Abstract
Grass lignins contain substantial amounts of p-coumarate (pCA) that acylate the side-chains of the phenylpropanoid polymer backbone. An acyltransferase, named p-coumaroyl-CoA:monolignol transferase (OsPMT), that could acylate monolignols with pCA in vitro was recently identified from rice. In planta, such monolignol-pCA conjugates become incorporated into lignin via oxidative radical coupling, thereby generating the observed pCA appendages; however p-coumarates also acylate arabinoxylans in grasses. To test the authenticity of PMT as a lignin biosynthetic pathway enzyme, we examined Brachypodium distachyon plants with altered BdPMT gene function. Using newly developed cell wall analytical methods, we determined that the transferase was involved specifically in monolignol acylation. A sodium azide-generated Bdpmt-1 missense mutant had no (<0.5%) residual pCA on lignin, and BdPMT RNAi plants had levels as low as 10% of wild-type, whereas the amounts of pCA acylating arabinosyl units on arabinoxylans in these PMT mutant plants remained unchanged. pCA acylation of lignin from BdPMT-overexpressing plants was found to be more than three-fold higher than that of wild-type, but again the level on arabinosyl units remained unchanged. Taken together, these data are consistent with a defined role for grass PMT genes in encoding BAHD (BEAT, AHCT, HCBT, and DAT) acyltransferases that specifically acylate monolignols with pCA and produce monolignol p-coumarate conjugates that are used for lignification in planta.
Keywords: BAHD acyltransferase, biomass, Brachypodium distachyon, DFRC method, grass, lignin, lignin acylation, NMR, thioacidolysis
Introduction
Lignocellulosic biomass from grasses could serve as a major feedstock for the generation of liquid biofuels and contribute more than half of the envisioned one billion tons of biomass available in the USA each year (Perlack et al., 2005; Carroll and Somerville, 2009; U.S. Department of Energy, 2011). Lignocellulosic biomass typically refers to the harvestable plant portions not consumed by humans (e.g., for grasses, the stems and leaves) and is primarily composed of secondary cell walls that consist of the polysaccharides cellulose and hemicelluloses embedded within the heterogeneous phenolic polymer lignin (Pauly and Keegstra, 2008). Lignin polymer ‘backbones’ in grasses are composed of guaiacyl (G) and syringyl (S) units (derived from the monolignols coniferyl alcohol and sinapyl alcohol, respectively), together with a lesser amount of p-hydroxyphenyl (H) units (derived from p-coumaryl alcohol), interconnected by β–O–4-aryl ether and other inter-unit bonds, and cross-linked to hemicelluloses by ferulate (Ralph et al., 1995, 2004; Boerjan et al., 2003; Zhang et al., 2009; Lapierre, 2010).
C3 grasses such as rice, wheat, rye, oat, and bamboo, and C4 grasses such as corn, switchgrass, and sorghum, all have lignin polymers that are partially acylated by p-coumarate ‘appendages’ and, as more recently shown, by acetate (Ralph, 2010; Ralph and Landucci, 2010; del Río et al., 2012). It is now well established that such lignin acylation results from in planta lignification using pre-acylated monolignols (Ralph et al., 1994; Lu and Ralph, 1999, 2002, 2008; Hatfield et al., 2009; Ralph, 2010; Withers et al., 2012); in the case of p-coumarate, this process is via coniferyl and sinapyl p-coumarate ester conjugates 3 (Figure1). As the p-coumarate acylates the monolignol γ-OH, the resulting lignin is partially γ-p-coumaroylated (c.f., 5, Figure1) (Ralph et al., 1994; Ralph, 2010).
Figure 1.
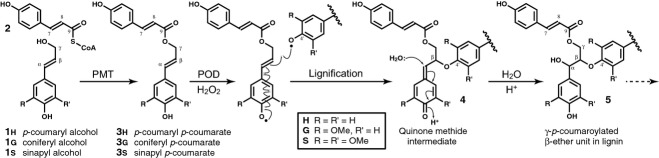
Pathway for the acylation of monolignols 1 by p-coumarate, via its CoA derivative 2, to form monolignol p-coumarate conjugates 3, and an example (using the favored β–O–4-coupling, following rearomatization of the quinone methide intermediate 4) of how the monolignol moiety undergoes radical coupling reactions during lignification resulting in lignin units such as 5 in which p-coumarate acylates the γ-position of lignin unit side-chains; monolignol p-coumarate conjugates 3 can also β–5-couple (with a G or H unit) or β–β-couple with a monolignol 1 or another monolignol p-coumarate 3. Further lignification via coupling with additional monomers can etherify the phenolic unit of 5, via either 5–β- (when the phenolic ring is H or G) or 4–O–β-coupling.
Although the role for these conjugated monolignols remains somewhat a mystery, there is evidence that they are synthesized in the cell and exported to the wall along with the traditional monolignols 1, coniferyl and sinapyl alcohols (along with smaller amounts of p-coumaryl alcohol) where they undergo the radical coupling and cross-coupling reactions, mainly by so-called endwise polymerization in which the monomer radical couples to the growing polymer radical (Ralph, 2010). It was initially puzzling but is now clear why the p-coumarate moieties, as phenolic entities, remain as free-phenolic pendant units on the polymer and do not themselves undergo radical coupling reactions. Although they readily form radicals, such radicals preferentially undergo radical transfer reactions with phenols that produce more stable phenolic radicals, such as the monolignols themselves and the guaiacyl and syringyl phenolic end-units on the growing polymer, i.e., they undergo radical transfer reactions rather than participating in radical coupling (Takahama et al., 1996; Ralph et al., 2004; Hatfield et al., 2008; Ralph, 2010). This process has been demonstrated in a model system that involves the typically slow peroxidase-H2O2-mediated dehydrodimerization of sinapyl alcohol that is significantly sped up in the presence of catalytic levels of a p-coumarate ester (Ralph et al., 2004; Hatfield et al., 2008). Therefore, the polymerization, whether from the monolignols 1 or from the p-coumaroylated monolignols 3, is solely via the monolignol moiety.
Current preliminary evidence, and more extensive evidence with monolignol acetates, shows that acylation does not greatly affect the coupling propensity of the monolignol moiety, i.e., radical coupling produces the same array of coupling products in approximately the same ratios whether the monolignols are acylated or not (Lu and Ralph, 2008). The one difference is that, for the product of monolignol β–β-coupling or dehydrodimerization, the post-coupling rearomatization pathways are necessarily different because the γ-OH is no longer available for internal trapping of the quinone methide intermediate.
Recently, we identified from rice the putative monolignol transferase gene, OsPMT. Heterologously expressed OsPMT had all the properties anticipated for a p-coumaroyl-CoA:monolignol transferase (PMT) enzyme responsible for p-coumaroylation of monolignols (Figure1) (Withers et al., 2012). The enzyme produced, with favorable kinetics, monolignol p-coumarate conjugates 3 from p-coumaroyl-CoA 2 and p-coumaryl, coniferyl, and sinapyl alcohols 1h,1g, and 1s (Hatfield et al., 2009; Withers et al., 2012). In this paper, we demonstrate the veracity of the hypothesized in planta action by misregulation of the closest gene homolog in Brachypodium, and furthermore demonstrate its selectivity for p-coumaroylation of monolignols and, thereby lignin, over the arabinoxylans that are also partially p-coumaroylated in grasses.
Results and Discussion
It has been shown recently that bacterially expressed and purified OsPMT protein could acylate monolignols with p-coumarate (pCA) to produce monolignol-pCA ester conjugates (Withers et al., 2012). We hypothesized that, in planta, this enzyme is therefore responsible for biosynthesizing the monolignol p-coumarate conjugates that become incorporated into the lignin polymer by oxidative radical coupling reactions (Figure1). To validate the in planta activity of PMT, we employed a reverse genetic approach, using the model grass Brachypodium distachyon (Brachypodium).
To identify the likely Brachypodium PMT gene, the OsPMT (LOC_Os01g18744.1) amino acid sequence was used as a query in a BLASTP search of the Brachypodium proteome (http://www.gramene/org/; http://mips.helmholtzmuenchen.de/plant/brachypodium/). The predicted protein Bradi2g36910 was found to have the highest sequence homology to OsPMT, sharing 63% amino acid sequence identity and 75% similarity. Recently published phylogenetic trees that contained both Brachypodium and rice BAHD acyltransferases showed that Bradi2g36910 is the most closely related Brachypodium protein to OsPMT (Bartley et al., 2013; Molinari et al., 2013).
As the Bradi2g36910 coding sequence was only partially supported by published expressed sequence tag sequences (Vogel et al., 2006), we generated and sequenced Bradi2g36910 cDNA clones (GenBank accession no. HG421450), from Brachypodium inbred line Bd21-3 stem tissue transcripts, finding the open reading frame (ORF) sequence to be 100% identical to the annotated gene model Bradi2g36910.1.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was performed on transcripts that had been isolated from various plant tissues, and revealed that Bradi2g36910 (hereafter referred to as BdPMT) was expressed in all organs tested (spikelet, stem internodes, leaves, and root), with highest expression levels in the spikelet (Figure2a). Co-expression analysis using PlaNet (http://aranet.mpimp-golm.mpg.de/index.html), a web-based platform that predicts transcriptomic co-expression networks based on temporal and spatial gene expression data (Mutwil et al., 2011; Harrington et al., 2012), revealed that BdPMT expression clustered with those of other predicted lignin biosynthetic genes including Phenylalanine Ammonia Lyase (PAL; Bradi3g49250 and Bradi3g49260), Cinnamoyl-CoA Reductase (CCR; Bradi3g36887), and Caffeic acid O-Methyltransferase (COMT; Bradi3g16530) (Table S1).
Figure 2.
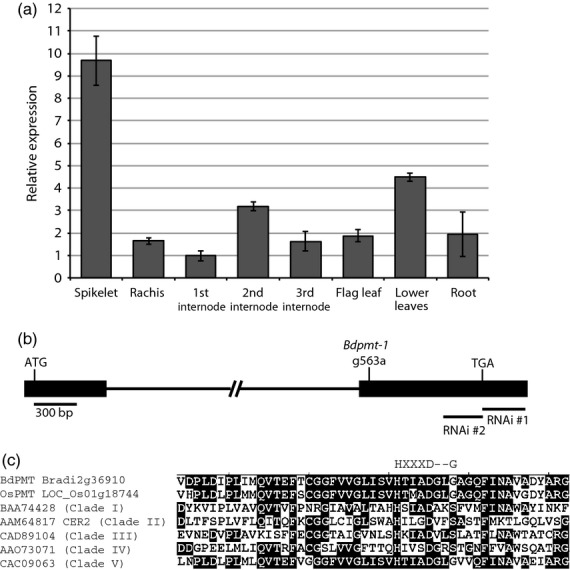
BdPMT expression levels, locations of mutations, and sequence homologies. (a) BdPMT transcript levels in WT Brachypodium tissues, as determined by qRT-PCR. Columns represent means (n = 3), bars, standard deviations. Means were normalized to the first internode mean transcript level value, which was set to one. (b) Diagram of the BdPMT gene Bradi2g36910 (drawn to scale; 300 bp scale bar). Solid rectangles represent the two exons, connected by a 4009 bp intron. Labeled are relative locations of translational start/stop codons, Bdpmt-1 transition mutation, and the RNAi constructs. (c) Alignment of BdPMT polypeptide sequences containing the HXXXD motif with sequences from BAHD acyltransferases representing each of the five clades as delineated by phylogenetic analysis (D'Auria, 2006). Consensus amino acids are boxed in black. The HXXXD motif is delineated along with the position of the G that was mutated in the Bdpmt-1 mutant allele. References for the acyltransferases (delineated by accession numbers) are in D'Auria (2006).
To study how altered BdPMT expression and function affects cell wall composition and structure as well as plant growth, we generated Brachypodium BdPMT RNA interference (RNAi) as well as overexpression (OX) lines. We also identified a Brachypodium BdPMT mutant in a sodium azide-mutagenized population generated by the Institut National de la Recherche Agronomique (INRA; Dalmais et al., 2013).
For the RNAi approach, two different hairpin constructs were generated, using BdPMT coding sequences that were chosen based on their relatively low homologies to other Brachypodium genes (Figure S1). RNAi construct #1 (BdPMT RNAi #1) consisted of a 301 base pair fragment derived from the 3′ untranslated region (UTR) of the BdPMT coding sequence, whereas BdPMT RNAi #2 consisted of a 258 base pair fragment derived from coding sequence positioned just upstream of the translational termination site (Figure2b). These constructs were transformed into embryo-derived callus using Agrobacterium tumefaciens, from which transgenic plants were regenerated on hygromycin selection media. qRT-PCR analysis was performed on transcripts that had been isolated from the stems of T1 and T2 generation plants from four independent transgenic lines, revealing average BdPMT transcript level reductions of 53 to 92%, i.e., residual levels of 47% to just 8% (Table S2).
The Brachypodium Bdpmt-1 mutant was isolated serendipitously during a Targeting Induced Local Lesions in Genomes (TILLING) screen for another gene (Bouvier d'Yvoire et al., 2013; Dalmais et al., 2013). One of the lines isolated in that screen had cell wall compositional changes that were consistent with those expected for a pmt mutant (described below). Subsequent DNA sequencing of the BdPMT gene in that line (hereafter named Bdpmt-1) revealed a nucleotide base substitution at position 563 of the Bradi2g36910 ORF (guanine replaced by adenine), resulting in codon 188 of the 450-codon ORF changed to encode aspartic acid instead of glycine (G188D). Incidentally, this base substitution disrupts a NarI restriction enzyme site, which allows one to track the mutation with a Cleaved Amplified Polymorphic Sequence (CAPS) marker (Neff et al., 2002).
The mutated codon in Bdpmt-1 is positioned three codons downstream of that encoding aspartic acid in the highly conserved HXXXD motif, which is considered the catalytic center of BAHD acyltransferases (Figure2c) (St-Pierre and De Luca, 2000; D'Auria, 2006). Altering or deleting the HXXXD motif in BAHD acyltransferases was found to greatly reduce enzyme activities of those proteins (Suzuki et al., 2003; Bayer et al., 2004; D'Auria, 2006). A scan of the amino acid sequences of 20 known and putative BAHD acyltransferases that fall into five clades based on phylogenetic analysis (D'Auria, 2006) revealed that proteins that belonged to Clade V (to which BdPMT belongs) all have a glycine residue at the position mutated in Bdpmt-1. Proteins in clades I to IV have a serine (S), glycine (G), or threonine (T) at that position (Figure2c). The amino acids S, G, and T all have small and uncharged side-chains, whereas the aspartic acid (D) that substitutes for glycine in the Bdpmt-1 mutant protein has a negatively charged and slightly larger side-chain (Betts and Russell, 2003).
As with BdPMT RNAi plants, Bdpmt-1 mutant plants grew in a similar way to wild-type (WT) and empty vector transgenic control plants grown side by side under our controlled growth chamber conditions, with no obvious morphological differences (Figures3a,b and S2).
Figure 3.
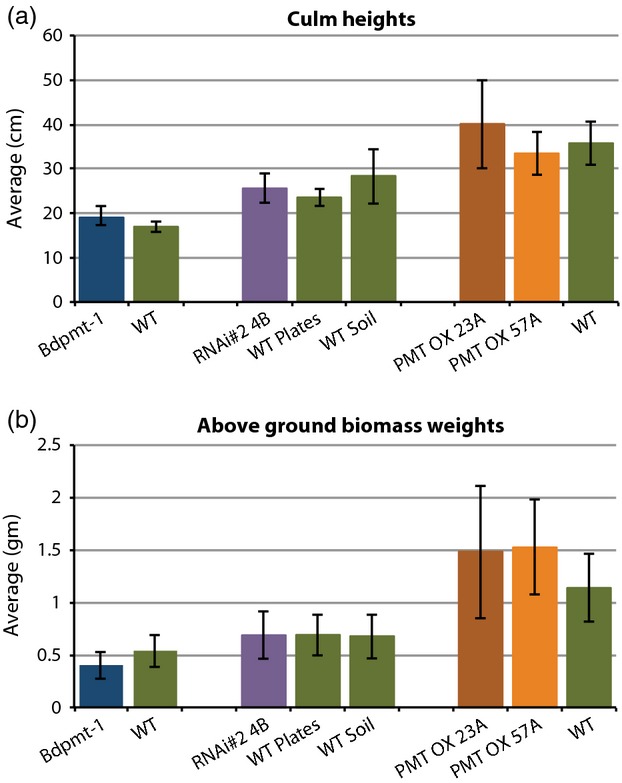
Comparisons of tallest culm heights (a) and total above-ground biomass weights (b) of senesced mutant and wild type (WT) plants grown under long days. Columns represent means ± standard deviations. Samples grouped together in the graphs represent plants grown side by side. Note that subtle growth chamber and/or epigenetic differences affect Brachypodium average plant growth, so only plants grown side by side and having similar seed histories should be compared with each other. 11 < n < 63.
To determine if cell wall biomass from the aforementioned BdPMT mutants had reduced levels of ester-linked pCA, as was our hypothesis, pulverized senesced stems were solvent-extracted then subjected to mild alkaline hydrolysis. Figure4(a) and Table S2 show that Bdpmt-1 mutant plant cell walls had nearly an 80% reduction in the amount of released pCA. Stem cell walls from all four RNAi lines also had reduced amounts of released pCA compared with WT, with the relative levels of reduction coinciding with the relative reductions in BdPMT transcript levels. For example, line 4B stems had the highest levels of reduction in pCA and BdPMT transcripts (51% and 92%, respectively) whereas line 8A stems had the lowest levels of reduction (3 and 53%, respectively). The 3% pCA reduction from line 8A plant cell walls was not significantly different from that of the WT, whereas the 14% pCA reduction from line 1A cell walls was significantly different; line 1A stems had a 58% reduction in BdPMT transcripts. These data suggest that the 53% reduction in BdPMT transcript levels represents the approximate threshold at which significant cell wall compositional changes occur.
Figure 4.
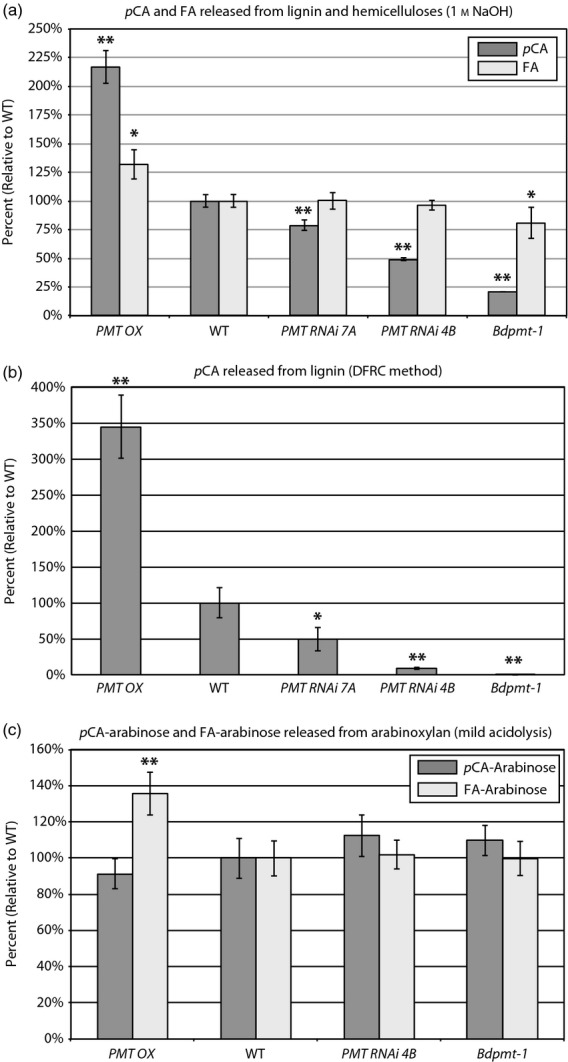
Graphs showing the amounts of pCA, FA, pCA-Arabinose, and FA-arabinose released from senesced stem biomass lignin and hemicelluloses (a), lignin only (b), or arabinoxylan only (c), using the treatments denoted in the graph titles. Columns represent mean percents relative to wild type (WT), with mean WT amounts normalized to 100%. Bars denote standard deviations. Asterisks denote statistically significant differences compared with WT plants that were grown alongside, as determined by Student's t-tests, where * represents P < 0.02 and ** represents P < 1 × 10−4. PMT OX denotes plants from transgenic BdPMT OX lines 23A, 57A, 64A, and 79A. See Tables S2 and S3 for actual values, n ≥ 3.
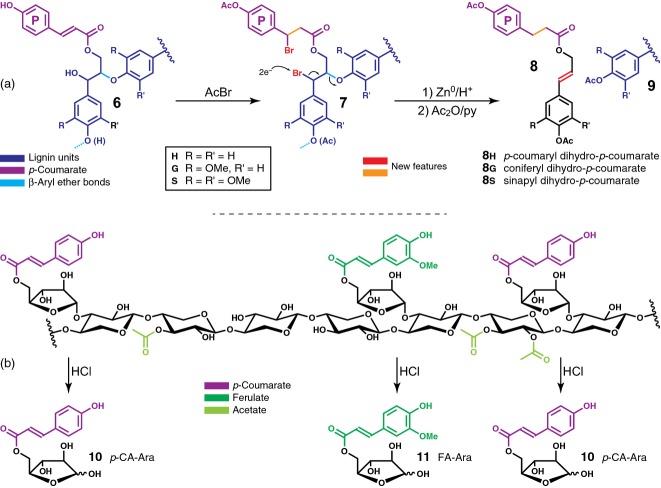
Analysis of p-coumarate levels is complicated by the fact that, unlike ferulate in Brachypodium mature stems, p-coumarate acylates both cell wall hemicelluloses (more specifically, arabinoxylans) and lignin (Mueller-Harvey et al., 1986). Although mild alkaline hydrolysis clearly detected reduced amounts of pCA liberated from alkali-treated Bdpmt-1 and BdPMT RNAi cell wall biomass compared with WT (Figure4a and Table S2), it remained unclear whether that reduction was due to a decrease of the pCA ester levels on lignin and/or on arabinoxylan. To unambiguously parse apart these possibilities, we employed two methods: (1) derivatization followed by reductive cleavage (DFRC; Lu and Ralph, 1997, 1998c, 1999), which cleaves β-aryl ether bonds in lignin while leaving ester bonds intact, thereby releasing diagnostic lignin-derived coniferyl and sinapyl dihydro-p-coumarate conjugates 8 (Figure5a); and (2) a newly developed mild acidolysis method that allows for the purification and quantification of pCA-arabinose 10 and FA-arabinose 11 ester moieties originating from arabinoxylans (Figure5b).
Figure 5.
Schematic for analytical methods used to delineate between hydroxycinnamates, p-coumarate and ferulate, on lignin versus on polysaccharides. (a) The derivatization following by reductive cleavage (DFRC) method cleaves lignin ethers while leaving the γ-esters intact, releasing quantifiable, lignin-diagnostic acylated units, the monolignol dihydro-p-coumarate ester conjugates 8. (b) Mild hydrolysis cleaves arabinose units from arabinoxylans and, at least in large part, leaves esters intact, therefore releasing 5-O-p-coumaroyl arabinose 10 and 5-O-feruloyl arabinose 11. Free p-coumaric and ferulic acids are also released and detected/quantitated; the p-coumarate may derive from either its ester on lignin or arabinoxylan, but compounds 10 and 11 are specifically derived from acylated arabinoxylan.
The DFRC method releases monolignol units and retains their γ-acylation by pCA, (conjugates 8 in Figure5a), products that are obviously exclusively from lignin. In WT Brachypodium, in which the lignin has an approximately 60:40 S:G ratio, the pCA was found on the syringyl units with no detectable amount of the pCA on guaiacyl units (Figure S3). Biomass stem tissue samples from Bdpmt-1, BdPMT RNAi line 4B and line 7A plants were found to have 0% (<0.5%), 8, and 50% residual lignin pCA levels, respectively (Figure4b and Table S3). The Bdpmt-1 mutant allele appears to be essentially a null, yielding a negligible amount of lignin-derived pCA. These data strongly suggest that BdPMT is solely responsible for placing pCA on stem lignin in Brachypodium.
Using a mild acidolysis method (Figures5b and S4), we quantified pCA-arabinose 10 and FA-arabinose 11 released from stem tissue biomass; these moieties were derived exclusively from the hemicellulose arabinoxylan. The mild acidolysis method released essentially the same amounts of pCA-arabinose and FA-arabinose from the Bdpmt-1 and BdPMT RNAi line 4B stem cell wall samples as was released from the WT samples (Figure4c and Table S3). This result signifies that another enzyme, or other enzymes, and not BdPMT, acylates arabinosyl units with pCA and FA, one of those enzymes perhaps being the Brachypodium orthologue to the rice OsAT10 enzyme (Bartley et al., 2013). Taken together with the DFRC results, these data unambiguously establish that BdPMT is responsible for the acylation of lignin with pCA in planta and is not involved in the incorporation of pCA into hemicelluloses.
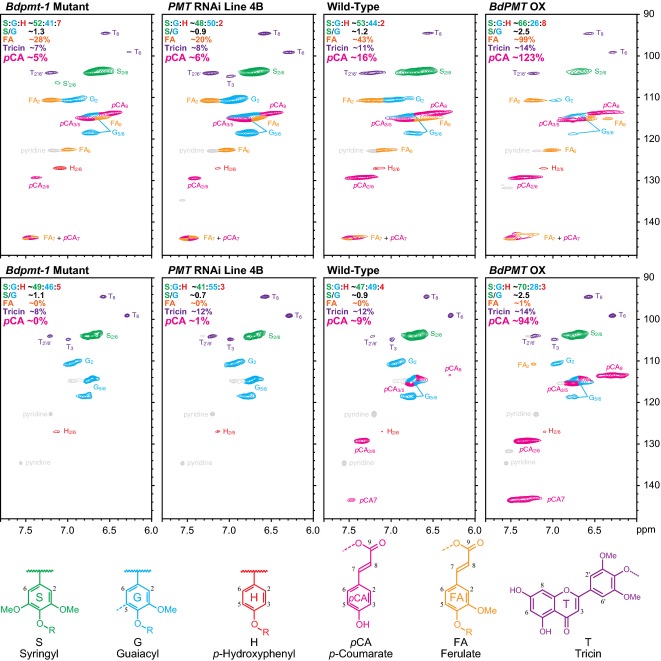
In order to obtain a more detailed picture of the cell wall compositional and structural differences between Bdpmt-1, BdPMT RNAi, and WT plants, whole cell walls of senesced stems were analyzed by gel-state 2D nuclear magnetic resonance (NMR) (Kim and Ralph, 2010). Figure6, top row, shows the spectral aromatic fingerprints obtained by this method, profiling S, G, and H lignin backbone units as well as pendent moieties, pCA, FA, and tricin, in the cell wall; the latter mobile units on the periphery of polymer chains are over-represented due to their slower spin relaxation than units in the polymer backbone. As with total saponified hydroxycinnamate measurements, these spectra, being from the whole wall material, reflect the total levels of pCA and FA in the wall, i.e., without distinction between those components on lignins versus polysaccharides.
Figure 6.
Solution-state HSQC NMR of ball-milled plant cell walls (top row) and cellulolytic enzyme lignin (CEL) treatment (bottom row) in DMSO-d6/pyridine-d5.From left to right are the Bdpmt-1 mutant, BdPMT downregulated line 4B (PMT RNAi line 4B), wild-type and BdPMT overexpressed (BdPMT OX). Sample peaks have been color-coded according to the lignin substructures below the plots. The percentages and ratios reported for each spectrum are integrals relative to total H+G+S aromatic units.
Compared with WT, and consistent with the alkaline treatment analysis results detailed above, the correlation peaks for pCA are clearly reduced in Bdpmt-1 and BdPMT RNAi line 4B, whereas those of FA appear unchanged. In addition, the peaks for syringyl units are relatively lower in line 4B and Bdpmt-1 cell walls compared with the WT, whereas those for guaiacyl units and p-hydroxyphenyl units appear relatively unchanged, denoting a relative decrease in S lignin in line 4B cell walls and a reduced S/G units ratio. This altered S/G units ratio result was corroborated by thioacidolysis data, in which we found Bdpmt-1 as well as BdPMT RNAi plant cell walls had reduced S/G units ratios compared with the WT (Table1).
Table 1.
Lignin analyses for wild-type (WT) and BdPMT-misregulated Brachypodium plants: lignin content and composition for extract-free senesced stems as determined by Klason and thioacidolysis methods
| Genotype | Lignin content (%) | Relative frequency of lignin-derived thioacidolysis monomers | ||
|---|---|---|---|---|
| % H | % G | % S | ||
| WT | 16.85 ± 0.07 | 3.5 ± 0.2 | 31.8 ± 0.0 | 64.8 ± 0.1 |
| Bdpmt-1 | 16.71 ± 1.60 | 4.6 ± 0.2 | 36.7 ± 3.4 | 58.8 ± 3.2 |
| WT | 18.53 ± 0.16 | 4.6 ± 0.1 | 42.9 ± 1.1 | 52.5 ± 1.2 |
| RNAi#2 4B | 18.48 ± 0.39 | 4.9 ± 0.1 | 45.8 ± 1.2 | 49.4 ± 1.3 |
| RNAi#2 7A | ND | 6.0 ± 0.9 | 45.7 ± 1.6 | 48.3 ± 2.1 |
| WT | 17.12 ± 0.38 | 4.3 ± 0.2 | 35.8 ± 0.7 | 59.9 ± 0.9 |
| BdPMT OX | 13.20 ± 0.26* | 3.4 ± 0.4* | 24.9 ± 2.4* | 71.8 ± 2.6* |
Values are means ± standard deviation (SD) from individually analyzed plants (n ≥ 3). Asterisks denote statistically significant differences (Student's t-test) compared with the wild type (WT) values at P < 0.0001. For the lignin content analysis (Klason method), UBIprom:GUS transgenic plants were used as WT.
The lignin-bound pCA (and FA moieties, if there were any) are more clearly delineated after enzymatic digestion of the wall polysaccharides using crude polysaccharidases (Chang et al., 1975; Wagner et al., 2007). The resulting ‘milled wood enzyme lignins’ (MWELs) are lignin-enriched (retaining essentially the entire lignin component), and are devoid of the hemicellulosic polysaccharides that are also acylated with pCA and FA, i.e., most or all of the hydroxycinnamates remaining are on the lignin component only. NMR spectra of these cellulolytic enzyme lignins (MWELs) compellingly confirm the conclusions made from examining the saponification, polysaccharide-specific, and lignin-specific analyses above. Consistent with ferulates’ solely acylating wall arabinoxylans, signals that corresponded to ferulate are absent in both WT and mutant lines, with the possible exception of a trace in the overexpression line (BdPMT OX), which bears further scrutiny. p-Coumarates remain, but at a lower level, because those acylating arabinoxylans are no longer in this fraction and only those acylating lignin remain. Comparisons with the WT material indicated that 11, and 0% residual levels (compared with the WT) remain in BdPMT RNAi line 4B and the Bdpmt-1 mutant. Such data are in line with the reduced levels in these materials determined by the lignin-specific DFRC method. The data from both analysis methods therefore compellingly demonstrate that the PMT activity is associated solely with p-coumaroylation of lignin and not arabinoxylan.
To more fully explore how PMT influences lignin biosynthetic fluxes and cell wall composition, we generated transgenic plants that overexpressed BdPMT in the form of an ENHANCED YELLOW FLUORESCENT PROTEIN (EYFP):BdPMT fusion protein, with expression driven by the maize UBIQUITIN promoter and intron; these lines are hereafter referred to as BdPMT OX. qRT-PCR analysis of T2 generation plants from two independent transformation events (initially chosen as having high expression based on EYFP fluorescence and western blot analysis. Figure S5a) revealed BdPMT transcript levels to be, on average, 35-fold or more higher than in the WT (Table S2). In our climate-controlled growth chambers, BdPMT OX plants grew in a similar manner to that of the WT and attained comparable culm heights and above-ground biomass weights (Figures3a,b and S5b,c). However, for five lines with the highest BdPMT expression, including BdPMT OX lines 57A and 67B but not 23A, a small percentage of the plants were stunted. This stunted phenotype displayed high penetrance (was commonly observed) when BdPMT OX plants were grown in a greenhouse, where growth conditions were variable and perhaps more challenging to the plants (Figure S5d).
In growth chambers as well as in the greenhouse, BdPMT OX leaf collars commonly became blackened (never observed with WT) and leaf blades grew along the stems, whereas WT leaf blades grew at about 60° angles from the stems (Figure S5e,f). The blackened collar phenotype might be due to a pathogen infection, deposition of a phenolic compound, and/or spontaneous necrosis. We note that all of the gene expression experiments and cell wall analyses described herein were performed on BdPMT OX plants that grew to be the same sizes as WT plants grown side by side in climate-controlled chambers.
Whereas BdPMT OX and WT stem cross-sections revealed no morphological differences and no collapsed vascular bundle cells (Figure7a–d), BdPMT OX stem section tissues stained considerably less with both phloroglucinol (Weisner test) and Mäule reagent compared to WT (Figure7a–d). Phloroglucinol stains cinnamaldehyde end groups of lignin, whereas Mäule reagent is used as a diagnostic of S lignin units (Lin and Dence, 1992). Consistent with these observed staining differences, Klason lignin analysis revealed that senesced BdPMT OX stems contained, on average, 23% less lignin than WT stems (Table1). This reduced lignin was corroborated by gel-state 2D NMR analysis (Figure6), and may be due to excessive BdPMT activity drawing p-coumaroyl-CoA away from monolignol biosynthesis, thereby lowering the amounts of monolignols available for polymerization. We speculate the above-noted stunted growth phenotype observed with the highest overexpressing BdPMT OX plants might be due to lignin amounts being reduced below levels necessary for normal plant growth.
Figure 7.
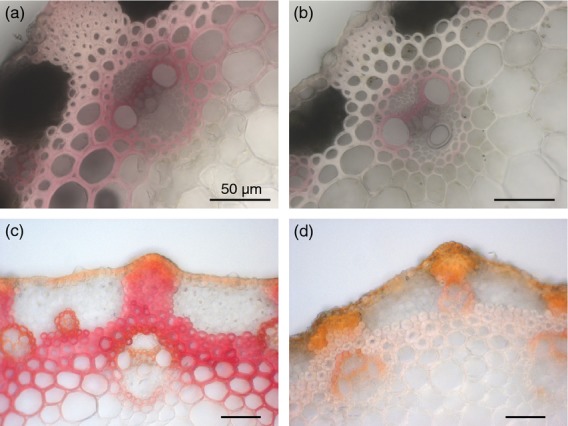
Stem section staining to visualize lignin levels and/or composition. BdPMT OX stem sections (b,d) have considerably lighter phloroglucinol (a,b) and Mäule Reagent (c,d) staining compared with wild type (WT) (a,c), which is consistent with relatively less cell wall lignin. Scale bars represent 50 μm.
Using the above-mentioned mild alkaline hydrolysis, cell wall pCA levels were found to be more than two-fold higher in BdPMT OX plants stem biomass compared with the WT (Figure4a and Table S2). As expected, the increased amounts of pCA were determined to be exclusively due to higher pCA levels on lignin (3.4-fold higher, Table S3 and Figure S3) and not on polysaccharides (Figure4b,c and Table S3), reinforcing our conclusion that BdPMT exclusively catalyzes the acylation of monolignols. With the increased level of pCA, a small but notable amount of acylation on the guaiacyl subunits was observed (Figure S3); p-coumaroylation was, however, found to favor S over G units (approximately 99:1).
FA levels were found to be as much as 40% higher in BdPMT OX stem biomass compared with WT. These higher FA levels mirrored the slightly lower FA levels detected in the Bdpmt-1 loss of function mutant (Figure4a and Table S2). At least part of the relative cell wall FA content increase can be explained as being a consequence of the reduced amounts of lignin in BdPMT OX plant stems; ferulate in the cell wall is on polysaccharides (arabinoxylans), so an increase in the ratio of arabinoxylan to lignin would lead to a relative FA level increase. It seems unlikely that these altered FA levels are due directly to altered BdPMT activity, given that in vitro studies of OsPMT substrate specificity revealed that the PMT enzyme utilizes the thioester feruloyl-CoA only poorly as a substrate (Withers et al., 2012). We cannot rule out the possibility that the expression and/or activity of a currently unknown transferase responsible for feruloylating arabinosyl units on arabinoxylans may be altered in the BdPMT mutants, or that PMT-related flux alterations in the phenylpropanoid biosynthetic pathway result in altered FA availability and/or incorporation.
To determine how the BdPMT misexpression-related cell wall alterations affect biomass digestibility, we first subjected the WT and BdPMT OX extract-free senesced stems to simple cellulolysis assays, as previously described (Berthet et al., 2011). The susceptibility of stem cell walls to enzymatic hydrolysis was roughly evaluated by the induced weight loss. Comparing BdPMT OX to WT, we observed a 33% higher weight loss upon hydrolysis with a commercial cellulase preparation containing both cellulase and hemicellulase activities (mean weight loss from BdPMT OX samples 45.6 ± 1.9, versus 34.3 ± 1.2 for WT samples; P < 0.0001; n = 5). This result was consistent with an expectation that BdPMT OX biomass would have improved digestibility owing to a significant reduction in cell wall lignin (Chapple et al., 2007; Chen and Dixon, 2007; Fu et al., 2011).
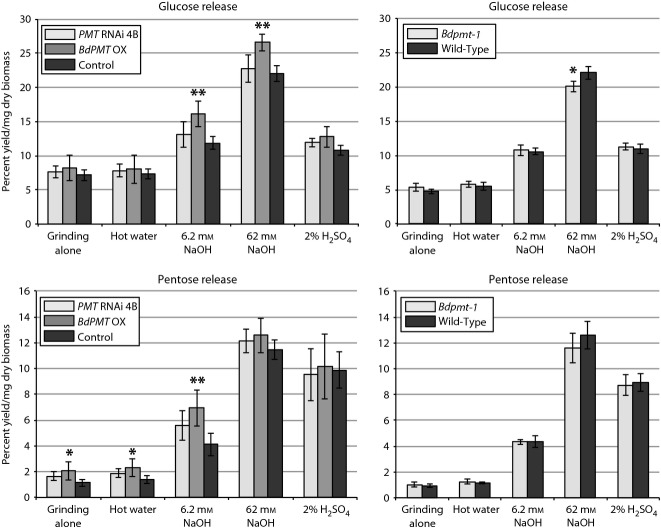
As a more encompassing evaluation of cell wall degradability, we subjected pulverized stem biomass to a range of pretreatments and enzyme hydrolysis using a high-throughput lignocelluloses digestibility assay (Santoro et al., 2010). With the exception of 62 mm NaOH, these methods were designed to be mild and incomplete so as to identify possible differences in rates of biomass digestion. We found BdPMT OX senesced stem tissue to consistently have higher cell wall polysaccharides sugar release compared with the controls, in particular with base pretreatments (Figure8; on average, 21 to 36% more glucose and 10% to 69% more pentose sugars released with 62 and 6.2 mm NaOH, respectively). By contrast, the amounts of sugars released from BdPMT RNAi and Bdpmt-1 tissues were not consistently different from controls and suggested that the abolishment of pCA on lignin has limited if any effects on stem biomass digestibility (Figure8).
Figure 8.
Digestibility of stem biomass, comparing BdPMT OX and BdPMT RNAi line 4B to transgenic control plants (UBIprom:GUS), and Bdpmt-1 to wild-type.The average amounts of glucose (top two charts; non-cell wall glucose subtracted from totals) and pentose sugars (bottom two charts) released per mg of senesced ground biomass using the various pretreatments along with enzyme hydrolysis are shown. Vertical bars represent standard deviations. Asterisks represent Student's t-test (*P < 0.03; **P < 0.005). n = five biological replicates and three technical replicates.
Conclusion
Diagnostic analytical methods provide compelling support for our contention that the PMT gene BdPMT identified in Brachypodium, a homolog of the OsPMT gene in rice, is specifically involved in the acylation of monolignols with p-coumarate. Its upregulation roughly doubles the total pCA and more than triples the pCA on lignin level. Conversely, RNAi downregulation results in pCA total levels down to as low as 50%, with approximately 10% on lignin, whereas a PMT mutant likely to be a null, with approximately 24% total pCA, has zero (or at least <0.5%) on lignin. In all cases, pCA levels on arabinoxylan remain statistically unchanged. Thus, we appear to have finally identified and authenticated the first transferase, PMT, involved in acylating monolignols. BdPMT OX plant biomass was found to have about 23% less lignin compared with WT and a higher S:G lignin ratio; BdPMT loss of function mutants had a lower S:G lignin ratio. These data together suggest that PMT integrally affects phenylpropanoid biosynthetic fluxes in grasses.
Experimental Procedures
Transgenic plant generation
Two BdPMT RNAi constructs were generated by PCR amplification of Bradi2g36910 coding sequences from Brachypodium accession Bd21-3 purified genomic DNA using the primer pairs listed in Table S4. The PCR products were restriction digested and cloned into both the BamHI–AscI and KpnI–SpeI sites of the pStarling vector (http://www.plantindustry.csiro.au/RNAi/vectors.htm). The resulting hairpin cassettes were removed as NotI fragments and ligated into the pWBVec8 binary vector backbone (Wang et al., 1998).
The BdPMT OX construct (Zea mays Ubiqitin-1 promoter:EYFP:BdPMT) was generated by performing reverse transcription PCR (RT-PCR) on Bd21-3 stem RNA using oligo(dT)18, then amplifying the 1.35 kb Bradi2g36910 ORF. The PCR product was digested with BamHI–XhoI, cloned into pENTR2B, then Gateway (Invitrogen, http://www.lifetechnologies.com/us/en/home/brands/invitrogen.html) cloned into a modified pH7WGY2 (Karimi et al., 2002) binary vector that had the 35S promoter replaced with the Zea mays Ubiquitin-1 promoter plus intron 1 from pStarling (Christensen and Quail, 1996). All constructs were sequenced to validate accuracy.
Plants harboring the BdPMT RNAi or BdPMT OX construct were regenerated from Bd21-3 embryonic callus tissue transformed using Agrobacterium tumefaciens strain AGL-1, as described by Vogel and Hill (2008); media were supplemented with 40 U ml−1 hygromycin B (Phytotechnology Laboratories, http://www.phytotechlab.com/).
Transgenic BdPMT RNAi plants were identified and confirmed by both seed selection on hygromycin and PCR amplification of RNAi construct portions from leaf genomic DNA using the ExtractNAmp Plant PCR kit (Sigma-Aldrich, http://www.sigmaaldrich.com/united-states.html) and the following primers: pStarling_F1 + pStarling_R1 and pStarling_F2 + pStarling_R2 flanking BdPMT RNAi DNA in the sense and antisense orientations, respectively.
BdPMT OX plants were identified by tracking EYFP fluorescence from the EYFP:BdPMT fusion protein using a Leica MZ8 fluorescence dissecting scope. The EYFP:BdPMT protein in these plants was found to be the expected size based on western blot analysis of stem tissue protein extracts detected with an anti-GFP antibody (1:2500 dilution, Allele Biotech catalogue # ABG-MP-MMGFP10; secondary antibody: 1:5000 goat anti-mouse IgG-HRP, Thermo-Fisher catalog # 31430, http://www.thermofisher.com/global/en/home.asp).
Seed sterilization and plant growth
Seeds were surface sterilized and plated on selective plates (1.5% agar/one half-strength Murashige and Skoog salts (Caisson Laboratories, http://www.caissonlabs.com/)/40 U ml−1 hygromycin B), stratified for 3 days (4°C, dark), then moved to a growth chamber (22°C, 16 h light) until hygromycin selection was apparent.
Plants were grown in a 50:50 mix of SunGro Rediearth and MetroMix 510 soil in 4 inch pots in growth chambers (20 h light:4 h dark photoperiod, 22°C, 50% humidity). Control plants were either WT Bd21-3 seedlings plated on non-selective plates or planted directly in soil, or hygromycin-selected plants harboring a Zea mays Ubiquitin-1 promoter with intron driving GUSPlus (Cambia Labs, http://www.cambia.org/daisy/cambialabs/home.html) in pWBVec8. The UBIprom:GUS control was used for the Klason lignin analysis and the digestibility analyses.
Quantitative RT-PCR (qRT-PCR) and RNA-seq
Total stem RNA was extracted from the top two internodes of transgenic and control plants by grinding in liquid nitrogen, followed by RNA extraction using the Plant RNeasy Extraction Kit (Qiagen Ltd, http://www.qiagen.com/) following the manufacturer's protocol. 0.5 μg of DNase I-treated (Fermentas, http://www.thermoscientificbio.com/fermentas/) RNA was reverse transcribed using oligo(dT)18 in a 20 μl MMLV (Promega, http://www.promega.com/) reaction volume.
BdPMT expression analysis was performed by qRT-PCR using a DyNAmo HS SYBR Green qPCR kit (Thermo-Fisher), cDNA samples diluted to 10 ng μl−1 and primers listed in Table S4. Thermal cycling conditions: 95°C for 6 min; 40 cycles of 95°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec; followed by a dissociation stage 95°C for 15 sec, 60°C for 30 sec, 95°C for 15 sec. PCR reactions were analyzed in triplicate by an Applied Biosystems 7300 Real-Time PCR system (http://www.lifetechnologies.com/us/en/home/brands/applied-biosystems.html). Relative expression ratios of target genes were determined by normalizing to WT samples using an efficiency calibrated model (Pfaffl, 2001; Equation 1 in Yuan et al., 2006); BdUBC18 (Bradi4g00660) or EF1α (Bradi1g06860) were used as reference genes. Primer efficiency (E), defined as 2−(1/slope), was calculated from log2 cDNA concentration versus Ct value plot slopes of serially diluted cDNA samples across treatment groups (Yuan et al., 2006).
For RNA-Seq analysis, Poly(A) RNA and libraries were quality-checked before 2 × 100 bp sequencing with an Illumina HiSeq2000 instrument (Illumina, Inc., http://www.illumina.com/). Gene expression data were computed using the CLC Genomics Workbench version 6.5. Reads were trimmed and filtered on quality with the Trim Sequences algorithm (Limit: 0.05, Maximum ambiguities: 2). RPKM was generated by aligning expressed sequence tags (ESTs) to Gene annotations using the RNA-Seq Analysis algorithm for annotated sequence. (Parameters: Similarity 0.8; Length fraction 0.9). Genome sequence and annotations were downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/bioproject/PRJNA74771). Genome sequence, annotation, and expression data were stored in an Oracle relational database and displayed using a custom web application.
Stem sectioning, phloroglucinol and Mäule reagent staining
Hand-cut stem sections from the first internode (1 cm below first node below spikelet) of plants 39 days after planting were made with a Teflon-coated razor blade (GEM, Ted Pella). Sections were stained 15 min in acid phloroglucinol (2% (w/v) phloroglucinol/20% (v/v) ethanol/2.4 m HCl), mounted on microscope slides, and then photographed using a Leica DMRBE microscope with brightfield illumination. Care was taken to stain each section the same duration of time before imaging. Mäule method analysis was performed as in Bouvier d'Yvoire et al. (2013).
Digestibility assays
Digestibility assays were performed as in Santoro et al. (2010). Ball mill-pulverized senesced culms that had spikelets and leaves at leaf collars removed were analyzed. Free glucose amounts in the undigested biomass were determined by extracting the glucose from ground biomass with distilled water and quantifying using a glucose oxidase/peroxidase method (K-GLUC, Megazyme, http://www.megazyme.com/) following manufacturer's instructions with some modifications as described (Santoro et al., 2010). Free glucose values were subtracted from the glucose values obtained after the chemical and enzymatic treatments so as to have digestibility values that reflected sugars released exclusively from cell wall polysaccharides. Free glucose values did not need to be subtracted for NaOH-treated samples, as those treatments degraded any free glucose in the ground biomass before enzyme hydrolysis. No detected glucose originated from starch, as the hydrolytic enzyme mixes contained no amyloglucosidase or α-amylase activity.
The cellulolysis protocol was performed as described in Berthet et al. (2011).
Thioacidolysis method
The thioacidolysis protocol was performed as described in Bouvier d'Yvoire et al. (2013). Lignin-derived thioacidolysis monomers were identified by gas chromatography–mass spectrometry as their trimethylsilylated derivatives.
Mild alkaline hydrolysis of extract-free senesced stems
The determination of ester-linked pCA and FA was via mild alkaline hydrolysis, following procedures published previously (Ralph et al., 1994; Bouvier d'Yvoire et al., 2013).
DFRC method to quantitate pCA specifically bound to lignin
The quantification of lignin-bound pCA was performed using the DFRC method, which is capable of cleaving lignin ethers while retaining esters (Lu and Ralph, 1997, 1998a,b, 1999).
Dried extract-free whole cell walls (20-40 mg) were treated with acetyl bromide in acetic acid (5 ml, 1:4 v/v) at 50°C for 3 h. The benzyl (α-carbon) brominated lignin solution was evaporated to dryness on a rotary evaporator (water bath at 50°C). The dry film was suspended in 2 ml absolute ethanol and the ethanol was evaporated. The dry sample was then immediately dissolved in a mixture of 1,4-dioxane:acetic acid:water (5 ml, 5:4:1 by volume), and a stir bar and zinc dust (150 mg) was added to the flask with immediate rapid stirring.
The reaction was stirred for 1 h at room temperature and then quenched with saturated ammonium chloride. The sample was spiked with 50 μg diethyl 5,5′-diferulate diacetate and the organics were extracted with dichloromethane (4 × 15 ml). The combined organic fraction was dried over sodium sulfate, filtered, and the solvent was removed under vacuum. The residue was treated with a mixture of acetic anhydride and pyridine (4 ml, 1:1 v:v) at room temperature overnight. Solvents were removed on a rotary evaporator. The crude product was loaded on a Supelco Supelclean LC-SI SPE tube (Sigma-Aldrich part #505048) with DCM (3 × 1 ml), eluted using a mixture of 1,4-dioxane:ethyl acetate (5 ml, 1:1 v:v), and concentrated to dryness. The dry film was dissolved in DCM (1 ml) and analyzed on a GC-MS (Shimadzu QP2010 Plus) using a 0.25 μm × 0.25 mm ×30 m DB-1701 (Agilent) column with He as a carrier gas (0.85 ml min−1, 10:1 split ratio).
The column temperature was held at 150°C for 1 min then increased at 20°C min−1 to 300°C and held for 24 min. The injector was set at 250°C, the ion source was set at 260°C and the detector voltage was 0.99 kV. The detector was run in selective ion monitoring (SIM) mode, acquiring three masses per target compound: coniferyl dihydro-p-coumarate diacetate (the diacetate of 8g, Figure5a) m/z: 370, 163, 131; sinapyl dihydro-p-coumarate diacetate (the diacetate of 8s, Figure5a) m/z: 400, 193, 161; diethyl-5,5′-diferulate diacetate (DEDF) m/z: 484, 442, 350. Authentic standards of 8g,8s, and DEDF were used to generate a six-point calibration curve, with the DEDF used as an internal reference.
Analysis of arabinoxylan-bound pCA and FA
Mild acidolysis was carried out on 5–10 mg of extractive-free sample using 2 ml dioxane/water 0.2 m HCl that contained 0.05 mg C21 internal standard in a Teflon-lined screwed cap tube overnight at 50°C on a carousel. Then, 2 ml water was added and samples were extracted with 3 × 4 ml EtOAc. The combined organic extracts were dried over Na2SO4 and then concentrated to approximately 0.5–1 ml. For GC-MS analyses, a 10 μl aliquot of the sample solution was silylated with 100 μl BSTFA + 10 μl pyridine for 1 h at 50°C before injection into a GC-MS system (Supelco, www.sigmaaldrich.com/Supelco) methylsilicone 15 m × 0.32 mm × 0.25 μm film thickness column, using He (1.5 ml min−1 flow rate) as the carrier gas, and with the following temperature program: from 45–180°C at 30°C min−1, then from 180–280°C at +3°C min−1. Quantitative determinations were made from ion chromatograms reconstructed at m/z (57 + 71 + 85) for the internal standard C21 (heneicosane), at (308 + 293) for pCA-TMS, at (338 + 323) for FA-TMS, and at 219 for pCA-Ara-TMS, the per-TMS ether of 5-O-p-coumaroyl arabinose 10, and at 249 for FA-Ara-TMS, the per-TMS ether of 5-O-feruloyl arabinose 11.
2D HSQC NMR analysis of cell wall components
The preparation of the extract-free whole cell walls and the MWELs was performed as described previously (Lu and Ralph, 2003; Wagner et al., 2007; Kim and Ralph, 2010)). Ball-milled, extract-free whole cell wall samples (40 mg) were prepared in a 5 mm NMR tube and suspended/swelled in dimethyl sulfoxide (DMSO)-d6/pyridine-d5 ‘100%’ (4:1, 500 μl).
The MWEL was prepared from ball-milled material (50–70 mg) as described previously (Chang et al., 1975; Wagner et al., 2007). The material was digested at room temperature with 2 mg of crude cellulases (Cellulysin; Calbiochem), in pH 5.0 acetate buffer (30 ml) on a shaker for 3 days. The samples were then centrifuged (Sorval biofuge primo; 10 016 g for 10 min), and decanted. The cellulase digestion was then repeated. After decanting the second batch of acetate buffer, the sample was rinsed with reverse osmosis water (3 × 40 ml) and the sample lyophilized. The resulting brown powder (5–10 mg) was transferred to a 5 mm NMR tube and dissolved in DMSO-d6/pyridine-d5 ‘100%’ (4:1, 500 μl).
HSQC NMR spectroscopy was performed as described previously (Kim and Ralph, 2010; Mansfield et al., 2012) on a Bruker BioSpin (Rheinstetten, Germany) Avance 700 MHz NMR spectrometer equipped with an inverse gradient 5 mm TXI 1H/13C/15N cryoprobe. The central DMSO solvent peak was used as internal reference (δc 39.5, δH 2.49 ppm). Peak assignments for syringyl (S), guaiacyl (G), and p-hydroxyphenyl (H) lignin units, as well as p-coumarate (pCA), ferulate (FA), and tricin (T), were made by comparison with previously assigned spectra (Lu and Ralph, 2003; Marita et al., 2003; Kim et al., 2008; Kim and Ralph, 2010; Ralph and Landucci, 2010; Mansfield et al., 2012; del Rio et al., 2012; del Río et al., 2012; Rencoret et al., 2013).
Conflicts of interest
The authors have no conflict of interest to declare.
Acknowledgments
We thank Frédéric Legée for performing the Klason lignin analyses, Cliff Foster for performing thioacidolysis analyses, Hoon Kim for his help with gel-NMR methods, Nick Thrower for help processing RNA-Seq datasets, and Stephen Lutgen, Heather Welch, and Michael Krzyskowski for prepping tissue samples. We thank Marek Mutwil and Staffan Persson for providing access to AraNet Brachypodium co-expression tools before their publication. This work was supported by the Department of Energy's Great Lakes Bioenergy Research Center (Department of Energy, Biological and Environmental Research, Office of Science grant no. DE–FC02–07ER64494).
Supporting Information
Clustal W alignments of the nucleotide sequences comprising BdPMT RNAi constructs.
Figure S2. Time course growth measurements of Bdpmt-1 (squares) and WT (circles) plants in soil.
Figure S3. Partial GC-SIM traces of the diacetylated monolignol conjugates generated by DFRC treatment of extractive-free senesced stems from (top to bottom) BdPMT overexpressed (BdPMT OX), wild-type (WT), BdPMT downregulated line 7A (PMT RNAi line 7A), BdPMT downregulated line 4B (PMT RNAi line 4B), and Bdpmt-1 mutant.
Figure S4. Partial GC-MS traces of phenolics recovered by mild acidolysis of extractive-free senesced stems from wild-type (WT) and BdPMT misregulated lines (Bdpmt-1 mutant and BdPMT overexpressor BdPMT OX).
Figure S5. Western blot analysis of BdPMT OX expression, and growth phenotypes of BdPMT OX plants.
Classes of genes predicted by PlaNet to be co-expressed with Bradi2g36910.1 (BdPMT).
Table S2. Impact of misregulating the BdPMT gene on BdPMT transcript levels and on the amount of p-coumaric acid (pCA) and ferulic acid (FA) released by mild alkaline hydrolysis.
Table S3. Amount of DFRC-released p-coumaric acid (pCA) and amounts of pCA, ferulic acid (FA), p-coumaroylated arabinose (pCA-Ara) and feruloylated arabinose (FA-Ara) released by mild acidolysis of extract-free senesced stems.
Table S4. Primer sequences and qRT-PCR amplification efficiencies.
References
- Bartley LE, Peck ML, Kim SR, et al. Overexpression of a BAHD acyltransferase, OsAt10, alters rice cell wall hydroxycinnamic acid content and saccharification. Plant Physiol. 2013;161:1615–1633. doi: 10.1104/pp.112.208694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A, Ma XY, Stockigt J. Acetyltransfer in natural product biosynthesis - functional cloning and molecular analysis of vinorine synthase. Bioorg. Med. Chem. 2004;12:2787–2795. doi: 10.1016/j.bmc.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for Geneticists. Chichester, UK: John Wiley and Sons; 2003. pp. 289–316. [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Ann. Rev. Plant Biol. 2003;54:519–549. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Bouvier d'Yvoire M, Bouchabke-Coussa O, Voorend W, et al. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 2013;73:496–508. doi: 10.1111/tpj.12053. [DOI] [PubMed] [Google Scholar]
- Carroll A, Somerville CR. Cellulosic biofuels. Annu. Rev. Plant Biol. 2009;160:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- Chang H-M, Cowling EB, Brown W, Adler E, Miksche G. Comparative studies on cellulolytic enzyme lignin and milled wood lignin of sweetgum and spruce. Holzforschung. 1975;29:153–159. [Google Scholar]
- Chapple C, Ladisch M, Meilan R. Loosening lignin's grip on biofuel production. Nat. Biotechnol. 2007;25:746–748. doi: 10.1038/nbt0707-746. [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Dalmais M, Antelme S, Ho-Yue-Kuang S, et al. A TILLING platform for functional genomics in Brachypodium distachyon. PLoS ONE. 2013;8:e65503. doi: 10.1371/journal.pone.0065503. (65501-65510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl Acad. Sci. USA. 2011;108:3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington MJ, Mutwil M, Barriere Y, Sibout R. Molecular biology of lignification in grasses. Adv. Bot. Res. 2012;61:77–112. [Google Scholar]
- Hatfield RD, Ralph J, Grabber JH. A potential role of sinapyl p-coumarate as a radical transfer mechanism in grass lignin formation. Planta. 2008;228:919–928. doi: 10.1007/s00425-008-0791-4. [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Marita JM, Frost K, Grabber JH, Lu F, Kim H, Ralph J. Grass lignin acylation: p-coumaroyl transferase activity and cell wall characteristics of C3 and C4 grasses. Planta. 2009;229:1253–1267. doi: 10.1007/s00425-009-0900-z. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim H, Ralph J. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org. Biomol. Chem. 2010;8:576–591. doi: 10.1039/b916070a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J, Akiyama T. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6. Bioenergy Res. 2008;1:56–66. [Google Scholar]
- Lapierre C. Determining lignin structure by chemical degradations. In: Heitner C, Dimmel D, Schmidt JA, editors. Lignin and Lignans–Advances in Chemistry. Boca Raton, FL: CRC Press/Taylor & Francis Group; 2010. pp. 11–48. [Google Scholar]
- Lin SY, Dence CW. Methods in Lignin Chemistry. Heidelberg: Springer-Verlag; 1992. [Google Scholar]
- Lu F, Ralph J. The DFRC method for lignin analysis. Part 1. A new method for β-aryl ether cleavage: lignin model studies. J. Agric. Food Chem. 1997;45:4655–4660. [Google Scholar]
- Lu F, Ralph J. The DFRC method for lignin analysis. Part 3. NMR studies. J. Wood Chem. Technol. 1998a;18:219–233. [Google Scholar]
- Lu F, Ralph J. The DFRC method for lignin analysis. Part 2. Monomers from isolated lignins. J. Agric. Food Chem. 1998b;46:547–552. doi: 10.1021/jf970676m. [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. Efficient ether cleavage in lignins: the “DFRC” method as a basis for new analytical methods. In: Lewis NG, Sarkanen S, editors. Lignin and Lignan Biosynthesis. Washington, DC: American Chemical Society; 1998c. pp. 294–322. [Google Scholar]
- Lu F, Ralph J. Detection and determination of p-coumaroylated units in lignins. J. Agric. Food Chem. 1999;47:1988–1992. doi: 10.1021/jf981140j. [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. Preliminary evidence for sinapyl acetate as a lignin monomer in kenaf. Chem. Commun. 2002:90–91. doi: 10.1039/b109876d. [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. Non-degradative dissolution and acetylation of ball-milled plant cell walls; high-resolution solution-state NMR. Plant J. 2003;35:535–544. doi: 10.1046/j.1365-313x.2003.01817.x. [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. Novel tetrahydrofuran structures derived from β–β-coupling reactions involving sinapyl acetate in Kenaf lignins. Org. Biomol. Chem. 2008;6:3681–3694. doi: 10.1039/b809464k. [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J. Whole plant cell wall characterization using solution-state 2D-NMR. Nat. Protoc. 2012;7:1579–1589. doi: 10.1038/nprot.2012.064. [DOI] [PubMed] [Google Scholar]
- Marita JM, Vermerris W, Ralph J, Hatfield RD. Variations in the cell wall composition of maize brown midrib mutants. J. Agric. Food Chem. 2003;51:1313–1321. doi: 10.1021/jf0260592. [DOI] [PubMed] [Google Scholar]
- Molinari HB, Pellny TK, Freeman J, Shewry PR, Mitchell RA. Grass cell wall feruloylation: distribution of bound ferulate and candidate gene expression in Brachypodium distachyon. Front. Plant Sci. 2013;4:50. doi: 10.3389/fpls.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Harvey I, Hartley RD, Harris PJ, Curzon EH. Linkage of p-coumaryl and feruloyl groups to cell wall polysaccharides of barley straw. Carbohydr. Res. 1986;148:71–85. [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- Perlack RD, Wright LL, Turhollow AF, Graham RL, Stokes BJ, Erbach DC. Biomass as feedstock for a biomass and bioproducts industry: the technical feasibility of a 1 billion ton annual feedstock supply. ORNL/TM-2005/66. Oak Ridge, TN: Oak Ridge Natl. Lab./US DOE/USDA; 2005. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2004–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J. Hydroxycinnamates in lignification. Phytochem. Rev. 2010;9:65–83. [Google Scholar]
- Ralph J, Landucci LL. NMR of Lignins. In: Heitner C, Dimmel DR, Schmidt JA, editors. Lignin and Lignans; Advances in Chemistry. Boca Raton, FL: CRC Press (Taylor & Francis Group); 2010. pp. 137–234. [Google Scholar]
- Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H-JG. Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J. Am. Chem. Soc. 1994;116:9448–9456. [Google Scholar]
- Ralph J, Grabber JH, Hatfield RD. Lignin-ferulate crosslinks in grasses: active incorporation of ferulate polysaccharide esters into ryegrass lignins. Carbohydr. Res. 1995;275:167–178. [Google Scholar]
- Ralph J, Bunzel M, Marita JM, Hatfield RD, Lu F, Kim H, Schatz PF, Grabber JH, Steinhart H. Peroxidase-dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem. Revs. 2004;3:79–96. [Google Scholar]
- Rencoret J, Ralph J, Marques G, Gutiérrez A, Martínez ÁT, del Rio JC. Structural characterization of the lignin from coconut (Cocos nucifera) coir fibers. J. Agric. Food Chem. 2013;61:2434–2445. doi: 10.1021/jf304686x. [DOI] [PubMed] [Google Scholar]
- del Rio JC, Rencoret J, Prinsen P, Martínez ÁT, Ralph J, Gutiérrez A. Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 2012;60:5922–5935. doi: 10.1021/jf301002n. [DOI] [PubMed] [Google Scholar]
- del Río JC, Prinsen P, Rencoret J, Nieto L, Jiménez-Barbero J, Ralph J, Martínez ÁT, Gutiérrez A. Structural characterization of the lignin in the cortex and pith of elephant grass (Pennisetum purpureum) stems. J. Agric. Food Chem. 2012;60:3619–3634. doi: 10.1021/jf300099g. [DOI] [PubMed] [Google Scholar]
- Santoro N, Cantu SL, Tornqvist CE, Falbel TG, Bolivar JL, Patterson SE, Pauly M, Walton JD. A high-throughput platform for screening milligram quantities of plant biomass for lignocellulose digestibility. Bioenergy Res. 2010;3:93–102. [Google Scholar]
- St-Pierre B, De Luca V. Evolution of acyltransferase genes: origin and diversification fo the BAHD superfamily of acyltransferases involved in secondary metabolism. In: Romeo JT, Ibrahim R, Varin L, De Luca V, editors. Recent Advances in Phytochemistry. Evolution of Metabolic Pathways. Amsterdam: Elsevier; 2000. pp. 285–315. Vol. 34. [Google Scholar]
- Suzuki H, Nakayama T, Nishino T. Proposed mechanism and functional amino acid residues of malonyl-CoA: anthocyanin 5-O-glucoside-6’’’-O-malonyltransferase from flowers of Salvia splendens, a member of the versatile plant acyltransferase family. Biochemistry. 2003;42:1764–1771. doi: 10.1021/bi020618g. [DOI] [PubMed] [Google Scholar]
- Takahama U, Oniki T, Shimokawa H. A possible mechanism for the oxidation of sinapyl alcohol by peroxidase-dependent reactions in the apoplast: enhancement of the oxidation by hydroxycinnamic acids and components of the apoplast. Plant Cell Physiol. 1996;37:499–504. [Google Scholar]
- Perlack RD, Stokes BJ, editors. U.S. Department of Energy. U.S. Billion-Ton Update: Biomass Supply for a Bioenergy and Bioproducts Industry. Oak Ridge, TN: Oak Ridge National Laboratory; 2011. p. 227. (Leads)). ORNL/TM-2011/224. [Google Scholar]
- Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 2008;27:471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D. EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor. Appl. Genet. 2006;113:186–195. doi: 10.1007/s00122-006-0285-3. [DOI] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr KM, Nanayakkara B, Te Kiri L. Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA: shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc. Natl Acad. Sci. 2007;104:11856–11861. doi: 10.1073/pnas.0701428104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M-B, Li Z-Y, Matthews P, Upadhyaya N, Waterhouse P. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic. 1998;461:401–407. [Google Scholar]
- Withers S, Lu F, Kim H, Zhu Y, Ralph J, Wilkerson CG. Identification of a grass-specific enzyme that acylates monolignols with p-coumarate. J. Biol. Chem. 2012;287:8347–8355. doi: 10.1074/jbc.M111.284497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinforma. 2006;7:85–97. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Lu F, Sun R, Ralph J. Ferulate-coniferyl alcohol cross-coupled products formed by radical coupling reactions. Planta. 2009;229:1099–1108. doi: 10.1007/s00425-009-0894-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clustal W alignments of the nucleotide sequences comprising BdPMT RNAi constructs.
Figure S2. Time course growth measurements of Bdpmt-1 (squares) and WT (circles) plants in soil.
Figure S3. Partial GC-SIM traces of the diacetylated monolignol conjugates generated by DFRC treatment of extractive-free senesced stems from (top to bottom) BdPMT overexpressed (BdPMT OX), wild-type (WT), BdPMT downregulated line 7A (PMT RNAi line 7A), BdPMT downregulated line 4B (PMT RNAi line 4B), and Bdpmt-1 mutant.
Figure S4. Partial GC-MS traces of phenolics recovered by mild acidolysis of extractive-free senesced stems from wild-type (WT) and BdPMT misregulated lines (Bdpmt-1 mutant and BdPMT overexpressor BdPMT OX).
Figure S5. Western blot analysis of BdPMT OX expression, and growth phenotypes of BdPMT OX plants.
Classes of genes predicted by PlaNet to be co-expressed with Bradi2g36910.1 (BdPMT).
Table S2. Impact of misregulating the BdPMT gene on BdPMT transcript levels and on the amount of p-coumaric acid (pCA) and ferulic acid (FA) released by mild alkaline hydrolysis.
Table S3. Amount of DFRC-released p-coumaric acid (pCA) and amounts of pCA, ferulic acid (FA), p-coumaroylated arabinose (pCA-Ara) and feruloylated arabinose (FA-Ara) released by mild acidolysis of extract-free senesced stems.
Table S4. Primer sequences and qRT-PCR amplification efficiencies.