Abstract
The exosome is a conserved multi-subunit ribonuclease complex that functions in 3′ end processing, turnover and surveillance of nuclear and cytoplasmic RNAs. In the yeast nucleus, the 10-subunit core complex of the exosome (Exo-10) physically and functionally interacts with the Rrp6 exoribonuclease and its associated cofactor Rrp47, the helicase Mtr4 and Mpp6. Here, we show that binding of Mtr4 to Exo-10 in vitro is dependent upon both Rrp6 and Rrp47, whereas Mpp6 binds directly and independently of other cofactors. Crystallographic analyses reveal that the N-terminal domains of Rrp6 and Rrp47 form a highly intertwined structural unit. Rrp6 and Rrp47 synergize to create a composite and conserved surface groove that binds the N-terminus of Mtr4. Mutation of conserved residues within Rrp6 and Mtr4 at the structural interface disrupts their interaction and inhibits growth of strains expressing a C-terminal GFP fusion of Mtr4. These studies provide detailed structural insight into the interaction between the Rrp6–Rrp47 complex and Mtr4, revealing an important link between Mtr4 and the core exosome.
Keywords: nuclear exosome, RNA degradation, X-ray crystallography, yeast genetics
Introduction
Most cellular ribonucleic acids are transcribed in the nucleus as larger precursor molecules that are then processed to produce mature, functional RNAs. The maturation of ribosomal RNAs (rRNAs), small nuclear RNAs and nucleolar RNAs (snRNAs and snoRNAs) requires the trimming of the extended 3′ end of their nascent transcripts and the elimination of the excised RNA fragments (reviewed in Bernstein & Toth, 2012). These processes involve a complex of ribonucleases known as the RNA exosome (Allmang et al, 1999a). In addition to 3′ end processing, the exosome functions in RNA turnover and surveillance pathways (reviewed in Schmid & Jensen, 2008; Schaeffer et al, 2011; Chlebowski et al, 2013). In the nucleus, it mediates the turnover of precursor transfer RNAs (pre-tRNAs) and precursor messenger RNAs (pre-mRNAs) (Gudipati et al, 2012). The nuclear exosome also swiftly eliminates misprocessed tRNAs (Kadaba et al, 2004) and pre-mRNAs (Bousquet-Antonelli et al, 2000; Hilleren et al, 2001) as well as cryptic unstable transcripts (CUTs) generated by antisense and intergenic transcription (Wyers et al, 2005; Davis & Ares, 2006; Neil et al, 2009). In the cytoplasm, the exosome participates in the turnover of mature mRNAs (Anderson & Parker, 1998) and in quality-control pathways that eliminate defective mRNAs with premature stop codons (Mitchell & Tollervey, 2003) or without a stop codon (van Hoof et al, 2002).
The exosome core complex consists of ten subunits that are evolutionarily conserved and are essential in yeast (Allmang et al, 1999b). Nine of these subunits form a catalytically inactive barrel-like structure (Exo-9) (Liu et al, 2006; Dziembowski et al, 2007) that threads RNA substrates to the tenth subunit, Rrp44 (also known as Dis3) (Bonneau et al, 2009; Malet et al, 2010; Wasmuth & Lima, 2012; Makino et al, 2013a; Liu et al, 2014). Rrp44 is bound at the bottom of the Exo-9 barrel and contains a processive 3′–5′-exoribonuclease site and an endonuclease site (reviewed in Schneider & Tollervey, 2013; Makino et al, 2013b). While the Exo-10 core is found in both the nucleus and the cytoplasm, physical and genetic interactions have linked the exosome core to several cofactors that have specific subcellular localization (reviewed in Schneider & Tollervey, 2013). In the cytoplasm, the exosome functions together with the Ski complex, a multi-subunit assembly centered at the helicase Ski2 (Anderson & Parker, 1998; Araki et al, 2001; Halbach et al, 2013). In the yeast nucleus, the exosome is associated with a set of conserved proteins that include Rrp6 (known as PM/Scl-100 in humans), Rrp47 (also known as Lrp1 in yeast and as C1D in humans), Mpp6 and Mtr4 (also known as Dob1 in yeast) (reviewed in Butler & Mitchell, 2011).
Rrp6 contains a 3′–5′-exoribonuclease site. In contrast to Rrp44, the Rrp6 nuclease functions in a distributive manner and stalls when encountering structured RNA sequences (Briggs et al, 1998; Burkard & Butler, 2000; Liu et al, 2006; Januszyk et al, 2011). Rrp6 binds the exosome directly, near the top of the Exo-9 barrel (Cristodero et al, 2008; Makino et al, 2013a; Wasmuth et al, 2014). Although in vitro Rrp6 and Rrp44 can bind Exo-9 independently of each other, Exo-9 binding interconnects the enzymatic properties of the two ribonucleases (Liu et al, 2006; Wasmuth & Lima, 2012). The interplay between Rrp6 and Rrp44 also emerges from in vivo studies. During the maturation of 5.8S rRNA, Rrp44 degrades the 3′ end of the precursor to leave a processing intermediate that is then trimmed to the final product by Rrp6. This intermediate features a 3′ extension of 30 nucleotides (Briggs et al, 1998), a length that corresponds to the size of the internal channel of Exo-10 (Bonneau et al, 2009; Makino et al, 2013a). Rrp6 can carry out the last processing step even when separated from Exo-10 (Callahan & Butler, 2008).
Rrp47 and Rrp6 interact in vitro (Stead et al, 2007) and in vivo (Mitchell et al, 2003; Synowsky et al, 2009). Consistently, depletion of Rrp47 leads to defects in RNA processing and degradation that are similar to those observed in rrp6Δ strains (Mitchell et al, 2003; Peng et al, 2003). The presence of Rrp6 protects Rrp47 from degradation in yeast and conversely Rrp47 stabilizes Rrp6 (Feigenbutz et al, 2013a,b; Stuparevic et al, 2013). Knockout of either Rrp6 or Rrp47 is synthetically lethal with the absence of Mpp6, another factor that functions in the maturation of 5.8S rRNA, the degradation of CUTs and pre-mRNA surveillance (Milligan et al, 2008). The human orthologue of Mpp6 has been shown to interact with PM-Scl100–C1D (Rrp6–Rrp47) in co-immunoprecipitation experiments, and also with human Mtr4 (Schilders et al, 2007). Mtr4 is a Ski2-related RNA helicase (Jackson et al, 2010; Weir et al, 2010; Halbach et al, 2012) and is essential for viability in yeast (de la Cruz et al, 1998). Mtr4 is required for Rrp6-dependent and Rrp6-independent functions of the nuclear exosome (Jackson et al, 2010; Klauer & van Hoof, 2013) and is also part of the TRAMP complex (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). In human cells, Mtr4 has been shown to associate with either Mpp6 (Schilders et al, 2007) or Rrp6 (Lubas et al, 2011). In yeast, all genetic data suggest a close association between Mtr4 and the exosome, but no direct interaction has been reported thus far. In this work, we dissected the interaction network of the nuclear cofactors of the yeast exosome in vitro and identified the structural basis for how Rrp6 and Rrp47 assemble in a complex that directly recruits Mtr4.
Results
Saccharomyces cerevisiae Rrp6–Rrp47 recruits Mtr4 to the exosome
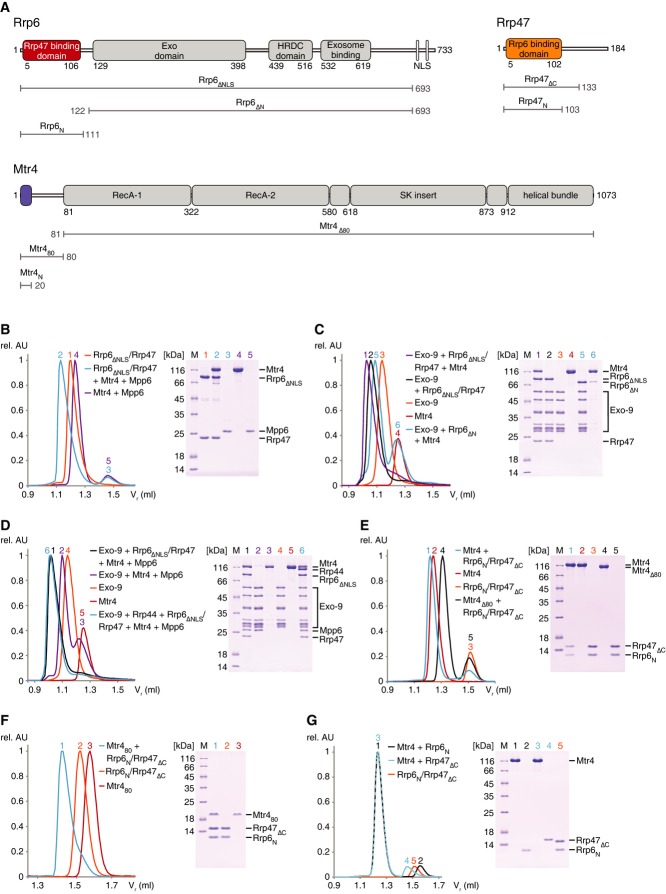
The domain organization of nuclear cofactors of the S. cerevisiae exosome is in several cases known from previous structural studies or can be extrapolated from sequence analysis (Fig1A). The Rrp6 exoribonuclease is a modular protein of 733 residues. The Rrp6 N-terminal region (so-called PMC2NT) mediates the interaction with Rrp47 and is expected to be a folded domain (Stead et al, 2007). The central region encompasses the exoribonuclease (Exo domain), which includes the catalytic DEDD site and the regulatory HRDC domain (Midtgaard et al, 2006). The C-terminal region consists of mainly low-complexity sequences and contains both the Exo-9-binding segment and two nuclear localization signals (NLSs) (Callahan & Butler, 2008; Makino et al, 2013a). Rrp47 (184 residues) has an N-terminal domain that binds Rrp6 and a C-terminal low-complexity region rich in positively charged residues (Costello et al, 2011) (Fig1A). The Mtr4 helicase (1073 residues) contains a low-complexity N-terminal region of 80 residues followed by a DExH helicase core characterized by an insertion domain also known as the arch domain (Jackson et al, 2010; Weir et al, 2010) (Fig1A). Mpp6 (186 residues) is a small basic protein without recognizable domains.
Figure 1. Direct interactions of the nuclear cofactors of the yeast exosome.
- A Schematic representation of the domain arrangement of the nuclear exosome cofactors from Saccharomyces cerevisiae used in this study. Gray-filled rectangles denote domains whose structures are known from previous studies: the central region of Rrp6 with the exonuclease (Exo) and HRDC domains (Midtgaard et al, 2006), the exosome-binding domain in the C-terminal region of Rrp6 (Makino et al, 2013a) and the entire helicase region of Mtr4 (Jackson et al, 2010; Weir et al, 2010). Colored rectangles highlight the N-terminal domains of Rrp6, Rrp47 and Mtr4 visualized in the structure of the ternary complex reported here. Truncation mutants engineered for the biochemical and structural analysis are indicated.
- B–G Size-exclusion chromatography assay to assess formation of protein complexes. Purified samples (as indicated) were incubated and co-injected on an analytical size-exclusion column (Superdex 200 Increase 3.2/300, GE Healthcare, exclusion volume 0.8 ml). On the left are the overlays of the chromatograms (rel. AU and Vr denote relative absorbance and retention volume of the proteins, respectively). On the right are the Coomassie-stained SDS–PAGE gels with samples from the corresponding peak fractions. The detailed analysis of the chromatography profiles is shown in Supplementary Table S1.
We recombinantly expressed and purified different versions of these proteins, incubated them in different combinations and analyzed the mixtures using size-exclusion chromatography to dissect their direct interactions (Fig1B–G). In the case of Rrp6, we engineered a version of the protein that spans from the Rrp47-binding domain to the exosome-binding domain (Rrp6ΔNLS), as the inclusion of the very C-terminus resulted in an unstable sample that was quickly degraded. Rrp6ΔNLS co-eluted with full-length Rrp47 (Fig1B, peak 1 in the size-exclusion chromatography profile on the left and lane 1 in the corresponding Coomassie gel on the right). Rrp6ΔNLS–Rrp47 did not interact with Mpp6 (Fig1B, peaks and lanes 2 and 3). Instead, Rrp6ΔNLS–Rrp47 interacted with full-length Mtr4 (Fig1B, peak and lane 2). Interestingly, Mtr4 required the Rrp6ΔNLS–Rrp47 complex to bind to Exo-9 (Fig1C, compare peak and lane 1, with peaks and lanes 4 and 5). Rrp6 lacking the N-terminal Rrp47-binding domain (Rrp6ΔN) co-eluted with Exo-9 but lost most of the binding to Mtr4 (Fig1C, peaks and lanes 5 and 6). The other nuclear exosome cofactor, Mpp6, interacted with Exo-9 both in the presence and in the absence of Rrp6ΔNLS–Rrp47 (Fig1D, peak and lane 1, compare with peak and lane 2). Finally, Exo-9, Rrp6ΔNLS, Rrp47, Mtr4, Mpp6 and Rrp44 co-eluted in a single peak that corresponds to the 14-subunit nuclear exosome (Fig1D, peak and lane 6). These data indicate that S. cerevisiae Rrp6, Rrp44 and Mpp6 can bind directly, independently and concomitantly to Exo-9, while Mtr4 is recruited to the exosome mainly by binding to Rrp6–Rrp47.
The N-terminal domains of Mtr4, Rrp6 and Rrp47 form a ternary complex in vitro
Consistent with previous studies (Stead et al, 2007; Costello et al, 2011; Dedic et al, 2014), we observed an interaction between the N-terminal regions of Rrp6 (Rrp6N, residues 1–111) and Rrp47 (Rrp47ΔC, residues 1–133) (Fig 1A). Rrp6N and Rrp47ΔC co-eluted with Mtr4 in size-exclusion assays (Fig1E, peak and lane 1). Next, we assessed which part of Mtr4 is recognized by Rrp6–Rrp47. Rrp6N and Rrp47ΔC co-eluted with the N-terminal region of Mtr4 (Mtr480) (Fig1F, lane and peak 1), while no interaction was detected with the helicase domain (Mtr4Δ80) (Fig1E, peaks and lanes 4 and 5). In isolation, neither Rrp6N nor Rrp47ΔC interacted with Mtr4 (Fig1G, peaks and lanes 1, 2 and 3, 4, respectively), suggesting that both proteins are required for binding. Finally, the interaction is conserved across species, as the S. cerevisiae Rrp6N–Rrp47ΔC complex formed a complex with the N. crassa Mtr4 orthologue, FRH (Supplementary Fig S1A). Sequence analysis showed that only the first 20 residues of the N-terminal region of Mtr4 are evolutionarily conserved ( 4Fig C). Rrp6N–Rrp47ΔC indeed formed a ternary complex with an Mtr41-20 peptide (Mtr4N) (Supplementary Fig S1B). As a note, the cytoplasmic Ski2 helicase does not contain an analogous N-terminal sequence, and consistently, the cytoplasmic exosome complex does not contain Rrp6 and Rrp47. Finally, using limited proteolysis experiments, we could narrow down the Rrp6-binding domain of Rrp47 even further to residues 1–103 (Rrp47N) (Fig 1A and Supplementary Fig S1C).
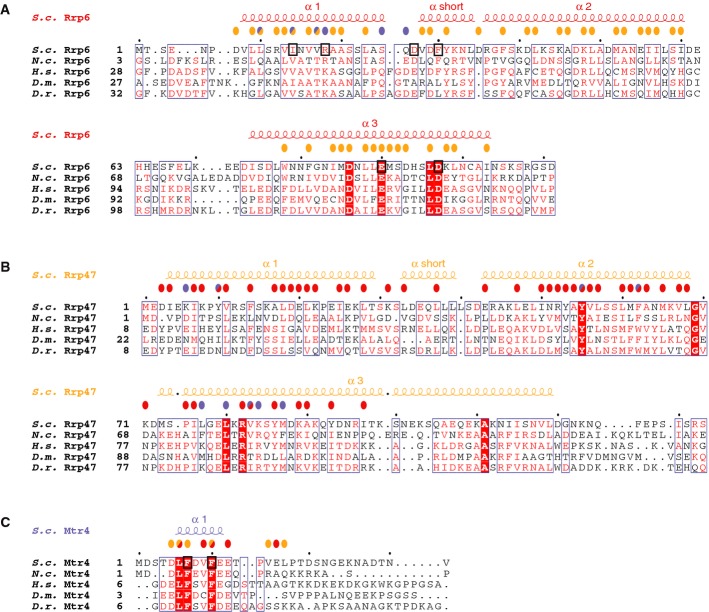
Figure 4. Structure-based sequence alignments of Rrp6N, Rrp47N and Mtr4N.
- A–C The alignments of Rrp6 (A), Rrp47 (B) and Mtr4 (C) include orthologues from the representative species Saccharomyces cerevisiae (S.c.), Neurospora crassa (N.c.), Homo sapiens (H.s.), Drosophila melanogaster (D.m.) and Danio rerio (D.r.), based on a comprehensive alignment. The secondary structure elements are shown above the sequences. Conserved residues are highlighted in color. Colored circles above the sequences identify residues involved in the interaction with Rrp6 (red circles), with Rrp47 (orange circles) and with Mtr4 (blue circles). Circles of two colors indicate residues involved in interactions with two partners in the ternary complex. Residues targeted for mutagenesis are highlighted with a black square.
Structure determination of the Rrp6N–Rrp47N–Mtr4N complex
Alone, Rrp47ΔC eluted as an apparent oligomer in size-exclusion chromatography (Fig 1G, peak 4), consistent with previous reports of its oligomeric nature when in isolation (Feigenbutz et al, 2013b). Upon co-expression, however, the Rrp6N–Rrp47ΔC complex eluted with a predominant peak corresponding to the expected molecular weight of a 1:1 complex (Fig1G, peak 5). Rrp6N–Rrp47ΔC crystallized in a tetragonal space group with three independent binary complexes in the asymmetric unit (Supplementary Fig S2A). The structure was determined by combining phases from single-wavelength anomalous dispersion (SAD) experiments using crystals containing tantalum bromide and crystals of selenomethionine-substituted protein. The model has been refined to 2.35 Å resolution with an Rfree of 22.4%, Rfactor of 18.4% and good stereochemistry (Table1).
Table 1.
Crystallographic statistics
| Data set | Rrp6N–Rrp47ΔC native | Rrp6N–Rrp47ΔC Ta6Br14 | Rrp6N–Rrp47ΔC Se-Met | Rrp6N–Rrp47N–Mtr4N native |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P 41 2 2 | P 41 2 2 | P 41 2 2 | P 3 1 2 |
| Unit cell (a, b, c in Å) | 98.4, 98.4, 208.0 | 98.6, 98.6, 205.3 | 97.9, 97.9, 207.7 | 142.7, 142.7, 63.5 |
| Wavelength (Å) | 0.873 | 1.255 | 0.979 | 1.000 |
| Resolution range (Å) | 88.95–2.35 (2.43–2.35) | 88.85–5.19 (5.80–5.19) | 48.96–3.59 (3.94–3.59) | 47.42–2.40 (2.49–2.40) |
| Unique reflections | 43613 (4107) | 4311 (1169) | 12424 (2850) | 28898 (2918) |
| Completeness (%) | 99.8 (97.8) | 99.6 (98.6) | 99.6 (98.5) | 99.6 (96.5) |
| Multiplicity | 12.9 (12.7) | 11.7 (11.6) | 41.8 (42.9) | 17.3 (17.2) |
| Mean I/ σ (I) | 16.6 (1.6) | 14.7 (3.5) | 20.3 (8.6) | 25.7 (1.7) |
| R-merge | 0.151 (1.955) | 0.140 (0.837) | 0.278 (0.644) | 0.094 (2.088) |
| R-pim | 0.043 (0.562) | 0.043 (0.254) | 0.043 (0.098) | 0.023 (0.510) |
| CC1/2 | 0.999 (0.586) | 0.998 (0.879) | 0.998 (0.972) | 1.000 (0.591) |
| Refinement | ||||
| R-work (%) | 18.37 | 20.07 | ||
| R-free (%) | 22.40 | 24.26 | ||
| Rmsd bonds (Å) | 0.008 | 0.009 | ||
| Rmsd angles (°) | 1.04 | 1.29 | ||
| Average B-factor | 65.53 | 69.5 | ||
| Ramachandran favored (%) | 99 | 99 | ||
| Ramachandran outliers (%) | 0 | 0 | ||
Values in parentheses correspond to the highest resolution shell.
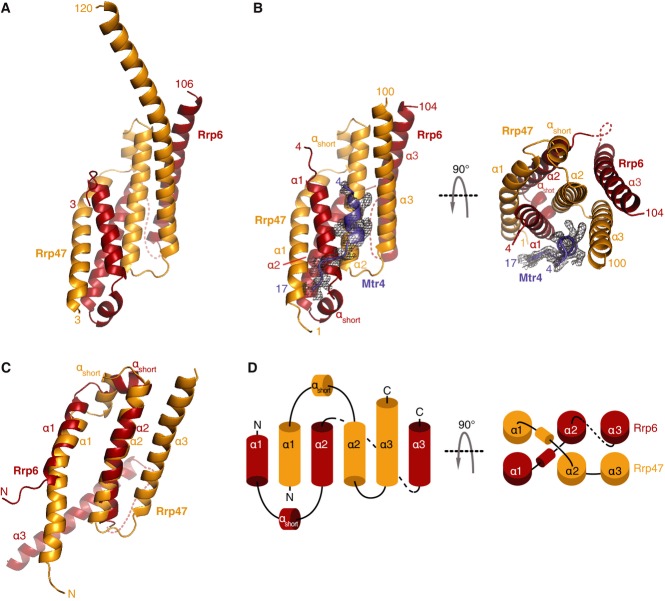
Based on the atomic model of the binary Rrp6N–Rrp47ΔC complex (Fig 2A) and on the limited proteolysis experiments (Supplementary Fig S1C), we trimmed the C-terminus of Rrp47 further and crystallized a ternary complex of Rrp6N, Rrp47N and Mtr4N. Rrp6N–Rrp47N–Mtr4N crystallized in a merohedrally twinned trigonal space group with three independent copies of the complex in the asymmetric unit (Supplementary Fig S2B). The structure was determined by molecular replacement using the coordinates of Rrp6N–Rrp47ΔC in combination with anomalous dispersion from yttrium ions that were required for crystallization and mediated lattice contacts. The model has been refined to 2.4 Å resolution with an Rfree of 24.3%, Rfactor of 20.1% and good geometry (Table1). The atomic models of Rrp6 and Rrp47 are very similar in the three ternary complexes in the asymmetric unit and are also very similar when compared to the structure of the binary complex (root mean square deviation (rmsd) of 1.35 Å over 184 α-carbon atoms). The main difference is that in one of the three copies of Rrp6N–Rrp47ΔC, twenty more residues of the C-terminal helix of Rrp47 are well ordered as a result of lattice contacts with a symmetry-related molecule (compare Fig 2A and B, left panel). The atomic model of Mtr4 shows well-defined electron density from residue 4 up to residue 17 (Fig2B and Supplementary Fig S2C).
Figure 2. Structure of the yeast Rrp6N–Rrp47N–Mtr4N ternary complex.
- Structure of the Rrp6N–Rrp47ΔC binary complex shown in cartoon representation with Rrp6 in red and Rrp47 in orange. The N- and C-terminal residues are indicated. This and all other cartoon drawings were generated with PyMOL (http://www.pymol.org/).
- Structure of the Rrp6N–Rrp47N–Mtr4N ternary complex shown in cartoon representation in two orientations related by a 90° rotation about the horizontal axis. The orientation of the ternary complex in the left panel is the same as that of the binary complex in (A). The secondary structure elements are labeled. A disordered region in the structure is indicated as a dotted line. The electron density for Mtr4N (Fo-Fc, contoured at 2.0 σ in PyMOL) and the model built into this density (blue) are shown.
- Superposition of the atomic models of Rrp6N and Rrp47N, showing the similarity of their secondary structure elements (labeled as in B).
- Topological diagram of the secondary structure elements of Rrp6N and Rrp47N. The left and right panels correspond to the views of the structure in the left and right panels of (B), respectively.
The Rrp6–Rrp47 interaction: an intertwined structure
The Rrp6N–Rrp47N complex has a compact α-helical fold (Fig2B). At the secondary structure level, Rrp6N and Rrp47N are remarkably similar. They both consist of three long α-helices (α1, α2 and α3) with a short α-helix (αshort) between α1 and α2. The stretch of Rrp6N encompassing the first helix-turn-helix (α1–αshort–α2) can be superposed to the equivalent stretch of Rrp47N (rmsd of 1.99 Å over 39 αC atoms) (Fig2C). The main topological difference between the two proteins is that helix α3 is oriented antiparallel to α2 in the case of Rrp47N, while it points in the opposite direction in the case of Rrp6N (Fig2C and topology in D).
The secondary structure elements of Rrp6N and Rrp47N are highly intertwined (Fig2B and D). The first helix-turn-helix of Rrp6N interdigitates with the first helix-turn-helix of Rrp47N, forming a heterodimeric 4-helix bundle (Figs2D and 3A). The α3 helices of Rrp6N and Rrp47N pack against the side of the bundle that is lined by the α2 helices (Figs2D and 3B). Evolutionarily conserved residues of the α1 and α2 helices form an extensive hydrophobic core in the center of the bundle (Figs3A, 4A and B). The interactions between the α2 and α3 helices are also extensive, apolar and conserved (Figs3B, 4A and B). The interaction of Rrp6N with Rrp47N buries 5,560 Å2 (i.e., more than 33%) of the surface area of the two proteins. The complex appears to be further stabilized by inter-molecular salt bridges present on the outer surface of the heterodimer. Finally, the structure of the binary complex shows that the C-terminal helix of Rrp47 protrudes out of the globular core of Rrp6N–Rrp47ΔC, extending about 30 Å into the solvent (Fig2A). The C-terminal region of Rrp47 that has been reported to interact with proteins involved in snoRNP assembly (Costello et al, 2011) would likely extend even further.
Figure 3. Rrp6N and Rrp47N form a highly intertwined structural module.
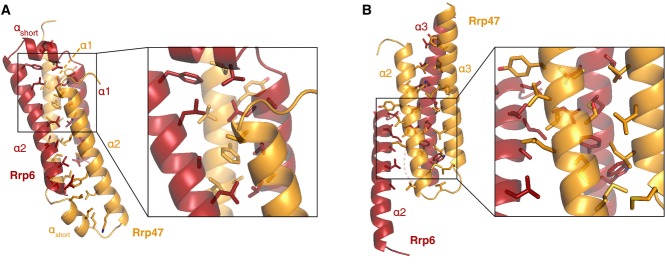
- Intermolecular contacts between the α1 and 2 helices (from both Rrp6N and Rrp47N), which form the center of the helical bundle. On the right is a zoom-in with a subset of the extensive Van der Waals interactions shown on the left.
- Intermolecular contacts between helices α2 and α3 (from both Rrp6N and Rrp47N), which are at the side of the bundle. A zoom-in view of part of the interactions is shown on the right.
Mtr4 binds Rrp6–Rrp47 via evolutionarily conserved interactions
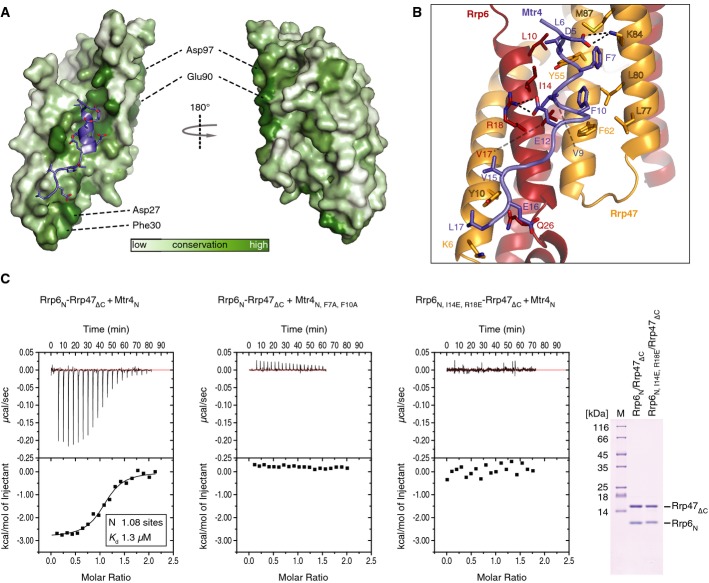
The overall structure of Rrp6N–Rrp47N resembles a turn of a superhelix with a conserved concave surface that is formed by the α1 helix of Rrp6 and the α2 and α3 helices of Rrp47 and that provides the binding site for Mtr4N (Figs 2B and 5A). Mtr4N binds as a short α-helix (residues 6–11), with extended segments at both ends. Apolar residues of Mtr4 (Leu6, Phe7, Val9, Phe10, Val 15 and Leu17) contact hydrophobic residues of Rrp6 (Leu10, Ile14 and Val17) and Rrp47 (Tyr10, Tyr55, Phe62, Leu77, Leu80 and Met87) (Fig5B). In addition, Mtr4 Glu12 forms a salt bridge with Rrp6 Arg18, while Mtr4 Asp5 interacts electrostatically with Lys84 of Rrp47. We tested the effect of mutating a set of the conserved interacting residues in in vitro binding assays. Isothermal titration calorimetry (ITC) experiments showed that Rrp6N–Rrp47ΔC bound to the Mtr4N peptide with a Kd of 1.3 μM, but no binding to an Mtr4N F7A, F10A mutant was detected (Fig5C, left and central panel). Conversely, an Rrp6N I14E, R18E–Rrp47ΔC mutant showed no binding to wild-type Mtr4N by ITC (Fig5C, right panel). We note that there are also additional conserved residues, including Asp27 and Phe30 in the Rrp6 αshort helix and Rrp6 Glu90 and Asp97 on the convex surface of the superhelix (Fig5A). These residues do not contact Mtr4N in the structure, and consistently, their mutation did not affect binding to Mtr4N in ITC experiments (Supplementary Fig S3A).
Figure 5. Mtr4N binds at an evolutionarily conserved surface groove of Rrp6N–Rrp47N.
- Surface representation of Rrp6N–Rrp47N colored according to sequence conservation (corresponding to the comprehensive alignments used for Fig4). The complex is shown in two orientations related by a 180° rotation about the vertical axis. The concave surface of Rrp6N–Rrp47N is viewed on the left (with an orientation corresponding to that in Fig2B, left panel) where Mtr4 is shown with the helix in cartoon representation. Peripheral patches of conserved residues of Rrp6 not involved in the interaction with Mtr4 are also indicated.
- Close-up view of the interactions of Mtr4N with Rrp6N and Rrp47N. Interacting residues are shown in stick representation and labeled.
- Isothermal titration calorimetry (ITC) experiments of the Mtr4N peptide with Rrp6–Rrp47. The MicroCal cell was filled with Rrp6N–Rrp47ΔC complex at 50 μM, and the Mtr4 peptide was injected at 500 μM concentration consecutively in 2 μl volumes. The left panel shows binding with the proteins used in the structure determination. Shown in the inset are the number of calculated binding sites (N) and the dissociation constant (Kd), as calculated with the program Origin. The middle and right panels show the corresponding ITC experiments with structure-based mutations. A Coomassie-stained gel with the wild-type and mutant complexes used in these experiments is shown on the right.
Impact of Rrp6N–Rrp47ΔC on RNA binding and degradation
The concave surface of Rrp6N–Rrp47N that binds Mtr4N is highly positively charged (Fig6A). Given these electrostatic properties and previous reports that Rrp47 binds RNA (Stead et al, 2007), we tested whether the interaction between Rrp6 and Rrp47 might also serve as an RNA-binding site (at least in the absence of Mtr4). Since the structure of the N-terminal region of Rrp6 is expected to be unfolded in the absence of Rrp47, we compared Rrp61–518–Rrp47ΔC with Rrp6122–518. We measured RNA-binding affinities by fluorescence anisotropy using fluorescein-labeled poly(A)35 or poly(U)30 RNAs and using a catalytic mutant of Rrp6 (Asp296 to Asn). In these experiments, we did not detect RNA binding to Rrp6N–Rrp47ΔC (Fig6B), suggesting its positively charged concave surface is a protein–protein interaction site for Mtr4N rather than an RNA-binding site. Rrp6122–518, D296N and Rrp61–518, D296N–Rrp47ΔC bound RNA with a similar affinity in the low micromolar range (Fig6B). In RNA degradation assays, the Rrp61–518–Rrp47ΔC complex showed somewhat lower activity as compared to that of Rrp6122–518 (Fig6C), indicating that the Rrp6N–Rrp47ΔC module subtly downregulates the enzymatic properties of the Rrp6 ribonuclease. Although the rationale for this effect is currently unclear, similar observations have been recently reported (Barbosa et al, 2014; Dedic et al, 2014).
Figure 6. Impact of Rrp6N–Rrp47N on ribonuclease activity.
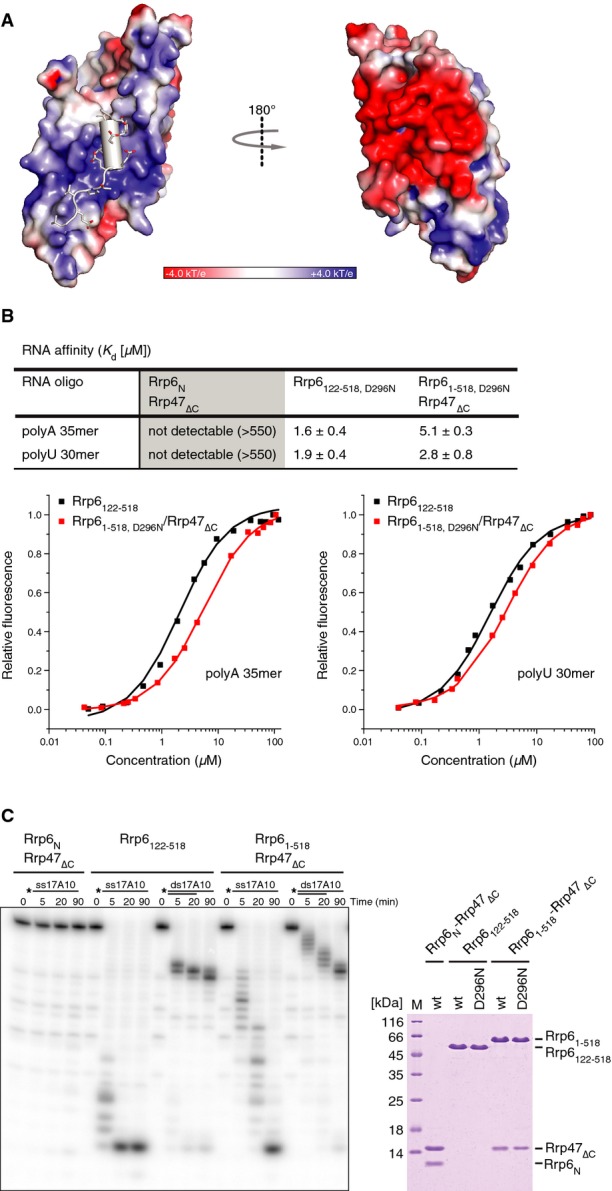
- Surface representation of Rrp6N–Rrp47N colored according to electrostatic potential (blue for electropositive and red for electronegative). Mtr4N is shown in gray, with negatively charged residues in a stick representation. The molecule is viewed in the same orientations as in Fig5A.
- Quantitative measurements of RNA-binding affinities in solution by fluorescence anisotropy using fluorescein-labeled homopolymeric substrates. The data were fitted to a binding equation describing a single-site binding model to obtain the dissociation constants (Kd). The best fit was plotted as a solid line. The Kds and their corresponding errors are the mean and standard deviation of a minimum of 3 independent experiments and are compiled in the table (top panel).
- Nuclease activity of the indicated Rrp6 and Rrp6–Rrp47 constructs toward single-stranded RNAs and duplex RNAs with 3′ overhangs. Substrates were designed to have a 17-base pair GC-rich duplex (ds17), corresponding to the 3′ end of tRNATyr (Vincent & Deutscher, 2006; Lorentzen et al, 2008), and a 3′ overhang of 10 adenine nucleotides (A10). The RNAs were 5′-end-labeled with [γ-32P]ATP, and the reaction products were resolved on 20% denaturing PAGE and visualized with a phosphorimager. A Coomassie-stained gel with the wild-type and nuclease-deficient mutant proteins used in the RNase activity and RNA affinity assays (B) is shown on the right.
We carried out a set of degradation assays of Rrp6ΔNLS–Rrp47ΔC in the presence of Mtr4, with and without the other subunits of the nuclear exosome complex (Supplementary Fig S3B). We first tested a double-stranded substrate with a short 3′ overhang (10 nucleotides) that from previous work is known to be inaccessible to the Rrp44 exoribonuclease when in the context of Exo-9 (Bonneau et al, 2009) (Supplementary Fig S3B, upper panel). We also tested a double-stranded substrate with a long 3′ overhang (35 nucleotides) that is accessible to the processive exoribonuclease activity of Rrp44 (Supplementary Fig S3, lower panel). We found that the Rrp6–Rrp47 degradation properties on these substrates were not affected by the presence of Mtr4 (Supplementary Fig S3B). Although we saw no significant effect of Mtr4 on the degradation of these substrates by either Rrp6 or Rrp44, we caution that it is possible that the helicase domain of Mtr4 might operate in the context of more complex RNA structures.
Structure-based mutations in the Rrp6 N-terminal domain result in 5.8S rRNA processing defects in vivo
Loss-of-function rrp6 and rrp47 mutants show strong defects in the 3′ processing of 5.8S rRNA and box C/D snoRNAs (Briggs et al, 1998; Allmang et al, 1999a; Mitchell et al, 2003). These mutants accumulate the 5′ external transcribed spacer (5′ ETS) fragment that is released during early processing of the pre-rRNA transcript, as well as truncated fragments of U3, 5S and snR13 as a result of impaired RNA surveillance processes (Briggs et al, 1998; Allmang et al, 1999a; Mitchell et al, 2003). To address the importance of the interaction between Mtr4 and the Rrp6–Rrp47 heterodimer in vivo, we designed specific rrp6 and rrp47 mutants based on the structure of the Rrp6N–Rrp47N–Mtr4N complex and analyzed the levels and integrity of the above RNAs in these mutants by Northern blot hybridization. Mutations were generated in the N-terminal region of Rrp6 and Rrp47 to either block the interaction with Mtr4 in vitro (rrp6I14E,R18E, see Fig5) or alter other conserved surface residue pairs (rrp6D27R,F30R, rrp6E90R,D97R, rrp47Y55A,S59Y, rrp47L77E,L80E or rrp47A111Y,I115Y).
The rrp6I14E,R18E, rrp6D27R,F30R and rrp6E90R,D97R mutants showed an accumulation of the 3′ extended ‘5.8S +30’ species and a defect in the degradation of the 5′ ETS fragment (see Fig 7A, lanes 4–6). The phenotype was stronger for the rrp6D27R,F30R mutant and weaker with the rrp6I14E,R18E mutant, but reproducible (see also Supplementary Fig S4A). In contrast, no clear effect was seen on the 3′ maturation of the snR38 snoRNA or the accumulation of degradation fragments from U3, snR13 or 5S rRNA in these mutants (denoted with asterisks in Fig 7A). These data are consistent with a partial loss of Rrp6 function in the rrp6I14E,R18E, rrp6D27R,F30R and rrp6E90R,D97R mutants. All rrp6 alleles complemented the temperature-sensitive growth phenotype of an rrp6Δ mutant (Fig7B), were expressed comparably to the wild-type protein and had no effect on the expression level of Rrp47 (Fig7C). Northern analyses of the rrp47 mutants revealed no strong phenotypes, although a weak but reproducible accumulation of the 5.8S + 30 fragment was observed for the Y55A, S59Y and L77E, L80E mutants (Supplementary Fig S4B). We concluded that mutation of conserved surface residues in the Rrp6 N-terminal domain, in particular D27 and F30, results in a clear defect in 5.8S rRNA maturation. It is currently unclear why conserved surface residues of Rrp6–Rrp47 that are not involved in Mtr4N binding are important for function, but it is possible that they are involved in additional interactions within the nuclear exosome complex or with the substrate ribonucleoprotein particle.
Figure 7. Structure-based mutations in the Rrp6 N-terminal domain result in 5.8S rRNA processing defects in vivo.
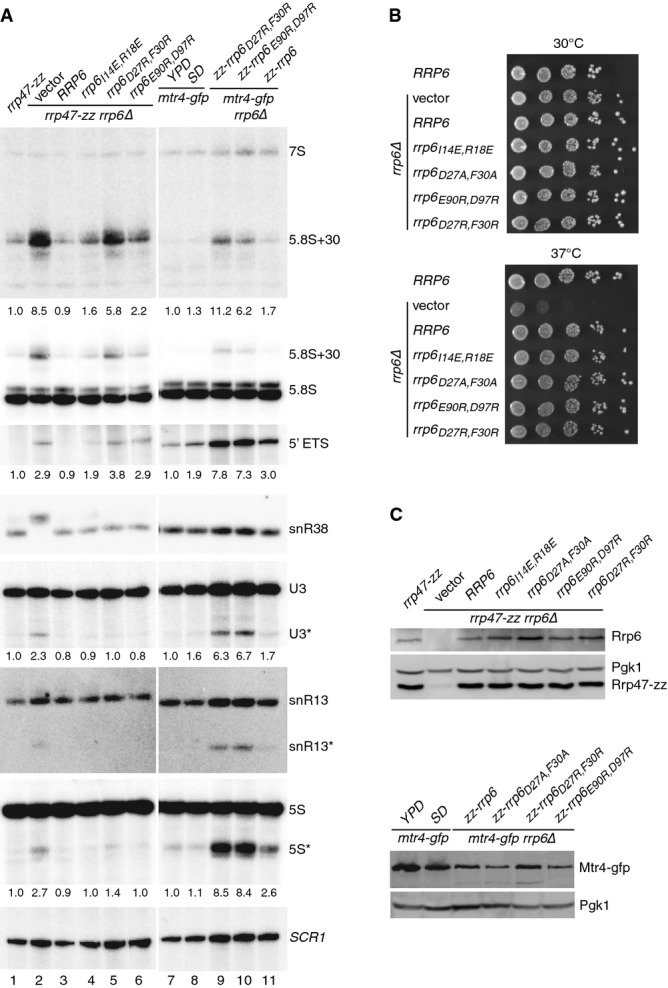
- Northern blot analyses of RNA from rrp6, mtr4-gfp and mtr4-gfp rrp6 double mutants. Strains were grown in selective minimal medium, unless indicated otherwise. Consecutive hybridizations of a single blot are shown; panels shown for lanes 1–6 and 7–11 are juxtaposed from a single image. The major RNAs detected by each probe are indicated on the right. Asterisks indicate truncated RNA fragments of U3, snR13 and 5S. The amount of ‘5.8S+30’ RNA, the 5′ ETS fragment and the major U3 and 5S degradation fragments in each mutant (indicated beneath the appropriate panel) is expressed relative to the rrp47-zz strain in lanes 1–6 and to the mtr4-gfp strain during growth in YPD in lanes 7–11.
- Spot growth assays of rrp6 mutants. Serial dilutions of pre-cultures were spotted onto selective minimal medium plates and incubated at 30 or 37°C. The plates were photographed after 3 days.
- Western analyses of rrp6 mutants. (Upper panels) Expression levels of Rrp6 and the Rrp47-zz fusion protein in the rrp6 mutants. (Lower panels) Mtr4–gfp fusion protein expression levels in the mtr4-gfp rrp6 double mutants. Pgk1 levels were analyzed as a loading control in each case.
Mtr4 and Rrp6 mutants show synergistic effects in vivo
C-terminally tagged Mtr4 fusion proteins can support cell growth (Ghaemmaghami et al, 2003; Huh et al, 2003). Indeed, we observed no clear growth defect for an mtr4-gfp strain when compared to an isogenic wild-type strain. Northern analyses of RNA isolated from the mtr4-gfp strain during growth in either minimal or rich medium did not show significant accumulation of the 5.8S + 30 fragment or the 3′ extended forms of snoRNAs that are characteristic of rrp6Δ mutants (Briggs et al, 1998; Allmang et al, 1999a). However, the Northern analyses showed defects in the degradation of other structured RNAs, with a clear accumulation of the 5′ ETS fragment that has been previously shown to accumulate upon Mtr4/Dob1 depletion (de la Cruz et al, 1998) (see Fig7A, lanes 7 and 8). These results indicated that the presence of the C-terminal GFP protein partially compromises the function of Mtr4, without causing a general affect on Rrp6-dependent processing or degradation pathways. We therefore generated an rrp6Δ allele in the haploid mtr4-gfp strain and tested for genetic interactions between the mtr4-gfp allele and the rrp6 mutants.
Attempts to delete the RRP6 gene in the mtr4-gfp strain directly were unsuccessful, but correct integrants were isolated when the mtr4-gfp strain had been transformed with a URA3 plasmid encoding a functional Rrp6 fusion protein (Allmang et al, 1999b). Notably, deletion of the RRP6 gene was also achieved in mtr4-gfp strains expressing rrp6D27R,F30R and rrp6E90R,D97R variants of the Rrp6 fusion protein but not the rrp6I14E,R18E mutant. To determine whether the mtr4-gfp rrp6I14E,R18E double mutant is synthetic lethal, wild-type and rrp6I14E,R18E mutant alleles were subcloned into the LEU2 plasmid pRS415 and transformed into the mtr4-gfp rrp6Δ plasmid shuffle strain and the resulting transformants were tested for growth on medium containing 5-fluoroorotic acid (5 FOA). While transformation with the wild-type RRP6 gene gave rise to viable colonies on 5 FOA medium, no growth was observed for the mtr4-gfp rrp6Δ strain after transformation with a plasmid encoding the rrp6I14E,R18E mutant or the cloning vector (Fig8A). We concluded that the mtr4-gfp strain is dependent upon Rrp6 for cell growth.
Figure 8. Disruption of the Rrp6/Mtr4 interaction in yeast blocks growth of mtr4-gfp strains.
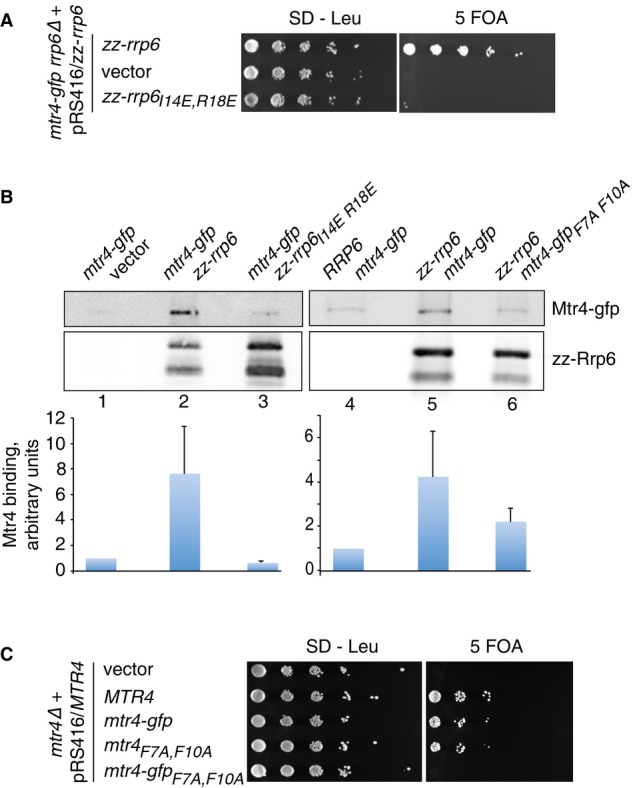
- Plasmid shuffle assay on an mtr4-gfp rrp6Δ strain expressing zz-Rrp6. Transformants harboring LEU2 plasmids encoding wild-type zz-Rrp6, the I14E, R18E variant or the vector alone were cultured in selective medium and then spotted onto medium lacking leucine or containing 5 FOA. Plates were incubated at 30°C and photographed after 3 days.
- Pull-down assays of zz-Rrp6 with Mtr4-gfp. (Left panels) Pull-downs of zz-Rrp6 in an mtr4-gfp strain harboring the vector (lane 1), expressing zz-tagged wild-type Rrp6 (lane 2) or the I14E, R18E variant (lane 3). The amount of Mtr4-gfp associated with Rrp6 is expressed relative to the negative control and is normalized to the Rrp6 levels in the eluates. The mean values of assays on two independent biological replicates are shown. Error bars indicate the experimentally observed range. (Right panels) Pull-downs of zz-Rrp6 in a wild-type strain expressing the Mtr4-gfp fusion protein or the F7A, F10A mutant. A wild-type strain was transformed with plasmids expressing either non-tagged or zz-tagged Rrp6 and Mtr4-gfp or the F7A, F10A mutant and analyzed by Western blot as above.
- mtr4 plasmid shuffle assay. An mtr4Δ mutant strain harboring a URA3 plasmid containing the wild-type MTR4 gene was transformed with LEU2 plasmids encoding mtr4-gfp, mtr4F7A,F10A or mtr4-gfpF7A,F10A mutants, the wild-type MTR4 gene or the vector. Strains were grown up in selective medium and then spotted onto medium lacking leucine (SD-Leu) or containing 5 FOA. The plates were incubated at 30°C and photographed after 3 days.
To determine whether the Rrp6/Mtr4 interaction in yeast is blocked by the rrp6I14E,R18E mutation, pull-downs were performed on lysates from mtr4-gfp strains expressing plasmid-encoded, zz epitope-tagged wild-type or mutant Rrp6 fusion proteins (in addition to the endogenously encoded Rrp6). Mtr4-gfp was bound to the wild-type zz-Rrp6 protein, but not the I14E, R18E mutant (Fig8B, left panel). We then addressed whether mutation of the N-terminal region of Mtr4 also causes a block in the Rrp6/Mtr4 interaction in vivo. Immobilized zz-Rrp6 protein retained the wild-type Mtr4-gfp protein, whereas binding of the mtr4-gfpF7A,F10A mutant was only slightly above background levels (Fig8B, right panel). These data support the conclusion that the structurally defined Rrp6N–Rrp47N–Mtr4N complex forms the principal interaction between Rrp6 and Mtr4 in yeast.
If the synthetic lethal phenotype observed for the mtr4-gfp rrp6I14E,R18E mutant (Fig8A) is due to loss of interaction between Mtr4 and Rrp6, a strong synergistic effect would be predicted upon introduction of the F7A, F10A mutation in the mtr4-gfp mutant. Indeed, both the mtr4-gfp and the mtr4F7A,F10A mutants grew comparably to the wild-type strain, whereas the strain expressing an Mtr4-gfp fusion protein bearing the F7A, F10A mutation was nonviable (Fig8C). We concluded that the strong synergistic effects observed in strains expressing a C-terminal Mtr4-gfp fusion protein in combination with either the Rrp6 I14E, R18E mutation (that impairs the interaction with the N-terminus of Mtr4) or the Mtr4 F7A, F10A mutation (that impairs the interaction with the N-terminal domains of Rrp6–Rrp47) are due to loss of binding. The very C-terminus of Mtr4 is embedded within the base of the DExH core of the helicase, where the 3′ end of an RNA substrate is expected to emerge after unwinding (Jackson et al, 2010; Weir et al, 2010). Collectively, these data suggest that the base of Mtr4 is also engaged in interactions within the nuclear exosome complex and that linking the C-terminus of Mtr4 to a GFP protein weakens this interaction.
Northern analyses of the viable mtr4-gfp rrp6D27R,F30R and mtr4-gfp rrp6E90R,D97R double mutants revealed a strong synergistic block in the degradation of some RNAs, including the 5′ ETS fragment and truncated fragments of U3, snR13 and 5S rRNA (Fig7A, lanes 9 and 10). In contrast, the defect in 5.8S rRNA maturation seen in the rrp6D27R,F30R or rrp6E90R,E97R mutants was not exacerbated in the mtr4-gfp rrp6 double mutants. This suggests that mtr4-gfp rrp6 mutants may be defective in RNA surveillance mechanisms, rather than 5.8S rRNA or snoRNA processing.
Discussion
The interaction of Rrp6 and Rrp47 is known to stabilize the individual proteins in vivo and to influence their function in exosome-mediated RNA processing and turnover pathways. The molecular basis for these effects, however, has remained unclear. Here, we show that the N-terminal domains of Rrp6 and Rrp47 assemble into a globular heterodimer with an elaborate architecture formed by intertwined pairs of α-helices. The interlocked Rrp6N–Rrp47N structure explains why heterodimer formation leads to stabilization and to functional interdependence. In isolation, the individual proteins are expected to be partially unfolded and aggregated. As a 1:1 complex, they form a composite molecular surface for the direct recruitment of the N-terminal region of Mtr4, which binds via a set of evolutionarily conserved interactions. Together with the observation that Rrp6 and Rrp47 assemble only after they are independently imported into the nucleus (Feigenbutz et al, 2013b), these results rationalize how the cell might avoid the untimely recruitment of Mtr4 to the exosome in the cytoplasm.
The Rrp6N–Rrp47N heterodimer does not undergo significant conformational changes upon Mtr4N binding and thus appears to function as a rather rigid platform. The structural analysis and in vivo data suggest that this platform also contains docking sites for other interaction partners whose identities remain to be explored. It is also likely that Rrp6, Rrp47 and Mtr4 are engaged in additional, albeit weaker, contacts in the context of the nuclear exosome complex. Although we did not observe direct interactions between Rrp6–Rrp47 and Mpp6 and between Mtr4 and Mpp6, as has been reported for the human orthologues (Schilders et al, 2007), it is possible that the yeast proteins might engage in analogous contacts but without the high affinity required for detection in the in vitro reconstitution assays we used. Indeed, synthetic lethal mutations in S. cerevisiae support the presence of redundant interactions among the yeast cofactors of the nuclear exosome (Milligan et al, 2008; Garland et al, 2013 and Fig8A). The presence of additional weak interactions would also rationalize why Mtr4 would be able to carry out at least part of its functions in vivo in the absence of Rrp6 and Rrp47, as can be inferred from the severity of the corresponding knockout studies (Briggs et al, 1998; de la Cruz et al, 1998; Mitchell et al, 2003). The emerging picture is that Rrp6, Rrp47, Mpp6 and Mtr4 assemble together with Exo-10 with a combination of high-affinity interactions and additional intermolecular contacts to form a functional nuclear complex (Fig9). Understanding how the nuclear exosome complex is structured and how it coordinates the multiple catalytic activities of core components and cofactors awaits future studies.
Figure 9. Model of the nuclear exosome complex.
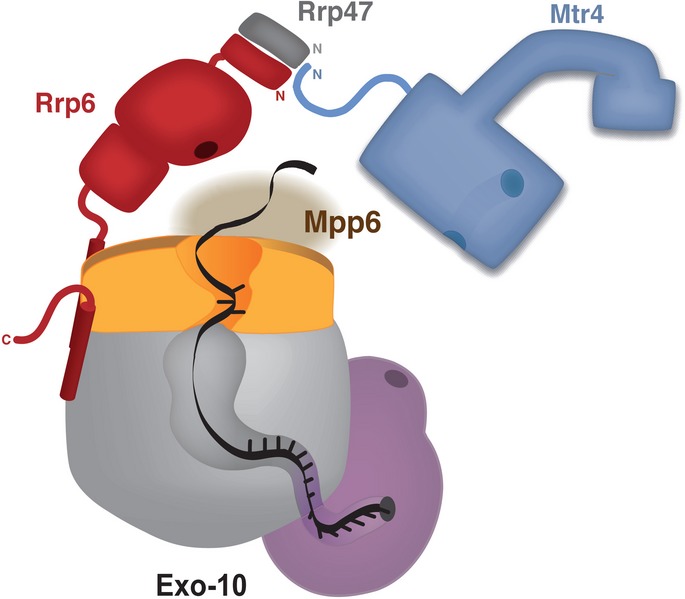
The schematic representation shows the architecture of the yeast nuclear exosome based on current structural and biochemical information. The exosome core (Exo-10) is depicted with the RNase PH-like subunits in gray, the S1/KH subunits in orange and Rrp44 in pink (based on the structure reported in Makino et al, 2013a). RNA is shown in black, threading through the internal channel and reaching the exoribonuclease site of Rrp44 (highlighted with a circle). The endonuclease site in the PIN domain of Rrp44 is also highlighted with a circle. Rrp6 is shown in red, with the exoribonuclease site highlighted as a circle. The exoribonuclease of Rrp6, depicted based on Midtgaard et al, 2006; is positioned near the exosome-binding domain of Rrp6, as determined by Makino et al, 2013a;. Rrp47 is in gray while Mtr4 is in blue. The channel in the helicase core and the insertion domain of Mtr4 are indicated in the model (according to the structures reported by Jackson et al, 2010; Weir et al, 2010). The N-terminus of Mtr4 binds at the interface between Rrp6 and Rrp47. The N-terminal interaction regions of Rrp6, Rrp47 and Mtr4 are denoted by N. Mpp6 is shown tentatively at the top of Exo-10, as its binding is not dependent on Rrp44.
Materials and Methods
Protein purification
Saccharomyces cerevisiae Mtr4 full-length, Mtr4Δ80 and the Neurospora crassa Mtr4 orthologue FRH were expressed and purified according to the protocol in Weir et al (2010). Exo-9 and Rrp44 were expressed and purified as in Makino et al, 2013a. All other S. cerevisiae proteins purified in this study were expressed recombinantly using E. coli BL21-Gold (DE3) pLysS cells (Stratagene) grown in TB medium and induced overnight at 18°C. Rrp6N, Rrp6ΔN and Mtr480 were expressed as His-tagged proteins. Rrp6N–Rrp47N, Rrp6N–Rrp47ΔC and Rrp6ΔNLS–Rrp47 complexes were co-expressed such that the Rrp6 constructs bear an N-terminal His tag while the Rrp47 constructs are untagged. The proteins were purified using cobalt-based (or nickel for Mtr480) affinity chromatography, followed by cleavage of the His tag with the appropriate protease (human rhinovirus 3C protease for Rrp6N, Rrp6N–Rrp47N, Rrp6N–Rrp47ΔC, tobacco etch virus (TEV) protease for Mtr480, Rrp6ΔNLS–Rrp47 and small ubiquitin-like modifier (SUMO) protease for Rrp6ΔN). Protease-treated Rrp6ΔN was loaded on the affinity column a second time to remove uncleaved species. After affinity purification, all samples (with the exception of Mtr480) were subjected to anion exchange chromatography (HiTrap Q HP, GE Healthcare). Rrp6ΔN and Rrp6ΔNLS–Rrp47 were further purified over a HiTrap Heparin Sepharose HP column (GE Healthcare).
Size-exclusion chromatography (SEC) on a Superdex 200 or Superdex 75 column (GE Healthcare) was performed as a final step of purification for all proteins. Rrp6N and complexes of this construct were finally purified in buffer A (20 mM Tris pH 7.5, 100 mM NaCl) supplemented with reducing agents. An additional 10% glycerol was added to the size-exclusion buffer when purifying Rrp6N and Rrp6ΔNLS–Rrp47. Mtr480 was purified in buffer A supplemented with an additional 50 mM NaCl and 2 mM DTT. Size exclusion for Rrp6ΔN was performed in buffer B (20 mM MES pH 6.0, 250 mM NaCl, 10% glycerol, 2 mM DTT). Rrp6 mutants were verified by DNA sequencing and purified using the protocol for the wild-type protein. Yeast Rrp47ΔC and Mpp6 were expressed as recombinant GST-tagged (3C- or TEV protease cleavable, respectively) proteins in conditions similar to those described above. The proteins were purified by affinity chromatography on Glutathione Sepharose resin (Clontech). Tag cleavage was followed by ion-exchange chromatography (HiTrap SP Sepharose HP column, GE Healthcare) and SEC (in buffer A supplemented with 1 mM DTT and 10% glycerol) for Rrp47ΔC. Tag-cleaved Mpp6 was purified over a HiTrap Heparin Sepharose HP column (GE Healthcare) as the final step.
Crystallization and structure determination
Crystals of yeast Rrp6N–Rrp47ΔC were grown at 20°C by sitting-drop vapor diffusion from drops formed by equal volumes of protein (at 18 mg/ml in size-exclusion buffer comprising 20 mM Tris, 100 mM NaCl and 1 mM DTT) and of crystallization solution (1.8 M (NH4)2SO4, 125 mM NaCl and 100 mM Na-cacodylate pH 5.8). For heavy-atom derivatization, native Rrp6N–Rrp47ΔC crystals (grown in 2.05 M (NH4)2SO4, 125 mM NaCl and 100 mM Na-cacodylate pH 6.4) were soaked for 15 min in crystallization solution supplemented with 1 mM Ta6Br14 prior to cryoprotection. Optimized Se-Met-derivatized crystals were obtained in 2.0 M (NH4)2SO4, 200 mM NaCl and 100 mM Na-cacodylate pH 6.2. Crystals were cryoprotected with crystallization solution supplemented with 24% glycerol for the native crystals and 17.5% glycerol for the Ta soaked and Se-Met-derivatized crystals prior to cryo-cooling and data collection.
Yeast Rrp6N–Rrp47N complex was mixed with a 1.5-fold molar excess of Mtr4N (synthesized peptide (H)-MDSTDLFDVFEETPVELPTK-(NH2); D20K substitution for synthesis strategy and solubility reasons) and 5 mM YCl3 and incubated at room temperature for 10 min. Crystals of the Rrp6N–Rrp47N–Mtr4N complex were grown at 20°C by sitting-drop vapor diffusion from drops formed by equal volumes of complex (at 27 mg/ml in size-exclusion buffer comprising 20 mM Tris, 100 mM NaCl and 0.5 mM TCEP) and crystallization solution (12% PEG 1000, 0.1 M imidazole pH 7.5 and 0.125 M calcium acetate). Crystals were cryoprotected in 19% PEG 1000, 0.1 M imidazole pH 7.5, 0.125 M calcium acetate, 5 mM YCl3 and 12% glycerol and supplemented with 0.5 mM Mtr4N peptide. Data were collected at the ID23-2 beamline of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) and at the PXII and PXIII beamlines of the Swiss Light Source (SLS) (Villigen, Switzerland) and processed using XDS (Kabsch, 2010) and Aimless (Evans & Murshudov, 2013).
The Rrp6N–Rrp47ΔC structure was solved at low resolution (5.2 Å) by SAD with SHELX (Sheldrick, 2008) and HKL2MAP (Pape & Schneider, 2004) using the anomalous Ta signal. Identifiable α-helices were manually placed with Coot (Emsley et al, 2010) and used as a starting model for SAD-MR by exploiting the anomalous Se signal using the program PHENIX AutoSol (Terwilliger et al, 2009). After manual chain tracing, the model was completed with Coot and refined against the native data using phenix.refine (Afonine et al, 2012). The Rrp6N–Rrp47N–Mtr4N structure was solved by SAD-MR using Phaser (McCoy et al, 2007) with parts of the Rrp6N–Rrp47ΔC model as a search model and AutoSol using the anomalous signal from yttrium. Merohedral twinning generated by a twofold axis perpendicular to a crystallographic threefold axis of the trigonal space group became apparent by the poor quality of the electron density for one of the three copies of the complex in the asymmetric unit. The twin law (-h,-k,l) and twinning fraction (0.5, perfect twin) was determined using phenix.xtriage (Adams et al, 2010). After manual tracing of the Mtr4N sequence, the model was completed using Coot and refined against twinned data using phenix.refine. Several Yttrium ions form clusters surrounded by electron density that likely corresponds to a negatively charged loop of Rrp6 (residues 63–73). In the Rrp6N–Rrp47ΔC structure, the same loop has only partial electron density and was not modeled. Although it is possible that in two copies of the binary complex, this loop is engaged with another molecule in the asymmetric unit, this is a crystal lattice artifact as it occurs neither in the third copy nor in the ternary complex.
Size-exclusion chromatography assay
Equimolar amounts of purified proteins as indicated (500 pmol for Fig1C and D, 700 pmol otherwise) were diluted in a total injection volume of 25 μl in SEC buffer A supplemented with 2 mM DTT. Samples were incubated for 1 h on ice to allow complex formation. Increase in particle size upon complex formation was assayed by comparing the retention volumes in SEC on a Superdex 200 Increase 3.2/300 (GE Healthcare). Composition of the SEC peak fractions were analyzed by SDS–PAGE and visualized by Coomassie staining (percentage of the SDS–PAGE depended on protein sample size).
Fluorescence anisotropy
Fluorescence anisotropy measurements were performed with a 5′-6-carboxy-fluorescein (6-FAM)-labeled poly(A)35 or poly(U)30 RNA at 20°C in 50 μl reactions on a Genios Pro (Tecan). The RNA was dissolved to a concentration of 10 nM (1 nM for Rrp6N, D296N–Rrp47) and incubated with Rrp6 or the Rrp6–Rrp47 complexes at different concentrations in a buffer containing 20 mM Tris pH 7.5, 100 mM NaCl and 1 mM DTT. The excitation and emission wavelengths were 485 nm and 535 nm, respectively. Each titration point was measured three times using ten reads with an integration time of 40 μs. The data were analyzed by nonlinear regression fitting using the BIOEQS software (Royer, 1993).
Isothermal titration calorimetry (ITC)
Rrp6N–Rrp47ΔC wild-type and mutant proteins were dialyzed overnight in the same buffer that was used to dissolve the lyophilized Mtr4N peptide (20 mM HEPES, pH 7.5, 100 mM NaCl, 0.5 mM TCEP). ITC experiments were carried out at 20°C with a iTC-200 MicroCal calorimeter (GE healthcare). The MicroCal cell was filled with Rrp6N–Rrp47ΔC at 50 μM concentration and stirred at 800 rpm. For each titration, Mtr4N was injected into the cell 20 times in 2 μl volumes per injection at the same intervals of time (4 min). The concentration of Mtr4N in the syringe (500 μM) was 10 times the concentration of the protein sample in the cell. The released heat was obtained by integrating the calorimetric output curves and was corrected for the effect of dilution by subtraction of the value of the last injection as background. As control for all ITC measurements, the injectant was titrated into buffer. The Kd values and binding ratios were calculated with the Origin (V7) software supplied with the calorimeter. We used the same protocol to measure the Kd of the Rrp6 and the Mtr4 mutants.
Nuclease assay
The exonuclease activity assay in Fig6C was carried out at 30°C in a buffer containing 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM magnesium diacetate, 10% (v/v) glycerol, 0.1% (v/v) NP-40 and 1 mM DTT. The reactions contained protein at a final concentration of 2 nM, while the concentration of RNA substrates was 200 nM. Substrates were verified by native gel electrophoresis. Two-microliter aliquots from a 10 μl total reaction volume were taken at indicated time points and quenched by addition of 14 μl loading dye consisting of 10 mM EDTA, 0.1% (w/v) bromophenol blue and 0.1% (w/v) xylene cyanol FF in formamide. The ‘0’ time point was taken before adding the protein. Reaction products were boiled for 5 min immediately before being resolved on a 20% acrylamide gel containing 8 M urea and visualized by phosphorimaging.
Plasmids and yeast strains
Yeast expression plasmids either encoding an N-terminal zz fusion of Rrp6 under the control of the RRP4 promoter (Allmang et al, 1999b), or containing a genomic clone of the RRP6 gene and lacking the CEN6 element from the vector backbone (Feigenbutz et al, 2013a), have been reported previously. Expression of the N-terminal zz-Rrp6 fusion protein from the RRP4 promoter is comparable to the endogenous expression level of the C-terminal TAP-tagged protein (Stead et al, 2007). The RRP47 genomic clone used in this study is described in Costello et al (Costello et al, 2011). The MTR4 construct pAv675 (Jackson et al, 2010) was kindly provided by Ambro van Hoof (University of Texas Health Science Center, Houston). The construct encoding mtr4-gfp was cloned in yeast by homologous recombination, using HindIII linearized pAv675 (after deletion of the HindIII polylinker site) and a PCR amplicon encompassing the mtr4-gfp::HIS3 allele. The isolated plasmid was confirmed by sequencing the mtr4/GFP junction along the complete length of the PCR product. Point mutations were introduced into the RRP6, RRP47 and MTR4 ORFs in these constructs by site-directed mutagenesis using the Quikchange kit (Agilent Technologies) and validated by sequence analysis. Plasmid inserts encoding the N-terminal epitope-tagged wild-type RRP6 and rrp6I14E,R18E mutants were subcloned into pRS415 (Stratagene) for the plasmid shuffle assay. The rrp6Δ::KANMX4 allele was amplified by PCR and integrated into the mtr4-gfp strain by homologous recombination after initial transformation with plasmids encoding zz-Rrp6 fusion proteins. Correct integrants were identified by PCR amplification of genomic DNA.
The mtr4-gfp strain (Huh et al, 2003) was obtained from Invitrogen. The mtr4 plasmid shuffle strain yAv1151 (Jackson et al, 2010) was kindly provided by Ambro van Hoof. Isogenic wild-type and rrp6Δ strains were obtained from Euroscarf (University of Frankfurt, Germany). The rrp47-zz and rrp47-zz rrp6Δ strains have been previously reported (Mitchell et al, 2003). Yeast strains were routinely cultured at 30°C in SD selective minimal medium (2% glucose, 0.5% ammonium sulfate, 0.17% yeast nitrogen base) supplemented with appropriate amino acids and bases or in YPD medium (2% glucose, 2% peptone, 1% yeast extract). Spot growth assays were performed on selective solid minimal medium or medium containing 5-fluoroorotic acid (5 FOA) using tenfold serial dilutions of freshly saturated pre-cultures. Plates were photographed after incubation for 3 days.
RNA analyses
Total cellular RNA was isolated from strains harvested during early log growth and resolved by electrophoresis through 8% polyacrylamide gels containing 50% urea. After transfer to Hybond N+ membranes (GE Healthcare), the RNA was hybridized with 5′-[32P]-labeled oligonucleotide probes complementary to the ITS2 region of the pre-rRNA transcript (tgagaaggaaatgacgct), 5.8S rRNA (gcgttgttcatcgatgc), the 5′ ETS region of the pre-rRNA (cgctgctcaccaatgg), snR38 (gagaggttacctattattacccattcagacagggataactg), U3 (ttcggtttctcactctggggtac), snR13 (caccgttactgatttggc), 5S (ctactcggtcaggctc), SCR1 (aaggacccagaactaccttg) and U6 (atctctgtattgtttcaaattgaccaa). Hybridized blots were placed under phosphor storage screens, and the data were captured using a Personal Molecular Imager (Bio-Rad). Nonsaturated images were adjusted for signal level and window using ImageJ (NIH, USA). RNA hybridization signals were quantified using ImageJ and normalized to the expression level of 5S rRNA.
Protein analyses
For protein expression analyses, total cellular protein was prepared by alkaline/SDS lysis followed by TCA precipitation. Extracts were resolved by SDS–PAGE, transferred to Hybond C extra membranes (GE Healthcare) and incubated with a rabbit anti-Rrp6 antiserum (Mitchell et al, 2003) or mouse anti-Pgk1 (clone 22C5D8, Life Technologies) primary antibody, followed by either goat anti-rabbit (A4914, Sigma) or goat anti-mouse (1706516, Bio-Rad) HRP-conjugated secondary antibodies. The Mtr4-gfp fusion protein was detected using the anti-GFP antibody. The Rrp47-zz fusion protein was detected directly using the PAP antibody conjugate (P1291, Sigma). ECL images were captured using a G:Box iChemi XL system (Syngene) and adjusted for the signal level and window using ImageJ.
For the pull-down experiments, yeast were lysed in 50 mM HEPES pH 7.4, 50 mM KCl, 5 mM MgCl2, 10% glycerol and 1 mM PMSF. Lysates were clarified by centrifugation at 13,000 g for 30 min, normalized for A280 units and passed through ∼200 μl IgG sepharose fast flow beads (GE Healthcare). The beads were washed 5 times with 1 ml wash buffer (50 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT), and the retained material was eluted with 0.5 M acetic acid. Eluates were resolved through 10% SDS–PAGE gels and analyzed by Western blotting, as described above.
Data deposition
The coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 4WFC for the Rrp6N–Rrp47ΔC structure and 4WFD for the Rrp6N–Rrp47N–Mtr4N structure.
Acknowledgments
We would like to thank the Max Planck Institute of Biochemistry (MPIB) Core Facility for synthesizing the peptides used in the study and for mass spectrometry analysis; the MPIB Crystallization Facility for crystallization screenings; and the beamline scientists at the SLS and the ESRF for assistance with data collection. We also thank Marc Baumgärtner, Tatjana Krywcun and Petra Birle for technical assistance and members of our laboratories for useful discussions and critical reading of the manuscript. This study was supported by the Max Planck Gesellschaft, the European Commission (ERC Advanced Investigator Grant 294371 and Marie Curie ITN RNPnet) and the Deutsche Forschungsgemeinschaft (DFG SFB646, SFB1035, GRK1721, FOR1680 and CIPSM) to EC and by a research grant from the Wellcome Trust (08836/Z/09/Z) to P.M. We thank David Tollervey (University of Edinburgh) for the yeast Rrp6 antiserum and Ambro van Hoof (University of Texas Health Science Center, Houston) for mtr4 strains and plasmids.
Author contributions
BS carried out the structural analyses; BS, DM and SF, the in vitro biochemical analyses; CB, the biophysical analyses; and MF and PM, the in vivo experiments. BS, EC and PM wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Table S1
Supplementary Information
Review Process File
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999a;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ –> 5′ exonucleases. Genes Dev. 1999b;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa RL, Legrand P, Wien F, Pineau B, Thompson A, Guimarães BG. RRP6 from Trypanosoma brucei: crystal structure of the catalytic domain, association with EAP3 and activity towards structured and non-structured RNA substrates. PLoS One. 2014;9:e89138. doi: 10.1371/journal.pone.0089138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J, Toth EA. Yeast nuclear RNA processing. World J Biol Chem. 2012;3:7–26. doi: 10.4331/wjbc.v3.i1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Burkard KT, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Mitchell P. Rrp6, rrp47 and cofactors of the nuclear exosome. Adv Exp Med Biol. 2011;702:91–104. doi: 10.1007/978-1-4419-7841-7_8. [DOI] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: the exosome. Biochim Biophys Acta. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Costello JL, Stead JA, Feigenbutz M, Jones RM, Mitchell P. The C-terminal region of the exosome-associated protein Rrp47 is specifically required for box C/D small nucleolar RNA 3′-maturation. J Biol Chem. 2011;286:4535–4543. doi: 10.1074/jbc.M110.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristodero M, Böttcher B, Diepholz M, Scheffzek K, Clayton C. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol. 2008;159:24–29. doi: 10.1016/j.molbiopara.2007.12.012. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedic E, Seweryn P, Jonstrup AT, Flygaard RK, Fedosova NU, Hoffmann SV, Boesen T, Brodersen DE. Structural analysis of the yeast exosome Rrp6p-Rrp47p complex by small-angle X-ray scattering. Biochem Biophys Res Commun. 2014;450:634–640. doi: 10.1016/j.bbrc.2014.06.032. [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenbutz M, Garland W, Turner M, Mitchell P. The exosome cofactor Rrp47 is critical for the stability and normal expression of its associated exoribonuclease Rrp6 in Saccharomyces cerevisiae. PLoS One. 2013a;8:e80752. doi: 10.1371/journal.pone.0080752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenbutz M, Jones R, Besong TMD, Harding SE, Mitchell P. Assembly of the yeast exoribonuclease Rrp6 with its associated cofactor Rrp47 occurs in the nucleus and is critical for the controlled expression of Rrp47. J Biol Chem. 2013b;288:15959–15970. doi: 10.1074/jbc.M112.445759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland W, Feigenbutz M, Turner M, Mitchell P. Rrp47 functions in RNA surveillance and stable RNA processing when divorced from the exoribonuclease and exosome-binding domains of Rrp6. RNA. 2013;19:1659–1668. doi: 10.1261/rna.039388.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Seraphin B, Steinmetz LM, Jacquier A, Libri D. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell. 2012;48:409–421. doi: 10.1016/j.molcel.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–1577. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauer AA, van Hoof A. Genetic interactions suggest multiple distinct roles of the arch and core helicase domains of Mtr4 in Rrp6 and exosome function. Nucleic Acids Res. 2013;41:533–541. doi: 10.1093/nar/gks1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Bratkowski MA, Liu X, Niu C-Y, Ke A, Wang H-W. Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nat Struct Mol Biol. 2014;21:95–102. doi: 10.1038/nsmb.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013a;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- Makino DL, Halbach F, Conti E. The RNA exosome and proteasome: common principles of degradation control. Nat Rev Mol Cell Biol. 2013b;14:654–660. doi: 10.1038/nrm3657. [DOI] [PubMed] [Google Scholar]
- Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, Saibil HR, Lorentzen E. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–942. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan L, Decourty L, Saveanu C, Rappsilber J, Ceulemans H, Jacquier A, Tollervey D. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol Cell Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, Mitsakakis N, Ryan OW, Datta N, Jojic V, Pal C, Canadien V, Richards D, Beattie B, Wu LF, Altschuler SJ, et al. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919–933. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- Royer CA. Improvements in the numerical analysis of thermodynamic data from biomolecular complexes. Anal Biochem. 1993;210:91–97. doi: 10.1006/abio.1993.1155. [DOI] [PubMed] [Google Scholar]
- Schaeffer D, Clark A, Klauer AA, Tsanova B, van Hoof A. Functions of the cytoplasmic exosome. Adv Exp Med Biol. 2011;702:79–90. doi: 10.1007/978-1-4419-7841-7_7. [DOI] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJM. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013;38:485–493. doi: 10.1016/j.tibs.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Stead JA, Costello JL, Livingstone MJ, Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuparevic I, Mosrin-Huaman C, Hervouet-Coste N, Remenaric M, Rahmouni AR. Cotranscriptional recruitment of RNA exosome cofactors Rrp47p and Mpp6p and two distinct Trf-Air-Mtr4 polyadenylation (TRAMP) complexes assists the exonuclease Rrp6p in the targeting and degradation of an aberrant messenger ribonucleoprotein particle (mRNP) in yeast. J Biol Chem. 2013;288:31816–31829. doi: 10.1074/jbc.M113.491290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synowsky SA, van Wijk M, Raijmakers R, Heck AJR. Comparative multiplexed mass spectrometric analyses of endogenously expressed yeast nuclear and cytoplasmic exosomes. J Mol Biol. 2009;385:1300–1313. doi: 10.1016/j.jmb.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol Cell. 2012;48:133–144. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth EV, Januszyk K, Lima CD. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014;511:435–439. doi: 10.1038/nature13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Table S1
Supplementary Information
Review Process File