Abstract
Winter wheat requires a period of low temperatures to accelerate flowering (vernalization). This requirement could make winter wheat more vulnerable to elevated global temperature via insufficient vernalization. All known vernalization genes are cloned according to qualitative variation in vernalization requirement between spring and winter wheat, but the genes controlling quantitative variation for more or less vernalization requirement among winter wheat cultivars remain unknown. We report here that the gene for the vernalization requirement duration in winter wheat was cloned using a BC1F2:3 population that segregated in a 3:1 ratio of early-flowering plants and late-flowering plants after vernalization for 3 weeks. The positional cloning of the gene for vernalization requirement duration demonstrated that this trait is controlled by TaVRN-A1 at the protein level. The Ala180 in vrn-A1a, encoded by the dominant allele for 3–week vernalization, was mutated to Val180 in vrn-A1b, encoded by the recessive allele for 6–week vernalization. Further studies with in vitro protein pull-down assays and immunoprecipitation analyses indicated that the mutated Val180 in vrn-A1b protein decreased the ability to bind with TaHOX1 (the first homeobox protein in Triticum aestivum). The direct binding of TaVRN-A1 and TaHOX1 proteins was confirmed in the nucleus of living plant cells by bimolecular fluorescence complementation (BiFC) analyses. The TaHOX1 gene was found to be upregulated by low temperatures, and to have a significant genetic effect on heading date, suggesting that TaHOX1 functions in the flowering pathway in winter wheat.
Keywords: vernalization requirement duration, VRN–1, homeobox protein, winter wheat, protein interaction, Triticum aestivum
Introduction
Triticum aestivum (wheat, 2n = 6x = 42, AABBDD) is cultivated across more land area than any other grain crop. Qualitatively, wheat cultivars are classified as two general types: winter wheat, with a variable low-temperature requirement for a proper flowering time (vernalization) and thus successful grain reproduction, and spring wheat, without this requirement (Chouard, 1960; Pugsley, 1971; Amasino, 2004). Quantitatively, winter wheat cultivars are classified as three types according to the low temperature duration required to reach the vernalization saturation point or to achieve the maximum vernalization effect: a weak winter type that is stimulated to flower by brief exposure to low temperature (for <2 weeks); a semi-winter type that requires 2–4 weeks of cold exposure for flowering; and a strong winter type that requires more than 4 weeks of cold exposure for timely flowering (Crofts, 1989).
Vernalization usually occurs at temperatures of <8°C (Chouard, 1960; Pugsley, 1971; Crofts, 1989; Amasino, 2004). Recent studies have shown that the average global surface air temperature rose by 0.5°C in the 20th century, and is projected to continue its increase by roughly 3°C or 5°C by the end of the 21st century (Kerr, 2007; Semenov and Halford, 2009). As various simulation models have shown, winter wheat is more vulnerable to increasing temperatures during winter seasons because of its low temperature requirement to ensure proper flowering time and successful seed reproduction (Miglietta et al., 1995; Humphreys et al., 2006). A shortened duration at low temperature, caused by global warming, could result in a failed or insufficient vernalization in winter wheat. The cloning of genes for quantitative vernalization requirement durations in winter wheat cultivars would provide new knowledge and molecular tools to breed novel cultivars to adapt to changing climates.
Three vernalization genes have been cloned from wheat by using a positional cloning approach, VRN1 (Yan et al., 2003), VRN2 (Yan et al., 2004a) and VRN3 (Yan et al., 2006), and each of them was cloned in the context of growth habit as a discrete trait of winter and spring types in diploid wheat and Hordeum vulgare (barley). VRN1 (=AP1) is a central promoter for spring cultivars to flower without vernalization (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003), and dominant Vrn-A1 alleles originated from mutations in the promoter or the first intron of the wild recessive vrn-A1 in hexaploid wheat (Yan et al., 2004b; Fu et al., 2005). A recent study reported that the increased copy number of TaVRN-A1 resulted in an increased requirement for vernalization in winter wheat (Díaz et al., 2012). VRN2 (=ZCCT1) is a flowering repressor, and a recessive vrn2 allele was caused by a point mutation at the conserved CCT domain or complete deletion of the wild dominant Vrn2 in diploid wheat (Yan et al., 2004a). VRN3 (=FT1) is another flowering promoter, and the early-flowering plants carry Vrn3 alleles originating from mutations in the promoter or the first intron in wheat and barley (Yan et al., 2006). The genetic and molecular mechanisms to account for the qualitative difference in the three genes between the two divergent types of wheat are not expected to explain the quantitative variation in vernalization requirement duration among winter wheat cultivars, because all winter wheat cultivars are supposed to have the same winter allele for each of the three cloned vernalization genes: recessive vrn1, recessive vrn3 and dominant Vrn2 alleles (Pugsley, 1971; Tranquilli and Dubcovsky, 2000). No exception was reported to invalidate this genetic model, when numerous cultivars/germplasm from different wheat species varying in ploidy level were screened using molecular markers for each of the three genes (Yan et al., 2003, 2004a,b; Fu et al., 2005; Bonnin et al., 2008; Zhang et al., 2008; Santra et al., 2009; Chen et al., 2010).
The molecular mechanism underlying quantitative vernalization requirement in a winter type of Arabidopsis is not yet known, but could be explained by three models. The first model is that vernalization results in a quantitative reduction in FLC mRNA levels, which negatively correlates with flowering time (Michaels and Amasino, 1999; Sheldon et al., 1999). The second model is that the transcript level of FLC is upregulated or downregulated by multiple genes, including VIN3 and FRI in the vernalization pathway, as well as GI, CO and FT in the photoperiod pathway (Reeves and Coupland, 2001; Levy et al., 2002; Corbesier et al., 2007; Heo and Sung, 2011). The signals from these pathways are integrated at FLC to induce AP1 for flowering. The last model is that quantitative vernalization requirement is modulated by the accumulation of the Polycomb-based epigenetic silencing complexes and histone modifications at the FLC gene (Angel et al., 2011), and the quantitative modulation of Polycomb silencing is associated with natural variation in the sequence of FLC (Coustham et al., 2012). Previous studies have demonstrated that vernalization has evolved different mechanisms between winter wheat and Arabidopsis (Yan et al., 2003). The gene(s) responsible for various vernalization requirement durations in winter wheat should be cloned from a population generated using two winter wheat cultivars with different vernalization saturation points.
In previous studies, we developed a population of recombinant inbred lines (RILs), generated from a cross between two winter wheat cultivars: ‘Jagger’ and ‘2174’. When different sets of the same population were tested in field, the RIL population was segregated in stem elongation, heading date and physiological maturity (Chen et al., 2009). In this previous study, a polymerase chain reaction (PCR) marker developed for allelic variation in exon 4 of TaVRN-A1 was found to have genetic association with a major quantitative trait locus (QTL) for the phenotypes, but it was not known whether TaVRN-A1 caused the QTL in winter wheat. If TaVRN-A1 was indeed responsible for this QTL, it could have different mechanisms in regulating the developmental process in winter wheat (as opposed to spring wheat). Otherwise, another gene at the TaVRN-A1 locus should be responsible for the QTL regulating developmental process in winter wheat. In the present study, we tested this RIL population under thermal- and photo-controlled glasshouse conditions, and found a major QTL for vernalization requirement duration. We then generated a large backcross population and cloned the first gene for vernalization requirement duration in winter wheat.
Results
The discovery of a major gene for vernalization requirement duration
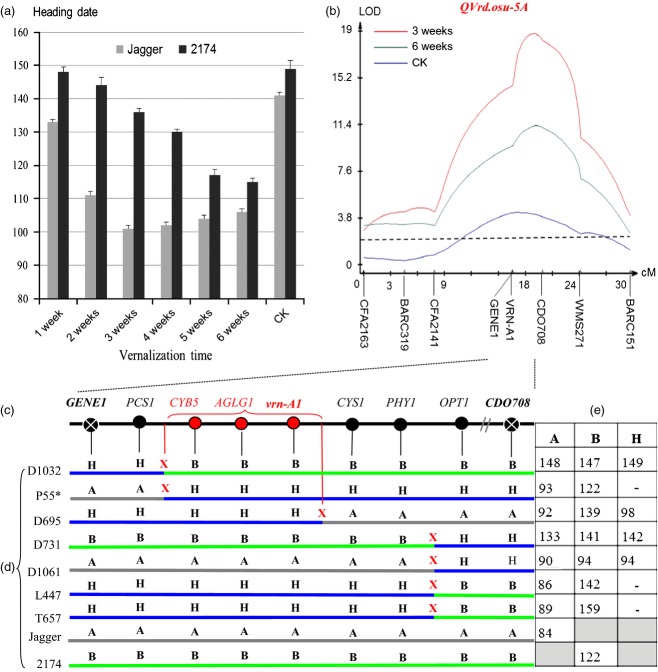
We tested the two parental lines, Jagger and 2174, for variation in vernalization requirement duration to reach the vernalization saturation point or gain the maximum vernalization effect on heading under long-day conditions (16–h day/8–h night). Jagger required 3 weeks at 4°C to reach the maximum vernalization effect on heading, whereas 2174 required 6 weeks under the same conditions (Figure1a). When vernalized for 3 weeks, Jagger flowered 101 days after planting but 2174 flowered 136 days after planting. When vernalized for 6 weeks, Jagger flowered 106 days after planting but 2174 flowered 115 days after planting. A major QTL for vernalization requirement duration that segregated in the mapping population of Jagger × 2174 RILs was mapped to the long arm of chromosome 5A in a genomic region encompassing the vrn-A1 locus (Figure1b), and this QTL was designated QVrd.osu-5A. The QVrd.osu-5A locus explained 63.4% (logarithm of the odds, LOD = 18.8) of the total phenotypic variation in the population vernalized for 3 weeks, 43.2% (LOD = 11.3) in the population vernalized for 6 weeks and 20.9% (LOD = 4.3) in the control population with no vernalization.
Figure 1.
Genetic and physical maps of the gene for QVrd.osu-5A.(a) Differential effects of vernalization requirement duration on heading date between ‘Jagger’ and ‘2174’. The plants were vernalized for a varying number of weeks (1–6 weeks), and non-vernalized plants were used as the control (CK). Heading date was the date of emergence of the first spike of each plant. The values represent mean heading date (n = 8), and error bars indicate standard errors.(b) Mapping of the QVrd.osu-5A locus. Two sets of the Jagger × 2174 RIL populations were vernalized for 3 and 6 weeks, and one set of the same population was not vernalized as a control (CK). The horizontal dotted line represents a threshold value of 2.5 LOD, logarithm of the odds.(c) The physical location of the QVrd.osu-5A locus.(d) The critical crossovers in the QVrd.osu-5A locus. ‘A’ represents the Jagger allele, ‘B’ represents the 2174 allele and ‘H’ represents heterozygosity at the given marker locus; ‘X’ indicates the recombinant site.(e) Days to heading of each critical recombinant plant. The detailed statistical analyses on the phenotype of the recombinant plants are provided in Table S1.
A recombinant inbred line, RIL23, carrying the Jagger vrn-A1 allele, was backcrossed with the parental line 2174 to generate a BC1F2 population, in which the QVrd.osu-5A locus was heterozygous but each of PPD-D1 and VRN-D3 were fixed at the homozygous allele. The latter two genes showed significant effects on heading date in the Jagger × 2174 RIL population when tested in the field (Chen et al., 2009). The resulting BC1F2 population was used to test genetic effects of the QVrd.osu-5A locus on vernalization requirement in winter wheat.
Ninety plants of the RIL23 × 2174 BC1F2 population were tested for heading date when the population was vernalized for 3 weeks. On average, 24 plants homozygous for the Jagger vrn-A1 allele headed at 110 days after planting, 20 plants homozygous for the 2174 vrn-A1 allele headed at 138 days and 46 plants heterozygous at vrn-A1 headed at 118 days. In order to avoid confusion of gene/allele in the terminology (Vrn-A1a, Vrn-A1b, Vrn-A1c, Vrn-A1d and Vrn-A1e were designed for the dominant or spring alleles; Yan et al., 2004b), the Jagger vrn-A1 allele is hereafter designated as vrn-A1a and the 2174 vrn-A1 allele is designated as vrn-A1b, and TaVRN-A1 rather than Tavrn-A1 is used for both vrn-A1a and vrn-A1b.
The 70 plants either homozygous or heterozygous for the Jagger vrn-A1a allele for early heading showed a significant difference from the 20 plants homozygous for the 2174 vrn-A1b allele for late heading (P < 0.001). The observed segregation ratio between the earlier-heading and later-heading groups was not significantly different from a 3:1 ratio (χ2 = 0.37, d.f. = 1, P = 0.54), and fits with a one-gene model. We genotyped a total of 6410 BC1F3 plants derived from BC1F2 plants heterozygous at vrn-A1 using PCR markers for GENE1 and CDO708 that flanked the gene responsible for QVrd.osu-5A (Figure1c), and the progeny of the resulting seven critical recombinant plants that exhibited a crossover between PCS1 and OPT1 (Figure1d) were tested for heading date after vernalization for 3 weeks. The results clearly showed that each of the progeny population was segregated in a 3:1 ratio of plants that headed early and plants that headed later, indicating the dominance of the vrn-A1a allele for early heading over the vrn-A1b allele for late heading (Figure1e; see Table S1).
The positional cloning of QVrd.osu-5A
We determined the locations of crossovers using PCR markers for nine genes at the QVrd.osu-5A locus (see Figures S1–S9). Subsequently, the gene responsible for QVrd.osu-5A was positioned within a genomic region flanked by PCS1 and CYS1, and included three candidate genes: CYB5, AGLG1 and vrn-A1. Gene expression results indicated that the vernalization requirement duration was not associated with any of the candidate genes at the transcriptional level (see Figure S10), suggesting that this trait was controlled by a candidate gene at the protein level.
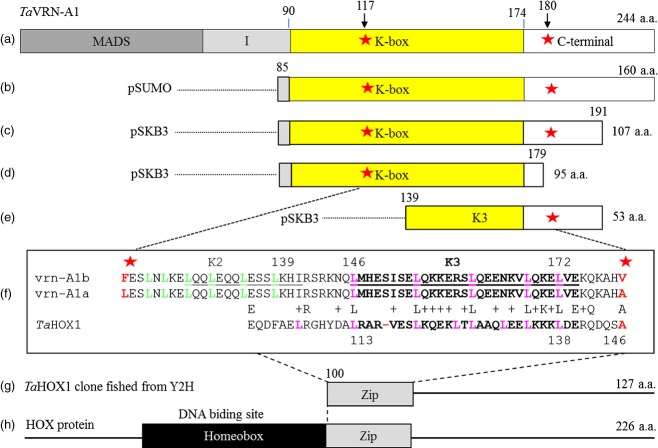
No difference was observed in the CYB5 protein encoded by the two alleles, eliminating the possibility that CYB5 is responsible for QVrd.osu-5A. One amino acid substitution occurred in AGLG1 but the substituted amino acids are similar, serine in Jagger and alanine in 2174, and AGLG1 may be involved in spike differentiation but not in flowering time (Yan et al., 2003). In contrast, two amino acid substitutions occurred in vrn-A1, and the substituted amino acids are very different. In vrn-A1, one single nucleotide polymorphism (SNP) occurred in exon 4, producing a leucine (Leu) at position 117 in vrn-A1a, encoded by the Jagger allele, but a phenylalanine (Phe) at the same position in vrn-A1b, encoded by the 2174 allele (Figure2a). The second SNP occurred in exon 7, producing an alanine (Ala) at position 180 in vrn-A1a, but a valine (Val) at the same position in vrn-A1b (Figure2a). The vrn-A1 is a MADS-box protein (Mandel et al., 1992), and the Leu117/Phe117 substitution occurred within the conserved K–box at positions 89–174, whereas the Ala180/Val180 substitution occurred in the divergent C terminal. In addition, VRN1 functions in the vernalization pathway in wheat (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003); therefore, we further tested and validated the hypothesis that allelic variation in the vrn-A1 protein is responsible for QVrd.osu-5A.
Figure 2.
Interacting site of TaVRN-A1 and TaHOX1 proteins.(a) The locations of the conserved domains in TaVRN-A1 and two altered sites in the amino acid sequence are indicated with red stars.(b–e) Four TaVRN-A1 fragments covering either one or two mutations were expressed to test differential interactions with TaHOX1.(f) Sequence comparison between the TaVRN-A1 and TaHOX1 proteins. Leu117/Phe117 or Ala180/Val180 residues are highlighted in red, and the conserved leucine residues are highlighted in pink.(g) The cDNA of TaHOX1 fished out from the Y2H library was expressed to test protein interactions with TaVRN-A1.(h) Positions of conserved HD and Zip domains in TaHOX1 proteins in plants.
A differential interaction of vrn-A1a and vrn-A1b with TaHOX1
To determine whether the protein properties of vrn-A1a or vrn-A1b have been altered by the presence of the two mutations, we first expressed vrn-A1a and vrn-A1b proteins of full length using the pSUMO vector. However, after the pSUMO portion containing 6xHIS-Tag was removed, vrn-A1b was almost insoluble, whereas vrn-A1a was soluble. The decrease in vrn-A1b protein solubility could result in altered function. To investigate if vrn-A1a or vrn-A1b has different interactions with any protein partners, we used vrn-A1a as bait to screen a yeast-two hybrid (Y2H) library generated from Jagger.
Positive clones identified from the Y2H library included TaSOC1 (T. aestivum wheat suppressor of overexpression of constans 1) and TaVRT2 (T. aestivum wheat vegetative to reproductive transition 2), both of which have physical interactions with other MADS proteins because of the presence of a K–box in these proteins (Mandel et al., 1992; Yang et al., 2004; Kane et al., 2005). Strong interactions between TaVRN-A1 and TaSOC1 and between TaVRN-A1 and TaVRT2 were detected in our pull-down assays and immunoprecipitation analyses (Figure3), but no significant difference was found in the interaction of vrn-A1a and vrn-A1b with TaSOC1 or TaVRT2, suggesting that the interaction of TaVRN-A1 with TaSOC1 or TaVRT2 was not directly involved in controlling vernalization requirement duration in winter wheat.
Figure 3.
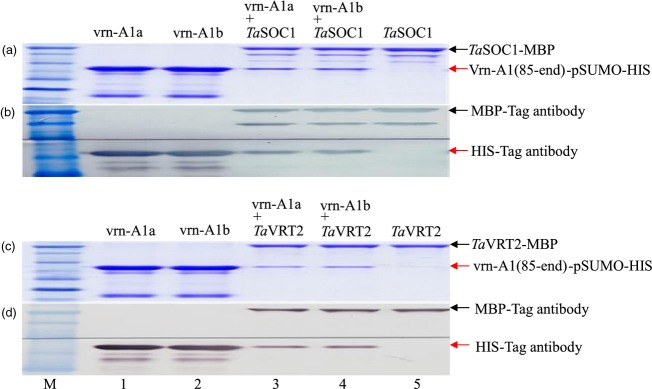
In vitro interactions of TaVRN-A1 with TaSOC1 and TaVRT2 proteins.(a, c) Pull-down assays of TaVRN-A1 with TaSOC1 and TaVRT2 interactions.(b, d) Protein immunoprecipitation analyses of TaVRN-A1 with TaSOC1 and TaVRT2. TaVRN-A1(85–end) with TaSOC1 (a and b) and TaVRT2 (c and d). TaVRN-A1 was detected by anti-HIS antibody, and TaSOC1 or TaVRT2 was detected by anti-MBP antibody: lane 1, purified Jagger vrn-A1a-HIS-tag; lane 2, purified 2174 vrn-A1b-HIS-tag; lane 3, interaction of TaVRN-A1a-HIS-tag with TaSOC1-MBP-tag or TaVRT2-MBP-tag; lane 4, interaction of TaVRN-A1b-HIS-tag with TaSOC1-MBP-tag or TaVRT2-MBP-tag; lane 5, purified TaSOC1-MBP-tag or TaVRT2-MBP-tag; M, protein marker. Arrowheads represent the expressed or interacting proteins. Six independent reactions for each interaction were performed, and one representative reaction is shown.
We discovered, however, that a protein TaHOX1 not only interacted with TaVRN-A1 but also interacted differently with vrn-A1a and vrn-A1b. Three independent cDNA clones identified from the Y2H library were predicted to encode 127 amino acids [100–226 aa (end)] of the TaHOX1 protein (Figure2g). In pull-down assays, both vrn-A1a and vrn-A1b proteins interacted with TaHOX1, but the vrn-A1b protein showed a visible decrease in its interaction ability with TaHOX1 (Figure4a). To determine whether the Leu117/Phe117 or Ala180/Val180 substitution in the TaVRN-A1 proteins caused the differential interaction with TaHOX1, three additional protein fragments for each of vrn-A1a and vrn-A1b were expressed. These fragments included: (i) the Leu117/Phe117 and Ala180/Val180 substitutions together (Figure2b, C) (ii) the Leu117/Phe117 substitution (Figure2d) alone; and (iii) the Ala180/Val180 substitution alone (Figure2e). Comparative in vitro interaction studies indicated that vrn-A1a and vrn-A1b protein fragments with both substitutions differed in their ability to interact with TaHOX1 (Figure4b), as did the vrn-A1a and vrn-A1b protein fragments including only the Ala180/Val180 substitution (Figure4c). However, the vrn-A1a and vrn-A1b protein fragments including only the Leu117/Phe117 substitution interacted similarly with TaHOX1 (Figure4d). The decreased interactive ability of vrn-A1b with TaHOX1 in pull-down assays was confirmed by protein immunoprecipitation analyses (Figure4e,f). We concluded the Ala180/Val180 substitution accounted for the differential interactions of vrn-A1a and vrn-A1b with TaHOX1.
Figure 4.
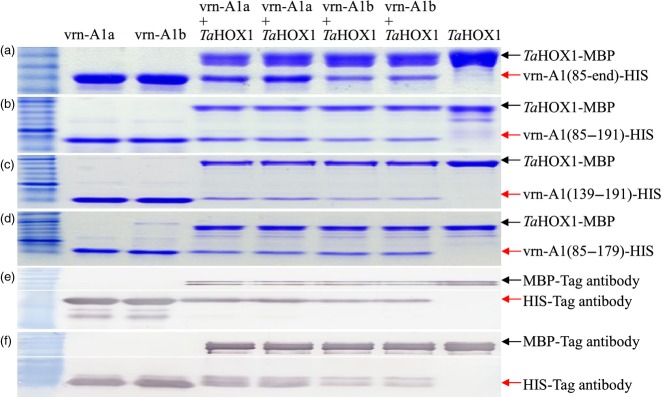
In vitro interaction between TaVRN-A1 and TaHOX1 proteins.(a–d) Pull-down assays of TaHOX1-MBP and TaVRN-A1 interactions. In vitro interaction between TaHOX1(100–end) and TaVRN-A1(85–end) (a), TaVRN-A1(85–191) (b), TaVRN-A1(139–191) (c), and TaVRN-A1(85–179) (d).(e, f) Protein immunoprecipitation analyses of TaHOX1 and TaVRN-A1. TaHOX1-MBP was detected by anti-MBP antibody (the upper panel, e and f). TaVRN1-HIS(85–end) and TaVRN-A1-HIS(139–191) were detected by anti-HIS antibody (the lower panel, e and f). Lane 1, purified Jagger vrn-A1a-HIS-tag; lane 2, purified 2174 vrn-A1b-HIS-tag; lanes 3 and 4, interaction of TaVRN-A1a-HIS-tag and TaHOX1-MBP-tag; lanes 5 and 6, interaction of TaVRN-A1b-HIS-tag and TaHOX1-MBP-tag; lane 7, purified TaHOX1-MBP-tag; M, protein marker. Arrowheads represent expressed or interacting proteins. Six independent replicates were performed for each interaction, and two replicates are shown to indicate consistency among replicates.
The TaHOX1 protein found in this study has the same structure of the homeodomain (HD) and the leucine zipper domain (Zip) (Figure2h) as HOX proteins reported in animals (Ariel et al., 2007), and TaHOX1 and TaVRN-A1 shared five leucine residues present in the Zip domain (Figure2f). It is likely that the physical interaction between TaHOX1 and TaVRN-A1 is attributable to the presence of the zipper in the two proteins; however, the Ala180 in vrn-A1b represented the same amino acid residue as TaHOX1, but Val180 caused a mismatch between vrn-A1b and TaHOX1 (Figure2f). This mismatch explained the decreased ability of vrn-A1b to interact with TaHOX1, indicating the Ala180/Val180 substitution outside the K–box in TaVRN-A1 was involved in its physical interaction with TaHOX1.
We confirmed that TaVRN-A1 and TaHOX1 proteins exhibited direct binding in living cells by using a transient expression system in tobacco leaves (Lu et al., 2010). The full-length TaVRN-A1 was expressed by pEG101-YFP vector, and enriched yellow fluorescent signals were detected predominantly in the nucleus (Figure5b). The full-length TaHOX1 was not expressed by the pEG101-YFP vector or by a series of bimolecular fluorescence complementation (BiFC) vectors (Waadt et al., 2008); therefore, we expressed a protein fragment, TaHOX1(1–180), to localize the protein in living cells. When TaHOX1-YFP(1–180) alone was expressed, enriched yellow fluorescent signals were detected only in the nucleus (Figure5b). Then, we cloned TaVRN-A1 into pEG201-YN vector and TaHOX1(1–180) into pEG202-YC vector and analyzed in vivo protein interactions by BiFC. When TaVRN-A1-YN and TaHOX1(1–180)-YC were simultaneously expressed in the same cell, yellow fluorescence was observed in the nucleus with a confocal microscope (Figure5c,e). We observed that when TaVRN-A1-YN and TaHOX1(100–end)-YC were simultaneously expressed in the same cell, TaVRN-A1 and TaHOX1 had a very strong interaction (Figure5f). Furthermore, TaHOX1(100–end) showed more interacting fluorescence with vrn-A1a than vrn-A1b in multiple images on a large scale. These results were consistent with the results for the differential in vitro interactions of TaHOX1(100–end) with vrn-A1a and vrn-A1b.
Figure 5.
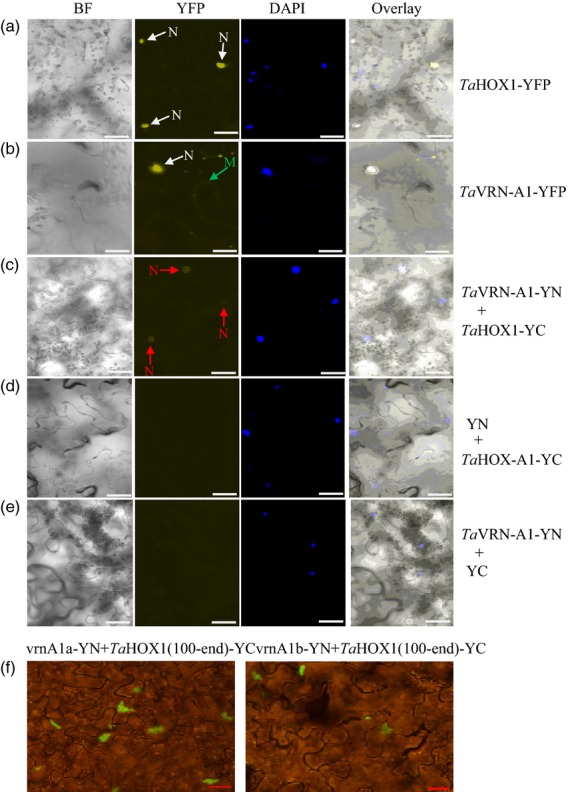
Subcellular localization and in vivo interaction of TaVRN-A1 and TaHOX1 proteins.(a) The subcellular location of TaHOX1(1–180)-YFP protein expressed by pEG101 in the nucleus (N) of living cells in Nicotiana tabacum (tobacco) leaves.(b) The subcellular location of TaVRN-A1(full-length)-YFP protein expressed by pEG101 predominantly in the nucleus (N) as well as partly on the cytoplasm membrane (M).(c) Bimolecular fluorescence complementation (BiFC) analysis of TaVRN-A1 and TaHOX1(1–180) proteins. The yellow fluorescent proteins (YFPs) resulted from the in vivo interaction between TaVRN-A1-YN and TaHOX1(1–180)-YC in the nucleus are arrowed ‘N’.(d–e) No YFP was observed in a negative control of the co-transformation of pEG202-YC with TaVRN-A1-pEG201-YN (d) or pEG201-YN with TaHOX1-pEG202-YC (e). The YFPs resulting from the in vivo interaction between TaVRN-A1-YN and TaHOX1(1–180)-YC in the nucleus are arrowed by ‘N’. (d, e) No YFP was observed in a negative control of the co-transformation of pEG202-YC with TaVRN-A1-pEG201-YN (d) or pEG201-YN with TaHOX1-pEG202-YC (e). YN, YFP fragment at the N-terminal end expressed from pEG201-YN vector; YC, YFP fragment at the C–terminal end expressed from pEG202-YC vector. Images were visualized under a confocal microscope.(f) BiFC analysis of TaVRN-A1 and TaHOX1(100–end) proteins. The green fluorescent proteins resulted from the in vivo interaction between TaVRN-A1-YN and TaHOX1(100–end)-YC. YN, YFP fragment at the N–terminal end expressed from pEG201-YN vector; YC, YFP fragment at the C–terminal end expressed from pEG202-YC vector. Images were taken under a fluorescent microscope. TaHOX1(100–end) was also tested for interaction with the vectors as negative controls, and no interaction signal was observed (data not shown).
The upregulation of TaHOX1 by low temperature and its genetic effects on heading date
We isolated the complete TaHOX1 gene from Jagger (JQ915061) and 2174 (JQ915062) for functional characterization. Using RT-PCR, we found that the TaHOX1 transcripts in leaves were greatly upregulated, and remained at a high level during vernalization (Figure6a). This result indicated that TaHOX1 had a similar expression pattern as the recessive vrn1 allele characterized in previous studies (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). TaHOX1 and TaVRN-A1 were concomitantly upregulated by low temperature, indicating that they functioned in the flowering pathway.
Figure 6.
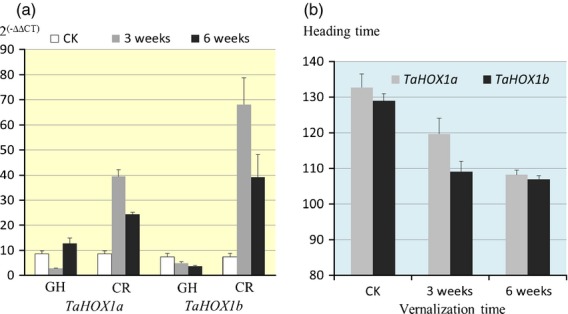
Expression profiles of TaHOX1 and its genetic effect on flowering time.(a) Expression pattern of TaHOX1 in the glasshouse (GH) and in the cold room (CR). Transcript levels of TaHOX1a (the Jagger allele) and TaHOX1b (the 2174 allele) in leaves of vernalized plants for 3 and 6 weeks, as well as non-vernalized plants (CK) are shown using the values calculated by the 2(−ΔΔCT) method, where CT is the cycle threshold, and actin was used as an endogenous control. The primers for TaHOX1 and actin are provided in Table S2. The values represent mean expression levels (n = 15–20), and the error bars indicate standard errors.(b) Genetic effect of TaHOX1 on heading date. The heading date was from each line of the population (n = 96) that was vernalized for 3 or 6 weeks, or used for the control in the glasshouse (CK). The significant effects of the TaHOX1 gene on heading date were determined using one-way analysis of variance (anova). Error bars indicate standard errors.
We developed a PCR marker for one SNP (see Figure S11A), and mapped TaHOX1 to chromosome 6B using the Jagger × 2174 RIL population (see Figure S11B). A significant difference in heading date was observed between the Jagger TaHOX1a and the 2174 TaHOX1b alleles (120 versus 109 days, respectively) in the RIL population vernalized for 3 weeks (P < 0.05), but not in the RIL population vernalized for 6 weeks or the control population (Figure6b). The SNP in exon 1 resulted in a ‘leucine’ residue at position 99 in TaHOX1b and a ‘proline’ residue at the same position in TaHOX1a (Figure S12A). This point mutation in TaHOX1 did not affect its interaction with vrn-A1 (Figure S12B). In contrast, the higher TaHOX1 transcriptional level was associated with earlier heading, suggesting that the regulation of heading date by TaHOX1 was at the transcriptional level.
Discussion
All of the vernalization genes that were previously cloned are related to segregation between spring wheat and winter wheat, which can differ by as many as several months in phenotype (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003, 2004a, 2006). The phenotypic segregation in flowering time among winter wheat cultivars usually spans a few days when the plants are tested in the field (Chen et al., 2009, 2010). The present study allowed us to observe more than 1 month for flowering time segregated between the two winter wheat cultivars when a 3–week vernalization was performed under thermal- and photoperiodic-controlled conditions. This approach was used to separate the effect of low temperature in accelerating the developmental transition from the effect of low temperature in delaying the growth rate in a semi-winter cultivar that required only 3 weeks at low temperatures for full vernalization. If the semi-winter wheat cultivar was vernalized for 6 weeks, as has been performed in numerous studies, the over-vernalized plant to the prolonged low temperatures after 3 weeks would not be accelerated flowering, but would instead result in delayed flowering. Winter wheat, for which any requirement for vernalization has been fully satisfied, will have similar responses to ambient temperature as spring wheat (Snape et al., 2001). If the fully vernalized plants continue to stay at low temperatures, however, the developmental process of these over-vernalized plants will be delayed because of a longer phyllochron and a slower growth rate (McMaster, 2005). The phenotyping approach used in this study has facilitated the positional cloning of the major gene for vernalization requirement duration in winter wheat.
In a previous study, we found a C/T polymorphism in exon 4 that is responsible for the amino acid change at Leu117/Phe117 in TaVRN-A1 between the Jagger allele and the 2174 allele, and we developed a PCR marker for this polymorphism that showed an association with developmental variation in the Jagger × 2174 RIL population tested in the field (Chen et al., 2009). It was recently reported that TaVRN-A1 was duplicated in winter wheat cv. ‘Hereward’ (JF965397) but not in ‘Claire’ (JF965395) (Díaz et al., 2012). In the population of Claire × Hereward, plants homozygous for the Claire allele flowered earlier and plants homozygous for the Hereward allele flowered later, whereas heterozygotes had an intermediate flowering time; therefore, the authors concluded that the increased copy number of TaVRN-A1 in Hereward resulted in an increased requirement for vernalization (and thus late flowering). We found that at the polymorphic sites of both exon 4 and exon 7, Jagger has the same allele as Claire and 2174 has the same allele as Hereward. We also observed the duplication event of vrn-A1b in 2174 for more vernalization, but not in Jagger for less vernalization (Figure S13). Thus, on the surface, it seems that the two studies have consistent results and that the phenomenon that Diaz et al. described in their paper, that plants with an increased copy number of TaVRN-A1 have an increased requirement for vernalization, was correct. However, we affirm that a greater vernalization requirement in 2174 was not because 2174 has one more vrn-A1 copy than Jagger, but because Jagger produces a different vrn-A1 protein than 2174. Our results from several independent progeny populations clearly and consistently demonstrated the Jagger vrn-A1a allele for early flowering was dominant over the 2174 vrn-A1b allele for late flowering, with phenotypic segregation of 3:1 (Figure1e; Table S1). The two studies have come to different conclusions for several reasons. First, in the Diaz et al. study, vernalization was performed under short-day conditions (7°C, 8 h of light), and then the vernalized plants were grown under long-day conditions (18 h of light) in a glasshouse (no temperature condition was provided). In our study, however, vernalization was performed at 4°C with 16 h of light, and then the vernalized plants were moved to 25°C and 16 h of light. The same long day was set throughout the experiment in our study to avoid any disruption of photoperiod, which is well known to have significant effects on flowering in winter wheat (Dubcovsky et al., 2006; Wang et al., 2009). Second, in the Diaz et al. study a small mapping population of 96 F2 lines was used to test the association between the vrn-A1 copy number and flowering time. By contrast, we generated a backcross population and screened 6500 plants to find several lines that had crossovers at the vrn-A1 locus. The flowering time in the progeny of these recombinant lines was segregated by a single gene vrn-A1a, without any disruption by other genetic factors for this phenotype. Third, vrn-A1 is the promoter of flowering, as shown in wheat and barley (Danyluk et al., 2003; Murai et al., 2003; Trevaskis et al., 2003; Yan et al., 2003), and in Arabidopsis (Ng and Yanofsky, 2001). The dosage effect of two copies from such a flowering promoter should not result in a phenotype of a later flowering time, as suggested in the previous study (Díaz et al., 2012). In this study, we used the positional cloning strategy to prove that vrn-A1a in Jagger was dominant for early flowering, regardless of the function of the duplicated vrn-A1b in 2174. Therefore, we concluded that less vernalization requirement by Jagger resulted from its vrn-A1a protein form, or that more vernalization requirement in 2174 was not due to the presence of its two copies of vrn-A1b.
Sequence comparison analyses showed that the A genome of the diploid donor (Triticum urartu, GenBank accession DQ291016) and the tetraploid donor (Triticum turgidum, GenBank accession AY747598) had exactly the same cDNA sequence of vrn-A1a as Jagger, suggesting that the vrn-A1a allele in Jagger is the wild type. The two amino acids, Phe117 and Val180, in vrn-A1b in cultivar 2174 were mutated during the domestication and adaptation of hexaploid wheat. Previous studies revealed that the recessive vrn-A1 allele for winter growth habit is the wild type, and the dominant Vrn-A1 allele for spring growth habit was mutated via insertions or deletions in the promoter or the first intron (Yan et al., 2004b; Fu et al., 2005). This study revealed that winter wheat cultivars Jagger and 2174 carried the same recessive vrn-A1 allele but encoded different vrn-A1 proteins that underlie various vernalization requirement durations. Altogether, these studies demonstrate that a single gene may evolve differential mechanisms to enable adaptation to multiple environmental conditions. Through one mechanism evolution occurred at the transcriptional level by mutations in the promoter or first-intron region of vrn-A1, producing Vrn-A1 that confers spring growth habit without vernalization requirement (Yan et al., 2004b; Fu et al., 2005). Through a second mechanism, the wild-type vrn-A1a allele for less vernalization requirement in semi-winter wheat cultivars like Jagger evolved at the protein level through mutations in the coding region, producing vrn-A1b for greater vernalization requirement in strong winter wheat cultivars like 2174. The proposed mechanism underlying the quantitative vernalization requirement in winter wheat differs from the mechanisms that explain the various vernalization requirement durations in winter types of Arabidopsis (Michaels and Amasino, 1999; Sheldon et al., 1999; Reeves and Coupland, 2001; Levy et al., 2002; Corbesier et al., 2007; Angel et al., 2011; Heo and Sung, 2011; Coustham et al., 2012).
TaHOX1 is an orthologue of rice homeobox HOX24 (NP_001047582.1) and Arabidopsis thaliana homeobox 7 (AtHB–7) (NP_182191.1). AtHB7 was characterized as a member of HD-Zip class I involved in responses to water and light stresses, and overexpression of AtHB7 resulted in the early flowering of transgenic Arabidopsis plants (Olsson et al., 2004), providing supporting evidence for the role of the HOX genes in the regulation of flowering time in plants. TaHOX1 is one of the HOX proteins known to function as an on–off switch in controlling development, including specialization of regional identities along the anterio/posterior axis in a wide range of phyla in animals (Manak and Scott, 1994), whereas vrn-A1 (=AP1) is one of the MADS-box proteins known to act as floral switches for the transition from vegetative to reproductive development in plants (Ng and Yanofsky, 2001). This study presented one example in which MADS and HOX proteins involving homeosis have a direct binding in higher plants.
In winter wheat, two models were established to explain the relationship between the three vernalization genes that have been cloned so far: the first model supposes that VRN3 positively regulates VRN1, which then negatively downregulates VRN2 (Yan et al., 2006); the second model hypothesizes that VRN1 activates its downstream gene VRN3, which then downregulates VRN2 (Shimada et al., 2009). In this study, we focused on the positional cloning of the gene for vernalization requirement duration in winter wheat. In the cloning population, flowering time segregated according to a single gene, vrn-A1, and no genetic effect was observed from VRN2 or VRN3. When vrn-A1 acts as a protein in either of the two models, it should appear as a TaVRN1-TaHOX1 protein complex because of a direct binding between them. TaHOX1 protein and TaVRN-A1 protein should also have direct interaction in spring wheat cultivars, although the role of the protein complex in regulating flowering time in spring wheat needs to be investigated.
Previous studies suggested that the gene for vernalization requirement duration was postulated as a sensor to code for a temperature-sensitive protein (Roberts, 1990; Amasino, 2004; Sung and Amasino, 2004). The findings from this study indicated that the vrn-A1 protein could function as such a sensor, to measure cold duration. Although the accumulation of vrn-A1a was accelerated by a 3–week vernalization duration, its activity reached a threshold for flowering. Compared with vrn-A1a, vrn-A1b needs a longer time at the low temperature to reach the same activity. The point mutation at Val180 of vrn-A1a is critical in retaining vrn-A1 activity. The TaHOX1 protein was suggested to function in the flowering pathway, based on its direct interaction with TaVRN-A1 and genetic association with flowering time; however, the role of TaHOX1 in regulating flowering time needs to be confirmed in transgenic plants, and further experiments are also needed to test if the TaVRN-A1-TaHOX1 protein complex is sensed by low temperature in winter wheat. The information on the role of TaVRN-A1 in controlling vernalization requirement duration is particularly useful for directing breeding cultivars for adaptation to global warming in future climate scenarios for winter wheat, which represents approximately 75% of the wheat grown worldwide.
Experimental procedures
Plant materials and vernalization experiments
Jagger and 2174 were initially grown in a glasshouse at 20–25°C under long days (LD, 16–h day/8–h night). At the fifth-leaf stage, the parental plants were moved into a cold room with 4°C and the LD photoperiod. The LD photoperiod was applied throughout this study to avoid any disruption from photoperiod effects on vernalization (Wang et al., 2009). The two parental lines were vernalized for varying numbers of weeks. The vernalized plants were returned to the glasshouse, whereas non-vernalized plants that continuously remained in the glasshouse were used as controls. The heading date for eight plants for each treatment was scored.
Three populations from the same Jagger × 2174 RILs were initially grown in the same glasshouse as used for the parental lines. At the fifth-leaf stage, two of these populations were vernalized for 3 and 6 weeks, respectively. The vernalized populations were returned to the glasshouse for comparison with the third population as a non-vernalized control. The heading date was scored for three plants of each line in these three populations.
Development of PCR markers for the genes flanking QVrd.osu-5A
A total of nine genes in the QVrd.osu-5A locus were converted to orthologous genes in chromosome 5A of hexaploid wheat tested in this study. PCR markers for each of nine genes at the QVrd.osu-5A locus were developed and used to determine the locations of critical crossovers in the recombinant plants (Figures S1–S9).
Allelic variation and expression of candidate genes for QVrd.osu-5A
Each of the two alleles for each of three candidate genes was sequenced. Two SNPs were found in CYB5 between the Jagger allele and the 2174 allele, 10 SNPs were found in AGLG1 between the Jagger allele and the 2174 allele, and 29 SNPs were found in vrn-A1 between the Jagger allele and the 2174 allele. Transcript levels for vrn-A1, AGLG1 and CYB5 were determined from the same cDNA sample collected from five plants for the vernalized and non-vernalized treatments, and actin was used as an endogenous control (see Figure S10). The specific primers for expression of these genes are listed in Table S2.
Identification of proteins interacting with vrn-A1
The ‘Matchmaker™’ Library Construction & Screening System (Clontech, http://www.clontech.com) was used to construct a Y2H ‘prey’ library for Jagger. RNA was extracted from pooled samples of young leaves and apices from vernalized plants for 1, 2 and 3 weeks, and the vrn-A1a was used as ‘bait’ to screen the ‘prey’ library. The constructing and screening procedures were described in a previous study in which 2174 was used as a host plant (Cao and Yan, 2013).
Protein pull-down assay
The complete vrn-A1a and vrn-A1b cDNA sequences were cloned into pSUMO vectors with an N–terminal 6XHIS-tag (Deng et al., 2008), and vrn-A1 fragments (85–end) were also cloned into the pSUMO vector. Three shorter fragments for each of vrn-A1a and vrn-A1b, encoding different mutations, were cloned into pSKB3 with an N–terminal 6XHIS-tag (Figure2c–e). The TaSOC1, TaVRT2 and TaHOX1 cDNA obtained from the Y2H library were cloned into the pMAL-c2X vector with a MBP-tag (New England BioLabs, http://www.neb.com). The primers used in cloning are listed in Table S3. The protein constructs were expressed in Escherichia coli (BL21 DE3). The expressed proteins from pSUMO and pSKB3 were dialyzed and purified using Ni-NTA column (QIAGEN, http://www.qiagen.com). The proteins fused with MBP-tag were dialyzed and purified with amylase column (New England BioLabs).
A protein fused with the MBP-tag (500 μg ml−1, 50 μl) was incubated with amylase resin (40 μl) for 1 h at 4°C in the column buffer, and the MBP-tag (500 μg ml−1, 50 μl) alone was used as a negative control. A protein fused with the 6XHIS-tag (500 μg ml−1, 100 μl) was added to the amylase resins and incubated for 4 h at 4°C. The interacting proteins in the resin were then washed with the column buffer between six and eight times, depending on when the protein tagged with the 6XHIS-tag was no longer observed in the negative control. The protein that bound to the resin was eluted by boiling the samples in 100 μl of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer for 5 min. The eluted protein samples (80 μl) were analyzed by 10% SDS-PAGE stained with Coomassie Brilliant blue.
Western blot was performed using the protocol previously described by Earley et al. (2006). Immunodetection was performed using primary antibodies (1:1000 dilution): anti-Maltose Binding Protein (MBP) (M1321; Sigma-Aldrich, http://www.sigmaaldrich.com) to detect HOX1-MBP, and anti-6XHIS (SAB4600048; Sigma-Aldrich) to detect TaVRN-A1-HIS, TaSOC1-HIS or TaVRT2-HIS. After washing, anti-Mouse IgG-Alkaline Phosphatase antibody (A3562; Sigma-Aldrich) was used as the secondary antibody. The membranes were washed three times before color was developed with Alkaline Phosphatase Conjugate Substrate Kit (170-6432; Bio-Rad, http://www.bio-rad.com).
Subcellular localization and in vivo interaction of TaVRN-A1 and TaHOX1 proteins
The complete TaVRN-A1 cDNA was cloned into pDONR207 with the BP cloning kit (Invitrogen, now Life Technologies, http://www.lifetechnologies.com), and then transferred to pEarleygate 101 (pEG101) using the LR cloning kit for subcellular localization of TaVRN-A1. TaVRN-A1 in pDONR207 was fused to the N–terminal 174 amino acid portion (1–174 aa) of YFP in the pEarleyGate201-YN vector (pEG201-YN) to test in vivo interaction with TaHOX1 [1–180 aa, 100–226 aa (end)] fused to the C–terminal amino acid portion (175–239 aa) of YFP in the pEarleyGate202-YC vector (pEG202-YC). Empty vectors were also used as negative controls for interaction with TaVRN-A1 or TaHOX1 proteins. Agrobacterium tumefaciens strains (GV3101) carrying the BiFC constructs were used together with the p19 strain for infiltration of Nicotiana benthamiana leaves (at 5 weeks old). Leaf discs were cut for BiFC for imaging 3 days after infiltration. Images were visualized under a confocal microscope (TCS SP2; Leica, http://www.leica-microsystems.com). The images (Figure5a–e) were also taken with a bright filter (BF) to indicate the background of the leaves infiltrated with A. tumefaciens carrying constructs, or with an ultraviolet filter to indicate the position of the nucleus stained with 4′,6–diamidino-2-phenylindole (DAPI). The overlay images align the locations of YFP with the DAPI-stained nucleus. Images (Figure5f) were taken under a fluorescent microscope (BX51; Olympus, http://www.olympus-global.com) with GFP filter to indicate the presence of fluorescent proteins. The scale bars in all images are 50 μm.
Acknowledgments
We thank J. Deng (Oklahoma State University) for his assistance with the development of the in vitro protein interaction systems, Y Cui (Agriculture and Agri-Food Canada) for providing pEarleygate201-YN and pEarleygate202-YC vectors and F. Zhang (Oklahoma State University) for his assistance with the development of the in vivo protein interaction systems in tobacco. We thank Y. Chen for technical assistance in generating backcross populations. This project was supported by the National Research Initiative Competitive Grants CAP project 2011-68002-30029 from the US Department of Agriculture (USDA) National Institute of Food and Agriculture and from the Oklahoma Center of Advanced Science and Technology (OCAST, PS07-032). This research project was partially funded by the Oklahoma Agricultural Experiment Station and the Oklahoma Wheat Research Foundation.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
PCR marker for GENE1.
Figure S2.PCR marker for PCS1.
Figure S3.PCR marker for CYB5.
Figure S4.PCR marker for AGLG1.
Figure S5.PCR marker for vrn-A1a.
Figure S6.PCR marker for CYS1.
Figure S7.PCR marker for PHY1.
Figure S8.PCR marker for OPT1.
Figure S9.PCR marker for CDO708.
Figure S10. Comparison of gene expression levels of three candidate genes for QVrd.osu-5A in Jagger versus 2174.
Figure S11. Mapping of TaHOX1.
Figure S12. Location of a point mutation in TaHOX1.
Figure S13. Copy number of TaVRN-A1.
Progeny test of critical recombinant plants for heading date.
Table S2. List of primers used for gene expression.
Table S3. List of primers used for protein expression.
References
- Amasino R. Vernalization, competence, and the epigenetic memory of winter. Plant Cell. 2004;16:2553–2559. doi: 10.1105/tpc.104.161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M. A polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Bonnin I, Rousset M, Madur D, Sourdille P, Dupuits C, Brunel D, Goldringer T. FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat. Theor. Appl. Genet. 2008;116:383–394. doi: 10.1007/s00122-007-0676-0. [DOI] [PubMed] [Google Scholar]
- Cao S, Yan L. Construction of a high-quality Yeast two-hybrid (Y2H) library and its application in identification of interacting proteins with key vernalization regulator TaVRN-A1 in wheat. BMC Res. Note. 2013;6:81–86. doi: 10.1186/1756-0500-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Carver BF, Wang S, Zhang F, Yan L. Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor. Appl. Genet. 2009;118:881–889. doi: 10.1007/s00122-008-0946-5. [DOI] [PubMed] [Google Scholar]
- Chen Y, Carver BF, Wang S, Cao S, Yan L. Genetic regulation of developmental phases in winter wheat. Mol. Breed. 2010;118:1339–1349. [Google Scholar]
- Chouard P. Vernalization and its relation to dormancy. Annu. Rev. Plant Physiol. 1960;11:191–238. [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. Quantitative modulation of Polycomb silencing underlies natural variation in vernalization. Science. 2012;337:584–587. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- Crofts HJ. On defining a winter wheat. Euphytica. 1989;44:225–234. [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JP, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR. Structure of the ROC domain from the Parkinson's disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc. Natl Acad. Sci. USA. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the photoperiod-B1 and vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum. PLoS One. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Fu D, Szucs P, Yan L, Helguera M, Skinner JS, Hayes P, Dubcovsky J. Large deletions in the first intron of the VRN1 vernalization gene are associated with spring growth habit in barley and polyploid wheat. Mol. Genet. Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- Humphreys MW, Yadav RS, Cairns AJ, Turner LB, Humphreys J, Skot L. A changing climate for grassland research. New Phytol. 2006;169:9–26. doi: 10.1111/j.1469-8137.2005.01549.x. [DOI] [PubMed] [Google Scholar]
- Kane NA, Danyluk J, Tardif G, Ouellet F, Laliberte JF, Limin AE, Fowler DB, Sarhan F. TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiol. 2005;138:2354–2363. doi: 10.1104/pp.105.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr RA. Climate change: global warming is changing the world. Science. 2007;316:188–190. doi: 10.1126/science.316.5822.188. [DOI] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science. 2002;297:243–246. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;60:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- Manak JR, Scott MP. A class act: conservation of homeodomain protein functions. Dev. Suppl. 1994:61–71. [PubMed] [Google Scholar]
- Mandel MA, Gustafsonbrown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene Apetala1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- McMaster GS. Phytomers, phyllochrons, phenology and temperate cereal development. J. Agric. Sci. 2005;143:137–150. [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglietta F, Tanasescu M, Marica A. The expected effects of climate change on wheat development. Global Change Biol. 1995;1:407–415. [Google Scholar]
- Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y. WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 2003;44:1255–1265. doi: 10.1093/pcp/pcg171. [DOI] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nature Rev. Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- Olsson AS, Engström P, Söderman E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004;55:663–677. doi: 10.1007/s11103-004-1581-4. [DOI] [PubMed] [Google Scholar]
- Pugsley AT. A genetic analysis of the spring-winter habit of growth in wheat. Aust. J. Agric. Res. 1971;22:21–31. [Google Scholar]
- Reeves PH, Coupland G. Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol. 2001;126:1085–1091. doi: 10.1104/pp.126.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DWA. Identification of loci on chromosome 5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome. 1990;33:247–259. [Google Scholar]
- Santra DK, Santra M, Allan RE, Campbell KG, Kidwell KK. Genetic and molecular characterization of vernalization genes Vrn-A1 Vrn-B1, and Vrn-D1 in spring wheat germplasm from the pacific northwest region of the USA. Plant Breed. 2009;128:576–584. [Google Scholar]
- Semenov MA, Halford NG. Identifying target traits and molecular mechanisms for wheat breeding under a changing climate. J. Exp. Bot. 2009;60:2791–2804. doi: 10.1093/jxb/erp164. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 2009;58:668–681. doi: 10.1111/j.1365-313X.2009.03806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape JW, Sarma R, Quarrie SA, Fish L, Galiba G, Sutka J. Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica. 2001;120:309–315. [Google Scholar]
- Sung SB, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Tranquilli GE, Dubcovsky J. Epistatic interactions between vernalization genes Vrn-Am 1 and Vrn-Am2 in diploid wheat. J. Hered. 2000;91:304–306. doi: 10.1093/jhered/91.4.304. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc. Natl Acad. Sci. USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008;56:505–516. doi: 10.1111/j.1365-313X.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Carver B, Yan L. Genetic loci in the photoperiod pathway interactively modulate reproductive development of winter wheat. Theor. Appl. Genet. 2009;118:1339–1349. doi: 10.1007/s00122-009-0984-7. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of wheat vernalization gene VRN1. Proc. Natl Acad. Sci. USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004a;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN1 promoter region in polyploid wheat. Theor. Appl. Genet. 2004b;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl Acad. Sci. USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fanning L, Jack T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2004;33:47–59. doi: 10.1046/j.0960-7412.2003.01473.x. [DOI] [PubMed] [Google Scholar]
- Zhang XK, Xia XC, Xiao YG, Dubcovsky J, He ZH. Allelic variation at the vernalization genes Vrn-A1 Vrn-B1 Vrn-D1 and Vrn-B3 in Chinese common wheat cultivars and their association with growth habit. Crop Sci. 2008;48:458–470. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR marker for GENE1.
Figure S2.PCR marker for PCS1.
Figure S3.PCR marker for CYB5.
Figure S4.PCR marker for AGLG1.
Figure S5.PCR marker for vrn-A1a.
Figure S6.PCR marker for CYS1.
Figure S7.PCR marker for PHY1.
Figure S8.PCR marker for OPT1.
Figure S9.PCR marker for CDO708.
Figure S10. Comparison of gene expression levels of three candidate genes for QVrd.osu-5A in Jagger versus 2174.
Figure S11. Mapping of TaHOX1.
Figure S12. Location of a point mutation in TaHOX1.
Figure S13. Copy number of TaVRN-A1.
Progeny test of critical recombinant plants for heading date.
Table S2. List of primers used for gene expression.
Table S3. List of primers used for protein expression.