Abstract
Spines are highly motile protrusions emerging from the dendritic shafts of neurons. The dynamics of these post-synaptic structures are ruled by actin filament turnover. However, our understanding of the mechanisms of actin polymerization in dendritic spines is quite ambiguous. A recent study by the Giannone laboratory (Chazeau et al, 2014) is now shedding some light on the peculiar features of actin polymerization in dendritic spines, which are distinct from the known canonical mechanisms.
See also: A Chazeau et al (December 2014)
Cell biologists live with dogmas, and one of the best kept ones is that actin polymerization in a cell should always occur from the plasma membrane where the fast incorporation of actin monomers into newly formed short filaments generates the force and meshwork to push out and stabilize the membrane. Understanding the mechanisms that regulate this process is fundamental for life since virtually any kind of cell motility relies on directed actin polymerization. As odd as it may sound, even the way we feel, what we memorize and what we might forget is ultimately controlled by actin polymerization in the synapses of neurons.
The complexity of actin polymerization in vivo is staggering as the collective work over more than 30 years has shown, and still controversial discussions are ongoing on the role of the participating protein complexes and actin-binding proteins, as well as the spatial organization of actin filaments. Without entering the disputable matter, it is generally accepted in the field that in areas of actin polymerization, such as the lamellipodia, nucleation promoting factors (NPFs) are activated at the plasma membrane (Lai et al, 2008), as well as actin nucleators such as the Arp2/3 complex, formins, or WH2 domain-containing nucleators (Campellone & Welch, 2010), which then promote de novo formation of actin filaments and incorporation of monomers into the growing filaments in order to exert protruding force (Rottner et al, 1999).
So far, so good.
What we tend to neglect though is that our current view on the spatial organization of actin polymerization in cells is biased by what cultured cells can actually perform on a two-dimensional stiff substrate. These conditions promote the formation of highly esthetic structures such as filopodia, lamellipodia, crowns—just to name some of the most studied. The beauty of this system is that it allows excellent imaging to dissect the spatiotemporal sequence of actin polymerization and to define the underlying structural filament meshwork. This has collectively added to our current picture of membrane-linked actin polymerization and the concept of retrograde F-actin flow from the cell membrane toward the cell center. This mechanism is unquestionably true for most membrane protrusions observed in cultured cells; however, it has remained unclear whether it also applies to actin dynamics in tissue-constrained cells and extremely small membrane compartments such as the dendritic spines.
In neurons, the post-synaptic terminals of most excitatory synapses consist of dendritic spines. They are tiny and highly motile membrane protrusions, emerging from dendritic shafts and representing a major entry point of neurotransmission in the brain. It has been long recognized that the predominant cytoskeletal content in spines is actin (Fifková & Delay, 1982), which has a dual function in shaping the spine and sustaining its motility during synaptic development and synaptic plasticity. Pathological alterations in spine shape have been linked to a growing number of disorders such as autism and mental retardation, moving actin dynamics in the focus of attention. The importance of actin dynamics in spines is further highlighted by the observation that spine size and shape are intimately linked to synaptic plasticity, LTP, and LTD.
So what is particular about actin in spines and why is an answer to this question not straightforward?
The main reason for the uncertainty concerning actin filament structure and the mechanisms of actin polymerization as well as the discussion whether retrograde F-actin flow also occurs in spines is due to the very small size of spines, having a variable diameter of about 300–600 nm. This is practically below the physical resolution limit of standard fluorescence microscopy, including confocal microscopy. Even EM studies turned out to be difficult. To follow single actin filaments and determine their polarity in the intricate dense network that fills such a small structure is technically challenging. Whether the actin filaments in spines are branched, linear, or a mixture of both is still debated. Recent experiments on cultured neurons support the view of a mixed filament structure as the most likely scenario (Korobova & Svitkina, 2010). However, it is important to keep in mind that currently there is no conclusive picture for the high-resolution structure of the actin cytoskeleton in a spine embedded in the tissue context.
Owing to the recent advances in imaging technologies, such as two-photon and super-resolution fluorescence microscopy, we have gained valuable insight into the cytoskeletal dynamics and organization of dendritic spines (Frost et al, 2010; Murakoshi et al, 2011; Bosch et al, 2014). In particular, super-resolution microscopy techniques, in the forms of photoactivation localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM), or stimulated emission depletion (STED), allowed to push the resolution limits to about 50–70 nm. This now opens the possibility to explore nanodomains such as dendritic spines, both fixed and live, in cultured neurons with unprecedented topographic precision.
This is the entry point of the article from Giannone and co-workers published in this issue of The EMBO Journal (Chazeau et al, 2014). Specifically, the Giannone laboratory has employed super-resolution fluorescence microscopy to show that Arp2/3-dependent actin polymerization in spines fundamentally differs from the canonical mechanism seen in lamellipodia. The authors studied the spatial organization and the dynamics of the Arp2/3 complex and its nucleation promoting factor, the WAVE complex, as well as nucleating formins, within dendritic spines of cultured hippocampal neurons.
Two major novel findings in this work substantially advance our understanding of actin dynamics in the spine and extend the current view of retrograde F-actin flow as a universal mechanism. Using single protein tracking photoactivation localization microscopy (sptPALM) allowed the authors to distinguish two sub-spine domains with different actin dynamics characteristics: (i) a first domain consisting of finger-like protrusions (peri-synaptic protrusions), emerging from the side of the post-synaptic density (PSD), which are driven by formin-dependent fast actin polymerization at their tips, similar to classical filopodia, and (ii) a second domain within the core of the spine, where actin polymerization is driven by Arp2/3 nucleation, from scattered nanodomains around the PSD scaffold—different from the classical Arp2/3 function at the membrane of a lamellipodium (see Fig1). This Arp2/3-dependent actin nucleation in spines produced only a slow and non-polarized actin subunit flow, suggesting that around the PSD, actin nucleation is dissociated from elongation and not directly involved in peri-synaptic protrusion and spine motility.
Figure 1. Comparison of actin nucleation and elongation mechanisms in the dendritic spine and the lamellipodium.
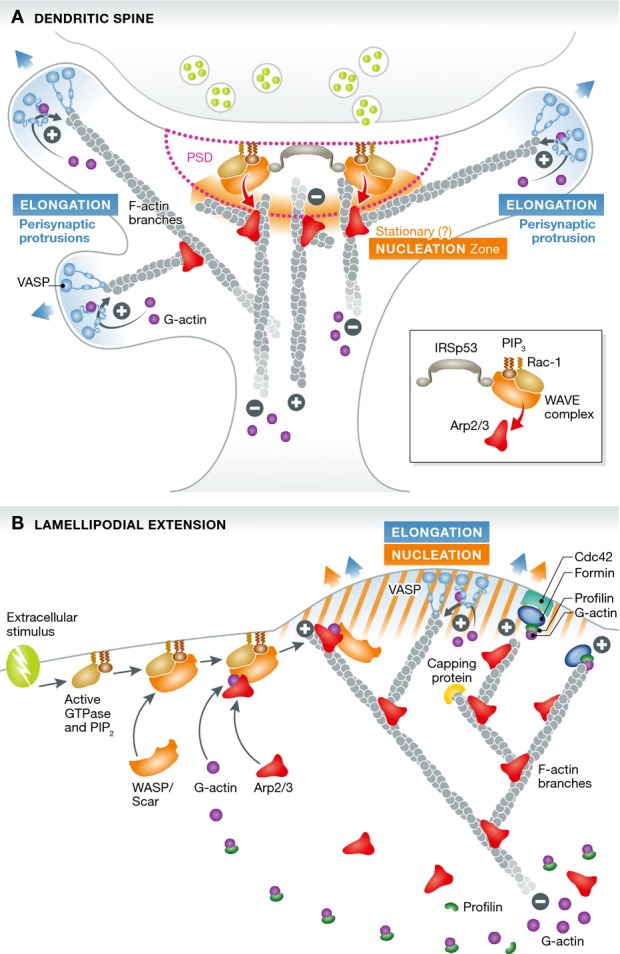
(A) In spines, the post-synaptic density functions as a hub for actin nucleation, concentrating signaling and regulatory factors (IRSp53, Rac1, WAVE complex) in order to activate the Arp2/3 complex in response to extra-synaptic signaling, recruiting it from a freely diffusible pool. Actin polymerization then proceeds in a slow and unpolarized fashion. Spine motility is ensured by protrusions stemming possibly by branched filaments on the sides of the PSD (peri-synaptic) in which formin-dependent elongation mechanisms at their tips and on the membrane generate a fast and polarized rear-flow of actin and exert the pushing force (from Chazeau et al). (B) In lamellipodia, both actin nucleation and rapid VASP- and formin-dependent elongation occur in synchrony at the membrane, where the pushing force is needed to extrude the lamellipodium, generating a fast and polarized rear-flow of actin and a complex branched actin network.
These findings support the interesting hypothesis that the PSD actually serves as a hub controlling actin nucleation and spine motility and structure (see also Bosch et al, 2014). According to this model, spine motility would depend on the elongation dynamics of the finger-like protrusions, which emerge from the Arp2/3-nucleated pool at the core of the spine. Importantly, with the same imaging approach, Giannone and co-workers showed that the WAVE complex (shown for three of the five subunits, Abi1, Nap and WAVE1), a known activator of the Arp2/3 complex, perfectly aligned with the PSD, which is in agreement with earlier biochemical findings in isolated synaptoneurosomes where the WAVE complex was only found tightly associated with the PSD (Pilo Boyl et al, 2007).
In summary, the work by Giannone and co-workers provides good evidence that actin nucleation in spines can also occur from the PSD and not only from the membrane. The WAVE complex as well as the BAR-domain-containing receptor IRSp53 remained bound to the PSD, even when all filamentous actin is depolymerized by latrunculin B treatment.
The implications of such an organization of actin polymerization are intriguing: It appears that the membrane juxtaposed to the PSD allows it to integrate extracellular signals via adhesion molecules, such as neuroligins, and neurotransmitter/receptor signaling cascades, such as the AMPA/NMDA-CaMKII pathway, and to translate them to actin nucleation, thereby coordinating spine motility and morphology in a highly synchronized way (Park et al, 2012; Chen et al, 2014).
The findings by Giannone and co-workers obviously stimulate a tail of interesting further questions. For example, it still needs clarification what type of actin network is actually formed in the spine. Since both branched and filamentous bundles have been observed in ultrastructural studies, it would be interesting to understand how the two different structures are coordinated at the molecular level and how filaments with different polarity are generated. One important aspect, which relates to this question and has not been addressed in the presented work, is the role of actin filament disassembly in spines by key regulators such as cofilin/ADF (Bosch et al, 2014). Another focus for future work should be the mechanisms of how the Arp2/3 and WAVE complex, as well as the formins, respond to physiological synaptic stimulation and how this translates into controlled actin polymerization.
In the best of all possible worlds, we would like to be able to study the actin cytoskeleton on a nanoscale level in vivo in the tissue context, combined with physiological cell stimulation. Given the rapidly evolving technical advances in microscopy over the past years, we can be considerably confident that this will not remain a dream.
Acknowledgments
PPB & WW are supported by Deutsche Forschungsgemeinschaft (DFG), CRC initiative 1089, SPP 1464, and the Bonner Forum Biomedizin (BFB).
References
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82:444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazeau A, Mehidi A, Nair D, Gautier JJ, Leduc C, Chamma I, Kage F, Kechkar A, Thoumine O, Rottner K, Choquet D, Gautreau A, Sibarita J-P, Giannone G. Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 2014;33:2745–2764. doi: 10.15252/embj.201488837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifková E, Delay RJ. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J Cell Biol. 1982;95:345–350. doi: 10.1083/jcb.95.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai FPL, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TEB, Dunn GA, Small JV, Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 2008;27:982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Chi S, Park D. Activity-dependent modulation of the interaction between CaMKIIα and Abi1 and its involvement in spine maturation. J Neurosci. 2012;32:13177–13188. doi: 10.1523/JNEUROSCI.2257-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo Boyl P, Di Nardo A, Mulle C, Sassoè-Pognetto M, Panzanelli P, Mele A, Kneussel M, Costantini V, Perlas E, Massimi M, Vara H, Giustetto M, Witke W. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]


