The native cellular microenvironment is complex, dynamic, and responsible for presenting both biomolecular (e.g., growth factors, cytokines) and biophysical (e.g., mechanical, structural) regulatory signals. While perhaps impossible to fully replicate the complexity of human tissues in engineered biomaterials, there is a specific need for combinatorial biomaterial platforms where combinations of biophysical and biomolecular cues can be rapidly screened. Such tools would enable investigations to identify hierarchies and synergies between multiple signals, and to determine the minimum constellation of cues necessary to induce a desired cellular phenotype. Advances in secretomic, genomic, and functional profiling as well as in systems biology could enable multi-scale analysis of interconnected signaling networks responsible for determining cell fate in response to complex extracellular cues. However, advances in biomaterials technology required to generate adaptable, three-dimensional microenvironments as test platforms to understand cell-material interactions on a similar scale have not yet been realized.
The growth of robotic and microfluidic technologies has enabled fabrication of a range of material arrays to screen the impact of biophysical and biomolecular regulators on embryonic[1] and adult stem cell[2] differentiation. While these studies provided important insights, most were performed with cells cultured on 2D hydrogel platforms. Recently, some progress has been made describing combinatorial arrays of 3D biomaterials to mimic elements of native tissue architecture. Such approaches have created combinatorial arrays of 3D macroporous synthetic polymeric scaffolds to explore the effects of polymer composition on MC3T3 preosteoblast adhesion and proliferation.[3] A separate approach using a gradient maker described a library of 3D poly(ethylene glycol)-based scaffolds with controlled mechanical properties, demonstrating selective osteogenic maturation of encapsulated MC3T3 cells in gels stiffer than 225 kPa.[4] Despite these advances, method for producing arrays of macroporous, naturally-derived polymeric scaffolds have yet to be described. Such a platform offers the potential to examine the combined impact of biophysical and biomolecular signals on regulating discrete cell behaviors. Further, the vast majority of the previously described 3D platforms require cells to be incorporated at fabrication, rather than having the ability to sequentially manipulate biomaterial structural, mechanical, and compositional properties, further incorporate discrete biomolecular signals, and then introduce cells.
Collagen-glycosaminoglycan (CG) scaffolds are naturally-derived polymeric biomaterials that have been used as regenerative templates for skin, peripheral nerves, conjunctiva and cartilage,[5] and as analogs of the native ECM to explore cell-matrix interactions in vitro.[6, 7] These scaffolds display high porosity (> 99%) and retain natural cell adhesion sites. Approaches have been described to tune structural and biomolecular properties at the macroscale via manipulation of the freeze-drying process used to fabricate these scaffolds. This makes this scaffold system particularly ideal for screening the impact of multiple regulators of cell activity in parallel. Notably, altering freezing conditions (rate, temperature, directionality) has been shown to impact scaffold pore size[8, 9] and anisotropy.[7] In addition to control over scaffold microstructure, our group has previously described methods to immobilize biomolecules within the scaffold in a ubiquitous[10] or spatially-restricted[11] manner to elicit specific cellular responses. Here we present a scalable, scaffold array platform that enables selective presentation of both biomolecular and structural cues at individually addressable nodes within the array, as well as its application to explore the combined impact of these signals on mesenchymal stem cell (MSC) bioactivity. While we have focused on CG materials here, the described freeze-drying fabrication approach should be amenable to other polymeric systems including chitosan,[12] silk,[13] and PLGA,[14] all of which have been used for generating large, macroscale biomaterials via lyophilization.
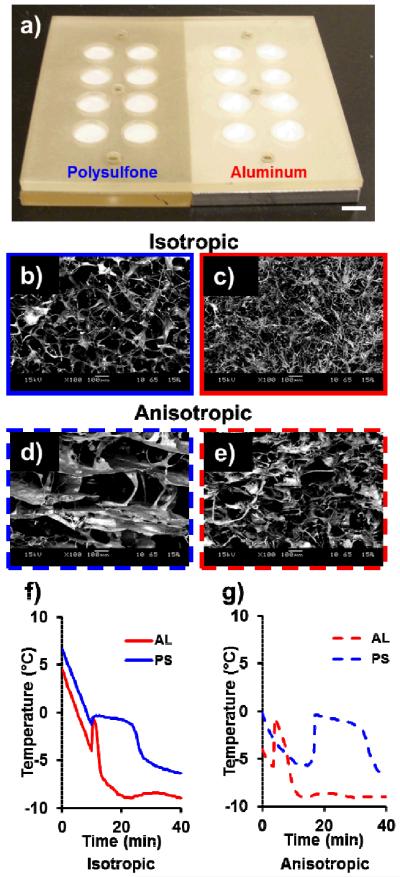
Inspired by the scalable architecture of the 96- and 384-well culture plate, we created a mold to enable fabrication of self-contained arrays of CG scaffolds with distinct variations in structural characteristics (e.g. pore size and anisotropy, Figure 1(a)) at unique nodes within the array. Pore size and shape are known regulators of cell behavior including adhesion, proliferation, and differentiation.[9, 15] The array was designed to be geometrically identical to a standard 96-well plate to enable interrogation of cell bioactivity within each scaffold node via existing microplate reader technologies. Each scaffold array was created via a polysulfone chip containing arrays of machined wells mounted on aluminum (AL) or polysulfone (PS) bases to regulate the solidification of the CG suspension in the array chip. With a thermal conductivity ~850 times higher than polysulfone, aluminum bottom wells displayed efficient heat transfer during freezing, shorter solidification times, and resultant smaller pores within the scaffold node. Additionally, we explored the use of constant versus directional cooling methods previously demonstrated for large scaffold constructs[7, 8] to create isotropic versus anisotropic pore geometries, respectively.
Figure 1.
(a) Scaffold array mold with disparate base thermal conductivities (kaluminum/kpolysulfone ≈ 850). SEM images of array scaffold microstructure with different base materials (aluminum; polysulfone) and freezing procedures (isotropic, constant cooling; anisotropic, directional cooling) reveal larger pores in the polysulfone groups and elongated, aligned pores in the anisotropic groups. (b) AL isotropic, (c) AL anisotropic, (d) PS isotropic, (e) PS anisotropic. Scale bars: 100 μm. Representative thermal profiles during solidification for CG scaffold arrays frozen under (f) isotropic or (g) anisotropic conditions. The ice crystal growth phase period is markedly shorter for AL scaffolds. Isotropic (constant) cooling conditions results in onset of solidification at about the same time for both base materials, but anisotropic (directional) freezing leads to later onset of solidification in less conductive PS group.
SEM micrographs (Figure 1(b–e)) and pore size quantification via stereology (Supplemental Table 1) showed a stark influence of both base thermal conductivity and freezing conditions on scaffold pore size and shape, respectively. As expected, pores were significantly larger in the PS groups compared to the corresponding AL groups (p < 0.05). SEM of the isotropic scaffolds supported observations made via stereology that the pores were relatively rounded. Additionally, while macro-scale anisotropic CG scaffolds can be fabricated via thermally-mismatched molds,[7] we were able to replicate this effect in the scaffold arrays via rapid cooling on a pre-cooled freeze-dryer shelf prior to lyophilization. Analysis of pore orientation angles from SEM images (Supplemental Figure 1) and light micrographs (Supplemental Figure 2) showed that the PS anisotropic group in particular had highly aligned pores.
We subsequently monitored suspension temperature profiles during lyophilization to compare the solidification processes in the scaffold array to previously reported macro-scale scaffolds. Directly following initial ice crystal nucleation, the temperature of the collagen suspension increased during an ice crystal growth phase, the length of which can be correlated to the average pore size of the scaffold. The solidification profiles shown in Figure 1(f–g) support the stereological pore analysis results with longer solidification times observed in the PS groups. Additionally, while constant (isotropic) cooling led to the onset of ice nucleation at about the same time in both the AL and PS groups (Figure 1(f)), directional (anisotropic) cooling led to delayed ice crystal nucleation in the PS group (Figure 1(g)).
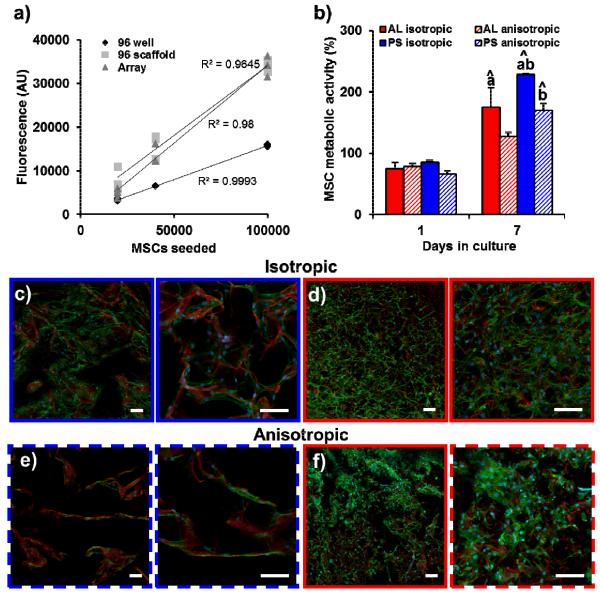
Next, we confirmed that the scaffold array could be integrated with conventional microplate reader technology to quantify cell bioactivity in each array node on-chip. Fluorescence spectrophotometry readouts of cell behavior were tracked over extended culture times. As a proof of concept, we compared the number of CellTracker Green® labeled human MSCs in three different groups: i) cells in suspension in a 96-well plate (96 well), ii) cells seeded on scaffold discs placed in a standard 96-well plate (96 scaffold), and iii) cells seeded on scaffold array nodes (Array). As expected, the average intensity for the array scaffolds, scaffold plugs, and cell-only controls increased linearly with increased cell seeding density (Figure 2(a)), validating the compatibility of the scaffold array setup with microplate technology. We also showed that all four array scaffold groups (AL, PS; isotropic, anisotropic) could support sustained MSC metabolic activity over a 7 day period, and that this response could be tracked on-chip, without the need for individual manipulation of each scaffold specimen within the array. Not surprisingly, MSC metabolic activity was significantly higher (p < 0.05) in the PS groups, likely due to increased scaffold permeability in the larger pores (Figure 2(b)).[16] MSCs within discrete nodes of the array were observed via confocal microscopy to attach and spread normally (Figure 2(c–f)). Paralleling previous observations in macro-scale scaffolds,[17] MSCs appeared to conform to the structural contact guidance cues provided by discrete struts (red) in the PS scaffold group with larger pores while MSCs were able to attach and spread over multiple pores in AL group nodes with significantly smaller pores.
Figure 2.
(a) MSCs labeled with CellTracker Green were seeded at three different amounts (0.2, 0.4, and 1 × 105) in solution (96 well), onto scaffolds placed in a 96 well plate (96 scaffold), or onto array scaffolds (Array). Linear increases in fluorescent signal with increased cell number were observed for each group, validating that quantitative cell metrics can be quantified directly within the array via standard microplate reader technology. (b) MSCs show sustained metabolic activity when cultured in array scaffolds. MSC metabolic activity is significantly higher in the larger pore PS groups compared to the corresponding AL groups. ^: significantly higher than day 1 value, a: significantly higher than corresponding anisotropic group, b: significantly higher than corresponding AL group. Confocal images of MSCs seeded within (c) PS isotropic, (d) AL isotropic, (e) PS anisotropic, and (f) AL anisotropic array groups. Pores are clearly larger in PS groups. MSCs conform to the scaffold microstructure more in the PS groups while spreading across multiple structural elements in the AL groups. Scale bars: 100 μm. Red: scaffold backbone (AlexaFluor® 633 carboxylic acid, succinimidyl ester), Green: actin (AlexaFluor® 488 phalloidin), Blue: nuclei (Hoechst 33258).
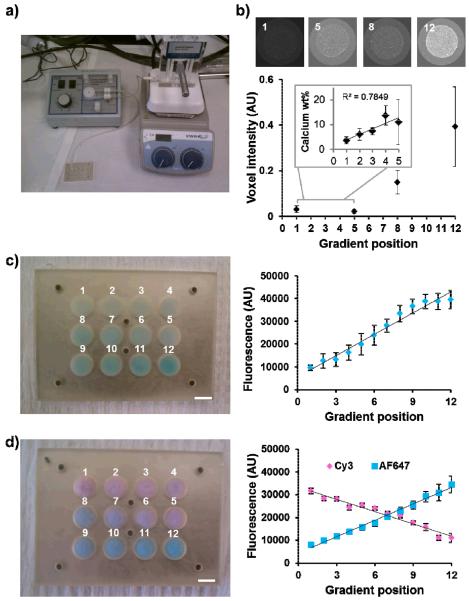
After confirming that scaffold arrays containing nodes with discrete microstructural properties could be fabricated and that subsequent cell response could be traced using plate reader technology, we used a commercially-available gradient maker to fabricate scaffold arrays containing node-to-node gradients in biomolecular signals (Figure 3(a)). Twelve-node scaffold arrays (AL base) were first fabricated with a gradient of fluorescently-labeled bovine serum albumin (BSA)-cyanine (Cy)5. Direct quantification of the fluorescent signal of the scaffold array nodes with the microplate reader showed incorporation of a linear gradient across the array (R2 = 0.97, Figure 3(c)). Opposing linear gradients of two model proteins were subsequently created by loading two different protein/CG solutions into the gradient maker with resultant linear gradients of Cy3-labeled BSA (R2 = 0.96) and AlexaFluor® 647-labeled streptavidin across the array (R2 = 0.99, Figure 3(d)). In addition to biomolecular signals, gradations of ECM components such as structural proteins and mineral content may also play a key role in the design of biomaterial analogs for a range of tissues such as orthopedic interfaces.[18] We therefore demonstrated a process to create array platform containing a gradient of mineral content across the array by mixing non-mineralized (CG) and mineralized (CGCaP) collagen suspensions via the gradient maker. MicroCT analysis and quantification of calcium content within discrete array nodes demonstrated the presence of a mineral gradient across the scaffold array (Figure 3(b)).
Figure 3.
(a) A gradient maker connected to a peristaltic pump was used to meter mixed CG suspensions to discrete nodes of the array mold. (b) Loading of the gradient maker with non mineralized and mineralized CG suspensions respectively results in the creation of mineral gradients as shown by microCT and calcium quantification (inset). (c) Monolithic gradient array containing a single linear gradient of Cy5-labeled BSA functionalized CG scaffolds. Direct plate reader quantification of Cy5 signal in array scaffolds demonstrates the creation of a linear biomolecular gradient. (d) Opposing linear gradients of Cy3-labeled BSA and AlexaFluor® 647-labeled streptavidin across a single scaffold array.
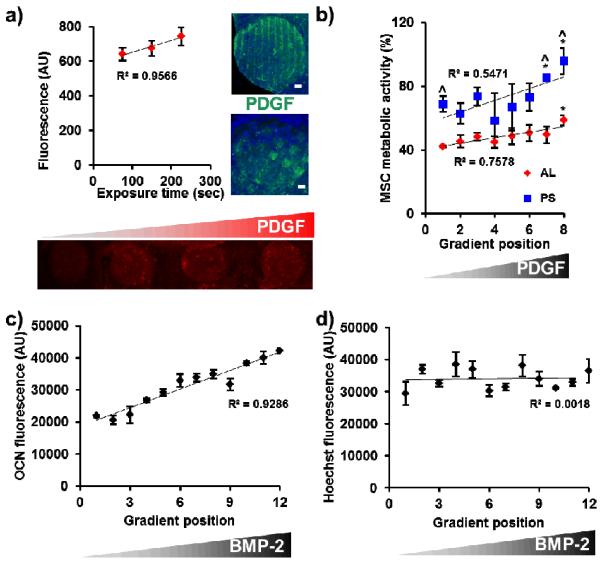
In addition to the simple incorporation of proteins and other ECM components into the gradient maker fabrication process, we demonstrated post-fabrication modification of discrete nodes with biomolecular signals. Here we used a benzophenone-based photolithography scheme, previously used to pattern biomolecules within large CG scaffolds.[11] We showed that linear gradients of covalently-immobilized platelet-derived growth factor BB (PDGF-BB) could be created within CG array nodes by simple variation of the UV exposure time during photolithographic patterning (R2 = 0.96, Figure 4(a)). We also demonstrated the creation of discrete patterns and shapes of immobilized PDGF-BB within individual array nodes (Figure 4(a)). Since PDGF-BB has known mitogenic and chemoattractant effects on MSCs, patterning of the factor within certain regions of the scaffold (e.g., the center) could be especially beneficial. We also used carbodiimide-mediated crosslinking chemistry to immobilize stepwise gradients of a model biomolecule (biotinylated concanavalin A) onto the array scaffolds (Supplemental Figure 3(a–b)). The resulting protein gradient was near-linear (R2 = 0.91, Supplemental Figure 3(c)). Maintenance of biomolecule activity following covalent immobilization was tracked by evaluating the effect of a linear gradient of PDGF-BB within AL and PS array scaffolds on MSC metabolic activity (Figure 4(b)). We observed a positive correlation between MSC metabolic activity and increasing PDGF-BB content in both the AL and PS groups. As expected we also saw higher activity in the larger pore PS groups, most likely due to improved nutrient biotransport associated with increased pore size.[16, 19]
Figure 4.
(a) Benzophenone photolithography was used to create a linear gradient of covalently-bound PDGF-BB across discrete nodes of a single array as a function of UV exposure time. Alternatively, discrete patterns of biomolecular signals (stripes, squares) can also be created within individual array nodes. Scale bars: 500 μm. Green channel: AlexaFluor® 647 anti-rabbit secondary antibody, blue channel: collagen autofluorescence. (b) PDGF-BB gradient directly incorporated across the array scaffolds drives increased MSC metabolic activity within both AL and PS groups. The larger pore PS scaffolds support higher levels of metabolic activity. ^: significantly higher than AL group at gradient position, *: significantly higher than gradient position 1. (c) BMP-2 peptide gradient incorporated across the array induces in a linear increase in osteocalcin expression (7 days) with (d) limited changes in MSC number (proliferation).
Finally, we used biomolecular gradients across the chip to induce specific biological responses. Here, a gradient of a modular peptide mimic of bone morphogenetic protein 2 (BMP-2)[20] was created to enhance MSC osteogenic differentiation across the array. MSC-seeded array scaffolds were cultured in osteogenic induction media for 7 days and then fixed, permeabilized, and incubated with an antibody against osteocalcin, a marker of osteogenesis. The osteocalcin antibody was then tagged with a fluorescently-labeled secondary antibody with osteocalcin-related fluorescent signal quantified directly within the scaffold nodes via a microplate reader. Cell number was quantified in parallel via Hoechst counterstaining. Notably, results demonstrated that increasing amounts of BMP-2 peptide incorporation resulted in trends towards increasing osteocalcin expression (Figure 4(c)) with little corresponding change in cell number (Figure 4(d)). This finding shows that biomolecules incorporated into scaffold arrays via gradient maker technology maintain activity and can elicit a specific biological response. Ongoing efforts will use this platform for monitoring of live cell behavior (proliferation, differentiation, etc.) in situ within the array setup using fluorescent reporters.
Strategies for combinatorial evaluation of biophysical and biochemical regulators of cell behavior are critical for the design of new biomaterial platforms. This work outlines a scalable approach to fabricate arrays of naturally-derived, three-dimensional collagen scaffolds while retaining the capacity to independently tune pore size, pore anisotropy, and the presentation of matrix or biomolecular cues node-to-node. Given recent findings from our lab that selective modification of CG scaffold structural properties can be used induce multi-lineage MSC differentiation in the context of orthopedic insertion repair,[21] but that incorporation of combinations of growth factors can lead to unexpected cell phenotypes,[10, 22] array-based platforms such as that described here may have significant potential for rapidly optimizing biomaterials for regenerative medicine applications. Further, this approach should be adaptable to the freeze-drying fabrication of other biopolymer systems and can be combined with existing patterning methods including photolithography and affinity-binding. Arrays were created with the geometry of a 96-well plate to enable integration with standard well-plate assay technologies. Eight and twelve node arrays were chosen for this study to demonstrate the capacity to create user-defined patterns of structural and biomolecular signals across a natural polymer-derived scaffold array. However, the flexibility of lyophilization-based fabrication suggests ease of scale-up to a wider range of array geometries (e.g., full 96 or 384 node arrays) in the future. Gradient maker technology was integrated to facilitate reproducible creation of linear biomolecular gradients across microstructurally-distinct nodes of a single CG scaffold array. We subsequently used gradients of biomolecular factors to drive cell proliferation and to regulate MSC fate. Scaffold arrays allow presentation of defined combinations of signals while tracing cell response node-to-node without having to physically remove discrete nodes from the array, hence facilitating parallel analysis of large numbers of scaffold specimens. Together, the material system described here should prove as a useful platform technology to aid studies attempting to decode the synergies in both nature and kinetics of microenvironmental factors necessary to regulate cell activity across a diverse range of tissue engineering and regenerative medicine applications.
Experimental Section
Extended procedures can be found in the Supporting Information.
Array scaffold fabrication
Scaffold arrays were fabricated in situ via freeze-drying. Arrays of wells (6.5 mm diameter; 96 well-plate geometry) were machined through polysulfone sheets (2 mm thick) that were subsequently mounted on removable bases made from aluminum and polysulfone (kaluminum/kpolysulfone ≈ 850). For some experiments, a monolithic array with 12 nodes and a single aluminum base was used. CG scaffolds within the array were composed of a suspension of type I collagen and chondroitin sulfate as described extensively[8, 23] while CGCaP suspension was derived from type I collagen, chondroitin sulfate, and calcium salts in phosphoric acid.[24] Suspension was pipetted into each node and a full scaffold array was fabricated via freeze-drying the suspension at a final temperature of −10°C via isotropic (constant) cooling or anisotropic (directional) cooling.
Cell culture within array scaffolds
Human bone marrow-derived mesenchymal stem cells (MSCs) were purchased from Lonza (Walkersville, Maryland) and cultured in low glucose DMEM with MSC-validated FBS (Invitrogen, Carlsbad, CA) and antibiotics (Invitrogen). MSCs were re-suspended at a concentration of 5 × 104 per 20 μL media respectively and seeded onto scaffold nodes using a previously validated static seeding method.[15]
Fluorescence quantification within array scaffolds via microplate reader
To quantify cell number on-chip, MSCs were stained with CellTracker™ Green CMFDA (Invitrogen) and added either directly onto array scaffolds, onto scaffolds placed within a 96-well plate, or in suspension in a 96-well plate. The array was placed on an inverted 96-well plate lid for reading on the plate reader. The average fluorescence for each sample was determined on-chip by taking 9 measurements at different locations in each node of the array (or individual well of the 96 well plate for control experiments), to account for heterogeneities in the scaffold groups. Subsequent analysis of cell activity within the arrays was performed similarly, though with different fluorescent antibodies.
Gradient scaffold fabrication
Scaffolds with linear biomolecular gradients were created using a gradient maker (CBS Scientific, San Diego, CA). The CG suspension was mixed with the biomolecule to be patterned then loaded into the gradient maker, with either CG suspension or CG suspension with a second biomolecule of interest loaded into the opposite chamber on the gradient maker. The linear mixture of the two suspensions created by the gradient maker was metered into the array using a mini-peristaltic pump (CBS Scientific), then freeze-dried as previously described.
Benzophenone photolithography
Benzophenone photolithography was used to localize biomolecules of interest within discrete nodes of the array, using methods previously described.[11] Briefly, the scaffold array was soaked in a solution of benzophenone-4-isothiocyanate in DMF to immobilize benzophenone groups to amines on the CG scaffold. Scaffolds were soaked in a PDGF-BB solution prior to patterning via UV irradiation (351.1–363.8, 20 mW/cm2) and thoroughly rinsed after patterning to remove non-immobilized protein.
Statistical analysis
One-way analysis of variance (ANOVA) followed by Tukey's HSD post-hoc test was performed on pore size and MSC metabolic activity data sets. Statistical significance was set at p < 0.05. At minimum of n = 3 independent scaffolds per group were used for all analyses. Error is reported as the standard error of the mean unless otherwise noted.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the SCS machine shop for construction of array molds and Prof. William Murphy (University of Wisconsin) for providing the BMP-2 peptide. This material is based upon work supported by the National Science Foundation under Grant DMR 1105300. Research reported in this publication was supported by NIH R03 AR062811 and R21 AR063331. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We are grateful for additional funding for this study provided by the Chemistry-Biology Interface Training Program NIH NIGMS T32GM070421 (SRC), NSF IGERT: Training the Next Generation of Researchers in Cellular & Molecular Mechanics and BioNanotechnology under Grant 0965918 (DWW), the Chemical and Biomolecular Engineering Dept. (BAH), and the Institute for Genomic Biology (BAH) at the University of Illinois at Urbana-Champaign.
Footnotes
Supporting Information Supporting Information is available online from the Wiley Online Library or from the author.
Literature Cited
- [1].Anderson DG, Levenberg S, Langer R. Nat. Biotechnol. 2004;22:863. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]; Flaim CJ, Teng D, Chien S, Bhatia SN. Stem Cells Dev. 2008;17:29. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]; Cosson S, Allazetta S, Lutolf MP. Lab Chip. 2013. [DOI] [PubMed] [Google Scholar]; Qi H, Du Y, Wang L, Kaji H, Bae H, Khademhosseini A. Adv Mater. 2010;22:5276. doi: 10.1002/adma.201002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nakajima M, Ishimuro T, Kato K, Ko IK, Hirata I, Arima Y, Iwata H. Biomaterials. 2007;28:1048. doi: 10.1016/j.biomaterials.2006.10.004. [DOI] [PubMed] [Google Scholar]; Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP. Nat Methods. 2011;8:949. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]; Woodruff K, Fidalgo LM, Gobaa S, Lutolf MP, Maerkl SJ. Nat Methods. 2013 doi: 10.1038/nmeth.2473. [DOI] [PubMed] [Google Scholar]
- [3].Yang Y, Bolikal D, Becker ML, Kohn J, Zeiger DN, Simon CG. Advanced Materials. 2008;20:2037. [Google Scholar]
- [4].Chatterjee K, Lin-Gibson S, Wallace WE, Parekh SH, Lee YJ, Cicerone MT, Young MF, Simon CG., Jr. Biomaterials. 2010;31:5051. doi: 10.1016/j.biomaterials.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yannas IV, Lee E, Orgill DP, Skrabut EM, Murphy GF. Proc Natl Acad Sci U S A. 1989;86:933. doi: 10.1073/pnas.86.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harley BA, Spilker MH, Wu JW, Asano K, Hsu HP, Spector M, Yannas IV. Cells Tissues Organs. 2004;176:153. doi: 10.1159/000075035. [DOI] [PubMed] [Google Scholar]; Yannas IV. Tissue and Organ Regeneration in Adults. Springer; New York: 2001. [Google Scholar]
- [6].Harley BA, Freyman TM, Wong MQ, Gibson LJ. Biophys. J. 2007;93:2911. doi: 10.1529/biophysj.106.095471. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harley BAC, Kim HD, Zaman MH, Yannas IV, Lauffenburger DA, Gibson LJ. Biophys. J. 2008;95:4013. doi: 10.1529/biophysj.107.122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Caliari SR, Harley BAC. Biomaterials. 2011;32:5330. doi: 10.1016/j.biomaterials.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].O'Brien FJ, Harley BA, Yannas IV, Gibson L. Biomaterials. 2004;25:1077. doi: 10.1016/s0142-9612(03)00630-6. [DOI] [PubMed] [Google Scholar]
- [9].Murphy CM, Haugh MG, O'Brien FJ. Biomaterials. 2010;31:461. doi: 10.1016/j.biomaterials.2009.09.063. [DOI] [PubMed] [Google Scholar]
- [10].Caliari SR, Harley BA. Tissue Eng Part A. 2013;19:1100. doi: 10.1089/ten.tea.2012.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Martin TA, Caliari SR, Williford PD, Harley BA, Bailey RC. Biomaterials. 2011;32:3949. doi: 10.1016/j.biomaterials.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Riblett BW, Francis NL, Wheatley MA, Wegst UGK. Advanced Functional Materials. 2012;22:4920. [Google Scholar]
- [13].Yan S, Zhang Q, Wang J, Liu Y, Lu S, Li M, Kaplan DL. Acta Biomater. 2013;9:6771. doi: 10.1016/j.actbio.2013.02.016. [DOI] [PubMed] [Google Scholar]
- [14].Ho M-H, Kuo P-Y, Hsieh H-J, Hsien T-Y, Hou L-T, Lai J-Y, Wang D-M. Biomaterials. 2004;25:129. doi: 10.1016/s0142-9612(03)00483-6. [DOI] [PubMed] [Google Scholar]
- [15].O'Brien FJ, Harley BA, Yannas IV, Gibson LJ. Biomaterials. 2005;26:433. doi: 10.1016/j.biomaterials.2004.02.052. [DOI] [PubMed] [Google Scholar]
- [16].Weisgerber DW, Kelkhoff DO, Caliari SR, Harley BAC. J Mech Behav Biomed Mater. 2013;28:26. doi: 10.1016/j.jmbbm.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jungreuthmayer C, Jaasma MJ, Al-Munajjed AA, Zanghellini J, Kelly DJ, O'Brien FJ. Med Eng Phys. 2009;31:420. doi: 10.1016/j.medengphy.2008.11.003. [DOI] [PubMed] [Google Scholar]
- [18].Schwartz AG, Pasteris JD, Genin GM, Daulton TL, Thomopoulos S. PLoS ONE. 2012;7:e48630. doi: 10.1371/journal.pone.0048630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].O'Brien FJ, Harley BA, Waller MA, Yannas IV, Gibson LJ, Prendergast PJ. Technol Health Care. 2007;15:3. [PubMed] [Google Scholar]
- [20].Lee JS, Wagoner-Johnson A, Murphy WL. Angew Chem Int Ed Engl. 2009;48:6266. doi: 10.1002/anie.200901618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Caliari SR, Harley BAC. Advanced healthcare materials. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Caliari SR, Harley BAC. Tissue Eng A. 2014 [Google Scholar]; Caliari SR, Harley BAC. Biomaterials. 2011;32:5330. doi: 10.1016/j.biomaterials.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yannas IV, Lee E, Orgill DP, Skrabut EM, Murphy GF. Proc Natl Acad Sci USA. 1989;86:933. doi: 10.1073/pnas.86.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harley BA, Lynn AK, Wissner-Gross Z, Bonfield W, Yannas IV, Gibson LJ. J Biomed Mater Res A. 2010;92:1066. doi: 10.1002/jbm.a.32361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.