Abstract
The Mre11–Rad50 nuclease–ATPase is an evolutionarily conserved multifunctional DNA double-strand break (DSB) repair factor. Mre11–Rad50's mechanism in the processing, tethering, and signaling of DSBs is unclear, in part because we lack a structural framework for its interaction with DNA in different functional states. We determined the crystal structure of Thermotoga maritima Rad50NBD (nucleotide-binding domain) in complex with Mre11HLH (helix-loop-helix domain), AMPPNP, and double-stranded DNA. DNA binds between both coiled-coil domains of the Rad50 dimer with main interactions to a strand-loop-helix motif on the NBD. Our analysis suggests that this motif on Rad50 does not directly recognize DNA ends and binds internal sites on DNA. Functional studies reveal that DNA binding to Rad50 is not critical for DNA double-strand break repair but is important for telomere maintenance. In summary, we provide a structural framework for DNA binding to Rad50 in the ATP-bound state.
Keywords: crystal structure, DNA double-strand break repair, homologous recombination, Mre11–Rad50, protein:DNA complex
Introduction
DNA double-strand breaks (DSBs) are highly genotoxic DNA lesions and result in cell death, genome instability, and gross chromosomal aberrations (Chen & Kolodner, 1999; Rothkamm & Lobrich, 2002). DSBs can be formed by ionizing radiation, genotoxic agents, or replicative stress, but are also introduced into the genome in a programmed manner during V(D)J recombination, meiosis, or yeast mating type switching (Costanzo et al, 2001; Longhese et al, 2009; Haber, 2012; Alt et al, 2013). Due to their highly genotoxic nature, DSBs require sensitive detection and repair. Eukaryotic cells react to DSBs through a very complex DNA damage response (DDR) that includes activation of DNA damage checkpoint kinases, chromatin modifications, cell cycle delay, and repair by non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Harper & Elledge, 2007; Lee et al, 2008; Jackson & Bartek, 2009; Stracker et al, 2013).
The evolutionarily conserved Mre11–Rad50 (MR) complex consists of the endo/exonuclease Mre11 and the ATP-binding cassette (ABC)-type ATPase Rad50. In eukaryotes, the complex is a critical factor in the early stages of DNA double-strand break repair and involved in the initial recognition and nucleolytic processing of DSBs (Williams et al, 2007; Stracker & Petrini, 2011). It contains the third component Xrs2 and is referred to as Mre11–Rad50–Xrs2 (MRX) in Saccharomyces cerevisiae and Mre11–Rad50–NBS1 (MRN) in mammals (Dolganov et al, 1996; Carney et al, 1998; Varon et al, 1998). MRN plays a decisive role in HDR (Bressan et al, 1999; Yamaguchi-Iwai et al, 1999) and NHEJ (Moore & Haber, 1996; Xie et al, 2009), meiosis (Moreau et al, 1999), in addition to telomere maintenance (Wilson et al, 1999; Tsukamoto et al, 2001; Reis et al, 2012), and the recruitment of DDR factors such as ATM/Tel1 (D'Amours & Jackson, 2001; Usui et al, 2001; Lee & Paull, 2005). MRN subunits are essential for embryonic viability, while hypomorphic mutations are implicated in severe human genetic disorders that are characterized by genome instability, cancer, and neurological aberration (Petrini, 2000). The bacterial Mre11 and Rad50 homologs are named SbcD and SbcC, respectively. The SbcCD complex helps to prevent gross chromosomal aberrations through degradation of hairpins at inverted repeats facilitating replication restart by recombination (Darmon et al, 2010).
The nuclease activities of MRN are required for DNA end processing and involve endonucleolytic cleavage as well as 3′–5′ exonucleolytic processing in the vicinity of DNA ends (Hopkins & Paull, 2008; Mimitou & Symington, 2008; Zhu et al, 2008; Cejka et al, 2010; Garcia et al, 2011; Shibata et al, 2013). The endonucleolytic cut made by MRN near DSBs, possibly in conjunction with 3′–5′ exonucleolytic degradation towards the DNA end (Garcia et al, 2011; Shibata et al, 2013), liberates covalently attached proteins, such as Spo11, at meiotic breaks and enables subsequent repair by HDR (Neale et al, 2005). A similar ability to remove proteins from DNA ends by introducing endonucleolytic cuts has been demonstrated for the bacterial MR (SbcCD) in vitro (Connelly et al, 2003).
The MR complex consists of a central Mre11 dimer and two Rad50 subunits. These assemble into a large elongated tetrameric complex with an ATP-regulated catalytic head, which binds and processes DNA, and long protruding Rad50 coiled-coil tails, which are able to tether DNA (Hopfner et al, 2000, 2001, 2002; de Jager et al, 2001). The Mre11 helix-loop-helix (HLH) motif C-terminal to the nuclease domain interacts with the base of Rad50's coiled-coil near its nucleotide-binding domain (NBD) and flexibly connects Rad50 to the nuclease dimer (Williams et al, 2011). In the absence of ATP, the two Rad50 NBDs in the MR complex are positioned on the outside of the Mre11 nuclease dimer and both the dsDNA-and metal-binding sites of Mre11 are accessible for DNA (Lammens et al, 2011). In the presence of ATP, however, the two Rad50 NBDs engage through sandwiching two ATP molecules (Hopfner et al, 2000). The engaged Rad50 NBD dimer binds into the Mre11 nuclease and DNA-binding cleft, thereby temporarily blocking the active site of Mre11 (Lim et al, 2011; Mockel et al, 2012).
While the ATP-free open conformation exhibits exonuclease activity, the ATP-bound closed conformation lacks, or has reduced, processive exonuclease activity (Herdendorf et al, 2011; Lim et al, 2011; Majka et al, 2012). The ATP-bound closed conformation still possesses endonuclease activity against ssDNA (archaeal system) (Majka et al, 2012), can clip off the terminal nucleotide on dsDNA in vitro (bacteriophage T4 system) (Herdendorf et al, 2011), or introduce an endonucleolytic cut near the DNA end (Connelly et al, 2003). Finally, eukaryotic MRN requires ATP binding to activate ATM and tether DNA (Lee et al, 2013; Deshpande et al, 2014).
Taken together, the available information suggests that ATP binding and hydrolysis switches MR/MRN between functional states: an open state in which Mre11 DNA binding at the nuclease active sites are accessible, and a closed state in which they are occluded. These different states provide the structural scaffold for the various functions of MR and are correlated to DSB processing on one hand and tethering as well as signaling on the other hand (Deshpande et al, 2014). To correlate the diverse functions with a structural mechanism, it is essential to understand how MR interacts with DNA in each functional state. While structural insights into dsDNA binding by the Mre11 dimer (Williams et al, 2008) may explain the interaction of DNA with the ATP-free open complex, no information is available about DNA binding to Rad50 in the ATP-bound closed conformation. This information is necessary to explain the structural basis for DNA tethering and—in the case of eukaryotic MRN—DSB signaling functions.
We report here the crystal structure of the Thermotoga maritima Rad50 nucleotide-binding domain (tmRad50NBD) in complex with the Rad50-interacting helix-loop-helix motif of Mre11 (tmMre11HLH), Mg2+, AMPPNP, and a dsDNA 15mer. The structural analysis reveals that DNA binds mainly to the N-terminal part of the Rad50 NBD (denoted ‘lobe I’) but forms additional contacts with the coiled-coil region (Fig1). An in vitro DNA-binding analysis validates the observed contacts, but also indicates that positively charged flanking residues contribute to DNA binding. In summary, our results provide a framework for the interaction of MR with DNA in different functional states and establish at the structural level that MR has at least two distinct DNA-binding sites, one on Rad50 in the ATP-bound form and one on Mre11 that is accessible after Rad50 ATP hydrolysis (Fig 5C).
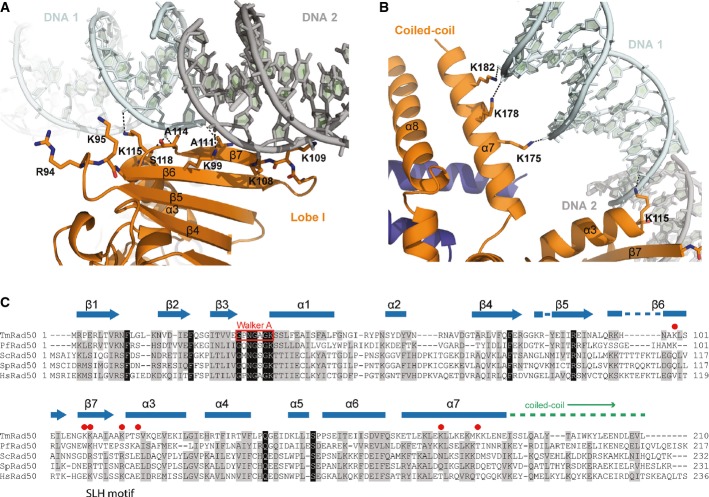
Figure 1. Structure of the tmRad50NBD–Mre11HLH–DNA complex.
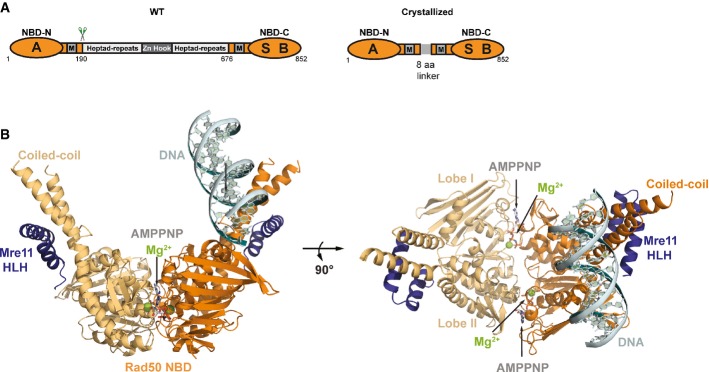
- Domain structure of wild-type Rad50 (WT, left) and the crystallized Rad50NBD construct (right). RAD50 contains a bipartite ATP-binding cassette-type nucleotide-binding domain (NBD, orange) consisting of N-terminal (NBD-N) and C-terminal (NBD-C) segments. The N-terminal segment harbors the Walker A motif (A); the C-terminal segment harbors the Walker B (B) and signature motifs (S). M: Mre11 binding sites. NBD-N and NBD-C are at the ends of a heptad-repeat segment that forms an antiparallel coiled-coil. The center of the heptad-repeat segment contains the Zn-hook dimerization motif.
- Ribbon representation with highlighted secondary structure of the nucleotide-binding domain (NBD) dimer of Rad50 (yellow and orange) in complex with the Mre11 C-terminal helix-loop-helix (HLH) motif, Mg2+-AMPPNP (Mg2+: green sphere, AMPNP: gray-color-coded sticks), and double-strand DNA (cyan ribbon and sticks) shown in two orientations. Rad50 dimerizes in the typical head-to-tail arrangement, sandwiching two Mg2+-AMPPNP moieties in the dimer interface. The DNA binds to a strand-loop-helix motif on one NBD of Rad50 and additional contacts are observed to the adjacent coiled-coil.
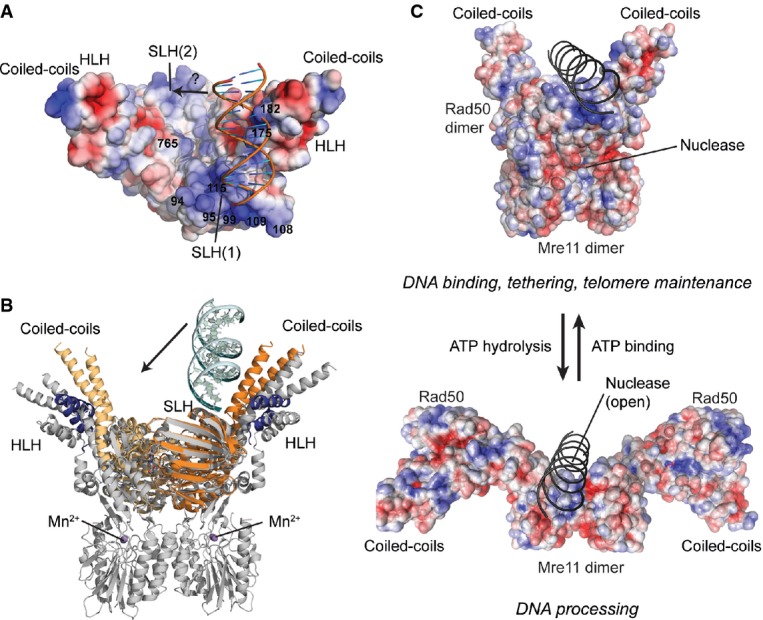
Figure 5. Framework for the interaction of DNA with Mre11–Rad50 functional states.
- Surface representation of tmRad50NBD–Mre11HLH with mapped electrostatic potential (blue: positive; red: negative). DNA is shown as cartoon. The SLH motifs and the surface in the groove between the coiled-coil domains carry a strong positively charged surface potential. A moderate shift of the bound DNA would enable additional contacts to the second SLH motif (see text).
- Superposition of the tmRad50NBD–Mre11HLH–DNA (color code of Fig1) and tmRad50NBD–Mre11FL (gray; PDB code 3THO) models. DNA binds to Rad50 on the opposite side of the Mre11 nuclease dimer and does not sterically compete with Mre11 binding.
- Model of DNA binding and its implication for the functions of the MRN complex. Top: the ATP-bound ‘closed’ conformation with DNA bound to Rad50 is implicated for DNA binding, tethering, and telomere maintenance functions. Upon ATP hydrolysis, Rad50 NBDs move away from each other, exposing the Mre11 active sites to allow for DNA processing.
Results
Structure of the Rad50NBD–Mre11HLH–DNA complex
We crystallized tmRad50NBD (residues 1–190 and 686–852 connected by GGAGGAGG linker) in complex with the C-terminal helix-loop-helix motif of tmMre11 (Mre11HLH residues 347–383), AMPPNP, Mg2+, and a 15mer dsDNA oligonucleotide in space group P1. The crystals diffracted to a limiting resolution of 2.7 Å and contained two (Rad50NBD)2–(Mre11HLH)2–(Mg2+-AMPPNP)2–DNA complexes in the asymmetric unit. Model building and refinement resulted in a final model with good R-factors and stereochemistry (Supplementary Table S1). Both complexes in the asymmetric unit are structurally very similar (Supplementary Fig S1C) (r.m.s.d. values for Cα atoms: 0.25 Å; r.m.s.d. values for DNA backbone phosphates: 0.35 Å) and the following description will be limited to one of these complexes.
Rad50NBD displays the dimer characteristic of ABC ATPases: the two Mg2+ ions and AMPPNP reside in the dimer interface and are sandwiched between the opposing Walker A/B and signature motifs (Hopfner & Tainer, 2003) (Fig 1; Supplementary Fig S1). The coordination of Mg2+ and AMPPNP in the Rad50NBD dimer interface is similar to that in several Rad50/SMC dimer structures in complex with ATP or non-hydrolysable analogs, indicating that the complex represents the typical ATP-bound form of Rad50/SMC proteins (Hopfner et al, 2000; Lammens et al, 2004; Williams et al, 2009, 2011; Lim et al, 2011; Mockel et al, 2012) (Supplementary Fig S1A). Proper ATP coordination is important in the context of the DNA complex reported here, because biochemical studies have shown that the ATP- or analogue-bound form of Rad50 is necessary for interaction with dsDNA (Raymond & Kleckner, 1993; Hopfner et al, 2000; Mockel et al, 2012).
The 15mer dsDNA binds in an orientation in which it is positioned between both coiled-coil arms of the Rad50 NBD dimer (Fig1) and interacts with both the NBD and the coiled-coil domain. On the basis of the number and distance of protein–DNA contacts, the main interaction site is located at the tip of the N-terminal part of Rad50 near the dimer interface (lobe I of the NBD fold). DNA binding is sequence independent and mediated by interactions between the protein main chain and side chain atoms and the sugar–phosphate backbone of the DNA. The DNA does not display significant deviations from the B-form. It protrudes at an approximately 45° angle relative to the Rad50 dimer axis and does not follow the twofold symmetry of the protein.
In lobe I, residues K115 and S118 bind to two consecutive phosphate moieties and additional interactions are made by the β7-strand main chain atoms of A111 and A114 (Fig 2A). Amino acids K99, K108, and K109 are located on the top and outer face of the lobe I β-sheets 6 and 7, respectively, and are positioned so that they may form additional interactions. These latter side chains are in close vicinity to a symmetry-related DNA molecule (DNA 2) that forms a quasi-continuous DNA helix with the directly bound DNA molecule (Fig2A; Supplementary Fig S1B).
Figure 2. Details of the Rad50NBD–Mre11HLH:DNA interface.
- Detailed view of the DNA interactions with the Rad50 NBD (orange) shown in ribbon representation with the contacting residues highlighted as sticks. DNA contacts involve interactions of the strand-loop-helix motif of the Rad50 dimer (residues K99, K108, K109, A111, A114, K115, and S118) with the sugar–phosphate backbone of the two DNA molecules (cyan and gray). The second DNA molecule, depicted in gray, belongs to the symmetry-related molecule and forms a quasi-continuous DNA strand in the crystal structure. Residues R94 and K95 are in close proximity of the DNA-binding region and evidently involved in DNA binding.
- Details of the DNA–protein contacts at the Rad50 coiled-coil region mediated by interactions of lysine residues K175, K178, and K182 with the DNA–phosphate backbone.
- Sequence alignment showing the conservation of residues involved in DNA binding.
We also observe a second, minor contact site for DNA in helix α7 of the Rad50 coiled-coil domain (Fig 2B). This second interaction site is mediated by three lysine–phosphate interactions that are approximately 3 Å distant between residues K175, K178, and K182 and the DNA. While residues K109 and K115 of the strand-loop-helix (SLH) motif are conserved across species (Fig2C), residues K175 and K182 of the coiled-coil domain are less conserved. Despite these two sites of interaction, the DNA molecule has considerably higher B-factor values compared with the neighboring protein regions, suggesting that the DNA is quite flexibly bound in the crystal lattice (Supplementary Table S1).
Comparison of tmRad50NBD–Mre11HLH structures with and without DNA reveals that the globular NBDs and their complexes are largely identical. Nevertheless, the coiled-coil domains and the Mre11 HLH motifs are repositioned by 2–3 Å relative to the apo structure (Supplementary Fig S2).
In summary, we identify here a SLH motif as the main DNA interaction site of Rad50 NBDs, with additional DNA contacts contributed by the coiled-coil region. Remarkably, the interactions are limited to internal sites within dsDNA and are apparently not directed toward the ends of DNA.
Biochemical analysis validates the positively charged DNA-binding groove
In order to validate the structurally identified DNA-binding sites on Rad50, we performed electrophoretic mobility shift assays (EMSAs) of several site-specific mutants of tmRad50NBD–Mre11HLH (Fig 3). In parallel, we analyzed the ATP- or AMPPNP-dependent dimer formation properties of wild-type and all examined mutant tmRad50NBD–Mre11HLH constructs through size-exclusion chromatography (Supplementary Fig S3).
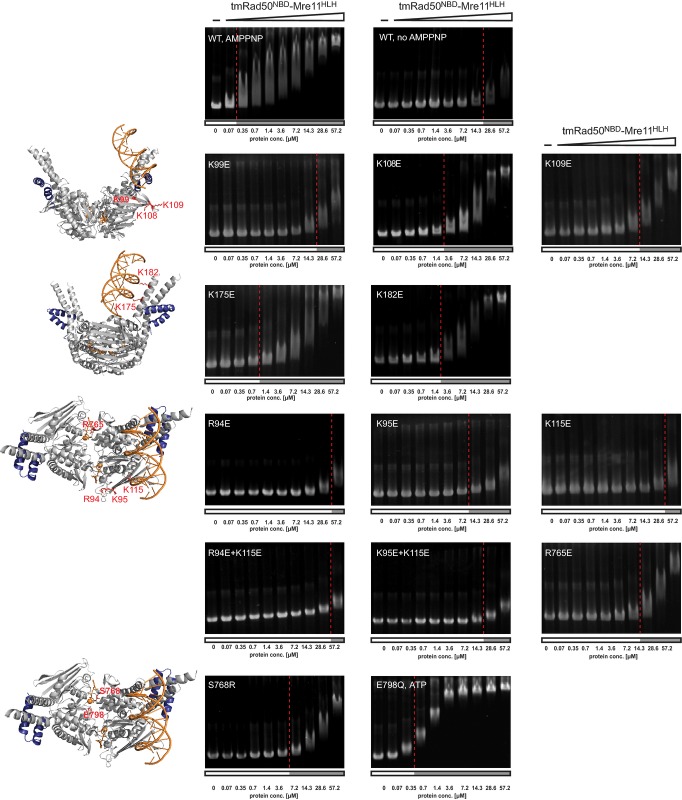
Figure 3. DNA-binding activity of the wild-type and mutant tmRad50NBD–Mre11HLH.
Shown are electrophoretic mobility shift assays of the ΦX174 RF II plasmid in the absence or presence of AMPPNP (or ATP if indicated) with increasing protein concentrations as indicated. For clarity, the positions of the mutations are marked in the structural cartoons left of the agarose gel images. The red lines mark the protein concentrations at which approximately half of the free DNA is shifted (see Supplementary Fig S4). K99E, K108E, K109E, K175E, K182E, and R765E mutants exhibit impaired DNA binding, whereas R94E, K95E, and K115E fail to bind DNA in these assays. ATP-induced structural changes and NBD dimer formation are important for the interaction with DNA as shown by the lack of DNA-binding activity in the absence of AMPPNP and of the non-dimerizing signature motif mutant S768R. In contrast, the Walker B mutation (E798Q) is proficient in DNA binding.
Consistent with previous observations, we find that tmRad50NBD–Mre11HLH is a ‘monomeric’ complex in the absence of ATP/AMPNP but forms a ‘dimeric’ complex (i.e. two tmRad50NBD–Mre11HLH protomers) in the presence of ATP/AMPPNP. The wild-type construct failed to form a stable dimer in the presence of ATP, likely because of ATP hydrolysis. Residue E798 in the Walker B motif positions and polarizes the attacking water molecule in the ATP hydrolysis site and the E-to-Q mutant can be used to ‘trap’ the ATP-bound state (see e.g. Lammens et al, 2004). In agreement with this, mutation of E798 to Q resulted in efficient dimer formation in the presence of ATP. Another notable mutation in this context is the signature motif mutation S768R that was previously shown to prevent ATP-induced engagement of the NBDs of Rad50 (Hopfner et al, 2000; Moncalian et al, 2004). Consistent with this observation, we observed no tmRad50NBD–Mre11HLH dimers even in the presence of AMPPNP. With the exception of S768R, all other mutants examined displayed wild-type-like dimer formation in size-exclusion chromatography, suggesting that the effects of these mutations on DNA binding are not due to aberrant AMPPNP-induced tmRad50NBD–Mre11HLH dimerization.
EMSAs performed with the ΦX174 RF II closed double-stranded DNA plasmid revealed that tmRad50NBD–Mre11HLH possesses robust DNA-binding activity only in the presence of AMPPNP (Fig 3; Supplementary Fig S4). Mutation of any of the three lysine residues (K115E, K175E, and K182E) involved in direct protein–DNA contacts affected DNA binding in vitro. Mutation of the coiled-coil residues K175E and K182E substantially reduced DNA binding, whereas the mutation K115E almost abolished DNA interactions in vitro. In agreement with our structural results, these data indicate that K115 on the tip of lobe I is the principal interaction site for DNA. Since its high B-factor indicates that the DNA is flexibly bound, we also mutated a number of conserved residues that flank these lysine residues. R94E and K95E are evidently also important for DNA binding, suggesting that the DNA-binding surface likely involves a more extended positively charged surface area than the direct contacts observed in the crystal structure. We also mutated the residue R765 in the center of the Rad50 dimer cavity to check whether the DNA may reach the symmetry-related binding site within the Rad50 dimer by traversing the positively charged Rad50 groove. The R765E mutation significantly diminished DNA binding without affecting dimer formation in vitro, lending support to the aforementioned hypothesis.
Finally, the Walker B motif ‘ATP trapping’ mutation E798Q showed robust DNA-binding ability in the presence of ATP. The slight reduction in affinity for DNA, compared to the wild-type protein in the presence of AMPPNP, could result from residual ATP hydrolysis of the E-to-Q mutant. The signature motif mutation S768R, however, substantially reduced DNA-binding activity in the presence of AMPPNP, suggesting that formation of an ATP/AMPPNP-bound NBD dimer of Rad50 is essential for robust DNA interaction (Fig3).
While these studies were performed with ΦX174 RF II plasmid DNA, binding of the wild-type protein was also analyzed with short dsDNA oligonucleotides. We observe that increasing the length of dsDNA from 30mer to 60mer substantially enhanced the affinity of tmRad50NBD–Mre11HLH for the DNA (Supplementary Fig S5A). This length-dependent increase in affinity is consistent with the structural finding that tmRad50NBD–Mre11HLH does not directly bind DNA ends but rather at internal sites of DNA. Taken together, the biochemical DNA-binding analysis confirms the importance of the structurally identified contact residues of Rad50 to the tip of NBD lobe I and suggests that the groove between the two coiled-coil domains, formed following ATP-dependent NBD engagement, provides a positively charged surface for DNA interaction ( 5Fig ; Supplementary Fig S6).
Analysis of the Rad50 DNA-binding site for DSB repair in Saccharomyces cerevisiae
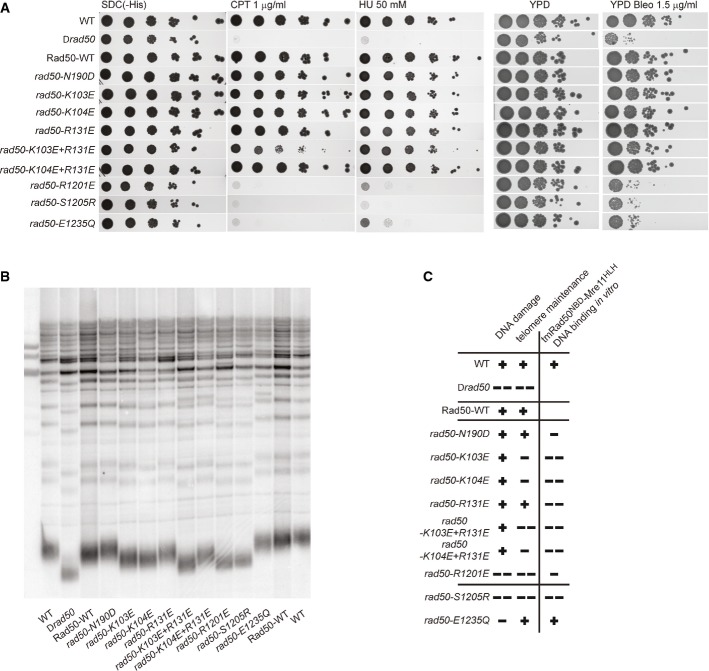
To further investigate the significance of the structurally observed DNA-binding site in the context of the eukaryotic MRN complex, we performed a mutational analysis of the equivalent residues in the S. cerevisiae Rad50 and determined the functional consequences with in vivo assays (Fig 4A and B). Western blot analysis of the mutants confirmed that all mutant proteins were expressed to wild-type levels (Supplementary Fig S6A). We first analyzed the viability and growth of S. cerevisiae transformants on topoisomerase I inhibitor camptothecin (CPT), ribonucleotide reductase inhibitor hydroxyurea (HU), and bleomycin-supplemented medium (Fig4A), as the Δrad50 strain grows poorly on media containing these DNA-damaging agents (D'Amours & Jackson, 2001). While plasmid-expressed wild-type Rad50 rescues the impaired DNA damage response of the Δrad50 strain, Rad50 carrying mutations in the signature motif (S1205ScR) and the Walker B motif E1235ScQ failed to rescue Δrad50, consistent with previous studies (Moncalian et al, 2004; Bhaskara et al, 2007). These mutations show the functional importance of ATP-binding-induced NBD engagement and ATP hydrolysis-induced NBD disengagement in the DSB repair activity of the MRN complex.
Figure 4. In vivo analysis of Rad50 mutations in Saccharomyces cerevisiae.
- Effects of rad50 mutants on S. cerevisiae survival in the presence of DNA-damaging agents. Plate survival assays show that R1201ScE, S1205ScR, and E1235ScQ mutants are deficient in the DNA damage response, comparable to the Δrad50 strain. K103ScE + R131ScE double mutant shows partially inhibited DNA damage response.
- Telomere maintenance assays show altered telomere metabolism for mutations or double mutations at the proposed Rad50 DNA-binding groove (K103ScE, K104ScE, K104ScE + R131ScE, R1201ScE) and the signature motif mutant S1205ScR. A notable exception is the Walker B mutant E1235ScQ, which is not proficient in ATP hydrolysis, suggesting that the ATP-bound, engaged Rad50 dimer is essential for the role of MRN at telomeres.
- Summary of S. cerevisiae phenotypic behavior in DNA repair and telomere maintenance compared with the effect of the corresponding tmRad50 mutations on DNA binding in vitro.
We also mutated the residues corresponding to R94, K95, and K115 in tmRad50NBD–Mre11HLH since these residues strongly affected DNA binding in vitro (Fig 3). The equivalent mutations K103ScE, K104ScE, and R131ScE had little influence on the DSB repair function of S. cerevisiae Rad50 (Fig4; Supplementary Fig S5B).
In the crystal structure, amino acids K99, K108, and K109 interact with a second DNA molecule forming a quasi-continuous DNA helix in the crystal lattice (Supplementary Fig S1B). This raised the question whether these residues are involved in the end-joining function of the MR complex. Indeed, two of these residues, K99 and K109, seem to be conserved in S. cerevisiae (Fig 2C) and the equivalent residues K110Sc and R125Sc have been mutated to glutamic acid to test the influence of these amino acids in a plasmid transformation assay in vivo (Supplementary Fig S6B). Whereas the Δrad50 strain showed substantially reduced transformant yields for NcoI-linearized pRS315-Kan plasmids relative to supercoiled plasmid transformation, the Rad50 point mutants K110ESc and R125ESc led to only insignificant reduction in plasmid recovery (Supplementary Fig S6B). This result argues against an involvement of these residues in end-joining processes in S. cerevisiae.
Rad50–DNA interactions are important for telomere length maintenance in Saccharomyces cerevisiae
MRN plays an important function in telomere maintenance, for example through activation or recruitment of the Tel1 kinase. We performed telomere maintenance assays to analyze the importance of the DNA-binding site on the Rad50 NBDs in this context. Interestingly, we observed a moderate reduction in telomere length in the case of K103ScE, K104ScE, and R131ScE, while the double mutant K103ScE+R131ScE at the SLH motif and the R1201ScE mutant at the center of the positive cleft substantially reduced the length of telomeres, almost equal to Δrad50 levels. This suggests that DNA binding by Rad50 at the SLH motif and possibly in the groove is critical for telomere length maintenance (Fig4B). Notably, while the signature motif mutant S1205ScR also substantially reduced telomere lengths, the Walker B mutant E1235ScQ is fully proficient in telomere maintenance. Together, this analysis suggests that MRN functions in telomere length maintenance in engaged conformation with DNA and ATP-bound NBDs.
Discussion
We report the first structural analysis of DNA binding to an SMC/Rad50/RecN family member of chromosome-associated ABC enzymes. Rad50 is the ATP-binding subunit of the Mre11–Rad50 (prokaryotes and phages) and Mre11–Rad50–Nbs1/Xrs2 (eukaryotes) complexes that are key genome maintenance factors in all kingdoms of life. Although considerable knowledge regarding the architecture of MR and MRN complexes has been acquired (reviewed in e.g. Schiller et al, 2014), the molecular mechanisms of MR/MRN complexes in replication-associated hairpin degradation, telomere maintenance, or DSB repair are still unclear.
Within MR/MRN, Rad50 is suggested to undergo a large structural change that is controlled by ATP binding and hydrolysis (Lammens et al, 2011). In the presence of ATP, the engaged Rad50 dimer sterically blocks the nuclease active site of Mre11 (Lim et al, 2011; Mockel et al, 2012), while following ATP hydrolysis and Rad50 disengagement, the Mre11 nuclease active sites become exposed (Lammens et al, 2011). Whereas Rad50 in its ATP-bound form apparently blocks the Mre11 DNA-binding and nuclease active sites, it becomes proficient for DNA binding in the presence of ATP. Therefore, ATP appears to switch MR/MRN between Mre11 and Rad50 DNA-binding modes.
Our structural analysis reveals how DNA interacts with MR in the Rad50 DNA-binding mode and, together with earlier studies on DNA binding by Mre11 (Williams et al, 2008), provides now a framework to understand DNA interaction of MR in different functional states. DNA predominantly binds to the strand-loop-helix (SLH) motif on the Rad50 NBD. This motif is located on the N-terminal lobe I of the conserved NBD fold. Lobe I harbors also the ATP-binding P-loop, while lobe II carries the signature motif and the coiled-coil protrusion. Besides the SLH motif, additional interactions are found between DNA and two lysine residues of the coiled-coil domain. These contacts of the coiled-coil domains to DNA are interesting in the context of DNA-induced mesoscale conformational changes between the two coiled-coil domains of MRN (Moreno-Herrero et al, 2005). Such a repositioning could be a direct consequence of DNA binding because of, for example, binding of K175 and K182 in the coiled-coil domain to DNA. On the other hand, the coiled-coil domains are notoriously flexible and the amount of their repositioning between apo- and DNA-bound Rad50NBD–Mre11HLH structures is also in the range of crystal lattice-induced differences, so further analysis is necessary to directly link DNA binding and coiled-coil movements. Nevertheless, binding of DNA to the coiled-coil domains is not unexpected and may proceed even further than the observed site in our truncated construct, because the coiled-coil domains are important for high-affinity DNA binding by human MRN (Lee et al, 2013).
The in vitro DNA-binding data indicate that although both interaction sites contribute to DNA binding, the residues within the SLH motif have a considerably larger impact in vitro (Fig3), suggesting this as the main DNA-binding motif in Rad50. Furthermore, we found that positively charged residues flanking the SLH motif R94 and R95 are also required for robust DNA binding, even though they do not directly contact the DNA in our crystal form. These observations suggest that DNA is recognized by lobe I on both sides of the Rad50 dimer, presumably through the extensive positive electrostatic surface potential across both NBDs (Fig5). Importantly, a comparison of the DNA-bound structure with the DNA-free structure of the bacterial MR complex (containing the nuclease domain of Mre11) confirms that there is no steric hindrance between Mre11 and DNA binding on Rad50 and shows that DNA binds to Rad50 on the opposite side of the Mre11 nuclease domain (Fig5B).
It was unexpected that DNA is bound only to one of the two NBDs, considering the internal symmetry of the Rad50 dimer and the biochemical data that clearly show that ATP-induced NBD dimer formation is important for DNA binding (Fig 3) (Hopfner et al, 2000; Mockel et al, 2012). How is ATP binding to Rad50 linked to DNA binding? One possibility is that we visualize an intermediate where DNA is bound to only one of the two SLH motifs of the NBD dimer. A shift of the DNA, taking the lobe I interaction with the SLH motif on one NBD as anchor point, would bring the DNA backbone into a suitable position to also bind the second SLH motif on the other NBD. Such a dual recognition on both SLH motifs requires at least 22-base-pair-long DNA consistent with the length dependence of the DNA binding (Supplementary Fig S5). Such a binding mode would also explain the strong effect of the R1201E mutant, which resides right at the surface of the NBD:NBD interface cleft. In vitro, this mutation still forms NBD dimers in the presence of ATP (tmRad50NBD R765E), so the effect of the mutation might proceed via perturbation of the electrostatics in the groove, but it could also affect the dimer structure in a way that DNA binding is compromised. ATP-dependent NBD dimer formation also leads to a strong positive electrostatic surface potential along the groove between the coiled-coils as previously noted (Hopfner et al, 2000; Lim et al, 2011; Mockel et al, 2012) (Fig 5; Supplementary Fig S7). Thus, NBD dimerization can contribute to robust DNA binding directly by forming a positively charged surface potential. Such a mechanism is consistent with our observation that robust in vitro binding of DNA to the NBDs is affected not only by mutations in directly interacting residues but also by mutations of basic side chains in the vicinity of the DNA in the structure. Finally, ATP binding induces structural changes between both lobes of a single NBD and repositions the SLH domain and the coiled-coil with respect to each other. Since both elements interact with DNA, ATP-triggered positioning of coiled-coil and SLH within a single NBD could be an important step for DNA recognition.
The SLH motif and its flanking residues turned out to play significant but divergent roles in the DNA damage response and telomere maintenance in S. cerevisiae. Single point mutations in the SLH motif had only minor effects on DNA repair functions in response to damaging agents as demonstrated by yeast survival assays. This suggests that the DNA repair pathway can still operate near wild-type physiological level, even if Rad50–DNA interactions are substantially weakened. It is also unlikely, given these minor consequences, that the nuclease activity of MRN is influenced by mutations in the Rad50 DNA-binding site. On the other hand, the strong negative effect of the signature motif mutant S2105ScR clearly argues against a role in which Rad50 merely blocks Mre11 nuclease prior to DNA processing, otherwise this mutation would not have such a strong effect. Likewise, imposing an ATP-bound dimer state through the Walker B motif E1235ScQ mutant abolishes DNA repair activities. We conclude that while DNA interactions to Rad50 appear to be less critical for the DNA repair activities of MRN, the ability to engage and disengage the NBDs in response to ATP binding and hydrolysis remains critical. We do not want to rule out, however, that mutating more than two residues in the identified DNA-binding surface, or combining these mutations with mutations in the coiled-coil domain will more substantially affect DNA repair activities. The coiled-coil domain is necessary for high-affinity DNA interactions—either directly or by promoting higher-order conglomerates—and might be sufficient to promote repair activities even when the DNA-binding site of the NBD is compromised (Lee et al, 2013).
This situation changes in a remarkable way in the context of telomere maintenance. The function of MRN in telomere maintenance includes the recruitment and activation of Tel1ATM (Hector et al, 2007), but also involves DNA-tethering activities (Reis et al, 2012). Here, the SLH mutations had much more severe effects, with the double mutant at the Rad50 DNA-binding site affecting telomere length nearly as much as a Rad50 deletion. Most notably, our analyses revealed that the SLH (K103E and K104E) and Walker B (E1235Q) motif mutations lead to a separation of function phenotype: while mutation of the SLH motif strongly impaired telomere maintenance but had little effect on repair activities, mutation of the Walker B motif resulted in the opposite behavior. Prohibiting, or at least severely delaying, ATP hydrolysis in Rad50 and consequently promoting formation of the closed complex do not influence telomere maintenance, while preventing ATP-induced NBD dimer formation through mutation of the signature motif has detrimental effects on telomere maintenance. Together, our analysis suggests that the Rad50 dimer with bound ATP and DNA is the functional state of MRN in the context of telomere maintenance. These data are consistent with findings that ATP binding but not hydrolysis is important for ATM activation by MRN (Lee et al, 2013; Deshpande et al, 2014).
We do not observe a direct molecular preference for a DNA end by Rad50 in our structural analysis. In general, it has been difficult to demonstrate a strong preference of MR/MRN for DNA ends, except in the case of scanning force microscopy studies, where the clustering of MRN complexes occurred preferentially at DNA ends (Chen et al, 2001; de Jager et al, 2001, 2002). Even in this case, one complex could bind to the end, while others may bind internal sites of DNA. In this regard, we analyzed the Rad50–Rad50 crystal lattice contacts on the quasi-continuous DNA as they might show how MR/MRN clusters on DNA. However, the lattice interactions would clash with the interfaces between Rad50 and Mre11 nuclease dimer in the Mre11–Rad50 complexes (Lim et al, 2011; Mockel et al, 2012), so it is unlikely that they represent interaction sites of MRN clusters on DNA.
With respect to DSB recognition, our results suggest that the affinity for DNA ends might reside in the Mre11 dimer, which can interact with one or two DNA ends or hairpins (Williams et al, 2008), or possibly in an yet to be characterized interplay between Mre11 and Rad50. It is also possible that binding of two DNA molecules via both SLH motifs in the Rad50 dimer and additional DNA:DNA contacts via DNA end stacking or overhangs with microhomologies can lead to DNA end recognition and tethering. These models will be addressed now in future studies. Nevertheless, DNA binding by Rad50 at internal sites could function in initial loading of the complex at or near DNA ends that are blocked by proteins such as Spo11 or Ku (Wasko et al, 2009; Bonetti et al, 2010; Mimitou & Symington, 2010; Garcia et al, 2011; Langerak et al, 2011; Sun et al, 2012), or at sites remote from breaks (Neale et al, 2005; Shibata et al, 2013).
Our data lead to a mechanistic model for MRN activity in which the ATP-bound ‘closed’ conformation of the complex recognizes internal sites of DNA via Rad50, yet also allows for the scaffolding and presumably ATM/Tel1 activation functions of the complex (Fig5C). Hereby, DNA binds between the coiled-coil domains and is suitably positioned to access the Mre11 dimer after ATP hydrolysis, which is suggested to switch MRN from the tethering and signaling mode to the ‘open’ DNA processing mode (Deshpande et al, 2014). In the open mode, MRN might also preferentially interact with DNA ends. It yet needs to be explained how these ATP-driven conformations communicate with the Rad50 coiled-coil structures and zinc hook, since heterozygous mutation of the zinc hook dimerization domains lead to increased ATM activation (Roset et al, 2014).
In general, the structure of the Rad50-DNA complex allows for a better understanding of the mechanism of Rad50/SMC/RecN/RecF-type chromosome-associated ABC ATPases as all these proteins contain an SLH-like secondary structure motif. SMC ATPases form dimers in the context of the cohesin, condensin, and SMC5/6 complexes (Hirano, 2006). For instance, a loop near the SLH motif is critical for DNA-stimulated ATPase activity in archaeal SMC NBDs (Lammens et al, 2004). Likewise, a lysine in the equivalent region is acetylated in eukaryotic SMC3 (part of the condensin complex) and results in the stable establishment of chromosome cohesion (Lee et al, 2008; Rolef Ben-Shahar et al, 2008; Unal et al, 2008), although the mechanistic role of this lysine acetylation still needs to be determined. On the basis of surface electrostatics, it was suggested that also the bacterial RecF and archaeal SMC proteins bind DNA at the NBD surface that contains the SLH motif (Lammens et al, 2004; Koroleva et al, 2007). Although the DNA binding mechanisms of these related chromosome-associated ABC ATPases each need to be determined experimentally, it is conceivable that DNA interacts with the ATP-bound NBD dimers in these complexes in a similar way as with the Rad50 NBDs.
Materials and Methods
Protein expression and purification
Rad50NBD–Mre11HLH from T. maritima was engineered and purified as described before (Lammens et al, 2011).
Crystallization and data collection
Crystals of tmRad50NBD–Mre11HLH–DNA were grown by hanging drop vapor diffusion method. 1 μl of protein–AMPPNP–DNA solution (12 mg/ml protein, 5 mM AMPPNP, 1.275 mM DNA) was mixed with 1 μl reservoir solution (150 mM D-maleic acid pH 6.5, 21% (v/v) PEG3350) and incubated over 400 μl reservoir solution at 20°C. DNA used for crystallization was prepared by annealing oligonucleotides 5′-GGTCGGTGACCGACC-3′ and 5′-GGTCGGTCACCGACC-3′. To this end, oligonucleotides were mixed at 1:1 molar ratio in annealing buffer (40 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2), preheated to 94°C, and cooled down to 4°C at the rate of 0.1°C/s. Prior to flash-freezing, crystals were transferred to cryoprotective condition containing 20% (v/v) glycerol. Native dataset collected at −170°C at wavelength 1.000020 Å at PXI beamline at Swiss Light Source (SLS, Villigen, Switzerland) was indexed and integrated with XDS (Kabsch, 1993). Crystals grew in the P1 space group with cell dimensions: a = 50.2 Å, b = 97.1 Å, c = 107.6 Å, α = 90.6°, β = 89.4°, γ = 98.3° and contained two dimer molecules per asymmetric unit.
Structure determination and refinement
The structure of tmRad50NBD–Mre11HLH–DNA was determined by molecular replacement phasing with PHASER (McCoy et al, 2007), using the AMPPNP–tmRad50NBD–Mre11HLH structure, determined in the absence of DNA, as a search model (PDB entry: 3QF7). The initial model was rebuilt manually in COOT (Emsley & Cowtan, 2004) and refined in PHENIX (Adams et al, 2002). At an early stage of manual building and refinement, a 15-bp DNA molecule was manually build into the Fo-Fc difference density. Further refinements included interactive cycles of bulk solvent corrections, overall B-value refinement, positional and individual B-value refinement, TLS refinement, and manual building. Prior to refinement, 5% of the reflections were randomly omitted to monitor the Rfree value. The Ramachandran statistics, calculated using Procheck (Lovell et al, 2003), of the final model are outliers (%): 0.5, allowed (%): 5.0, and favored (%): 94.5. The outliers are N713, which interacts with the Mre11 HLH motif and the DNA interacting residue K115. Statistics of data collection and model refinement are summarized in Supplementary Table S1. All figures of structural models were prepared with PyMOL (DeLano Scientific).
Electrophoretic mobility shift analyses
DNA-binding activity of tmRad50NBD–Mre11HLH (wild-type and mutants) was analyzed in electrophoretic mobility shift assay. Increasing amounts of dimerised protein (0, 71.5 nM, 357.5 nM, 715 nM, 1.43 μM, 3.57 μM, 7.15 μM, 14.3 μM, 28.6 μM and 57.2 μM) were incubated with 7.15 nM ΦX174 RF II plasmid DNA and 1 mM AMPPNP (ATP in case of E798Q mutant) in 5 mM Tris pH 7.8, 100 mM NaCl, 5 mM MgCl2 in a total volume of 20 μl for 15 min on ice. Reaction samples were then mixed with a loading buffer and separated in 0.5% agarose gel in TA buffer (40 mM Tris, 20 mM acetic acid) for 3.5 h at 80 V and 8°C. Protein–DNA complexes were stained with DNA-intercalating agent GelRed and visualized by UV Imaging System (Intas).
DNA-binding activity of wild-type tmRad50NBD–Mre11HLH was compared with 30mer and 60mer dsDNA. To this end, increasing amounts of dimerised tmRad50NBD–Mre11HLH (0, 125 nM, 250 nM, 375 nM, 625 nM, 1.25 μM, 2.5 μM, 5 μM and 12.5 μM) were incubated with 1 mM AMPPNP and 25 nM 30mer or 60mer fluorescently labeled dsDNA in 5 mM Tris pH 7.8, 100 mM NaCl, 5 mM MgCl2 in a total volume of 10 μl for 15 min on ice. Reaction samples were then mixed with a loading buffer and separated on 8% polyacrylamide native gels in TA buffer for 1–1.5 h at 100 V and 8°C. Protein–DNA complexes were visualized with a Typhoon System (Amersham Biosciences) using the green-excited (488 nm) fluorescence mode.
30mer and 60mer dsDNA were prepared by annealing oligonucleotides 6-FAM-5′-CCGGAAAGCATCTAGCATCCTGTCAGCTGC-3′ with 5′-GCAGCTGACAGGATGCTAGATGCTTTCCGG-3′ and 6-FAM-5′-GCTAATGCCGCGTGCCTTGTCTCACCTTCGATTTAGCATGGTATCAGCAGAGCAAGCCTC-3′ with 5′-GAGGCTTGCTCTGCTGATACCATGCTAAATCGAAGGTGAGACAAGGCACGCGGCATTAC-3′, respectively. To this end, oligonucleotides were mixed with 1.2 molar excess of fluorescently labeled oligonucleotide in annealing buffer (40 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2), preheated to 94°C, and cooled down to 4°C at the rate of 0.1°C/s.
Yeast complementation assay
Overnight cultures of S. cerevisiae transformed with empty pRS313 plasmid (W303-1a wild-type and W303-1a Δrad50 strains, kind gifts of Katja Strässer and Steve Jackson, respectively) or pRS313 carrying wild-type or mutated alleles of Rad50 (W303-1a Δrad50 strain) were diluted in deionised water to OD600 of 1. Serial tenfold dilutions were prepared, and 4 μl of each dilution was plated on SDC(-His)-agar or YPD medium supplemented with DNA-damaging agents: 1 μg/ml camptothecin (CPT), 50 mM hydroxyurea (HU), or 1.5 μg/ml bleomycin (Bleo). Cells were incubated for 72 h at 30°C. All experiments were performed in triplicates.
Western blot analysis
Trichloroacetate (TCA)-precipitated S. cerevisiae lysates were prepared as described before (Yaffe & Schatz, 1984) with modifications: Cells from 20 OD units of overnight culture were pelleted and lysed with 1.5 ml 0.2 M NaOH, 1% β-ME. Total protein was precipitated by addition of 150 μl 0.1% TCA. Protein pellet was resuspended in 25 μl twofold SDS–PAGE sample buffer. For analysis, protein extract corresponding to 5 OD units was resolved in 8% SDS–polyacrylamide gel, transferred onto nitrocellulose membrane, and immunodetected using standard Western blotting technique. Antibodies against S. cerevisiae Rad50 were a kind gift of John Petrini.
Analysis of telomere lengths
Telomere length in S. cerevisiae carrying wild-type and mutated Rad50 allele was analyzed as described before (Schiller et al, 2012).
Dimerization analysis by gel filtration chromatography
Dimerization of the monomeric tmRad50NBD–Mre11HLH complex was initiated by addition of AMPPNP (ATP in case of E798Q mutant). Protein was mixed to a final concentration of 20 mg/ml protein and 5 mM AMPPNP (or ATP) in dimerization buffer (5 mM Tris pH 7.8, 100 mM NaCl, 5 mM MgCl2) and incubated at 8°C. Dimerization was monitored by analytical gel filtration after 1, 5, 24, and 72 h after reaction start. Maximal dimerization was usually achieved after 1–5 h and not longer that 24 h. For practical reasons, protein was always dimerized for 24 h prior to in vitro activity assays.
Saccharomyces cerevisiae plasmid repair assay
The yeast strain used in this experiment was W303-1A Δrad50 transformed with empty pRS313 or pRS313 containing wild-type or mutated Rad50. Competent cells for yeast strains of the respective genotype were transformed with 5 μg of either supercoiled pRS315-Kan or NcoI-linearized pRS315-Kan plasmids by the method of Gietz and Schiestl (2007). Transformation reactions were then plated as serial dilutions onto selective media, and colonies were counted after plates had been incubated for 3–4 days. The relative transformation recovery after plasmid cleavage has been calculated by dividing the number of obtained transformants with the linearized plasmid by the number of transformants with intact plasmids.
Accession codes
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4W9M.
Acknowledgments
We thank Robert Byrne for discussions and comments on the manuscript. We thank the staff of the Swiss Light Source (Villigen) and European Synchrotron Radiation Facility (Grenoble) for support with data collection and analysis. We thank John Petrini for providing antibodies against S. cerevisiae Rad50, Steve Jackson and Katja Strässer for kind gifts of S. cerevisiae W303-1a Δrad50 and strain W303-1a wild-type strains, respectively. We are grateful to Brigitte Keßler for technical support. This work was supported by the European Research Council Advanced Grant ‘ATMMACHINE’, the German Research Council (SFBs 684, 646 and GRK1721) and the excellence cluster Center for Integrated Protein Science Munich to K-PH. AR acknowledges support by the International Max-Planck-Research School Molecular and Cellular Life Sciences and an Boehringer Ingelheim PhD Fellowship.
Author contributions
AR purified and crystallized the protein–DNA complex, built the atomic model, performed biochemical and yeast in vivo experiments, and participated in paper writing. KL performed and supervised the structure determination, model building, yeast in vivo experiment and participated in writing the paper. FUS performed yeast in vivo experiments and participated in writing the paper. CD obtained initial crystals. HF performed the yeast telomere maintenance and plasmid repair assays. K-PH designed and supervised the research and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://emboj.embopress.org
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table S1
Legends for Supplementary Figures
Review Process File
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara V, Dupre A, Lengsfeld B, Hopkins BB, Chan A, Lee JH, Zhang X, Gautier J, Zakian V, Paull TT. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell. 2007;25:647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Clerici M, Manfrini N, Lucchini G, Longhese MP. The MRX complex plays multiple functions in resection of Yku- and Rif2-protected DNA ends. PLoS One. 2010;5:e14142. doi: 10.1371/journal.pone.0014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst) 2003;2:795–807. doi: 10.1016/s1568-7864(03)00063-6. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 2001;15:2238–2249. doi: 10.1101/gad.208701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon E, Eykelenboom JK, Lincker F, Jones LH, White M, Okely E, Blackwood JK, Leach DR. E. coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol Cell. 2010;39:59–70. doi: 10.1016/j.molcel.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, Classen S, Guenther G, Russell P, Tainer JA, Paull TT. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JH. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garcia V, Phelps SEL, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature protocols. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Herdendorf TJ, Albrecht DW, Benkovic SJ, Nelson SW. Biochemical characterization of bacteriophage T4 Mre11-Rad50 complex. J Biol Chem. 2011;286:2382–2392. doi: 10.1074/jbc.M110.178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, Carney JP, Petrini JH, Tainer JA. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- Hopkins BB, Paull TT. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- de Jager M, Wyman C, van Gent DC, Kanaar R. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 2002;30:4425–4431. doi: 10.1093/nar/gkf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007;26:867–877. doi: 10.1038/sj.emboj.7601537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens A, Schele A, Hopfner KP. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr Biol. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, Strasser K, Hopfner KP. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Mejia-Ramirez E, Limbo O, Russell P. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 2011;7:e1002271. doi: 10.1371/journal.pgen.1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Mand MR, Deshpande RA, Kinoshita E, Yang SH, Wyman C, Paull TT. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J Biol Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATPgammaS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Bonetti D, Guerini I, Manfrini N, Clerici M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair (Amst) 2009;8:1127–1138. doi: 10.1016/j.dnarep.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Majka J, Alford B, Ausio J, Finn RM, McMurray CT. ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. J Biol Chem. 2012;287:2328–2341. doi: 10.1074/jbc.M111.307041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockel C, Lammens K, Schele A, Hopfner KP. ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Raymond WE, Kleckner N. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CC, Batista S, Ferreira MG. The fission yeast MRN complex tethers dysfunctional telomeres for NHEJ repair. EMBO J. 2012;31:4576–4586. doi: 10.1038/emboj.2012.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Roset R, Inagaki A, Hohl M, Brenet F, Lafrance-Vanasse J, Lange J, Scandura JM, Tainer JA, Keeney S, Petrini JH. The Rad50 hook domain regulates DNA damage signaling and tumorigenesis. Genes Dev. 2014;28:451–462. doi: 10.1101/gad.236745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Lobrich M. Misrepair of radiation-induced DNA double-strand breaks and its relevance for tumorigenesis and cancer treatment (review) Int J Oncol. 2002;21:433–440. [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, Mockel C, Schele A, Strasser K, Jackson SP, Hopfner KP. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Seifert FU, Linke-Winnebeck C, Hopfner KP. Structural studies of DNA end detection and resection in homologous recombination. Cold Spring Harb Perspect Biol. 2014;6:a017962. doi: 10.1101/cshperspect.a017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, Van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2013;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Roig I, Knobel PA, Marjanovic M. The ATM signaling network in development and disease. Front Genet. 2013;4:37. doi: 10.3389/fgene.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Lee KJ, Davis AJ, Chen DJ. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J Biol Chem. 2012;287:4936–4945. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Wasko BM, Holland CL, Resnick MA, Lewis LK. Inhibition of DNA double-strand break repair by the Ku heterodimer in mrx mutants of Saccharomyces cerevisiae. DNA Repair (Amst) 2009;8:162–169. doi: 10.1016/j.dnarep.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, Guenther G, SilDas S, Hammel M, Russell P, Tainer JA. ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat Struct Mol Biol. 2011;18:423–431. doi: 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, Tainer JA. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wilson S, Warr N, Taylor DL, Watts FZ. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, Kakazu N, Takeda S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table S1
Legends for Supplementary Figures
Review Process File