.
- Representative traces of mEPSC recordings from DIV18 hippocampal neurons. Cells transfected with GFP and 10 nM of the indicated anti-miRs were treated for 48 h with Ptx or vehicle and voltage clamped at −70 mV in the presence of tetrodotoxin and gabazine.
- Quantification of the median mEPSC frequencies from neurons treated as shown in (C). Data represent the average ± SD of a total of 15 cells per group collected from 4 independent experiments. P = 0.0125.
- Quantification of the median mEPSC amplitudes from neurons treated as shown in (C). Data represent the average ± SD of a total of 15 cells per group collected from 4 independent experiments.
MiR-134 differentially affects the surface expression of AMPA receptor subunits
We next sought to investigate the mechanism underlying the differential requirement of miR-134 in functional and morphological downscaling of excitatory synapses. Therefore, we considered differential regulation of AMPA receptor subunits as one possibility. GluA2 has been shown to be involved both in the regulation of synapse formation (Passafaro et al, 2003) and synaptic strength, whereas there is no evidence for an involvement of GluA1 in the former process. We therefore reasoned that in contrast to GluA2, GluA1 surface expression could be independent of miR-134, providing a potential explanation for the absence of defects in functional downscaling upon miR-134 inhibition. In this view, normal GluA1 internalization during miR-134 blockade would be sufficient to induce a functional downscaling of excitatory synaptic strength. Indeed, when we analyzed the response to chronic Ptx application of GluA1-containing AMPA receptor surface clusters, we observed a robust decrease in GluA1 cluster size in response to Ptx (Fig4A) that, contrary to GluA2-containing AMPA receptors, was independent of miR-134 activity (Fig4B). This result suggests that parallel and independent pathways control synaptic levels of the different subunit of the AMPA receptors during homeostatic plasticity, a miR-134-dependent pathway involved in the regulation of GluA2 and a miR-134-independent pathway responsible for GluA1. Two lines of evidence further support the hypothesis that GluA1 and GluA2 are independently regulated during homeostatic plasticity. First, a significant decrease in the total levels of GluA1 but not GluA2 protein was observed in Ptx-treated neurons compared to control conditions (Fig4C and D). Second, surface clusters formed by a YFP-GluA2 fusion protein, in which YFP is located on the extracellular surface and thus detectable by surface staining, did decrease in size in response to chronic Ptx in a miR-134-dependent fashion (Fig4E and F). Since the expression vector does not include the GluA2 3′UTR, this result further suggests that miR-134 indirectly regulates GluA2 levels by controlling the expression of a gene involved in the regulation of surface expression of this specific AMPA receptor subunit. Using time-lapse imaging with a photoactivable GFP-tagged GluA2 (paGFP-GluA2) construct (Tatavarty et al, 2013), we further obtained preliminary data suggesting that the Ptx-mediated acceleration of GluA2 removal from dendritic spines is also miR-134-dependent (Supplementary Fig S6). Taken together, different physiological responses appear to be associated with the control of GluA1 and GluA2 subunits. MiR-134-dependent control of GluA2 levels appears to be necessary for the regulation of synapse number during both the early phase of HSD and the later morphological downscaling of excitatory synapses, while GluA1 surface expression seems to be exclusively linked to the late phase of functional downscaling of synaptic strength.
Figure 4. Differential regulation of GluA1 and GluA2 AMPA receptor subunits during homeostatic synaptic depression.
- Representative images of DIV20 GFP-transfected neurons treated for 48 h with vehicle or Ptx and processed as shown in Fig1C to detect GluA1 surface clusters. Scale bar: 20 μm.
- Quantification of the size of GluA1-containing surface AMPA receptor clusters as shown in (A). The size of GluA1 surface clusters from 10 cells for each condition was quantified for each experiment. The average of 3 independent experiments ± SD is shown.
- GluA1 but not GluA2 protein expression decreases after 48 h of Ptx treatment. Protein extracts were prepared from DIV20 hippocampal neurons and analyzed by Western blot using antibodies directed against GluA1 and GluA2 or α-tubulin as a loading control. One representative blot is shown.
- Quantification of multiple experiments performed as in (C). The intensity of the GluA1 and GluA2 bands was normalized to the α-tubulin band from the same blot for each experiment. The average of 3 independent experiments ± SD is shown. P = 0.0078.
- miR-134 is required for the downscaling of recombinant GluA2 surface clusters. DIV18 hippocampal neurons transfected with 100 ng of YFP-GluA2 together with the indicated anti-miRs (10 nM) were treated for 48 h with Ptx and processed for surface staining using an anti-GFP antibody. Pictures of representative dendrites are shown for each condition. Scale bar: 20 μm.
- Quantification of the size of YFP-GluA2 clusters from neurons treated as in (E). The average ± SD of 3 experiments is shown (10 cells per condition). P = 0.0159 (YFP-GluA2 versus anti-miR-134), P = 0.0337 (anti-miR-134 versus anti-miR-control).
Source data are available online for this figure.
Pum2 is regulated by miR-134 under conditions of increased activity
To begin to address the mechanism by which miR-134 controls synapse number during HSD, we tested known targets of this miRNA (Pum2, Limk1, and Creb1) for regulation by increased network activity. We first investigated whether any of the targets are regulated by Ptx at the post-transcriptional level using destabilized luciferase 3′UTR reporter constructs. Only a reporter containing the 3′UTR of the RNA-binding protein Pumilio-2 (Pum2) reacted to Ptx in a manner consistent with miR-134-dependent regulation (Fig5A and B, and Supplementary Fig S2B and C). Pum2 is a known direct miR-134 target (Fiore et al, 2009) and represents an attractive candidate, since it regulates spine morphogenesis and is involved in the control of neuronal homeostasis in Drosophila and rodents (Mee et al, 2004; Vessey et al, 2010; Driscoll et al, 2013). The activity of a wild-type Pum2 reporter was significantly downregulated in DIV18 hippocampal neurons after 8 h of Ptx treatment, a time point corresponding to the peak of miR-134 activity (Figs5A and 1H and I). Accordingly, no regulation of the reporter was observed after 24-h Ptx treatment, a time point where no induction of miR-134 activity was observed. Together, our data are consistent with an involvement of miR-134 in the Ptx-mediated post-transcriptional downregulation of Pum2 expression. In further support for increased miR-134 activity as the cause of Pum2 repression, mutating the miR-134 binding site in the Pum2 3′UTR completely prevented Ptx-mediated downregulation of the reporter (Fig5A). Accordingly, anti-miR-mediated inhibition of miR-134 suppressed downregulation of the wild-type Pum2 reporter by Ptx, but had no effect on a reporter containing a mutated miR-134 binding site (Fig5B). Taken together, these results strongly argue that Pum2 expression is downregulated post-transcriptionally by miR-134 upon chronic activity.
Figure 5. Pum2 expression is regulated by miR-134 during homeostatic synaptic depression.
- Downregulation of a Pum2 luciferase reporter by Ptx requires a functional miR-134 site. Hippocampal neurons were transfected with 50 ng of a destabilized luciferase reporter gene bearing the wild-type 3′UTR of Pum2 (pGL4-Pest-Pum2-Long) or a reporter gene containing mutations in the seed region of the Pum2 miR-134 binding site (pGL4-Pest-Pum2-Long) and treated with Ptx at DIV18 for the indicated times. The ratio of normalized luciferase activities between Ptx-treated and untreated neurons is shown. Data represent the mean of three independent experiments ± SD. P = 0.0244.
- Downregulation of a Pum2 luciferase reporter by Ptx is miR-134-dependent. Hippocampal neurons were transfected with the indicated reporters and anti-miRs (8 nM) before Ptx or vehicle was applied for 8 h at DIV18. The average ± SD of 3 experiments is shown. P = 0.0006 (**), P = 0.0235 (*).
- Pum2 expression is differentially regulated by Ptx in the cell body and process compartment. Protein extract from the cell body (C) and process (P) compartment were prepared from DIV20 hippocampal neurons that were cultured in compartmentalized chambers and treated for 48 h with Ptx or vehicle. Extracts were analyzed by Western blot using antibodies directed against Pum2, Creb1, and α-tubulin as a loading control. Two different exposition times of the Pum2 detection are shown to demonstrate upregulation of Pum2 in cell bodies and downregulation in processes.
- Quantification of Creb1 signal intensities from multiple blots as shown in (C). The intensity of the band was normalized to the α-tubulin band from the same blot for each experiment. The average of three experiments ± SD is shown. P = 0.0400.
- Quantification of Pum2 signal intensities from multiple blots as shown in (C). The average of 3 experiments ± SD is shown. Pcell bodies = 0.0407; Pdendrites = 0.0064.
Source data are available online for this figure.
Both miR-134 and Pum2 mRNA were previously shown to be dendritically localized (Schratt et al, 2006; Vessey et al, 2006; Siegel et al, 2009), suggesting that at least part of the regulation could occur locally in dendrites. We further confirmed dendritic enrichment of the endogenous Pum2 isoform which contains the 3′UTR used in our luciferase reporter assay (Supplementary Fig S2D). In order to determine the site of regulation of Pum2 protein during HSD, we used a compartmentalized culture system that permits to physically separate cell bodies from neuronal processes, primarily consisting of dendrites (Bicker et al, 2013). Primary hippocampal neurons were cultured for 18 days in the chambers and then both compartments were incubated for 48 h with Ptx, since it was the time point we used in the majority of our functional experiments. Subsequently, protein extracts were prepared separately for both compartments and analyzed for the presence of endogenous Pum2 protein by Western blotting. In cell bodies, Ptx treatment significantly increased Pum2 protein levels, a result that is likely miR-134 independent and in agreement with previous results (Driscoll et al, 2013) (Fig5C, shorter exposition time lane, and E). These data are further supported by results obtained with whole-cell extracts, where we observed upregulation of Pum2 protein and mRNA by an 8-h Ptx treatment (Supplementary Fig S2E and F). Strikingly, the opposite effect was observed in dendrites, where Ptx significantly decreased Pum2 protein levels (Fig5C and E). The dendritic regulation of Pum2 by Ptx is specific, since another known miR-134 target, the activity-dependent transcription factor Creb1, was specifically downregulated in the cell body compartment and not detectable in dendrites. Together, these results suggest that miR-134 targets are downregulated by Ptx in different compartments and point to a possible dendritic function of the Pum2–miR-134 interaction in HSD.
Downregulation of Pum2 by miR-134 is necessary for homeostatic synapse elimination
Next, we investigated the functional relevance of the miR-134-dependent Pum2 regulation in the context of HSD. Therefore, we first assessed the effect of elevating Pum2 protein expression in Ptx-treated neurons on synapse elimination. Overexpression of Pum2, but neither of a constitutive active Creb1 (Creb-VP16) nor its inactive form (Creb-VP16m), did prevent synapse elimination induced by Ptx (Fig6A). Thus, Pum2 overexpression mimics miR-134 inhibition, consistent with a role for miR-134-dependent downregulation of Pum2 in HSD.
Figure 6. Pum2 downregulation is necessary for miR-134-dependent synapse elimination.
- Pum2 but not Creb1 overexpression prevents homeostatic synapse elimination. Hippocampal neurons were transfected with GFP alone or in combination with 100 ng of the indicated expression vectors. At DIV18 neurons were treated for 48 h with Ptx or vehicle, fixed and processed for immunofluorescence to detect PSD-95/synapsin I co-clusters. The average ± SD from 4 independent experiments (12 cells per experimental condition) is shown. P = 0.0349 (*); P = 0.0038 (**).
- Pum2 knockdown rescues the miR-134 loss-of-function phenotype in Ptx-treated hippocampal neurons. Neurons transfected with the indicated vectors and anti-miRs were processed as in Fig1E to detect PSD95/synapsin 1 co-clusters; representative images of Ptx-treated dendrites are shown. Scale bar: 20 μm.
- Quantification of synaptic co-cluster density from neurons treated as shown in (B). The average ± SD from 4 experiments (12 cells per experimental condition) is shown. P = 0.0078 (**); P = 0.0112 (*).
- Quantification of spine density from neurons treated as shown in (B). The average ± SD from 3 experiments is shown (8 cells per experimental condition). P = 0.0401 (GFP versus control-shRNA), P = 0.0433 (control-shRNA versus Pum2-shRNA).
To more definitely assess if Pum2 is indeed working downstream of miR-134 during homeostatic synapse elimination, we reduced Pum2 protein levels in the context of miR-134 inhibition using a previously described Pum2-shRNA (Fiore et al, 2009). We expected that the Pum2-shRNA should rescue defective synapse elimination induced by anti-miR-134. Consistent with our hypothesis, co-transfection of anti-miR-134 together with the specific Pum2-shRNA but not with a control-shRNA restored the reduction in both synapse and spine density induced by a 48-h Ptx treatment (Fig6B–D). Thus, Pum2 is a key component of the miR-134-dependent signaling pathway triggered by increased network activity. Interestingly, it appears that Pum2 levels have to be tightly regulated within a narrow physiological range during HSD, since Pum2 knockdown results in a phenotype similar to Pum2 overexpression (Supplementary Fig S7).
The miR-134–Pum2 regulation is upstream of the Plk2 pathway in HSD
Having determined that Pum2 regulation by miR-134 is necessary for excitatory synapses elimination, we next assessed whether this interaction is also important for the control of GluA2 surface levels during HSD. Downregulating Pum2 protein levels in the context of miR-134 inhibition restored downscaling of GluA2 receptor clusters after Ptx treatment, demonstrating that Pum2 mediates both aspects of miR-134-dependent HSD, synapse elimination and downscaling (Fig7A). We went on to identify Pum2-regulated mRNAs involved in the control of GluA2 surface levels. Therefore, we inspected the DoRiNA database (Anders et al, 2012) which contains genome-wide data for Pum2–RNA interactions obtained from a Pum2 PAR-CLIP study performed in HeLa cells (Hafner et al, 2010). By focusing on candidate genes involved in AMPA receptor trafficking, we detected a Pum2 CLIP tag in a highly conserved region the 3′UTR of the human Plk2 mRNA (Fig7B). Plk2 promotes homeostatic synaptic downscaling by targeting SPAR for degradation and by sequestering the GluA2-interacting protein NSF (Seeburg et al, 2008) (Evers et al, 2010). We therefore reasoned that downregulation of Pum2 by miR-134 could result in an activation of Plk2 pathways, thereby inducing GluA2 internalization. We first verified by UV cross-linking immunoprecipitation (CLIP) that the Plk2 mRNA is bound by Pum2 in DIV20 rat hippocampal neurons. The Pum2 antibody used for CLIP specifically immunoprecipitated endogenous Pum2 protein in neurons (Fig7C). Within the Pum2 immunoprecipitate, we observed a specific enrichment of Plk2 mRNA relative to control IgG immunoprecipitates as assessed by qPCR (Fig7D). To verify the specificity of this enrichment, we determined the levels of RNAs devoid of Pum2 CLIP tags in the Pum2 and IgG immunoprecipitates (U6, Limk1, GAPDH) using qPCR. The highly abundant GAPDH mRNA and the dendritically enriched LimK1 mRNA were readily detected in the input but neither in Pum2 nor IgG immunoprecipitates. The highly expressed non-coding RNA U6 was detectable in the Pum2 immunoprecipitate, but strongly depleted there compared to the IgG immunoprecipitate (Fig7D). Taken together, the absence (Limk1, GAPDH) or depletion (U6) of three different negative control RNAs from Pum2 immunoprecipitates supports the specificity of our highly stringent CLIP protocol.
Figure 7. Plk2 is a direct Pum2 target and functions downstream in Pum2-regulated downscaling of GluA2 surface receptors.
- Pum2 knockdown rescues defective Ptx-mediated downscaling of GluA2 surface receptors caused by miR-134 inhibition. Quantification of the average GluA2 surface cluster size in hippocampal neurons transfected with indicated shRNA constructs and anti-miR-134, depicted in principle as in Fig1C. The average ± SD from 3 experiments (10 cells per experimental condition) is shown. P = 0.0140 (GFP versus control-shRNA), P = 0.0139 (control-shRNA versus Pum2-shRNA).
- Position and conservation of the Pum2 CLIP tag in the 3′UTR of the human Plk2 mRNA according to Hafner et al (2010). The genomic location of the CLIP tag is indicated at the bottom (human assembly hg19). A sequence element fulfilling the Pum2 consensus binding site is highlighted with a dashed box.
- A polyclonal rabbit anti-Pum2 antibody specifically immunoprecipitates endogenous Pum2 protein. Western blot analysis for Pum2 using either anti-Pum2 (IgPum2) or control IgG immunoprecipitates. 10% of the input material before immunoprecipitation is shown for comparison.
- The Plk2 transcript is specifically enriched in the IgPum2 immunoprecipitates. qPCR analysis for the indicated genes using RNA purified from IgPum2 and IgG immunoprecipitates shown in (C). Values are shown as fold enrichment of IgPum2 RNA relative to IgG control RNA from 3 independent experiments ± SD, P = 0.0138.
- Pum2 overexpression interferes with Ptx-induced SPAR degradation. DIV18 hippocampal neurons transfected with 100 ng of GFP together with the indicated expression vectors (100 ng each) were treated for 48 h with Ptx and processed for immunostaining (red) using an anti-SPAR antibody. Photographs of representative dendrites are shown for each condition. Scale bar: 5 μm.
- Quantification of the average SPAR intensity of neurons treated as in (E). Values are plotted as ratio between Ptx- and vehicle-treated neurons. The average ± SD of three independent experiments is shown (8 cells for each experimental condition), P = 0.0003 (**), P = 0.0197 (*).
- Plk2 expression rescues defective downscaling of GluA2 surface receptors caused by Pum2 overexpression. Quantification of the average GluA2 surface cluster size in hippocampal neurons transfected with indicated expression vectors, depicted in principle as in Fig1C. The average ± SD from 3 experiments (10 cells per experimental condition) is shown. P = 0.0043 (Pum2-YFP versus GFP), P = 0.0022 (Pum2-YFP versus Pum2-YFP/Plk2), P = 0.022 (Pum2-YFP versus Creb-VP16 m).
Source data are available online for this figure.
Our CLIP results suggest that in unstimulated hippocampal neurons, Pum2 interacts with Plk2 mRNA, thereby potentially repressing Plk2 translation. If Pum2 indeed represses Plk2, one would expect that Pum2 expression should abolish Ptx-mediated degradation of the Plk2 target SPAR. We used a previously characterized SPAR antibody to determine endogenous SPAR levels in neurons using immunocytochemistry followed by confocal microscopy (Pak et al, 2001). In agreement with previous publications (Pak & Sheng, 2003; Seeburg et al, 2008), Ptx induced a significant decrease in the average intensity of the SPAR immunofluorescent signal in hippocampal neurons transfected with GFP alone or in combination with the Creb-VP16m control construct. In contrast, overexpression of Pum2-YFP completely prevented the decrease in SPAR levels induced by Ptx (Fig7E and F), consistent with the idea that the miR-134 target Pum2 regulates Plk2 activity during HSD. Lastly, we directly tested whether Pum2-dependent inhibition of Plk2 is functionally required for HSD. Therefore, we asked whether Plk2 expression was sufficient to reinstate the downscaling of GluA2 surface receptors in the context of Pum2 overexpression. As expected, downscaling of GluA2 surface cluster size induced by Ptx was abrogated upon Pum2-YFP expression. More importantly, GluA2 downscaling was fully reinstated when Pum2-YFP was overexpressed together with Plk2, consistent with Pum2 acting as an upstream inhibitor of Plk2 expression. GluA2 downscaling was also attenuated when Plk2 was overexpressed alone due to the already reduced levels of surface GluA2 in the absence of Ptx (Supplementary Fig S7B). Taken together, multiple lines of evidence suggest that Plk2 is an important downstream component in the miR-134–Pum2 regulatory pathway during HSD.
Discussion
HSD requires both synapse elimination and downscaling
We have characterized the temporal dynamics of the structural and functional synaptic rearrangements that underlie homeostatic plasticity following chronic activation of the neuronal circuit. We did uncover a mechanism of homeostatic synaptic depression that requires first the reduction of the densities of dendritic spines and synaptic puncta after 8 h of Ptx bath application. The remaining neuronal connections are then downscaled homogeneously if activation of the neuronal network persists for more than 24 h. Our results are in agreement with data from organotypic hippocampal slices where individual neurons chronically activated by an optogenetic approach undergo synaptic depression by decreasing both the frequency and amplitude of mEPSCs (Goold & Nicoll, 2010). We consider it unlikely that synapse elimination is due to developmental competition between synapses since at the time of our pharmacological treatments (DIV18–20) hippocampal cultures were already fully mature. It is well established that changes in postsynaptic parameters are accompanied by corresponding changes in presynaptic function (Burrone et al, 2002). Although our experimental setup strongly argues that postsynaptic miR-134 function is initially required for homeostatic synapse elimination and structural downscaling, it is conceivable that postsynaptic miR-134 regulates a retrograde signal that indirectly influences presynaptic functions. Future experiments will address a potential role of miR-134-dependent presynaptic rearrangements in certain aspects of HSD.
Molecular pathways involved in HSD
As indicated in the introduction, our current knowledge about molecules involved in HSD is relatively sparse. Calcium influx is likely to be the common starting event to eliminate and scale down synapses, since it is necessary for both synapse up- and downscaling (Ibata et al, 2008; Seeburg et al, 2008; Goold & Nicoll, 2010). In organotypic slices, synaptic depression requires somatic calcium influx and a transcriptional program activated by the CAMKK/CAMKIV pathway (Goold & Nicoll, 2010). In hippocampal neurons, synaptic downscaling activates also the Plk2/CDK2 pathway that further bifurcates in two downstream branches. First, Plk2 phosphorylates the AMPA receptor scaffolding protein SPAR, thereby inducing SPAR degradation and GluA1/2 internalization. Second, Plk2 associates with the GluA2-interacting protein NSF, thereby sequestering it away from GluA2 and rather specifically promoting GluA2 internalization (Seeburg & Sheng, 2008; Seeburg et al, 2008; Evers et al, 2010). We now show that miRNA-dependent regulation of mRNA translation is another important process regulating HSD insofar as miR-134 acts upstream of the Plk2 pathway via translational inhibition of Pum2, which in turn inhibits Plk2 expression. In contrast to transcription, translational pathways offer the possibility of spatial control in the cytoplasm, for example, within the synapto-dendritic compartment of neurons. The dendritic localization of miR-134 and Pum2 mRNA, as well as the selective dendritic downregulation of Pum2 protein expression by Ptx, provide support for a specific role of local protein synthesis in this form of homeostatic plasticity, similar to observations made from activity-deprived neurons (Sutton et al, 2006). Conclusive evidence for a role of dendritic miR-134 in the regulation of synapse-specific HSD will require further experiments with local manipulation of miR-134 activity. In this regard, it is interesting to note that Plk2 engages different homeostatic mechanisms in proximal versus secondary dendrites (Evers et al, 2010). This raises the possibility that specific aspects of Plk2 signaling are regulated post-transcriptionally by the miR-134 pathway in the dendritic compartment.
MiR-485 is the only other miRNA that has been previously implicated in the regulation of homeostatic synaptic plasticity (Cohen et al, 2011). However, based on the results from this study, there are several important differences between the function of miR-485 and miR-134. In contrast to miR-134, miR-485 already regulates synapse number in unstimulated neurons, suggesting that this miRNA has a more general function in synapse formation that is not limited to activity-dependent homeostatic synapse elimination. Accordingly, neurons in which miR-485 function was blocked maintained their ability to homeostatically reduce their number of synapses to a similar relative extent compared to control cultures. Thus, while miR-134 is clearly required for the induction and expression of homeostatic synaptic depression, miR-485 likely plays a more modulatory role in this process. In addition, miR-485 regulates the expression of a presynaptic synaptic vesicle protein (SV2A), which is more consistent with a presynaptic function of this miRNA. Therefore, it is possible that miR-134 and miR-485 control independently the pre- and postsynaptic functional and structural rearrangements necessary for this form of plasticity.
Differential regulation of AMPA receptor at synapses during HSD
The regulation of AMPA receptor accumulation at synapses as a central event of homeostatic plasticity is well established (Turrigiano, 2012). Our results with miR-134 inhibitors show that the GluA1 and GluA2 subunits are independently regulated during HSD and therefore likely control different aspects of this form of plasticity. Our results concerning GluA2 are most consistent with the idea that internalization of this subunit is necessary for early synapse elimination. A role of GluA2 in the regulation of synapse number is in agreement with the observation that the N-terminal domain of GluA2 is able to induce dendritic spine formation (Passafaro et al, 2003). Conversely, GluA1 internalization in response to activity occurs independently of miR-134, arguing that trafficking of this AMPA receptor subunit does not play a major role in synapse elimination. Rather, GluA1 seems to be specifically required for functional synaptic downscaling at later time points of HSD. Furthermore, control of GluA1 synaptic levels involves not only trafficking of this subunit but also regulation of cellular protein levels. The downstream signaling pathway components that differentially regulate GluA1 and GluA2 trafficking and whose activities are modulated by chronic network activation will have to be fully delineated in future studies. In particular, it will be interesting to see whether the Plk2–NSF interaction, which specifically regulates GluA2 internalization in a kinase-independent manner (Evers et al, 2010), is primarily affected by manipulations in miR-134 activity.
Pum2 is a key miR-134 target during HSD
Our results indicate that the RNA-binding protein Pum2 is a key miR-134 target during homeostatic plasticity. Intriguingly, Pum2 has been previously associated with the control of homeostatic plasticity in mammalian neurons (Driscoll et al, 2013). In this study, Pum2 was shown to be required for the homeostasis of membrane excitability, another important homeostatic plasticity mechanism. Consistent with our findings, Pum2 protein was upregulated by increased activity at the whole neuron level. Pum2 upregulation, which likely occurs in the neuronal soma independent of miR-134, was necessary to suppress translation of the voltage-gated sodium channel transcript Nav1.6 in order to decrease intrinsic excitability (Driscoll et al, 2013). On the other hand, we found that activity reduces Pum2 expression locally in the dendritic compartment, and it is tempting to speculate that this spatial control is important for both miR-134-dependent elimination of specific synapses and removal of GluA2-containing AMPA receptors from the surface of the neuronal membrane. In the future, it will be interesting to determine the relative contribution of somatic upregulation and dendritic downregulation of Pum2 to the stabilization of neuronal fire rates in response to chronically elevated activity.
(Patho)physiological role of miR-134
The physiological significance of miR-134-dependent regulation of HSD remains to be determined. MiR-134 has recently been shown to be necessary for the expression of LTP, a classical from of Hebbian plasticity, in the hippocampus (Gao et al, 2010). Since storing information within a circuit through differences in synaptic strength is no longer possible in the absence of homeostatic mechanisms that limit unrestrained potentiation (Abbott & Nelson, 2000), abnormal LTP in the absence of miR-134 might in fact be caused by the inability to express homeostatic plasticity. Defective homeostatic plasticity likely contributes in an important way to the etiology of epilepsy. For example, it has been shown that prolonged epileptiform activity in organotypic hippocampal slices triggers a homeostatic response that is crucial to reduce membrane excitability and the potentiation of synaptic strength (Seeburg & Sheng, 2008). Intriguingly, silencing miR-134 in vivo has neuroprotective and seizure suppressive effects during status epilepticus (Jimenez-Mateos et al, 2012). It will be interesting to determine to what extent the here described functions of miR-134 in homeostatic plasticity contribute to the beneficial effects of miR-134 inhibition in epilepsy.
Taken together, our results define a novel function of the miR-134–Pum2 pathway in homeostatic synaptic depression. More generally, they point to a prominent role of miRNA-regulated mRNA translation in neuronal homeostasis with important implications for activity-dependent neuronal development and neurological disorders.
Materials and Methods
DNA constructs
The vectors Pum2 3′UTR-luc wild type and mutant, pSuper—Pum2-shRNA, and control-shRNA have been described previously (Fiore et al, 2009). The expression vector YFP-GluR2 and PA-GFP-GluA2 were a kind gift of G. Turrigiano (Brandeis University, USA; Ibata et al, 2008; Tatavarty et al, 2013); Pum2-YFP expression vector is a kind gift of M. Kiebler (Ludwig-Maximilians-University, Munich, Germany; Vessey et al, 2006). CREB-VP16 and CREB-VP16m have been provided by M.E. Greenberg (Harvard Medical School, Boston, USA). The Plk2 expression vector (Plk2-myc) has been provided by M. Sheng (The Picower Institute for Learning and Memory, Cambridge, USA) and D.T. Pak (Georgetown University Medical Center, Washington, USA; Seeburg et al, 2008).
Cell culture and transfection of primary neurons
The culture and transfection of dissociated primary cortical and hippocampal neurons from embryonic day 18 (E18) Sprague–Dawley rats (Charles River Laboratories, Sulzfeld, Germany) were as described (Schratt et al, 2004).
For compartmentalized neuron cultures, dissociated hippocampal neurons were plated onto 1-μm pore and 30-mm diameter polyethylene tetra-phthalate (PET) membrane filter inserts (Millipore) that were matrix-coated with poly-L-lysine (Sigma-Aldrich) and Laminin (BD Biosciences) on the top and bottom (Poon et al, 2006).
Transfections of primary neurons at DIV13 using Lipofectamine 2000 (Invitrogen) were performed as described (Siegel et al, 2009). For stimulation, DIV18 neurons were treated either with picrotoxin (Ptx; 100 μm final concentration, Sigma) or solvent (ethanol absolute) for the indicated times.
Luciferase assay
Primary neurons were transfected in duplicates with 50 ng of pGL4 firefly reporter constructs and equal amounts of empty Renilla reporter as transfection control, alone or with 10 nM of the appropriate pLNA (Exiqon). Luciferase assays were performed 5 days post-transfection using the Dual-Luciferase reporter assay system (Promega) on the GloMax R96 Microplate Luminometer (Promega). P-values were calculated with Student's t-test (two-tailed, type 2) for one-way comparisons and with ANOVA followed by post hoc test (Student's t-test, two-tailed, type 2) for multi-way comparisons.
Immunofluorescence
For immunostaining of endogenous proteins, hippocampal neurons were fixed for 20′ at room temperature in paraformaldehyde/sucrose, rinsed in PBS, and sequentially incubated with primary antibody and Alexa-conjugated secondary antibodies (dilution 1:1,000), both diluted in 0.02% gelatin–0.5% Triton X-100–PBS. For surface staining of GluA2- and GluA1-containing AMPA receptors, the primary antibody was added to the culture medium prior to fixation and the neurons were incubated for 3 h at 37°C in 5% CO2. Four washes with Neurobasal medium were followed by fixation for 15′ in paraformaldehyde/sucrose and then the cultures processed as described above. The following primary antibodies were used: monoclonal anti-PSD-95 (1:300 dilution, Thermo Scientific), rabbit polyclonal anti-Synapsin 1 (1:500 dilution, Millipore), monoclonal anti-GluA2 (1.6 μg/ml final concentration, Millipore), rabbit polyclonal anti-GluA1 (2 μg/ml final concentration, Calbiochem), rabbit polyclonal anti-SPAR (kind gift of D.T. Pak). Alexa-546- and Alexa-647-conjugated secondary antibodies (1:2,000 dilution, Invitrogen) were used for detection.
Image analysis
All image analysis was performed with the scientist blinded to the experimental conditions. Images were taken with a confocal laser scanning microscope (Zeiss LSM 5 or Leica SP5) using a 64× objective at a resolution of 1,024 × 1,024 pixel corresponding to an image size of 144.72 × 144.72 μm (Leica Sp5) or 142.86 × 14286 μm (Zeiss LSM 5), the pinhole was set to 1 AU and the interval to 0.4 μm. For spine analysis, high-resolution z-stack images of GFP-positive neurons displaying pyramidal morphology for each condition were chosen. Spine volumes were subsequently analyzed with the ImageJ software as described (Schratt et al, 2006). Synapse density was determined by calculating the density of PSD-95/Synapsin-1 co-cluster in GFP-transfected neurons, using ImageJ following a described method (Paradis et al, 2007). For each independent experiment 12 neurons for condition were selected. The size of GluA1 and GluA2-containing cluster was determined after applying the threshold function to images in ImageJ and then using the analyzed particle function. Threshold was kept constant between experimental conditions, and particles with a size smaller than 0.1 μm2 were excluded from the analysis. The photoactivation protocol is described in the Supplementary Methods.
Statistics
Unless otherwise stated, three independent experiments were performed for each data set. The number of neurons analyzed for each experimental condition in each experiment is indicated in the respective figure legend. Error bars represent standard deviations. P-values were calculated with Student's t-test (two-tailed, type 2) for one-way comparisons and with ANOVA followed by post hoc test (Student's t-test, two-tailed, type 2) for multi-way comparisons.
Quantitative real-time PCR
Total RNA from cultured hippocampal and cortical neurons was isolated using mirVana miRNA isolation kit (Ambion). Genomic DNA contamination was eliminated with TURBO DNase (Ambion). RNA was reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad) according to manufacturer's instructions. Quantitative real-time PCR was performed with the StepOnePlus Real-Time PCR System (Applied Biosystems), using iTaq SYBR Green Supermix with ROX (Bio-Rad) for detection of mRNA and TaqMan MicroRNA Assay kits (Applied Biosystems) for detection of mature miRNAs.
Preparation of protein extracts and Western blotting
Protein extracts from compartmentalized chambers were prepared by gently scraping first the cell bodies and then the dendrites from the opposite faces of the membrane in warm 1× D-PBS using a plastic cell scraper (Costar® # 3010). The different cell fractions were transferred in 1.5-ml Eppendorf tubes on ice and spun down at 250 × g for 2 min at 4°C. The supernatant was removed, and the cells in each insert were lysed in 100 μl for modified RIPA buffer (50 mM Tris–HCl, 150 mM NaCl, 0.5% NP-40, 0.1% SDS, pH 7.4) containing Complete Protease Inhibitor Cocktail EDTA-free (Roche) for 20 min on ice. Samples were centrifuged at maximum speed for 5 min at 4°C, and the supernatant was collected and used for Western blotting after the measurement of protein concentration. Western blotting was performed as described previously (Siegel et al, 2009). The following primary antibodies were used: polyclonal rabbit anti-Pum2 (1:4,000 dilution, Novus Biologicals), rabbit polyclonal anti-tubulin (1:7,500 dilution, Cell Signaling), rabbit polyclonal anti-GluA1 (1:1,000 dilution, Thermo Scientific), rabbit anti-GluA2 (1:2,000 dilution, Millipore), monoclonal anti-Creb1 (1:1,000 dilution, Cell Signaling). Primary antibodies were recognized by either a horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (1:20,000; 401,315; Calbiochem) or an HRP-conjugated rabbit anti-mouse antibody (1:20,000; 402,335; Calbiochem). Secondary antibodies were detected by enhanced chemiluminescence with the ECL Plus Western Blotting Detection System (GE Healthcare).
UV cross-linking immunoprecipitation
A total of 5.6 million of cells were plated on two 6-well plates and treated at DIV5 with fluorodeoxyuridine (FUDR) to stop proliferation of non-neuronal cells. At DIV20 cells were cross-linked in 1× PBS on ice using an UV-Strata linker (Stratagene) at 400 mJ. Cells were lysed using 2.4 ml of lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 0.5% NP-40, 5 mM MgCl2, 1 mM DTT, Protease inhibitors (Roche), RNAsin Ribonuclease Inhibitor (Promega) for 10 min on ice under gentle shaking. Lysates were then centrifuged for 10 min at 4°C at 16,000 × g, and the supernatant was transferred to new tubes. Prior to immunoprecipitation, 7.5 μg of antibodies (rabbit anti-Pum2, Novus Biologicals; rabbit-IgG, Santa Cruz Biotechnology) was coupled to 25 μl each of protein-G Dynabeads (Invitrogen) by incubating for 60 min at room temperature in 500 μl of lysis buffer on an orbital shaker. Antibody/bead complexes were then washed three times with 1 ml lysis buffer. Ten percent of the lysates were stored for RNA and protein analysis and the remaining were divided equally between the antibody beads and incubated for 2 h at 4°C on an orbital shaker. The beads were washed three times with 1 ml of Washing buffer (lysis buffer with 500 mM NaCl) and two times in 1 ml of lysis buffer. After the last wash, the beads were split equally in two new tubes. Beads to be used for Western blot analysis were resuspended in 60 μl of lysis buffer/1× loading buffer, boiled for 5 min at 95°C, and the supernatant was collected. For RNA isolation, beads were resuspended in 50 μl of lysis buffer + 0.1% SDS and treated with 0.4 μg/μl Proteinase K (New England Biolabs) for 30 min at 50°C. Total RNA was purified using the mirVana RNA isolation kit according to the manufacturers’ instructions.
Electrophysiology
Miniature excitatory postsynaptic currents (mEPSCs) were recorded in whole-cell configuration of the patch clamp technique from DIV20 GFP-transfected rat spinal hippocampal neurons. For recordings, cover slips were constantly superfused with bath solution at RT, containing (in mM): 140 NaCl, 3 KCl, 2 CaCl2, 1 MgCl2, 15 glucose, 10 HEPES, pH 7.4, 315 mOsm. Pipette solution contained (in mM): 120 potassium gluconate, 15 KCl, 5 NaCl, 2 MgCl2, 10 EGTA, 10 HEPES, 4 MgATP, 0.1 NaGTP, pH: 7.25, 300 mOsm. EPSCs were isolated by tetrodotoxin (TTX, 1 μM) and gabazine (5 μM). Currents were recorded in voltage clamp mode at −70 mV using borosilicate glass pipettes (2–4 MΩ) and an EPC-7 amplifier (HEKA, Lambrecht, Germany). They were filtered at 3 kHz and digitized with a CED 1,401 interface (CED, Cambridge, UK; sample frequency: 20 kHz). Data were evaluated off-line with Spike7 software (CED, Cambridge, UK) using custom made routines. Numbers are given as mean ± SEM. After testing for normality, statistical significance was tested by Student's two-tailed t-test (GraphPad Software, San Diego, USA). Statistical significance of cumulative amplitude and interval distributions were tested by a two sample Kolmogorov–Smirnov test (OriginPro 8.6 Software; OriginLab Corporation, Northampton, USA).
Acknowledgments
We would like to thank M. Kiebler, G. Turrigiano, M. Sheng, D.T. Pak, and M.E. Greenberg for generously providing reagents. We thank P. Störchel and B. Honrath for cloning of the pGL4-PEST construct. The outstanding technical assistance of E. Becker, R. Gondrum, G. Jarosch, M.B. Kowalski, and T. Wüst is greatly acknowledged. This work was supported by grants from the European Research Council (ERC Starting Grant “NEUROMIR”) and the EU FP7 HEALTH (“EpimiRNA”) to G.S. A. A. is a recipient of a fellowship from the Medical Research Council (MRC).
Author contributions
RF and GS conceived the experiments and wrote the manuscript. RF performed and analyzed all experiments except electrophysiology. CS performed and analyzed the electrophysiology. KB and AD supervised electrophysiological recordings. MR and SB contributed biochemical and molecular biology data. AA performed photoactivation experiments.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Abbott LF, Nelson SB. Synaptic plasticity: taming the beast. Nat Neurosci. 2000;3(Suppl):1178–1183. doi: 10.1038/81453. [DOI] [PubMed] [Google Scholar]
- Anders G, Mackowiak SD, Jens M, Maaskola J, Kuntzagk A, Rajewsky N, Landthaler M, Dieterich C. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2012;40:D180–D186. doi: 10.1093/nar/gkr1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bicker S, Khudayberdiev S, Weiss K, Zocher K, Baumeister S, Schratt G. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013;27:991–996. doi: 10.1101/gad.211243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrone J, O'Byrne M, Murthy VN. Multiple forms of synaptic plasticity triggered by selective suppression of activity in individual neurons. Nature. 2002;420:414–418. doi: 10.1038/nature01242. [DOI] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci USA. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll HE, Muraro NI, He M, Baines RA. Pumilio-2 regulates translation of Nav1.6 to mediate homeostasis of membrane excitability. J Neurosci. 2013;33:9644–9654. doi: 10.1523/JNEUROSCI.0921-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers DM, Matta JA, Hoe HS, Zarkowsky D, Lee SH, Isaac JT, Pak DT. Plk2 attachment to NSF induces homeostatic removal of GluA2 during chronic overexcitation. Nat Neurosci. 2010;13:1199–1207. doi: 10.1038/nn.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Islas C, Wenner P. Spontaneous network activity in the embryonic spinal cord regulates AMPAergic and GABAergic synaptic strength. Neuron. 2006;49:563–575. doi: 10.1016/j.neuron.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengen KB, Lambo ME, Van Hooser SD, Katz DB, Turrigiano GG. Firing rate homeostasis in visual cortex of freely behaving rodents. Neuron. 2013;80:335–342. doi: 10.1016/j.neuron.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O'Tuathaigh C, Waddington JL, Prenter S, Delanty N, Farrell MA, O'Brien DF, Conroy RM, Stallings RL, DeFelipe J, Henshall DC. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck T, Keller GB, Jacobsen RI, Eysel UT, Bonhoeffer T, Hubener M. Synaptic scaling and homeostatic plasticity in the mouse visual cortex in vivo. Neuron. 2013;80:327–334. doi: 10.1016/j.neuron.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Kirov SA, Sorra KE, Harris KM. Slices have more synapses than perfusion-fixed hippocampus from both young and mature rats. J Neurosci. 1999;19:2876–2886. doi: 10.1523/JNEUROSCI.19-08-02876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei A, Nelson SB, Turrigiano GG. Selective reconfiguration of layer 4 visual cortical circuitry by visual deprivation. Nat Neurosci. 2004;7:1353–1359. doi: 10.1038/nn1351. [DOI] [PubMed] [Google Scholar]
- Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg DP, Sheng M. Activity-induced Polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J Neurosci. 2008;28:6583–6591. doi: 10.1523/JNEUROSCI.1853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Tatavarty V, Sun Q, Turrigiano GG. How to scale down postsynaptic strength. J Neurosci. 2013;33:13179–13189. doi: 10.1523/JNEUROSCI.1676-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. Cold Spring Harb Perspect Biol. 2012;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Vaccani A, Xie Y, Dahm R, Karra D, Kiebler MA, Macchi P. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di Leva F, Karra D, Thomas S, Kiebler MA, Macchi P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci USA. 2010;107:3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga CJ, Walsh MF, Turrigiano GG. Temporal regulation of the expression locus of homeostatic plasticity. J Neurophysiol. 2006;96:2127–2133. doi: 10.1152/jn.00107.2006. [DOI] [PubMed] [Google Scholar]
- Yu LM, Goda Y. Dendritic signalling and homeostatic adaptation. Curr Opin Neurobiol. 2009;19:327–335. doi: 10.1016/j.conb.2009.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.