Abstract
Mitochondrial research is experiencing a renaissance in part due to the recognition that these endosymbiotic descendants of primordial protobacteria appear to be pursuing their own biological agendas. Not only is mitochondrial metabolism required to produce most of the biochemical energy that supports their eukaryotic hosts (us), but mitochondria can actively (through apoptosis and programmed necrosis) or passively (through reactive oxygen species toxicity) drive cellular dysfunction or demise. The cellular mitochondrial collective autoregulates its population through biogenic renewal and mitophagic culling; mitochondrial fission and fusion, two components of mitochondrial dynamism, are increasingly recognized as playing central roles as orchestrators of these processes. Mitochondrial dynamism is rare in striated muscle cells, so cardiac-specific genetic manipulation of mitochondrial fission and fusion factors has proven useful for revealing non-canonical functions of mitochondrial dynamics proteins. Here, we review newly described functions of mitochondrial fusion/fission proteins in cardiac mitochondrial quality control, cell death, calcium signaling, and cardiac development. A mechanistic conceptual paradigm is proposed in which cell death and selective organelle culling are not distinct processes, but are components of a unified and integrated quality control mechanism that exerts different effects when invoked to different degrees, depending upon pathophysiological context. This offers a plausible explanation for seemingly paradoxical expression of mitochondrial dynamism and death factors in cardiomyocytes wherein mitochondrial morphometric remodeling does not normally occur and the ability to recover from cell suicide is severely limited.
Keywords: mitochondria, dynamics, apoptosis, autophagy
INTRODUCTION
Mitochondria are important because: 1. we need them to live; and 2. they can kill us
The heart is the most mitochondrial-rich mammalian organ. Hearts are therefore exquisitely dependent upon, and susceptible to, being damaged by mitochondria. Although the basic mechanisms of mitochondrial oxidative phosphorylation and ATP synthesis are conserved throughout evolution, adult cardiomyocyte mitochondria have morphological and functional properties that distinguish them from mitochondria of other mammalian cell types. It is therefore not clear how attributes of mitochondria from fibroblasts, neurons, and other cells relate to the unique biological context of cardiomyocytes. For example, a PubMed search of the term “mitochondrial dynamics” retrieves >3,500 scientific papers. Yet, those who have performed live-cell studies understand that “mitochondrial dynamism” seems oxymoronic when applied to adult hearts: cardiomyocyte mitochondria appear static, neither moving, fusing, nor dividing. Nevertheless, the proteins that mediate mitochondrial fusion and fission are highly expressed in hearts. Why is this? Hearts also possess a robust machinery for mitochondrial-mediated apoptotic and necrotic cardiomyocyte death, despite a limited capacity to regenerate or replace suicidal cardiomyocytes. Again, why?
Here, recent data that have advanced our understanding of non-canonical functioning of mitochondrial dynamics and death factors in cell pathways dedicated to surveillance and elimination of dysfunctional mitochondria are reviewed. Studies that have employed genetic manipulation to uncover unexpected accessory functions of pro-cell death, pro-fusion, and pro-fission proteins in pathways central to maintaining cardiomyocyte mitochondrial quality are interpreted in a broad context. Because adult cardiomyocytes are non-replicating, compact, and structurally static, the need to move and remodel mitochondrial networks in these cells is minimal. However, the requirement for mitochondrial ATP to fuel cardiac excitation contraction coupling is endless, and the necessity to detect and remove potentially toxic senescent or damaged mitochondria is continuous. For this reason, we propose that maintaining mitochondrial fitness through vigorous quality control may be the dominant normal function of mitochondrial dynamics and cell death factors in adult hearts.
Where do cardiomyocyte mitochondria come from? And where do they go?
The answers to these questions appear obvious from names applied to the relevant processes. Mitochondrial biogenesis means creation of new mitochondria, and mitophagy means eating mitochondria… alpha and omega; done. The quandary is that mitochondria are not born, live and die in the conventional sense. Rather, our mitochondria are literally immortal, having been passed down from our mothers, and from their mothers, all the way back to the figurative “mitochondrial Eve” 1. If one goes even further back in time, mitochondria were initially derived from independent protobacteria that invaded our primitive unicellular ancestors and established permanent residency as endosymbionts 2, 3. After a billion years, and despite having exported 99% of their genes to their hosts (i.e. to our nuclei), mitochondria retain key characteristic of their bacterial ancestors: 1) they have their own circular genomes encoding 13 electron transport complex (ETC) enzymes; 2) they possess replicative, transcriptional, and translational machinery necessary to sustain normal homeostatic functioning, organelle growth, and proliferation through replicative fission; and 3) they communicate with other members of the cellular mitochondrial pool through fusion-mediated exchange of DNA, proteins, and lipids.
Given this context, the answer to the first question is that an adult individual’s cardiomyocyte mitochondria are all descended from his or her embryonic cardiomyocyte progenitor cells. As the embryonic cells grew and proliferated, its resident mitochondria did the same. This represents de facto mitochondrial biogenesis; a perpetual cycle in which mitochondria import nuclear-encoded proteins and synthesize mitochondrial-encoded proteins and genomic components for individual organelle growth; so-called “new” organelles are periodically created via replicative fission (Figure 1a). The same biogenic process is used for homeostatic mitochondrial renewal in adult cardiomyocytes, and is dynamically regulated by physiological or pathological stress. Much has been learned about the genes that coordinate nuclear gene expression for mitochondrial biogenesis, especially PGC-1α and PPARγ (reviewed in 4), but the mechanisms by which mitochondria communicate with the nucleus to promote or suppress biogenic gene expression are poorly described.
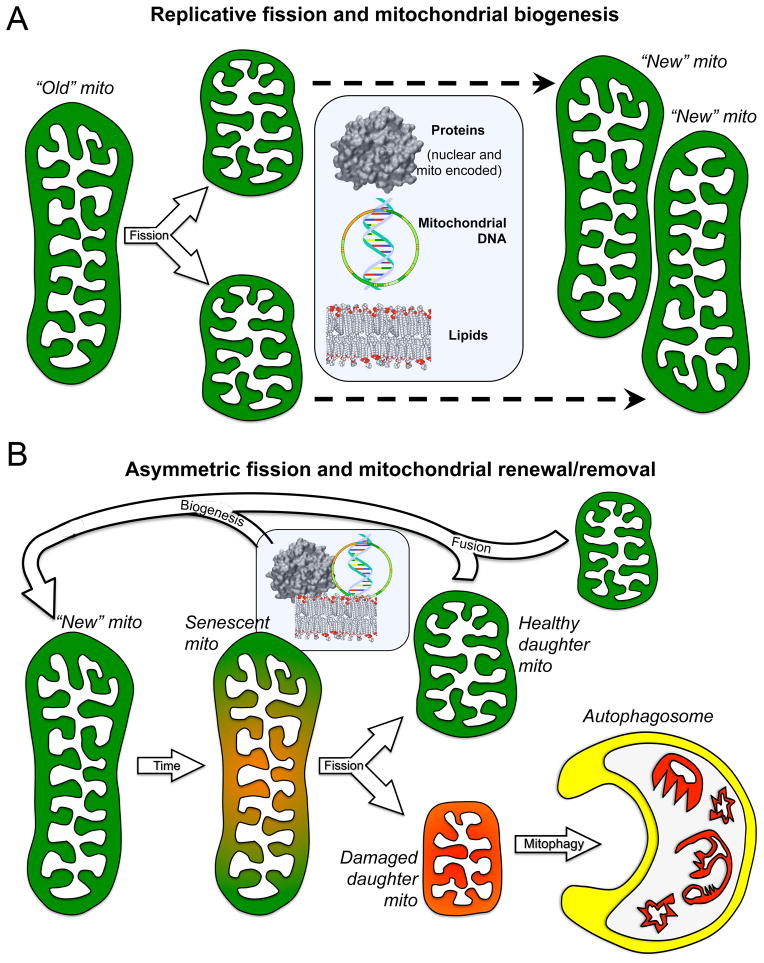
Figure 1. Consequences of replicative vs asymmetric mitochondrial fission.
A. Replicative fission of one healthy “old” parent mitochondrion produces two small healthy daughter organelles that incorporate biogenically produced protein, DNA, and lipids (central rectangle) to grow into “new” mitochondria. B. Asymmetric fission of a damaged or senescent mitochondrion produces one healthy daughter that fuses with other healthy organelles to regenerate the collective, and one severely damaged/depolarized (red) daughter that is rapidly eliminated by autophagosomal engulfment, thereby protecting the cell from mito toxicity and providing new recycled components for biogenic repair.
So, where do mitochondria go? The mitochondria collective of a given organism is essentially immortal as long as the host is viable, but individual mitochondria will sustain damage or eventually become senescent. Modest organelle damage is repaired through biogenic replacement of damaged components, or by fusion with and complementation by a healthy organelle (Figure 1b). Lethal damage of a magnitude or nature that precludes successful repair places the entire cellular mitochondrial pool at risk for contamination (because the consequence of fusion between a severely damaged and healthy mitochondrion is not a larger healthy organelle, but a larger damaged organelle with the potential to fuse with other healthy mitochondria, damaging them, and so on). We call fusion-mediated contamination of the cellular mitochondrial pool “mitochondrial contagion” 5. To prevent mitochondrial contagion, cells employ mitophagy to identify, functionally sequester, and remove severely damaged mitochondria (Figure 1b).
We have begun to unravel the cellular decision process to either repair or remove a damaged mitochondrion. Consider that the extent of mitochondrial damage will inevitably range across a continuum from mild to severe, but the decision to retain or remove a damaged organelle is categorical, i.e. thumbs up or thumbs down. Operationally, the cell must establish a threshold level of mitochondrial damage that will trigger mitophagic removal, while tolerating sub-threshold damage. Because sub-threshold organelle damage can nevertheless be toxic, mitochondria utilize the mechanism of asymmetric fission, integrating mitochondrial fission and mitophagy, to remove functionally compromised organelles before they are sufficiently impaired to do harm to the cell. In this situation, instead of normal replicative fission the senescent mitochondrion preferentially packages damaged DNA and proteins into one daughter organelle while directing undamaged components into the other. Asymmetric fission of a sublethally damaged parent mitochondrion thereby produces one healthy daughter and one damaged daughter. Since the dysfunctional components from the parent are enriched in just one of its daughters, this organelle can meet the threshold for removal by mitophagy, thus avoiding cell toxicity (Figure 1b).
These general concepts illustrate how mitochondrial fusion, fission, biogenesis, and mitophagy are operationally interrelated. The molecular mechanisms by which they interact to orchestrate homeostatic mitochondrial regeneration, renewal, and targeted removal are described in detail in the following sections.
Cardiomyocyte mitochondria, where fusion and fission have special meaning
Before digging into the molecular weeds of cardiac mitochondrial dynamism and quality control, it is worth noting that mitochondria in most mammalian cell types do not resemble the stubby ovoid organelles diagramed in high school textbooks. In cell types most frequently studied, such as fibroblasts, elongated mitochondria are integrated into a branching and highly interconnected cell-wide reticulum (Figure 2). These mitochondrial networks constantly undergo structural remodeling through fusion and fission; fusion promotes intra-network organelle communication and complementation/repair, while fission can isolate damaged mitochondrial components from the network prior to their functional sequestration and physical removal (see Figure 1b). Mitochondrial network fission is accelerated prior to cell mitosis, facilitating equal mitochondria distribution into daughter cells (reviewed in 6). Dissolution of the mitochondrial network during cell replication may also be necessary to accommodate structural changes in the parent, and then daughter, cells as they divide and reform.
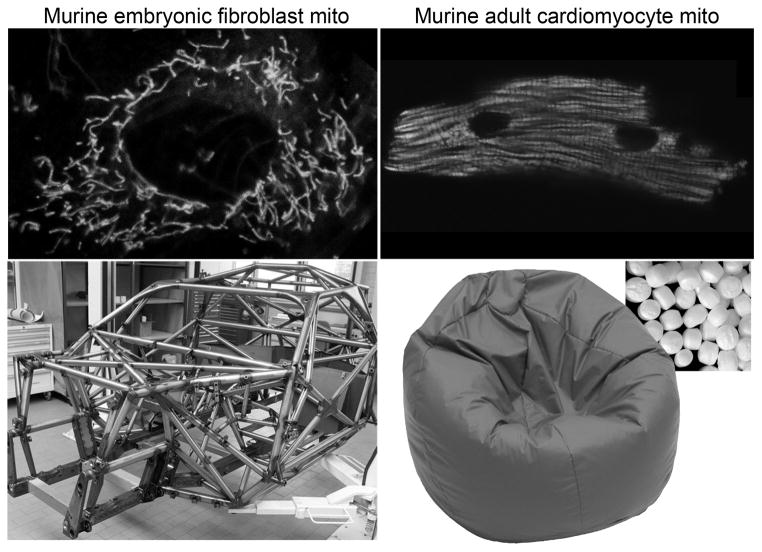
Figure 2. Structural differences between mouse fibroblast and adult cardiomyocyte mitochondria.
A. Top left is Mito-Tracker Green stained filamentous, interconnected mitochondria of a cultured murine embryonic fibroblast; below is the roll cage of a NASCAR racing car, specifically designed to withstand compressive forces. B. Distinct individual rounded GFP-labeled mitochondria on an isolated adult mouse cardiomyocyte; below is a bean bag (inset shows “bean” structure), specifically designed to be readily and reversibly deformable.
By comparison, in the adult cardiomyocytes of flies, mammals, and organisms that evolutionarily span the two, mitochondria exist largely as discrete rounded organelles packed together between the myofibrils (Figure 2). Adult cardiomyocyte mitochondria also cluster within the perinuclear and subsarcolemmal regions, but in no instance do they normally form interconnected networks. Thus, compared to non-myocytes, individual cardiomyocyte mitochondria appear “fragmented”.
In considering why cardiomyocyte (and skeletal muscle) mitochondria lack the connectivity that typifies most other cell types, a biomechanical explanation arises: a collection of individual cardiomyocyte mitochondria is intrinsically more deformable than a highly interconnected network. The real-world analogy is the interconnected components of the roll cage of a racing car, a structure specifically designed to resist deformation, compared to a bean bag that can readily and reversibly conform to almost any shape (Figure 2). Thus, mitochondrial fragmentation that is observed in most cells prior to mitosis not only enables partitioning of mitochondria to daughter cells, but promotes structural malleability necessary to accommodate plasticity in cell size and shape. If mitochondria in these cells maintained their basal interconnected morphometry then, like a roll cage, cell plasticity would be physically restricted. By extension, if a cardiomyocyte contained highly interconnected mitochondria, it might have to cyclically fragment and reform mitochondrial interconnections with each contraction and relaxation cycle, or expend sufficient energy to overcome the resistance produced by this internal elastic component. Consistent with a link between contraction and a fragmented mitochondrial structure, early embryonic cardiomyocytes exhibit a more network-like and interconnected mitochondrial morphometry, but acquire the typical mature individual ovoid structure later in development as the heart is increasingly called upon to generate circulatory flow 7. Therefore, mitochondrial networking is at least dispensable, and likely would be detrimental, to the normal pump functioning of adult cardiac myocytes.
Because mitochondrial networks do not exist in adult cardiac myocytes, and any role for mitochondrial fusion and fission in network remodeling is therefore irrelevant, genetic interdiction of mitochondrial fission and fusion in hearts has uncovered atypical functions of mitochondrial dynamics proteins. Protein mediators of mitochondrial fusion and fission are highly conserved across evolution, and the biomolecular events they evoke are well known 8, 9. Mitochondrial fission results from recruitment and directed multimerization of a dynamin superfamily GTPase, Dynamin-related protein 1 (Drp1). Drp1 oligomerizes in a head to toe configuration around the mitochondrial equator and constricts in a GTP-dependent manner, ligating the parent organelle into two daughters (Figure 3). The specific signals that stimulate Drp1 recruitment to pre-fission mitochondria are not fully understood, although contact points with ER play a role in cells with interconnected mitochondrial networks 10. Indeed, accumulating evidence supports roles for actin, myosin II, and the ER-associated protein inverted formin 2 acting upstream of Drp1 to direct its circumferential localization on mitochondria 11, 12.
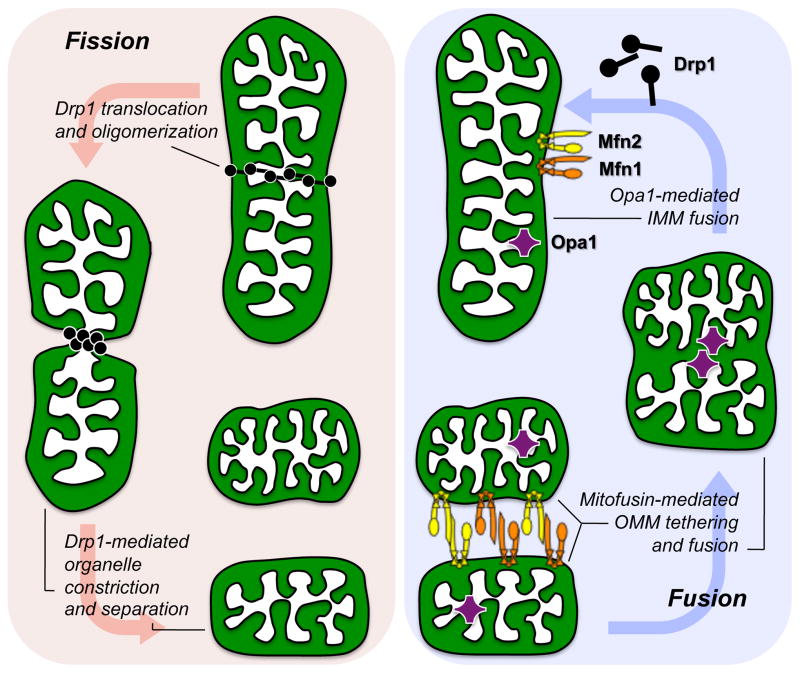
Figure 3. Molecular mechanism of mitochondrial fission and fusion.
The three molecular drivers of fission and fusion are schematically depicted as they would be associated with a normal mitochondrion. Replicative fission (left red panel) is initiated by recruitment of cytosolic Drp1 to the organelle, Drp1 oligomerization, and constriction of the parent into two daughters. Asymmetric fission uses the same mechanism. Fusion (right blue panel) requires initial Mfn1/Mfn2-mediated outer membrane tethering followed by fusion, and finally Opa1-mediated inner membrane fusion.
Mitochondrial fusion is somewhat more complex, requiring three distinct steps: tethering, outer membrane fusion, and inner membrane fusion. Tethering is the physical attachment between outer membranes of two mitochondria, and is a prerequisite to actual membrane fusion. Tethering can be mediated by either (or both) of two other dynamin superfamily GTPases, the mitofusins (Mfn) (so named because they also promote outer mitochondrial membrane fusion). If one considers the amino terminal GTPase domain of a Mfn protein to be its “head”, then its carboxyl terminal “tail” is comprised of an alpha helical heptad repeat that can homotypically (Mfn1-Mfn1 or Mfn2-Mfn2) or heterotypically (Mfn1-Mfn2) dimerize in trans (i.e. tail to tail) with other Mfn molecules on adjacent mitochondria. Tethering occurs via trans dimerization of the anti-parallel coiled coils of two mitofusin tails, and is GTPase-independent 13 (Figure 3). The physical nature of this interaction is much like that of Velcro hooks attaching to loops. Because the Mfn-Mfn tethering interaction is accomplished by hydrogen bonding, tethering (like Velcro binding) is fully reversible.
Mitochondrial tethering can be followed by physical fusion between the outer membranes of two tethered organelles. Fusion is also mediated by Mfn1 or Mfn2, but unlike tethering, is dependent upon GTP hydrolysis and is irreversible. The product of outer membrane fusion is an organelle having two distinct internal matrix structures, described as a “double yolk egg” 14 (Figure 3). This organelle configuration is not commonly observed because outer membrane fusion is normally followed immediately by inner membrane fusion mediated by yet another dynamin superfamily GTPase, Optic Atrophy 1 (Opa1) 15, 16 (Figure 3).
Mitochondrial fission and fusion occur constantly in cultured fibroblasts, and is readily observed using a variety of fluorescent techniques. For those who are interested, static and time-lapse moving images demonstrating typical mitochondrial fission/fusion dynamics in HeLa cells and mouse fibroblasts have been posted on his web-site by Dr. David C. Chan, one of the pioneering researchers into mechanisms of mitochondrial fusion (http://www.hhmi.org/research/mitochondrial-dynamics-development-and-disease). Genetic ablation or suppression studies in cells such as these wherein mitochondria are constantly repositioning and mutually interacting have proven that Drp1, Mfn1, Mfn2, and Opa1 are essential to normal mitochondrial network maintenance and remodeling. However, fluorescent live cell studies of adult Drosophila heart tubes found no evidence for intracellular mitochondrial movement or inter-organelle fusion or fission over several hours 17. Absence of measurable mitochondrial dynamism in adult cardiomyocytes raises the question of what secondary or ancillary functions mitochondrial dynamics factors might be playing in their apparently static mitochondria.
Cardiac mitochondria and ducks: calm on the surface, but working like mad underneath
As noted above, fibroblast mitochondria are obviously dynamic, whereas cardiomyocyte mitochondria might best be characterized as languid. It therefore seems incongruous that mitochondrial network remodeling fission/fusion proteins are abundant in normal adult cardiomyocytes. To more completely define atypical roles of these factors several groups independently began in vivo genetic dissections of mitochondrial fusion pathways in fly and mouse hearts.
The initial in vivo genetic manipulations of cardiac mitochondrial fusion factors were performed in the heart tubes of Drosophila fruit flies 17. Flies have a single mitofusin, called MARF (mitochondrial assembly regulatory factor), which is suppressible in a tissue-specific manner using transgenically-expressed RNAi 18. MARF RNAi was transgenically expressed in fly hearts under control of a tinman promoter driver (tinman is the Drosophila ortholog of the mammalian cardiomyocyte-specific differentiation factor, nkx2.5). Compared to controls, MARF-deficient fly cardiomyocytes had smaller mitochondria; cardiac MARF-deficient flies developed enlarged and hypocontractile heart tubes, i.e. a cardiomyopathy 17. Suppression of Drosophila Opa1 using the same tinman RNAi approach induced a somewhat different mitochondrial dysmorphology characterized by greater organelle heterogeneity, but a similar cardiomyopathy 17. These results provided the first evidence that (in flies at least) cardiomyocyte mitochondria must undergo fusion (because they became smaller as a consequence of suppressing MARF), and also demonstrated that MARF- and Opa1-dependent mitochondrial fusion is (for whatever reason) essential to normal heart functioning.
Higher organisms, including mammals, have two mitofusin proteins encoded by distinct genes. Germ-line ablation of either mitofusin gene in mice is embryonic lethal, likely due to adverse consequences on placental development 19. However, studies of embryonic fibroblasts derived from Mfn1, Mfn2, and Mfn1/Mfn2 double knockout mice revealed that: 1. Mfn1 and Mfn2 are almost entirely redundant in promoting mitochondrial tethering and outer membrane fusion; each can substitute when the other is absent 19. 2. Mfn2 is unique in its ability to localize to endoplasmic reticulum and tether this calcium-containing organelle to mitochondria; Mfn1 does not share this activity 20. 3. Absence of both mitofusins is compatible with cell viability, but mitochondria lacking both Mfn1 and Mfn2 are severely dysmorphic and partially depolarized 21. David Chan’s group worked around embryonic lethality of germ-line Mfn1 and Mfn2 ablation by creating the respective floxed allele mice, which have been used by multiple research teams to produce viable tissue-specific mouse genetic knockouts for Mfn1 and Mfn2. The Chan laboratory was the first to prove in vivo functional redundancy between the two Mfns and show that mitofusin activity is essential for normal brain and skeletal muscle development and function 22, 23. As detailed below, the same Mfn1 and Mfn2 floxed allele mouse models have been used in combination with cardiomyocyte-specific Cre lines to ablate Mfn1 and Mfn2 individually, and both Mfns in combination, in mouse hearts.
To date, Mfn1 and Mfn2 have been genetically deleted from mouse hearts using three different types of cardiomyocyte-specific Cre mouse lines. The genetic approach that most closely paralleled the Drosophila heart tube MARF knockdown study 17 utilized Cre expressed from an nkx2.5 knock-in allele. Like fly tinman, the mouse nkx2.5 gene is activated very early in heart development, conferring largely cardiac-specific gene ablation beginning in the cardiac crescent (~E7.5). Combined Mfn1 and Mfn2 ablation in the early embryonic heart was uniformly lethal, revealing an absolute requirement for mitochondrial fusion in normal cardiac development 24. Perinatal cardiac-specific ablation of both Mfn1 and Mfn2 using myh6-Cre also resulted in early lethality (by P16), with mitochondrial dysmorphology and matrix degeneration, impaired biogenic gene expression, and cardiomyopathy 25.
Early lethality with embryonic or perinatal Mfn1/Mfn2 ablation mandated an alternate approach to uncover the effects of perturbing mitochondrial fusion in adult hearts. Accordingly, we conditionally ablated the Mfn1 and Mfn2 genes using an estrogen receptor (ER)-Cre fusion protein expressed as a myh6-driven transgene. The ER-Cre transgene is activated in cardiomyocytes after birth, but Cre-mediated gene recombination requires administration of tamoxifen (or raloxifen) to induce nuclear Cre translocation. Using this system, mice carrying all five gene alleles required for conditional combined cardiac Mfn1/Mfn2 ablation were grown until 8 weeks of age, at which time Mfn1 and Mfn2 gene recombination was induced using pharmacological administration of estrogen receptor ligand. Concomitant ablation of Mfn1 and Mfn2 in the adult mouse heart induced mitochondrial fragmentation, cardiomyocyte respiratory defects, and rapidly progressive cardiac dilatation with lethal heart failure within 6–8 weeks 24. Based on the half-time for mitochondrial size reduction after combined Mfn1/Mfn2 ablation, we calculated that a complete mitochondrial fission-fusion cycle takes ~15 days in the adult mouse heart. The extremely slow rate of fusion and fission explains why cardiomyocyte mitochondria appear static in short-term studies. The conditional Mfn1/Mfn2 double cardiac knockout experiments provided the first evidence that mitochondrial fusion occurs in adult cardiomyocytes, and demonstrated that these two mitochondrial fusion proteins are necessary for mammalian cardiac health. However, this early work did not define either the essential physiological requirement for cardiac mitofusins or the underlying physiological defect that provokes cardiomyopathy when their expression is interrupted.
Regulation of calcium signaling by Mfn2 and mitochondrial death proteins
As introduced above, inter-mitochondrial tethering by mitofusins is mediated by reversible linking of the ends of filamentous structures attached to bodies requiring physical attachment (like Velcro); mitofusins have sticky antiparallel coiled-coils at their C-termini (whereas Velcro has plastic hooks and loops). An intrinsic characteristic of this tethering mechanism is that the tethers themselves are agnostic as to what they link: It doesn’t matter whether Velcro is on your ball peen hammer or David Letterman’s “Human Fly” jump suit 26, it will form attachments with and stick to its opposite number. This idea also applies to mitofusins; they tether mitochondria together because they are located on mitochondrial outer membranes. By extension, if mitofusins were present on other organelles they could tether those organelles to mitochondria. This hypothetical promiscuity in organelle bonding has not been described for Mfn1, which is exclusively mitochondrial. However, a fraction of Mfn2 has been localized to endoplasmic/sarcoplasmic reticulum (SR), as well as to mitochondria, in fibroblasts and cardiomyocytes 20, 27–29. Consequently, trans inter-molecular binding between ER/SR-localized Mfn2 and mitochondrial-localized Mfn1 or Mfn2 physically links these calcium-containing and ATP-producing organelles. Tethering of cardiomyocyte mitochondria to SR has functional implications because it creates protected appositional microdomains through which high concentrations of ionic calcium can be exchanged between organelles (without diffusing away into the cytosol). Cardiomyocyte-specific genetic ablation of Mfn2 demonstrated the existence of these microdomains, and showed how they enhance mitochondrial sensing and uptake of SR-derived calcium29, 30. Mfn2-mediated tethering of SR to mitochondria thus constitutes the sensory arm of a rapid response system that minimizes mitochondrial ATP depletion and reactive oxygen species (ROS) production when workload and metabolic demand are acutely increased. Because calcium can also be a potent stimulus for opening the mitochondrial permeability transition pore (MPTP) 31, the downside to physical tethering of SR and mitochondria is increased sensitivity of MPTP opening to SR-derived calcium 32. Indeed, cardiomyocyte-specific Mfn2 ablation protects the heart against a number of insults linked to calcium-mediated MPTP opening, including ischemia-reperfusion injury 28. Thus, under pathological conditions the molecular and functional features of Mfn2 that normally promote mitochondrial health (through fusion-mediated complementation) and that facilitate a rapid metabolic response to acute hemodynamic stress (through SR-mitochondrial calcium crosstalk) also predispose to MPTP-mediated cardiomyocyte death.
It is interesting that dual localization of factors to mitochondria and adjacent endo/sarcoplasmic reticulum, with resulting modulation of SR-mitochondrial calcium crosstalk, is a common characteristic of mitochondrial factors implicated in programmed cell death. For example, the pro-cell death mitochondrial Bcl2 family protein Nix (also known as Bnip3L) originally garnered interest because its transcriptional upregulation in cardiac hypertrophy induces apoptotic cardiomyocyte death 33–35. However, Nix also localizes to SR and modulates SR-mitochondrial calcium signaling that can evoke MPTP opening. Indeed, cardiomyocytes undergoing Nix-mediated cell death exhibit characteristics of both apoptosis and necrosis 36. By genetically targeting Nix either to mitochondria or to endo/sarcoplasmic reticulum we showed that Nix organelle localization, and not some intrinsic property of the protein, determines whether Nix provokes cell death via (mitochondria pathway) apoptosis or (endo/sarcoplasmic reticulum and MPTP-mediated) necrosis 37. Likewise, the closely related pro-apoptotic factor Bnip3, which mediates apoptotic cardiomyocyte death and adverse cardiac remodeling after myocardial infarction, 38 contributes to cardiac dysfunction by localizing to reticular structures and increasing sarcoplasmic reticulum-mitochondrial calcium transport 39, 40. This functional duality in factors regulating mitochondrial homeostasis and SR-mitochondrial calcium signaling is a common theme that emerges from cardiac-specific in vivo molecular dissections of outer membrane mitochondrial fusion and apoptosis proteins.
Mitochondrial fusion, calcium regulation, and cardiac development
Mitochondria are calcium rich; sufficiently calcium rich that mitochondrial disruption and release of calcium can produce dystrophic calcification, as observed in necrotic myocardium 41. Conversely, cardiac mitochondria can act as calcium sponges, taking up and buffering available free cytosolic calcium 42. As detailed below, a surprising role was recently discovered for mitochondrial calcium buffering in signaling pathways that orchestrate expression of genes that direct cardiomyocyte differentiation during embryonic cardiogenesis 7.
Mitochondrial structure and sub-cellular organization in embryonic cardiomyocytes and cardiomyocyte progenitor cells resemble those in fibroblasts (i.e. long reticular organelles organized into highly interconnected networks), rather than the short round individual organelles observed in fully differentiated adult cardiomyocytes. These developmental differences in mitochondrial morphometry lead to surprising functional interactions between mitochondrial network remodeling, mitochondrial calcium uptake, and embryonic cardiac development. Differentiating embryonic stem cell-derived cardiomyocytes normally have a copious number of perinuclear mitochondria arranged in reticular networks. Inhibiting mitochondrial fusion by suppressing either Mfn2 or Opa1 promotes network discontinuity and produces fragmented mitochondria that redistribute away from the nucleus and toward the cell membrane7. The atypical sub-sarcolemmal location of the fragmented mitochondria physically positions them to act as buffers for normal transmembrane (outside-in) calcium signaling, paradoxically increasing capacitative calcium entry and activating calcineurin that, in turn, increases notch signaling 7. Because notch suppression is a requisite for cardiomyocyte differentiation 43, 44, calcineurin-mediated notch activation in cardiomyocyte progenitor cells with defective mitochondrial fusion impairs their differentiation into cardiomyocytes (Figure 4). We determined the in vivo impact of impaired mitochondrial fusion on cardiomyocyte differentiation by genetically interrupting mitochondrial fusion (combined Mfn1 and Mfn2 ablation) in cardiomyocytes from the earliest stages of cardiac development (using nkx2.5-Cre) 7. Mfn1/Mfn2 double deficient hearts exhibited depressed Mef2c, Nkx2.5 and SRF-dependent gene expression, interruption of normal myocardial development, and lethal embryonic myocardial hypoplasia 7. The spectacular effects of induced mitochondrial dysmorphology on developmental cardiac gene expression and heart development provide a striking example of how these semi-autonomous cytosolic organelles can commandeer critical nuclear gene expression pathways to determine cell fate, and not just cell viability.
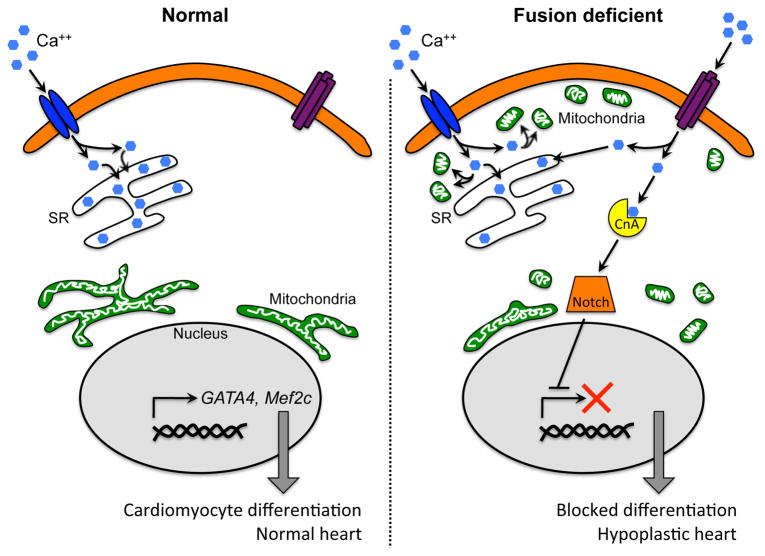
Figure 4. Mitochondrial fusion and control of cardiomyocyte differentiation/heart development.
Functional interactions between L-type calcium channels (LCC; blue), store-operated calcium channels (TRPC; purple), mitochondria (green), calcineurin A (yellow), Notch (orange), and developmental gene expression as conceived in cardiomyocyte progenitor cells. Left panel shows normal stem cell with fused peri-nuclear mitochondria in which LCC calcium signaling is normal and capacitative calcium entry is low. Right panel shows how mitochondrial fragmentation and sub-sarcollemmal redistribution disturbs LCC signaling through mitochondrial calcium uptake (“sponge”), invoking capacitative calcium entry that activates calcineurin and downstream Notch, repressing developmental gene expression.
Mitophagy, mitofusins, and mitochondrial quality control
A concept that emerges from experimental manipulation of mitochondrial fusion in hearts is that the critical pathophysiological roles of mitofusins derive more from higher order functions of organelle tethering and fusion than from structural mitochondrial remodeling per se. Thus, tethering of mitochondria to SR by Mfn2 is important for trans-organelle calcium signaling that normally regulates acute metabolic responsiveness 29, and under pathological conditions can evoke the mitochondrial permeability transition 28. Likewise, suppression of mitochondrial fusion provokes increased mitochondrial calcium buffering that can dysregulate developmental cardiac genes 7. Evidence is accruing that mitochondrial fusion factors also play central roles in mitophagic mitochondrial quality control. Indeed, since Mfn1 and Mfn2 serve functionally redundant roles in mitochondrial fusion, it is understandable how one of the redundant mitofusins may have evolved additional ancillary functions. Below we make the case that regulating mitophagy could be a primary function of Mfn2 in the heart.
One of the unexpected findings of early studies was that hearts with genetically engineered mitochondrial fusion defects were ineffective in clearing abnormal mitochondria 17, 24. Given high mitochondrial density in cardiomyocytes and the importance of removing damaged ROS-producing mitochondria before they become cytotoxic, it was expected that simple interruption of fusion-mediated organelle complementation (without concomitant suppression of biogenic renewal or mitophagic removal pathways) would impact organelle morphometry, but not mitochondrial fitness. However, the decrease in organelle size after complete Mfn ablation in mouse hearts is associated with markedly increased numbers of degenerated cardiac mitochondria during the early stages of the resulting cardiomyopathy 17,24. Ablation of Mfn2 alone in mouse hearts did not impede mitochondrial fusion (which was still induced by endogenous Mfn1 as evidenced by actual enlargement of Mfn2-deficient cardiomyocyte mitochondria 28, 29), but nevertheless provoked abnormal cardiomyocyte respiration and a slowly progressive chronic cardiomyopathy 45. The distinct mitochondrial and cardiac phenotypes of combined cardiac Mfn1/Mfn2 ablation compared to selective Mfn2 deficiency reveal different mechanisms for the resulting cardiomyopathies: Because abnormal mitochondria accumulated after selective Mfn2 ablation and combined Mfn1/Mfn2 ablation, but not after selective Mfn1 ablation, we hypothesized that Mfn2 is uniquely important for culling damaged mitochondria.
As introduced above, mitophagy or “eating mitochondria” is the central mechanism by which damaged mitochondria are removed to prevent cell damage. Two critical mediators of mitophagy are PINK1 and Parkin, encoded by genes in which loss of function mutations cause early-onset autosomal recessive Parkinson’s disease. The history of how conventional human genetic studies, gene manipulation in fruit flies and mice, and basic cellular research have all converged to reveal mitophagic dysfunction as the underlying cause of these heritable forms of Parkinson’s disease is a fascinating one, and the interested reader is referred to recent excellent reviews 46,47. For the purpose of understanding mitophagy signaling in the heart, PINK1 is a mitochondrial kinase that accumulates in damaged mitochondria and initiates the signal for translocation of the Parkin cytosolic E3 ubiquitin ligase specifically to damaged organelles 48. Parkin-mediated ubiquitination of outer membrane proteins on these damaged mitochondria attracts autophagosomes, thus initiating mitophagy (Figure 5). Mitochondria engulfed by an autophagosome are transferred to lysosomes and degraded, which minimizes cellular toxicity from reactive oxygen species that damaged mitochondria often produce and release. Interruption of PINK1-Parkin signaling disrupts mitophagy. Accordingly, accumulation of abnormal mitochondria in dopaminergic neurons is the pathological sine qua non of Parkinson’s disease linked to PINK1 and Parkin gene mutations. It is important to recognize that mitochondria can also be eliminated through non-selective macroautophagy 49 that is transduced though different signaling pathway(s) that are independent of PINK1 and Parkin. Because the terminal stages of both selective mitophagy and generalized mitochondrial autophagy proceed via autophagosomal mitochondrial engulfment and transfer to lysosomes, it can be challenging to differentiate between these processes.
Figure 5. The PINK1-Parkin mechanism of mitophagy.
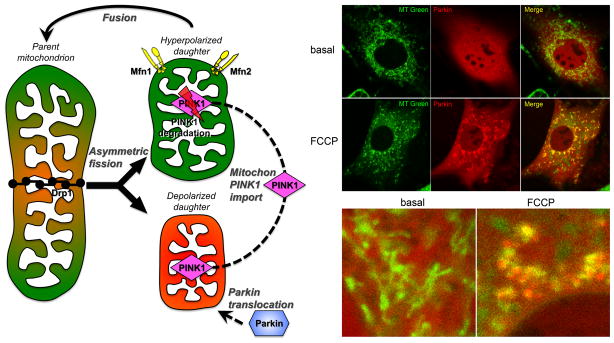
Left – Schematic diagram of PINK1-Parkin initiation of mitophagy signaling after asymmetric mitochondrial fission. Right – Confocal fluorescent images showing mcherryParkin (red) translocation from cytosol to mitochondria (MitoTracker green) after mitochondrial depolarization with the uncoupling agent FCCP. Parkin-containing mitochondria appear yellow in the merged image.
As Parkin is central to mitochondrial quality control in neurons, skeletal muscle, and perhaps cardiomyocytes 50,51, we examined the integrity of Parkin signaling in Mfn2-deficient mouse cardiomyocytes in which abnormal mitochondria appeared not to be properly eliminated, thus testing the hypothesis that genetic deficiency of Mfn2 interferes with normal mitophagy. Parkin was abundant in cardiomyocyte sarcoplasm, but did not translocate to depolarized mitochondria of Mfn2-deficient cardiomyocytes. By contrast, depolarization robustly stimulated Parkin translocation to mitochondria of wild-type and Mfn1-deficient cardiomyocytes 45.
Parkin recruitment to 45 and activation at 52, 53 mitochondria are obligatory steps for canonical mitophagy signaling. Re-localization of Parkin from cytosol to mitochondria results in Parkin-mediated ubiquitination of dozens of outer mitochondrial membrane proteins, including Mfn1 and Mfn2 54. It does not appear that Parkin exhibits much substrate selectivity for accessible mitochondrial proteins, but instead broadly paints the outer mitochondrial membrane with a coat of ubiquitin molecules. If Mfn2 plays a central role in the translocation of Parkin to damaged mitochondria, one would expect to see depressed mitochondrial polyubiquitination in Mfn2-deficient cardiomyocytes. This proved to be the case. Mitochondrial polyubiquitination and mitochondrial p62/sequestosome-1 localization (a marker of autophagosome recruitment) were impaired in Mfn2-deficient cardiomyocytes. Again, Mfn1 ablation had no effect on these Parkin-dependent mitophagy signaling events. From these results we concluded that Mfn2, but not Mfn1, is essential for attracting Parkin to damaged cardiomyocyte mitochondria, and inferred that its absence was interrupting mitophagy signaling.
The initiating signal in Parkin-mediated mitophagy is stabilization of PINK1 kinase 55, 56. Like hundreds of other nuclear-encoded mitochondrial proteins, PINK1 kinase is translated in the cytosol and the protein is then imported into mitochondria. Remarkably, upon gaining access to the mitochondria PINK1 is almost immediately degraded (likely by the mitochondrial protease presenilin-associated rhomboid-like protein [PARL] 57, 58), preventing PINK1 from having meaningful biological activity as a kinase (Figure 5). For this reason, although PINK1 mRNA is abundant in hearts and other tissues, PINK1 protein levels in normal mitochondria are so low as to stretch levels of detection. It is therefore functionally correct to consider a normal mitochondrion as being PINK1 suppressed or “knocked out”. However, mitochondrial damage or senescence sufficient to provoke depolarization interrupts normal PINK1 degradation. The consequence of this so-called mitochondrial dead-man switch is that PINK1 protein level increases only in damaged/depolarized mitochondria. Mitochondrial PINK1 activity is the proximate signal for Parkin translocation and activation 45, 52, 53, and is important to heart function 59. In the absence of an experimental manipulation such as forced overexpression that can overwhelm the normal degradative mechanisms 45, PINK1 stabilization only within depolarized mitochondria therefore recruits Parkin selectively to those damaged organelles.
The mechanism by which PINK1, a mitochondrial localized kinase, communicates with Parkin, a soluble cytosolic E3 ubiquitin ligase, was unclear. Like most kinases, PINK1 is fairly promiscuous and will phosphorylate any number of accessible proteins, including Parkin itself and ubiquitin 52, 53, 60, 61. We considered that co-localization of PINK1 and Mfn2 at the outer mitochondrial membrane 62 provided an opportunity for PINK1 to phosphorylate Mfn2, which our studies in Mfn2 knockout mice had shown was necessary for mitochondrial Parkin binding. We demonstrated that Mfn2 could be a PINK1 phosphorylation substrate by co-transfecting PINK1 and Mfn2 in cultured HEK293 cells. PINK1 decreased the electrophoretic mobility of Mfn2 in normal gels, and mobility retardation was exaggerated on PhosTag gels. We confirmed that Mfn2 is a PINK1 substrate using anti-phosphoserine antibodies, and further demonstrated overexpression of kinase active PINK1 (but not a kinase deficient PINK1 mutant) was sufficient to provoke Mfn2-Parkin binding in the absence of mitochondrial depolarization 45. To nail down the functional relevance of Mfn2 phosphorylation by PINK1 we used site-directed mutagenesis to map PINK1-mediated phosphorylation to Mfn2 amino acids to Thr111 and Ser442, located within the Mfn2 GTPase domain and just distal (i.e. C-terminal) to the first heptad repeat, respectively 45. Mutation of either of these two amino acids to non-phosphorylatable Ala depressed, but did not eliminate, Mfn2-Parkin binding. However, combinatorial mutation of both PINK1 phosphorylation sites to Ala (Mfn2 111/442 AA) completely abrogated Mfn2-Parkin binding. Finally, we performed the reciprocal experiment by mutating Mfn2 Thr111 and Ser442 to Glu (Mfn2 111/442 EE), thereby mimicking phosphorylation. The Mfn2 111/442 EE mutant was sufficient to bind Parkin in the absence of PINK1 kinase. Thus, pseudo-PINK1 phosphorylated Mfn2 is a constitutive mitochondrial Parkin receptor.
The functional interaction between PINK1, Mfn2, and Parkin (Figure 6) helps explain loss-of-mitochondrial quality control phenotypes induced by cell-type specific Mfn2 ablation in heart, brain, and liver. For example, tissue-specific Cre-mediated Mfn2 gene ablation in cardiomyocytes and neurons causes accumulation of abnormal mitochondria linked to a Parkin translocation defect 45, 63, and increased numbers of abnormal mitochondria are also observed after Mfn2 deletion from dopaminergic neurons 64 and hepatocytes 65 (although Parkin function was not directly evaluated therein).
Figure 6. Dual roles of Mfn2 in mitochondrial fusion and mitophagy.
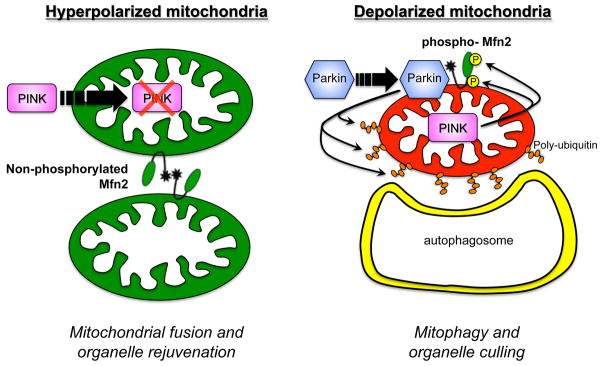
Left – Non-phosphorylated Mfn2 provokes tethering and fusion of normal (hyperpolarized) mitochondria. Right – PINK1-phosphorylated Mfn2 acts as a receptor that attracts Parkin to depolarized mitochondria (in which PINK1 protein is stabilized), initiating ubiquitylation of mitochondrial outer membrane proteins that recruits autophagosomes.
The insights gained from these studies have implications for future investigations of the PINK1-Mfn2-Parkin mitophagy pathway. For example, Parkin can be induced to translocate to the mitochondria of MEFs derived from (embryonic lethal) germ-line mitofusin deficient (Mfn1/Mfn2 knockout) mice 66. We believe this reflects developmental plasticity and induction of one or more alternate pathways leading to mitochondrial Parkin recruitment when the prototypical mitochondrial Parkin receptor (Mfn2) is genetically absent from the germ-line. It will be interesting to determine whether acute conditional Mfn2 deletion in tissue culture (e.g. using Mfn2 floxed allele MEFs with adeno-Cre) might avoid developmental plasticity and experimentally replicate the mitophagy-suppressing effects provoked by in vivo tissue-specific Mfn2 ablation. Likewise, conventional overexpression of Parkin does not intrinsically activate mitophagy (because in healthy cells Parkin resides almost entirely in the cytosol), but simply enhances the capacity to respond mitophagically to a mitochondrial damaging insult 67. It may be possible however to use the constitutive Parkin-binding Mfn2 T111/S442 EE mutant as a research tool to interrogate the effects of mitochondrial-specific Parkin activation on mitophagy in otherwise normal cells and test whether promiscuous Parkin-mediated mitophagy can have detrimental effects on mitochondria and cell fitness 68.
Generalized mitochondrial autophagy when selective mitophagy fails
Mitophagy might be considered as a smart bomb for eliminating damaged (and potentially damaging) mitochondria. PINK1 phosphorylation of Mfn2 acts as the targeting laser for the incoming Parkin/autophagosome bomb. The mitophagy smart bomb approach can eliminate potentially harmful organelles without collateral damage to healthy mitochondria in the same locale. However, if the need to eliminate damaging mitochondria is great, as when mitophagy is interrupted or there is generalized cell-wide mitochondrial dysfunction, the less selective approach of generalized autophagy can be invoked. This is analogous to using carpet bombing when you run out of smart bombs; the harmful mitochondria are removed at a cost of greater collateral damage. As described below, the evidence for compensatory autophagy induction when primary mitophagy fails (either because mitophagy is dysfunctional or is overwhelmed) is inferential, but accumulating data provide some direct support for partial homeostatic redundancy between mitophagy and mitochondrial autophagy.
Before defining the conditions under which mitophagy may be supplemented or supplanted by mitochondrial autophagy, it is worth noting the degree to which the two process overlap. Indeed, the differences between mitophagy and autophagy are all at the front end, i.e. at target acquisition. Both mitophagy and mitochondrial autophagy proceed through autophagosomal engulfment of the organelle and transfer to a degradative lysosome, followed by breakdown and recycling or export of constituent components. The downstream effectors of mitophagy and autophagy are therefore shared, which can be potentially confounding if assays are not performed and interpreted with care 69. By way of example, the normally cytosolic microtubule-associated protein 1A/1B-light chain 3 (LC3) protein is conjugated to phosphatidylethanolamine on autophagosomal membranes and forms LC3-II. The conversion of LC3-I to LC3-II is readily detected by a mobility shift on immunoblots, and is often used as a marker of ongoing autophagy. However, autophagosomal LC3-II is degraded by lysosomal hydrolases, and so its elimination can reflect increased autophagy. Conversely, increased LC3-II might indicate enhanced stimulation, but failure of autophagy to proceed normally through the lysosomal degradation stage. These vagaries indicate that, under steady-state conditions, immunoreactive LC3 is problematic as even a qualitative metric of autophagosomal flux. The same conundrum exists for other autophagosome-associated proteins like p62/sequestosome-1. Confocal fluorescence studies are helpful to localize LC3 or p62 at specific cellular components, but LC3 or p62 immunoreactivity in unfractionated cell or tissue homogenates cannot distinguish between selective mitophagy and non-selective mitochondrial autophagy.
There are some biochemical approaches to specifically detect and (semi-quantitatively) assay mitophagy in heart tissue. First, Parkin localization to mitochondria and polyubiquitination of mitochondrial proteins can be measured in mitochondrial fractions obtained by differential centrifugation of cardiac homogenates. Enrichment of Parkin and increased immunoreactive ubiquitin in cardiac mitochondrial fractions reflects activation of mitophagy signaling, although mitophagy per se will not proceed in the absence of critical downstream effectors. Mitophagy itself, i.e. the actual engulfment of mitochondria by an autophagosome, can be detected as the association of autophagosomal proteins, such as p62 and LC3, with mitochondria. Thus, if mitophagy signaling is increased and mitophagy proceeds intact through lysosomal degradation, one will observe increased Parkin, p62, and LC3-II in the mitochondrial-rich subcellular fraction (mitophagy-specific markers), but decreased p62 and LC3-II in cardiac homogenates (because of increased autophagic degradation). Conversely, if autophagy is generally increased without a specific mitophagy component, Parkin, p62, and LC3 levels in mitochondrial protein fractions may be unaltered, but autophagosomal markers will increase in the tissue homogenates. Finally, the visualization of mitochondria within autophagosomes or lysosomes, either by electron microscopy or a confocal visualization of co-localized fluorescent mitochondrial and lysosomal markers 70, 71, indicates that mitochondria are undergoing autophagosomal engulfment and lysosomal transfer that is the common pathway of organelle disposal for both Parkin-mediated mitophagy and non-specific mitochondrial autophagy; these imaging techniques do not discriminate between mitophagy and mitochondrial autophagy.
To explore the role of ROS in the upstream processes leading to mitochondrial clearance we examined markers of mitophagy and autophagy in Mfn2 deficient hearts72. ROS are frequently implicated as injurious factors released by damaged, dysfunctional, or senescent mitochondria, and can contribute to various forms of heart disease 73. Scavenging mitochondrial-derived ROS has therefore prevented or ameliorated experimental heart disease linked to mitochondrial dysfunction 74. Because accumulation of abnormal mitochondria is a hallmark of cardiomyocyte-specific mitofusin deficiency, we anticipated that ROS scavenging would prevent the cardiomyopathy evoked by cardiomyocyte-specific mitofusin ablation. While this was partially correct, we unexpectedly found that mitochondrial-derived ROS also play an important role in stimulating mitochondrial quality control pathways.
Our initial efforts to test ROS scavenging in cardiomyopathies provoked by mitofusin deficiency utilized cardiac-specific Drosophila Parkin and MARF RNAi models. Mammalian Mfn2 appears functionally synonymous with Drosophila MARF because expression of Mfn2 normalizes mitochondrial morphology and heart tube dysfunction induced in the fly by cardiomyocyte-specific expression of MARF RNAi 17. Accumulation of dysfunctional mitochondria after MARF suppression suggested that inadequate mitophagy might be contributing to the phenotype, as it does after Mfn2 ablation in mammalian hearts (vide supra). RNAi-mediated suppression of either MARF or Parkin in fly hearts provoked similar cardiomyocyte mitochondria pathology and heart tube abnormalities, showing that interruption of Parkin signaling is central to both of these fly cardiomyopathies 5, 45. We therefore reasoned that the mechanistic link between interruption of mitophagy, increased numbers of abnormal mitochondria, and cardiac failure could be increased ROS produced by accumulating damaged organelles; we tested this idea by expressing the potent anti-oxidant, superoxide dismutase (SOD), in Parkin- and MARF-deficient fly hearts. Consistent with the postulated role of ROS in the mitophagic cardiomyopathy induced by Parkin suppression, both mitochondrial SOD2 and soluble SOD1 completely prevented mitochondrial dysfunction and heart tube dilatation in Parkin RNAi fly hearts 5. Unexpectedly, SOD was minimally and only transiently protective in the cardiomyopathy caused by Drosophila MARF insufficiency 5, 17. These results supported our hypothesis that mitochondrial ROS-derived cytotoxicity is a major factor leading to cardiomyopathy induced by interdiction of Parkin-mediated mitophagy, but did not support the same role for ROS in cardiac MARF insufficiency. Thus, fly MARF appears to be important for mitochondrial fitness in ways that are independent of any role in Parkin-mediated mitophagy, likely by promoting mitochondrial fusion. However, another possibility raised by the absence of cardioprotection afforded by SOD in the cardiomyopathy provoked by fly mitofusin (MARF) insufficiency is that mitochondrial ROS can function as a signaling molecule in compensatory pathways75.
Because the single Drosophila protein MARF acts like both mammalian mitofusins, Mfn1 and Mfn2, the cardiac MARF RNAi fly pathophysiologically resembles the Mfn1/Mfn2 double cardiac knockout mouse: both mitochondrial fusion signaling and any related Parkin-mediated mitophagy are interrupted 17, 24. To better understand the roles that ROS can play within the Mfn2-Parkin mitophagy signaling axis, we returned to studies of the cardiac-specific Mfn2 null mouse. As recently reported 76, we detected increased endogenous catalase levels and observed increased ROS production by mitochondria isolated from Mfn2-deficient mouse hearts, constituting evidence for chronically elevated in vivo mitochondria-derived ROS. We determined how mitochondrial ROS impact organelle and organ dysfunction in cardiac-specific Mfn2 null mice by transgenically co-expressing mitochondria-directed catalase (mitoCAT, another potent anti-oxidant enzyme), and examining the compound genetic mice and their parent lines for ROS formation, mitochondrial function and integrity, and the cardiomyopathy that characteristically develops over a ~30 week period 76. We used two different mitoCAT mouse lines previously shown to protect against angiotensin II-induced cardiac hypertrophy and Gq-mediated cardiomyopathy 77: a β-actin driven CAT transgene expressed in hearts at high levels that we called hi-CAT, and a Cre-activated flox-stopped CAT transgene expressed in hearts at 10-fold lower levels that we called low-CAT 76. As expected, expression of either hi- or low-CAT alone was totally benign, and low-CAT expression completely rescued organ and organelle dysfunction in Mfn2 deficient hearts. This was associated with induction of macroautophagy likely evoked as a secondary mechanism for mitochondrial culling 49. The primary defect in Parkin-mediated mitophagy produced by Mfn2 deficiency was not affected by mCAT expression.
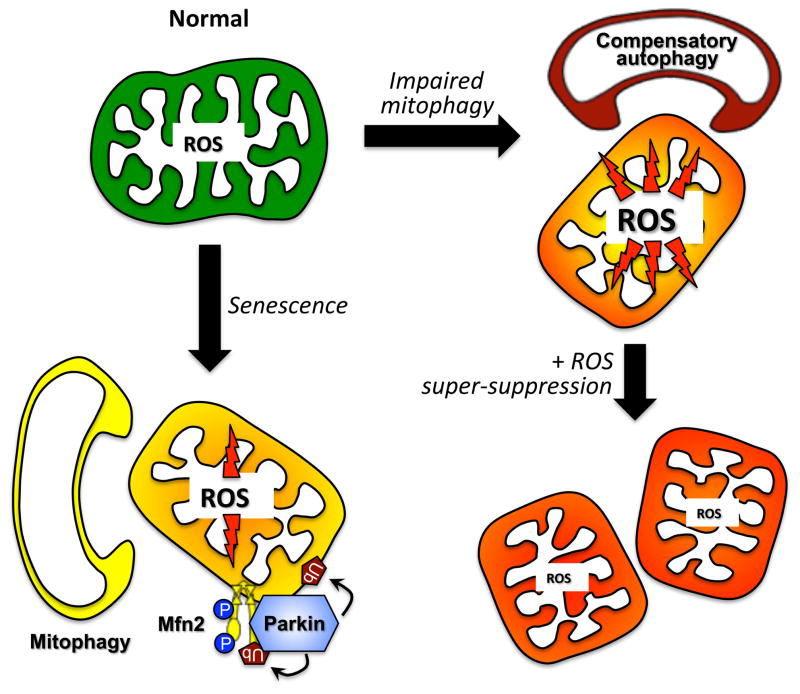
Given the benefits on the Mfn2 knockout mouse heart of ROS suppression with low-CAT, we were astonished that expression of hi-CAT in Mfn2-deficient hearts failed to improve either the mitochondrial abnormalities or the cardiomyopathy, despite super-suppressing mitochondrial ROS formation to levels well below normal 76. Failure of hi-CAT to rescue Mfn2 hearts was associated with failure to induce macroautophagy, i.e. with a breakdown in the putative secondary mitochondrial quality control mechanism. From these results we conclude that interruption of Parkin-mediated mitophagy in Mfn2 null mouse hearts leads to heart failure because damaged mitochondria that would normally be culled instead accumulate and form ROS. This initiates a vicious cycle leading to more mitochondrial damage and activation of compensatory mitochondrial quality control pathways that include generalized autophagy. Preventing toxic accumulation of mitochondria-derived ROS by expressing lower levels of mitochondrial-targeted catalase (lowCAT) interrupts this vicious cycle, improving mitochondrial health and diminishing the load on compensatory autophagy. On the other hand, failure of compensatory autophagy in Mfn2 null hearts co-expressing hi-CAT reveals that a threshold level of mitochondrial ROS is necessary to induce macroautophagy, as suggested in a different context (in vitro cardiomyocyte lipopolysaccharide toxicity) by Gottlieb and co-workers 72. Thus, super-suppression of ROS by hi-CAT impairs the back-up mechanism of autophagic mitochondrial culling which, together with ineffective mitophagy, induces two hits to the mitochondrial quality control apparatus and accelerates the generalized mitochondrial dysfunction and heart failure (Figure 7).
Figure 7. Role of mitochondrial ROS in mitochondrial autophagy signaling.
The primary mechanism for culling damaged or senescent mitochondria normally is Parkin-mediated mitophagy (left). When mitophagy is impaired, increased mitochondrial ROS acts as a signal to stimulate compensatory macroautophagy, resulting in Parkin-independent mitochondrial autophagy (right top). Super-suppression of mitochondrial ROS, as with highly expressed mitochondrial catalase, suppresses the ROS signal and compensatory mitochondrial autophagy, provoking further deterioration of the cell mitochondrial collective.
Mitoptosis: connecting cell death and mitochondrial quality control
Cardiomyocytes possess robust cell death programs. Regulated forms of cell death – apoptosis and necrosis – contribute critically to the evolution of myocardial infarction, and pathological cardiac remodeling, and heart failure (reviewed by Konstantinidis 78). The recent recognition that some protein mediators of mitochondria-dependent death pathways are also involved in mitochondrial dynamics has raised the intriguing possibility that cell death proteins perform a double-duty as regulators of mitochondria quality control.
Mitochondria are the organelle gatekeepers of the intrinsic pathways of apoptosis and necrosis. While apoptosis and necrosis signaling at the mitochondria is intertwined, the defining point of no return differs. For apoptosis, it is permeabilization of the outer mitochondrial membrane allowing the release into the cytosol of cytochrome c and other apoptogens that promote formation of the apoptosome and activation of caspases. In contrast, in necrosis, the sentinel event is opening of the MPTP in the inner membrane resulting in dissipation of the proton gradient that drives ATP synthesis and energetic catastrophe.
Bcl-2 proteins are the major regulators of outer mitochondrial membrane permeabilization in apoptosis (reviewed by Chipuk et al. 79). The BH3-only subclass of this family transduces various death signals from the periphery to Bax and Bak, which then promote outer mitochondrial membrane permeabilization. More precisely, the BH3-only proteins, Bim and truncated Bid (tBid), serve as the direct messengers that bind and conformationally activate Bax and Bak. The other BH3-only proteins are “sensitizers” that work by displacing Bim and tBid from anti-apoptotic family members such as Bcl-2 and Bcl-xL that serve as reservoirs to sequester Bim and tBid (Figure 8a). Conformational activation of Bax sends this protein from the cytosol, where it resides in healthy cells, to the mitochondria, where it inserts tightly into the outer mitochondrial membrane. Bak, on the other hand, is constitutively localized to the outer mitochondrial membrane, and the direct consequences of its conformational activation are less well understood. Once activated, Bax and Bak homo- and hetero-oligomerize to bring about outer mitochondrial membrane permeabilization through mechanisms that are poorly understood – perhaps pore formation.
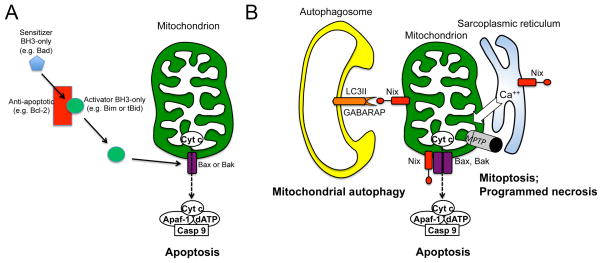
Figure 8. Multiple roles of Bcl2 proteins in selective mitochondrial destruction and generalized cell death.
A. Bax and Bak permeabilize the outer mitochondrial membrane after they undergo conformational changes induced by the direct binding of “activator” BH3-only proteins Bim or tBid. Anti-apoptotic Bcl-2 proteins, such as Bcl-2, sequester activator BH3-only proteins so that they are unavailable for binding to Bax or Bak. “Sensitizer” BH3-only proteins bind anti-apoptotic Bcl-2 proteins and displace Bim and tBid. B. The conventional role of Nix as a pro-apoptotic BH3-only factor that facilitates Bax/Bak-mediated cytochrome c release and caspase-mediated apoptosis is shown at the bottom center. To the right, SR-localized Nix increases SR calcium content and mitochondrial calcium cross-talk, inducing MPTP opening. When MPTP opening is selective, the result is mitoptosis, a non-mitophagic mechanism of mitochondrial culling. When MPTP opening is generalized, the cell dies from programmed necrosis. To the left, mitochondrial Nix is a receptor for autophagosomal proteins LC3 and GABARAP, targeting Nix-associated mitochondria for mitochondrial autophagy.
We previously discussed the BH3-only proteins Nix and Bnip3 in connection with their promoting cardiomyocyte death by apoptosis or necrosis depending on whether they are localized at the mitochondria or endo/sarcoplasmic reticulum respectively. The pathophysiological significance of Nix is that it is transcriptionally upregulated in cardiac hypertrophy and provokes cardiomyocyte death that mechanistically contributes to the transition from compensated hypertrophy to decompensated heart failure 33–35, while Bnip3 plays a parallel cell death promoting role in post-ischemic cardiac failure 38, 80. Recent evidence has uncovered additional roles for these proteins in mitophagy. Although genetic ablation of Nix or Bnip3 protects hearts against cardiomyocyte dropout and adverse remodeling (after pressure overloading or ischemia-reperfusion injury respectively), their combined deficiency in mouse hearts induces a progressive cardiomyopathy associated with accumulation of abnormal mitochondria 81. This observation uncovered a previously unsuspected role for Nix and Bnip3 in cardiomyocyte mitochondria quality control, and pointed to mechanistic connections between proteins involved in mitochondria-mediated cell death and mitophagy.
Consistent with this paradigm, data have emerged demonstrating that Nix and Bnip3 can each play dual roles as mediators of both programmed cell death (via apoptosis or necrosis) and programmed elimination of mitochondria (via mitophagy or autophagy). Nix is recruited specifically to damaged mitochondria and inserts itself into mitochondrial outer membranes. Its pro-apoptotic activity derives from facilitation of Bax/Bak-mediated outer mitochondrial membrane permeabilization that releases cytochrome c and activates the caspase cascade 37 (Figure 8b). Additionally, mitochondrial Nix can act as a chaperone to promote autophagosome recruitment by binding to γ-aminobutyric acid type A receptor-associated protein (a.k.a. GABARAP), which is an LC3 homologue critical for autophagosome maturation 82–84 (Figure 8b). The dual effects of Nix on apoptosis and mitophagy are perhaps most evident during erythrocyte (red blood cell) development and maturation from bone marrow precursor erythroblasts. Because retained mitochondria in highly oxygenated circulating red blood cells would produce damaging ROS and lead to intravascular hemolysis, the penultimate stage of erythrocyte formation wherein the erythroblast nucleus is extruded is also associated with the elimination of erythroblast mitochondria via developmentally programmed mitophagy. This developmental mitophagy is a Nix-driven and -dependent process: Nix is transcriptionally upregulated during the early stages of erythropoiesis and fine-tunes erythrocyte formation by apoptotically limiting erythroblast abundance 85. Then, during the terminal stages of erythropoiesis, Nix upregulation mediates mitophagic elimination of erythroblast mitochondria 86–88. Thus, within the same cell type Nix promotes either programmed cell or programmed organelle elimination, depending upon developmental context. Recent studies have uncovered a similar role for Bnip3 as an LC3-interacting mitophagy effector 89–92.
Another example of dual-functionality of a single factor in mitochondrial quality control and mitochondrial-mediated apoptosis pathways is the pro-fission protein, Drp1. Apoptosis is almost universally associated with mitochondrial fragmentation, i.e. a disproportionate increase in mitochondrial fission relative to fusion. In asymmetric mitochondrial fission that is integral to mitophagic quality control (see Figure 5), the central effector of apoptosis-associated mitochondrial fission is Drp1. Pharmacological inhibition of Drp1 with either mdivi 93–96 or P110 97 can protect against apoptosis and cardiac failure in ischemically, hemodynamically, or toxically damaged hearts. Importantly, recruitment of Drp1 to mitochondria during apoptosis is mediated by Bax and Bak 98, the same proteins responsible for outer mitochondrial membrane permeabilization in apoptosis. Thus, Bax and Bak provide a molecular link between apoptosis and fission, which in a different context, is a prerequisite for normal mitophagic quality control.
Recent work has unexpectedly extended the role of Bax to the regulation of mitochondrial-mediated necrosis 99, 100. Deletion of Bax and Bak 100, or Bax alone 101 in mice reduced infarct size following ischemia-reperfusion, and markedly decreased ultrastructural markers of cell necrosis. These effects are of similar magnitude to those resulting from deletion of cyclophilin D 102, 103, a mitochondrial matrix protein that regulates opening of the MPTP. In fact, the triple knockout of Bax/Bak/cyclophilin D does not result in greater cardioprotection compared to the double knockout of Bax/Bak, suggesting that Bax/Bak and cyclophilin reside in the same or overlapping pathways 100. Mechanistic analyses in Bax/Bak null fibroblasts and isolated adult cardiomyopcyte mitochondria showed that the absence of Bax decreases the sensitivity of the MPTP to opening in response to Ca2+. Bax produces necrosis through a mechanism distinct from its activation of apoptosis as oligomerization-defective Bax mutants that are unable to support apoptosis remain capable of supporting necrosis 99, 100.
What is the precise mechanism? This is not known with certainty. Some data suggest that Bax on the outer mitochondrial membrane is in complex with MPTP 99. Another mechanism that we have explored is that Bax facilitates necrosis through its previously recognized ability to promote mitochondrial fusion 100, 104. Interestingly, oligomerization-deficient Bax mutants, the same ones that support necrosis, are capable of driving mitochondrial fusion 105. Consistent with this model, other genetic manipulations that disable mitochondrial fusion, such as Mfn2 deficiency, also desensitize fibroblasts to Ca2+-induced MPTP opening. Conversely, restoration of the fused mitochondrial morphology by an independent means, blocking fission, restores the sensitivity of Ca2+-induced MPTP opening in Bax/Bak null or Mfn2-null fibroblasts. These observations suggest that there is something about the fused mitochondrial state, no matter how it is attained, that sensitizes to necrosis.
At present, we do not know with certainty exactly what features of fused mitochondrial are responsible for heightened sensitivity to cell death. There is evidence that fused mitochondria have more efficient diffusion of Ca2+ waves than less interconnected ones suggesting an explanation for increased sensitivity to MPTP opening 106. Another possibility worthy of consideration is that larger, interconnected mitochondria have deficits in quality control possibly by virtue of being less susceptible to mitophagy, i.e., Bax-driven fusion may be interfering with mitochondria quality control. While this model remains to be tested, there are some enticing connections that may come into play. First, Bax is a target for Parkin-mediated ubiquitination, the effect of which is to damp Bax translocation to the mitochondria in response to a death stimulus 107. Second, although the precise mechanism by which Bax promotes fusion is not understood, there appear to be physical and functional interactions between Bax and Mfn2 104, 105, 108, the latter at the nexus of fusion and mitophagy.
These examples illustrate the multifunctionality of Bcl-2 proteins, Mfn2, and other fusion and fission factors in the regulation of mitochondrial dynamics, mitophagy, and cell death – apoptotic and necrotic. Whereas the mechanisms that adjudicate outcome are not completely understood, the involvement of the same cast of characters argues for a unified and highly integrated model. One possibility is that outcome is determined more by the quantitative degree the same cast of players is activated. Thus, mitophagy employs Nix, Mfn2, and Drp1 to selectively eliminate certain damaged mitochondria, whereas the same constellation of factors interact cell-wide to initiate apoptosis. This paradigm is consistent with the concept that mitochondria and their host cells developed one or more mechanisms for organelle self-elimination, which has been designated “mitoptosis” 109. As originally described, mitoptosis progresses via caspase-independent, ROS-mediated activation of MPTP (i.e. programmed necrosis), and is distinct from canonical mitophagy 110. However, molecular cross-talk between apoptosis, programmed necrosis, and mitochondrial autophagy/mitophagy signaling at the level of Nix and Bnip3, Mfn2, and Drp1 indicates that these pathway distinctions probably lack biological relevance. Interactive signaling between mitochondrial quality control/mitoptosis pathways and programmed cell death pathways does, however, provide a plausible explanation for why apoptosis is so often observed in diseased hearts. We posit that these integrated processes constitute a stress response whose primary objective is to maintain mitochondrial quality control through the elimination of defective organelles. Only when this fails (e.g. severely damaging conditions) does the same machinery bring about cell death. One potential implication of this notion is that care must be taken in contemplating therapies to suppress cell death so as not to also disrupt long-term maintenance of normal mitochondrial fitness. Conversely, enhancing mitochondrial quality control is likely to be protective in conditions where programmed cell death contributes to disease progression 67, 107, 111, 112.
Acknowledgments
SOURCES OF FUNDING
Supported by NHLBI HL59888 (GWD) and HL60665 (RNK).
Nonstandard Abbreviations and Acronyms
- CAT
Catalase
- Drp1
Dynamin-related protein 1
- ER
Endoplasmic reticulum
- ETC
Electron transport complex
- LC3
microtubule-associated protein 1A/1B-light chain 3
- MARF
Mitochondrial assembly regulatory facto
- Mfn
Mitofusin
- MPTP
Mitochondrial permeability transition pore
- Opa1
Optic atrophy 1
- ROS
Reactive oxygen species
- SOD
Superoxide dismutase
- SR
Sarcoplasmic reticulum
Footnotes
DISCLOSURES
None
References
- 1.Lewin R. The unmasking of mitochondrial Eve. Science. 1987;238:24–26. doi: 10.1126/science.3116666. [DOI] [PubMed] [Google Scholar]
- 2.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 3.Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 5.Bhandari P, Song M, Chen Y, Burelle Y, Dorn GW., 2nd Mitochondrial contagion induced by Parkin deficiency in Drosophila hearts and its containment by suppressing mitofusin. Circ Res. 2014;114:257–265. doi: 10.1161/CIRCRESAHA.114.302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hales KG. Mitochondrial fusion and division. Nature Education. 2010;3:12. [Google Scholar]
- 7.Kasahara A, Cipolat S, Chen Y, Dorn GW, 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- 8.Chappie JS, Dyda F. Building a fission machine-structural insights into dynamin assembly and activation. J Cell Sci. 2013;126:2773–2784. doi: 10.1242/jcs.108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin EE, Detmer SA, Chan DC. Molecular mechanism of mitochondrial membrane fusion. Biochim Biophys Acta. 2006;1763:482–489. doi: 10.1016/j.bbamcr.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 14.Dorn GW., 2nd Mitochondrial dynamism and cardiac fate-a personal perspective. Circ J. 2013;77:1370–1379. doi: 10.1253/circj.cj-13-0453. [DOI] [PubMed] [Google Scholar]
- 15.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Z, Chochani M, McCaffrey JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorn GW, 2nd, Clark CF, Eschenbacher WH, Kang MY, Engelhard JT, Warner SJ, Matkovich SJ, Jowdy CC. MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ Res. 2011;108:12–17. doi: 10.1161/CIRCRESAHA.110.236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and Parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Liu Y, Dorn GW., 2nd Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papanicolaou KN, Kikuchi R, Ngoh GA, Coughlan KA, Dominguez I, Stanley WC, Walsh K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ Res. 2012;111:1012–1026. doi: 10.1161/CIRCRESAHA.112.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suddath C. A brief history of: Velcro. Time Magazine. 2010 Jun 15; [Google Scholar]
- 27.Dorn GW, 2nd, Scorrano L. Two close, too close: sarcoplasmic reticulum-mitochondrial crosstalk and cardiomyocyte fate. Circ Res. 2010;107:689–699. doi: 10.1161/CIRCRESAHA.110.225714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papanicolaou KN, Khairallah RJ, Ngoh GA, Chikando A, Luptak I, O’Shea KM, Riley DD, Lugus JJ, Colucci WS, Lederer WJ, Stanley WC, Walsh K. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol Cell Biol. 2011;31:1309–1328. doi: 10.1128/MCB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Csordas G, Jowdy C, Schneider TG, Csordas N, Wang W, Liu Y, Kohlhaas M, Meiser M, Bergem S, Nerbonne JM, Dorn GW, 2nd, Maack C. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca(2+) crosstalk. Circ Res. 2012;111:863–875. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorn GW, 2nd, Maack C. SR and mitochondria: calcium cross-talk between kissing cousins. J Mol Cell Cardiol. 2013;55:42–49. doi: 10.1016/j.yjmcc.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Brenner C, Moulin M. Physiological roles of the permeability transition pore. Circ Res. 2012;111:1237–1247. doi: 10.1161/CIRCRESAHA.112.265942. [DOI] [PubMed] [Google Scholar]
- 32.Papanicolaou KN, Phillippo MM, Walsh K. Mitofusins and the mitochondrial permeability transition: the potential downside of mitochondrial fusion. Am J Physiol Heart Circ Physiol. 2012;303:H243–255. doi: 10.1152/ajpheart.00185.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., 2nd Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syed F, Odley A, Hahn HS, Brunskill EW, Lynch RA, Marreez Y, Sanbe A, Robbins J, Dorn GW., 2nd Physiological growth synergizes with pathological genes in experimental cardiomyopathy. Circ Res. 2004;95:1200–1206. doi: 10.1161/01.RES.0000150366.08972.7f. [DOI] [PubMed] [Google Scholar]
- 35.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW., 2nd Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 36.Diwan A, Matkovich SJ, Yuan Q, Zhao W, Yatani A, Brown JH, Molkentin JD, Kranias EG, Dorn GW., 2nd Endoplasmic reticulum-mitochondria crosstalk in NIX-mediated murine cell death. J Clin Invest. 2009;119:203–212. doi: 10.1172/JCI36445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Lewis W, Diwan A, Cheng EH, Matkovich SJ, Dorn GW., 2nd Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc Natl Acad Sci U S A. 2010;107:9035–9042. doi: 10.1073/pnas.0914013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, Jones WK, Dorn GW., 2nd Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaanine AH, Gordon RE, Kohlbrenner E, Benard L, Jeong D, Hajjar RJ. Potential role of BNIP3 in cardiac remodeling, myocardial stiffness, and endoplasmic reticulum: mitochondrial calcium homeostasis in diastolic and systolic heart failure. Circ Heart Fail. 2013;6:572–583. doi: 10.1161/CIRCHEARTFAILURE.112.000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Li L, Liu H, Borowitz JL, Isom GE. BNIP3 mediates cell death by different pathways following localization to endoplasmic reticulum and mitochondrion. FASEB J. 2009;23:3405–3414. doi: 10.1096/fj.08-124354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korff S, Riechert N, Schoensiegel F, Weichenhan D, Autschbach F, Katus HA, Ivandic BT. Calcification of myocardial necrosis is common in mice. Virchows Arch. 2006;448:630–638. doi: 10.1007/s00428-005-0071-7. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan PG, Balke CW, Esser KA. Mitochondrial buffering of calcium in the heart: potential mechanism for linking cyclic energetic cost with energy supply? Circ Res. 2006;99:109–110. doi: 10.1161/01.RES.0000234949.61052.9f. [DOI] [PubMed] [Google Scholar]
- 43.Rones MS, McLaughlin KA, Raffin M, Mercola M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development. 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- 44.Nemir M, Croquelois A, Pedrazzini T, Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiol Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- 47.Lees AJ, Hardy J, Revesz T. Parkinson’s disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 48.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahlqvist G, Landin S, Wroblewski R. Ultrastructure of skeletal muscle in patients with Parkinson’s disease and upper motor lesions. Lab Invest. 1975;32:673–679. [PubMed] [Google Scholar]
- 51.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate Parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 54.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, Abramov AY, Wood NW. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, Knebel A, Alessi DR, Muqit MM. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiba-Fukushima K, Imai Y, Yoshida S, Ishihama Y, Kanao T, Sato S, Hattori N. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hasson SA, Kane LA, Yamano K, Huang CH, Sliter DA, Buehler E, Wang C, Heman-Ackah SM, Hessa T, Guha R, Martin SE, Youle RJ. High-content genome-wide RNAi screens identify regulators of Parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee S, Sterky FH, Mourier A, Terzioglu M, Cullheim S, Olson L, Larsson NG. Mitofusin 2 is necessary for striatal axonal projections of midbrain dopamine neurons. Hum Mol Genet. 2012;21:4827–4835. doi: 10.1093/hmg/dds352. [DOI] [PubMed] [Google Scholar]
- 64.Pham AH, Meng S, Chu QN, Chan DC. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum Mol Genet. 2012;21:4817–4826. doi: 10.1093/hmg/dds311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sebastian D, Hernandez-Alvarez MI, Segales J, et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc Natl Acad Sci U S A. 2012;109:5523–5528. doi: 10.1073/pnas.1108220109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee K, Lee MH, Kang YW, Rhee KJ, Kim TU, Kim YS. Parkin induces apoptotic cell death in TNF-alpha-treated cervical cancer cells. BMB Rep. 2012;45:526–531. doi: 10.5483/bmbrep.2012.45.9.104. [DOI] [PubMed] [Google Scholar]
- 69.Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy. 2007;3:154–157. doi: 10.4161/auto.3634. [DOI] [PubMed] [Google Scholar]
- 71.Krebiehl G, Ruckerbauer S, Burbulla LF, Kieper N, Maurer B, Waak J, Wolburg H, Gizatullina Z, Gellerich FN, Woitalla D, Riess O, Kahle PJ, Proikas-Cezanne T, Kruger R. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One. 2010;5:e9367. doi: 10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. Am J Physiol Heart Circ Physiol. 2009;296:H470–479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen YR, Zweier JL. Cardiac mitochondria and reactive oxygen species generation. Circ Res. 2014;114:524–537. doi: 10.1161/CIRCRESAHA.114.300559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dai DF, Chen T, Johnson SC, Szeto H, Rabinovitch PS. Cardiac aging: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal. 2012;16:1492–1526. doi: 10.1089/ars.2011.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 76.Song M, Chen Y, Gong G, Murphy E, Rabinovitch P, Dorn GW., 2nd Super-suppression of mitochondrial reactive oxygen species signaling impairs compensatory autophage in primary mitophatic cardiomyopathy. Circ Res. 2014;115:348–53. doi: 10.1161/CIRCRESAHA.115.304384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dai DF, Johnson SC, Villarin JJ, Chin MT, Nieves-Cintron M, Chen T, Marcinek DJ, Dorn GW, 2nd, Kang YJ, Prolla TA, Santana LF, Rabinovitch PS. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ Res. 2011;108:837–846. doi: 10.1161/CIRCRESAHA.110.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Konstantinidis K, Whelan RS, Kitsis RN. Mechanisms of cell death in heart disease. Arterioscler Thromb Vasc Biol. 2012;32:1552–1562. doi: 10.1161/ATVBAHA.111.224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Galvez AS, Brunskill EW, Marreez Y, Benner BJ, Regula KM, Kirschenbaum LA, Dorn GW., 2nd Distinct pathways regulate proapoptotic Nix and BNip3 in cardiac stress. J Biol Chem. 2006;281:1442–1448. doi: 10.1074/jbc.M509056200. [DOI] [PubMed] [Google Scholar]
- 81.Dorn GW., 2nd Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, Hersch N, Hoffmann B, Merkel R, Willbold D. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–698. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 83.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diwan A, Koesters AG, Odley AM, Pushkaran S, Baines CP, Spike BT, Daria D, Jegga AG, Geiger H, Aronow BJ, Molkentin JD, Macleod KF, Kalfa TA, Dorn GW., 2nd Unrestrained erythroblast development in Nix-/- mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc Natl Acad Sci U S A. 2007;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Glick D, Zhang W, Beaton M, Marsboom G, Gruber M, Simon MC, Hart J, Dorn GW, 2nd, Brady MJ, Macleod KF. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol. 2012;32:2570–2584. doi: 10.1128/MCB.00167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ishihara M, Urushido M, Hamada K, Matsumoto T, Shimamura Y, Ogata K, Inoue K, Taniguchi Y, Horino T, Fujieda M, Fujimoto S, Terada Y. Sestrin-2 and BNIP3 regulate autophagy and mitophagy in renal tubular cells in acute kidney injury. Am J Physiol Renal Physiol. 2013;305:F495–509. doi: 10.1152/ajprenal.00642.2012. [DOI] [PubMed] [Google Scholar]
- 92.Zhu Y, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, Novak I, Dikic I, Hamacher-Brady A, Brady NR. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gharanei M, Hussain A, Janneh O, Maddock H. Attenuation of doxorubicin-induced cardiotoxicity by mdivi-1: a mitochondrial division/mitophagy inhibitor. PLoS One. 2013;8:e77713. doi: 10.1371/journal.pone.0077713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, Tyagi SC. Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PLoS One. 2012;7:e32388. doi: 10.1371/journal.pone.0032388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 96.Sharp WW, Fang YH, Han M, Zhang HJ, Hong Z, Banathy A, Morrow E, Ryan JJ, Archer SL. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J. 2014;28:316–326. doi: 10.1096/fj.12-226225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Disatnik MH, Ferreira JC, Campos JC, Gomes KS, Dourado PM, Qi X, Mochly-Rosen D. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J Am Heart Assoc. 2013;2:e000461. doi: 10.1161/JAHA.113.000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karch J, Kwong JQ, Burr AR, Sargent MA, Elrod JW, Peixoto PM, Martinez-Caballero S, Osinska H, Cheng EH, Robbins J, Kinnally KW, Molkentin JD. Bax and Bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice. Elife. 2013;2:e00772. doi: 10.7554/eLife.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Whelan RS, Konstantinidis K, Wei AC, et al. Bax regulates primary necrosis through mitochondrial dynamics. Proc Natl Acad Sci U S A. 2012;109:6566–6571. doi: 10.1073/pnas.1201608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hochhauser E, Kivity S, Offen D, Maulik N, Otani H, Barhum Y, Pannet H, Shneyvays V, Shainberg A, Goldshtaub V, Tobar A, Vidne BA. Bax ablation protects against myocardial ischemia-reperfusion injury in transgenic mice. Am J Physiol Heart Circ Physiol. 2003;284:H2351–2359. doi: 10.1152/ajpheart.00783.2002. [DOI] [PubMed] [Google Scholar]
- 102.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, 2nd, Robbins J, Molkentin JD. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 103.Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- 104.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 105.Hoppins S, Edlich F, Cleland MM, Banerjee S, McCaffery JM, Youle RJ, Nunnari J. The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol Cell. 2011;41:150–160. doi: 10.1016/j.molcel.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 107.Johnson BN, Berger AK, Cortese GP, Lavoie MJ. The ubiquitin E3 ligase Parkin regulates the proapoptotic function of Bax. Proc Natl Acad Sci U S A. 2012;109:6283–6288. doi: 10.1073/pnas.1113248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cleland MM, Norris KL, Karbowski M, Wang C, Suen DF, Jiao S, George NM, Luo X, Li Z, Youle RJ. Bcl-2 family interaction with the mitochondrial morphogenesis machinery. Cell Death Differ. 2011;18:235–247. doi: 10.1038/cdd.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Skulachev VP. Programmed death phenomena: from organelle to organism. Ann N Y Acad Sci. 2002;959:214–237. doi: 10.1111/j.1749-6632.2002.tb02095.x. [DOI] [PubMed] [Google Scholar]
- 110.Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell Death Differ. 2005;12 (Suppl 2):1484–1489. doi: 10.1038/sj.cdd.4401780. [DOI] [PubMed] [Google Scholar]
- 111.Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, Abbas N, Gu WJ, Hirsch EC, Rooney T, Ruberg M, Brice A. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12:517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 112.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]