Abstract
Background
Risk prediction models that incorporate biomarkers and clinicopathologic variables may be used to improve decision-making post radical prostatectomy (RP). We compared two previously validated post-RP classifiers—the Cancer of the Prostate Risk Assessment post-Surgical (CAPRA-S) and the Decipher genomic classifier (GC)—to predict prostate cancer-specific mortality (CSM) in a contemporary cohort of RP patients.
Objective
To evaluate the combined prognostic ability of CAPRA-S and GC to predict CSM.
Design, Setting and Participants
A cohort of 1,010 patients at high risk of recurrence post-RP was treated at Mayo Clinic between 2000–06. High-risk was defined by any of: pre-operative PSA >20ng/mL, pathological Gleason score ≥8 or stage pT3b. A case-cohort random sample identified 225 patients (cases defined as patients who experienced CSM), among whom CAPRA-S and GC could be determined for 185.
Outcome Measurements and Statistical Analysis
The scores were evaluated individually and in combination using concordance (c)-index, decision curve analysis, re-classification, cumulative incidence, and Cox regression for prediction of CSM.
Results and Limitations
Among 185 men, 28 experienced CSM. The c-index for CAPRA-S and GC were 0.75 (95% CI 0.65–0.84) and 0.78 (95% CI 0.68–0.88), respectively. GC showed higher net-benefit on decision curve analysis but a score combining CAPRA-S and GC did not improve AUC after optimism-adjusted bootstrapping. In 82 patients stratified to high-risk based on CAPRA-S score ≥6, GC scores were likewise high-risk for 33, among whom 17 had CSM events. GC reclassified the remaining 49 men as low to intermediate-risk; among these men 3 CSM events were observed. In multivariable analysis, GC and CAPRA-S as continuous variables were independently prognostic of CSM, with hazard ratios of 1.81 (p<0.001, per 0.1 unit change in score) and 1.36 (p=0.05, per one unit change in score). When categorized into risk groups, the multivariable HR for high CAPRA-S scores (≥6) was 2.36 (p=0.04), and 11.26 (p<0.001) for high GC scores (≥0.6). For patients with both high GC and CAPRA-S scores, cumulative incidence of CSM was 45% at 10 years. The study is limited by its retrospective design.
Conclusions
Both GC and CAPRA-S were significant independent predictors of CSM. GC was shown to re-stratify many men classified as high-risk based on CAPRA-S ≥6 alone. Patients with both high GC and CAPRA-S risk scores were at markedly elevated post-RP risk for lethal prostate cancer. If validated prospectively, these findings suggest that integration of a genomic-clinical classifier may enable better identification of those post-RP patients who should be considered for more aggressive secondary therapies and clinical trials.
Introduction
Accurate risk stratification of prostate cancer, both at time of diagnosis and at other decision points, is essential to identify those at high risk of prostate cancer-specific mortality (CSM). These patients are most likely to benefit from aggressive multimodal therapy, and it is important to distinguish them from the larger majority of patients who are cured by surgery or are otherwise at low risk of CSM, who may be spared the potential impact of additive treatments. The Cancer of the Prostate Risk Assessment post-Surgical (CAPRA-S) score was developed in a multi-institutional, community-based cohort to predict biochemical recurrence and CSM following radical prostatectomy (RP) by incorporating pre-operative prostate specific antigen (PSA) levels and pathologic information into a straightforward, easy to use calculation of postoperative patient risk1. It has also been validated in another multi-institutional, socio-demographically and clinically diverse cohort, which confirmed its ability to predict both recurrence and CSM2.
Over the last decade, many studies have tried to address the unmet clinical need for predicting aggressive prostate cancer using genomic information3–7. The Decipher® prostate cancer genomic classifier (GC) risk prediction model was developed by investigators at Mayo Clinic and GenomeDx Biosciences to predict with high specificity, early metastasis post-RP4. Using oligonucleotide-microarray expression profiling of about 1.4 million markers in 545 tumors, machine learning algorithms were used to discover and validate a 22-marker gene expression signature of metastasis. The GC model measures the activity of genes implicated in proliferation, cell migration and adhesion, tumor motility, androgen-signaling and immune system evasion.8. In blinded validation studies in prospectively accrued cohorts9, the GC model demonstrated improved performance over any individual clinicopathologic variable or clinical prediction model for clinical metastasis (confirmed by radiographic bone and CT imaging) in post-RP10 and post-BCR11 patient cohorts.
In this study, we further examined the relationship between the CAPRA-S and GC scores for predicting CSM, from the time of RP. We aimed to determine whether integrated genomic and clinical risk prediction models may further improve risk prediction than either model alone.
Materials and Methods
Patient Population
Subjects were identified from a population of 1,010 men prospectively enrolled in the Mayo Clinic Department of Urology RP registry for prostatic adenocarcinoma from 2000–2006. This population was clinically high-risk, as defined by pre-operative prostate-specific antigen (PSA) level >20ng/mL, pathological Gleason score ≥8 or stage pT3b. Patients that received neo-adjuvant therapy or were diagnosed with metastatic disease or failed to achieve PSA nadir after surgery were excluded. Clinical staging for patients with D’Amico high-risk disease or preoperative PSA≥10 ng/mL underwent cross sectional imaging either with CT or MRI and bone scan to rule out the presence of metastatic disease pre-surgery. Data were collected from patients selected using a case-cohort approach, as this design allows inference measures (e.g., survival estimates, hazards) about the whole cohort without requiring assessment of all 1,010 patients12,13. The case-cohort design is most useful in analyzing time to failure in a large cohort in which the failure event is rare. The case cohort design included all CSM events and a random sample of the full cohort. Of the 1,010 men, 28 (3.0%) were documented to have died from prostate cancer (at median follow-up 6.9 years). A 20% random sample of the entire cohort was selected for the analysis, including 11 patients with CSM (cases). The remaining 17 cases, who were not selected by random sampling were also included for analyses (Figure S1).
Tissue and RNA Processing
Following histopathological review, total RNA was extracted and amplified from 4 to 6 4μm formalin-fixed, paraffin embedded primary prostatic adenocarcinoma tissue sections from the nodule with the highest Gleason score. Macrodissection was used to enrich for tumor cells. RNA was extracted and hybridized to Human Exon 1.0 ST GeneChips (Affymetrix, Santa Clara, CA), which profile coding and non-coding regions of the transcriptome as described previously10. Following exclusion for tissue unavailability and microarray quality control (n=38), 187 out of the 225 patients sampled from the cohort remained with GC scores, of whom 185 had complete clinicopathologic data for estimating CAPRA-S scores.
Classifier Assessment
We compared and integrated two previously validated post-RP classifiers: CAPRA-S and GC. CAPRA-S is a postoperative risk stratification model, based on standard clinical and pathological risk factors for predicting BCR and CSM. It was developed using the Cancer of the Prostate Strategic Urologic Research Endeavor (CaPSURE) registry, and validated in the Shared Equal Access Regional Cancer Hospital (SEARCH) database. CAPRA-S scores may be grouped into three validated groups: 0–2, 3–5, ≥614,15. GC is a 0 to 1 score developed using clinical metastasis post-RP as the primary endpoint8 and subsequently validated in an independent data set for prediction of clinical metastasis10. In some analyses, GC scores were categorized using previously defined cut-offs: <0.4, 0.4–0.6, >0.6 indicating low-, intermediate-, and high-risk, respectively10.
CAPRA-S and GC were combined into an integrated genomic-clinical classifier (GCC), using the parameter estimates derived from a Cox proportional hazard model for CSM. The predicted score is normalized to have values between 0 and 1, using the minimum and maximum predicted values that model could generate. It should be stressed that neither GC nor CAPRA-S was individually trained or refined, in this patient population. The GCC model was validated using a bootstrap valuation method that estimates the optimism of the area under the receiver-operating characteristic curve (AUC) in the training set to adjust the AUC for over-fitting, and estimate the expected AUC in a potential validation sets16. A total of 10,000 bootstrapped samples were run to determine the optimism, and the average optimism from those runs is used to adjust the AUC.
Statistical Analysis
The primary endpoint in this analysis was CSM. Discrimination was measured by the concordance (c) index. Survival analyses were weighted12,17 to estimate parameters in the full cohort. Decision curve analysis was used to estimate the net benefit across a range of threshold probabilities for CSM at 5 years post-RP18. Univariable and multivariable Cox proportional hazards models for case-cohort study designs19 were used to estimate hazard ratios for both continuous and categorical predictive scores. An adaptation of Firth’s method towards the Cox model to reduce the potential bias associated with small sample size was also used for univariable and multivariable analysis23,24. Interaction effects between prediction models and adjuvant therapy were evaluated by comparing the multivariable Cox model with interaction terms to the model without the interaction terms using the likelihood ratio test. Log-rank tests were used to assess the significance of differences in the Kaplan-Meier curves of patients in different risk groups. Cumulative incidence curves were constructed using Fine-Gray competing risks analysis to adjust for death by other causes20. Median follow-up times are estimates using the censoring distribution21. Statistical analyses were performed in R v3.0 and all statistical tests were two-sided using a 5% significance level.
Results
Performance of genomic and clinicopathologic risk models for predicting CSM
GC scores were available for 187 patients (28 cases; median follow-up 6.4 years). Complete clinical data required to calculate CAPRA-S scores was available for 185 patients (Table 1). Patients in this high-risk cohort experienced CSM with a median 4.8 years post-RP (interquartile range 3.2–6.6 years). Medians and ranges for CAPRA-S and GC were 5 (2 – 12) and 0.37 (0.01 – 0.99), respectively.
Table 1. Characteristics high-risk post-RP patients.
Adjuvant therapies (hormone or radiation) were those that were administered within 90 days of RP. Salvage therapies are those that were administered beyond 90 days of RP; although in this population the majority of patients had salvage therapy after BCR.
| Patients Characteristics | N | CSM N (group %) |
No-CSM N (group %) |
Pearson’s Chi-squared, Fisher exact or Wilcoxon (P) |
|---|---|---|---|---|
| Study Cohort | 185 | 28 | 157 | |
| Age | 0.93 | |||
| Median | 63 | 64 | 63 | |
| Range | (46, 78) | (48, 78) | (46, 78) | |
| Pre-operative Prostate-specific Antigen | 0.57 | |||
| <10 ng/mL | 103 | 14(50) | 89(57) | |
| 10–20 ng/mL | 51 | 9(32) | 42(27) | |
| >20 ng/mL | 31 | 5(18) | 26(16) | |
| Pathological Gleason Score | 3.14E-05 | |||
| ≤6 | 15 | 0(0.0) | 15(10) | |
| 7 | 91 | 6(21) | 85(54) | |
| ≥8 | 79 | 22(79) | 57(36) | |
| Seminal Vesicle Invasion | 61 | 15(54) | 46(30) | 0.02 |
| Positive Surgical Margin | 103 | 14(50) | 89(57) | 0.65 |
| Extra-capsular Extension | 79 | 20(72) | 59(38) | 0.002 |
| Lymph Node Involvement | 26 | 8(29) | 18(12) | 0.03 |
| CAPRA-S | 0.002 | |||
| <3 | 1 | 0(0.0) | 1(0.6) | |
| 3 – 5 | 102 | 8(29) | 94(60) | |
| >5 | 82 | 20(71) | 62(39.4) | |
| Decipher | 9.63E-06 | |||
| <0.4 | 100 | 7(25) | 93(59) | |
| 0.4–0.6 | 40 | 3(11) | 37(24) | |
| >0.6 | 45 | 18(64) | 27(17) | |
| Adjuvant1 Radiation Therapy | 18 | 2(7) | 16(10) | 1 |
| Adjuvant1 Androgen Deprivation Therapy | 39 | 2(7) | 37(24) | 0.08 |
| Salvage2 Radiation Therapy | 55 | 15(54) | 40(26) | 0.006 |
| Salvage2 Androgen Deprivation Therapy | 57 | 25(89) | 32(20) | 3.03E-12 |
| Follow-Up Time | 0.75 | |||
| Median (years) | 6.44 | 4.81 | 5.83 | |
| Range | (0.17, 10.15) | (1.48, 9.64) | (0.17, 10.15) |
Adjuvant therapy administered within 90 days post-RP,
Salvage therapy administered anytime after 90 days post-RP, majority after BCR
The AUC of CAPRA-S and GC as prediction models for prostate cancer specific mortality in comparison to individual clinicopathologic variables was compared using ROC analysis (Table S1). CAPRA-S and GC both have the highest AUCs, of 0.75 (95% CI 0.55–0.84) and 0.78 (95% CI 0.68–0.87), respectively. In univariable analyses, CAPRA-S, GC, pathological Gleason score, lymph node status, extracapsular extension, seminal vesicle and adjuvant androgen deprivation therapy (ADT) were statistically significant predictors of CSM (Table S3). In a multivariable analysis after adjusting for adjuvant treatment, the hazard ratio (HR) of CAPRA-S as a continuous variable was 1.37 for every unit increase (p=0.01), and the HR for GC was 1.84 for every 0.1 unit increase (p<0.001). No significant interaction terms between the prediction models and adjuvant treatment were observed (p>0.05, data not shown). Using a penalized approach to the Cox model appropriate for the small sample size and event rates in this study found similar results (Table 2). In exploratory analyses, when scores were dichotomized based on previously reported high-risk group cut-points, multivariable analysis showed that patients with high CAPRA-S scores (≥6) had an HR of 2.5 (p=0.07), and high GC scores (≥0.6) had an HR of 12.2 (p<0.001) for CSM (Table S3). The corresponding hazard ratios from the penalized model (Table 2) were 2.4 for high CAPRA-S and 11.3 for high GC. Kaplan-Meier plots using previously reported cut-points for the models show significant differences in CSM-free survival for CAPRA-S and GC risk groups (Figure S2A,B). Survival differences were virtually unchanged when excluding patients that received any form of adjuvant therapy (Figures S2C,D).
Table 2. Univariable and Multivariable Hazard Ratios of CAPRA-S, GC and standard clinicopathologic variables via Firth’s penalized likelihood method.
23.
CAPRA-S was included in the multivariable analysis as it is essentially a summary of all those variables. CAPRA-S is compared to GC separately in multivariable settings. Hazard ratios for GC are reported per 10% unit increase. Due to the high-risk nature of this cohort, pathologic GS is dichotomized (≥8) because there were too few GS 6 tumors to use it as a reference group. Estimation of the effect sizes in the multivariable model was performed using an adaptation of Firth’s method towards the Cox model to reduce the potential bias associated with small sample size24. Confidence intervals and p-values are unadjusted for the case-cohort design. Estimates of hazard ratios may be compared to those listed in Table S3.
| Univariable Analysis Using Firth’s Penalized Method** | |||
|---|---|---|---|
| Variable Treatment | Hazard Ratio (95 %CI) | p-value | |
| GC | 0.1 unit increments | 1.86 (1.54 – 2.29)** | p<0.001 |
| CAPRA-S | 1 unit increments | 1.45 (1.25 – 1.68)** | p<0.001 |
| Pathologic Gleason Score | ≥8 vs. 6–7 | 6.39 (2.80 – 16.80)** | p<0.001 |
| Lymph Nodes | Present | 3.70 (1.56 – 8.07)** | 0.004 |
| Extra Capsular Extension | Present | 3.21 (1.49 – 7.57)** | 0.003 |
| Seminal Vesicle Invasion | Present | 2.39 (1.14 – 5.04)** | 0.02 |
| Positive Margins | Present | 0.62 (0.30 – 1.31)** | 0.21 |
| Pre-operative PSA | Log2 | 1.00 (0.99 – 1.02)** | 0.33 |
| Adjuvant Therapy | Radiation | 0.74 (0.15 – 2.27)** | 0.63 |
| Androgen Deprivation | 2.80 (1.34 – 5.91)** | 0.006 | |
| Multivariable Analysis Using Firth’s Penalized Method** | |||
| As continuous/ordinal variables | |||
| Hazard Ratio (95 %CI) | p-value | ||
| GC | 0.1 unit increments | 1.81 (1.48 – 2.25)** | p<0.001 |
| CAPRA-S | 1 unit increments | 1.36 (1.13 – 1.65)** | 0.001 |
| Adjuvant Therapy | Radiation | 0.18 (0.03 – 0.70)** | 0.01 |
| Androgen Deprivation | 1.15 (0.48 – 2.68)** | 0.75 | |
| As risk Categories | |||
| Hazard Ratio (95 %CI) | p-value | ||
| GC | 0.4–0.6 (ref: <0.4) | 1.09 (0.26 – 3.77)** | 0.9 |
| >0.6 (ref: <0.4) | 11.26 (4.69 – 30.37)** | p<0.001 | |
| CAPRA-S | >5 | 2.36 (1.06 – 5.68)** | 0.04 |
| Adjuvant Therapy | Radiation | 0.56 (0.11 – 1.80)** | 0.36 |
| Androgen Deprivation | 1.55 (0.72 – 3.36)** | 0.26 | |
Confidence Intervals and p-values were not adjusted to account for the case-cohort design
Comparison of genomic and clinicopathologic model risk groups
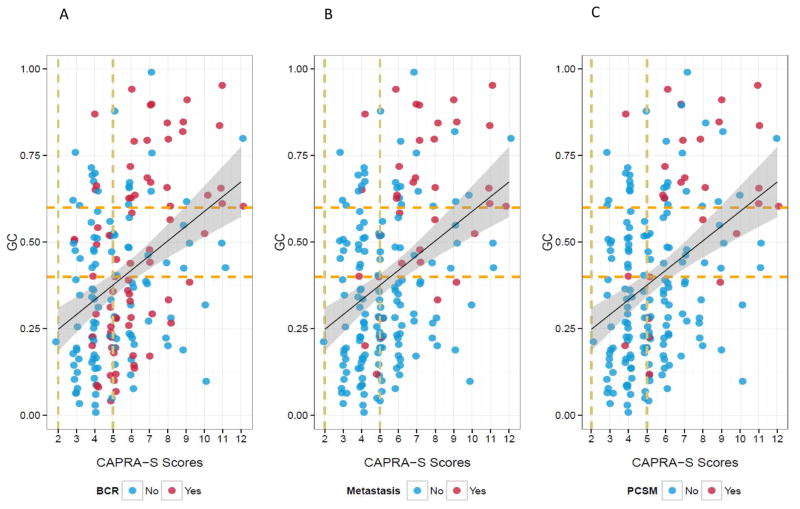
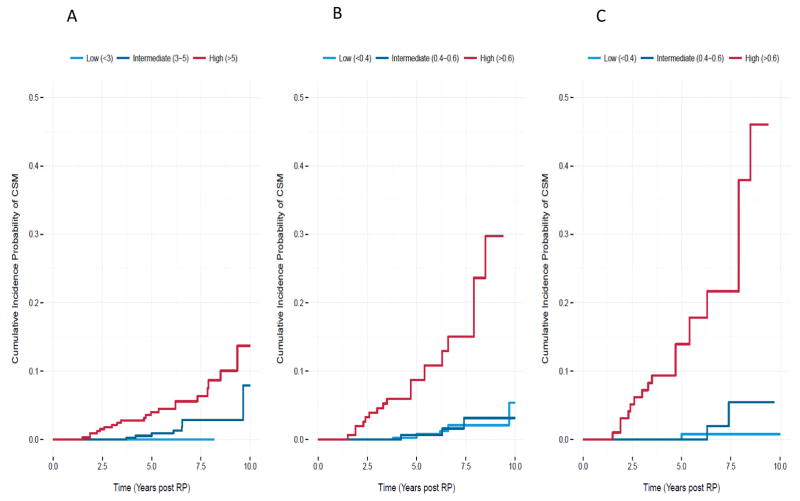
As expected, some correlation (although modest) between GC and clinical risk factors captured by CAPRA-S is observed (r-squared=0.38, p-value<0.0001) but the trend shows that patients with both high GC score and high CAPRA-S score are those most at risk to die from prostate cancer (Figure 1, Table S2). While patients with higher CAPRA-S score had multiple adverse pathologic features few that also had low GC (<0.4) scores developed metastasis or died of prostate cancer at this follow-up despite a high frequency of BCR. For the group of patients with high-risk CAPRA-S (≥6) or GC (≥0.6) scores, cumulative incidence of CSM at 10 years was estimated at 13% and 30%, respectively (Figure 2A–B). However, for patients with both high CAPRA-S and GC scores, the cumulative incidence of CSM was 45% at 10 years (Figure 2C). Further evidence that the models provide complementary information was found using Kaplan-Meier analysis of each models higher risk groups (Figure S3). Significant differences in CSM-free survival were observed when CAPRA-S risk groups were used to re-stratify GC > 0.6 (p=0.005) and GC risk groups to re-stratify CAPRA-S > 5 (p<0.001).
Figure 1. Agreement between GC and CAPRA-S scores.
The distribution of CAPRA-S and GC scores for A) Biochemical Recurrence, B) Clinical Metastasis, and C) Cancer Specific Mortality. The dashed vertical lines show the boundaries for low (≤2), intermediate (3–5) and high-risk (≥6) risk groups for CAPRA-S. The dashed horizontal lines mark the low (<0.4), intermediate (0.4 – 0.6) and high (≥0.6) GC scores as described previously. The solid black line and surrounded by a gray shadow demonstrates the regression line and its 95% confidence interval.
Figure 2.
Cumulative incidence of CSM for A) CAPRA-S, B) GC, and C) CAPRA-S high-risk stratified by GC. The cumulative probability of CSM increases with the CAPRA-S high risk when it is further stratified by GC.
The integrated genomic and clinicopathologic risk model identifies a very high risk subset of RP patients
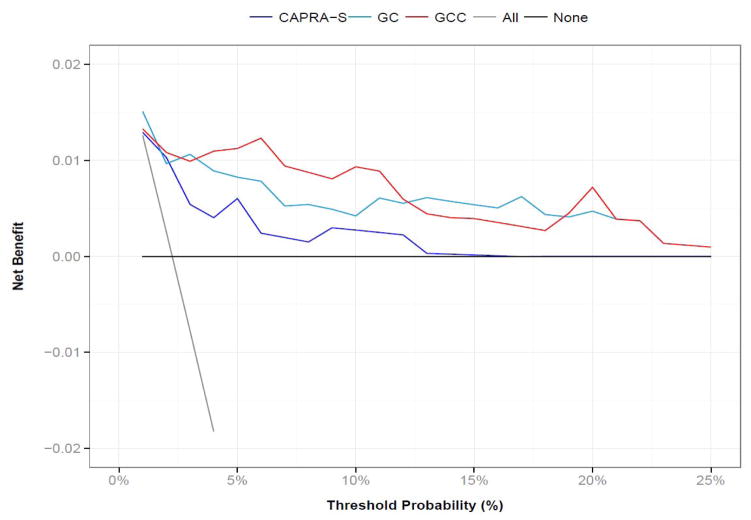
Next, GC and CAPRA-S were combined using Cox regression to generate an integrated genomic-clinical classifier (GCC). The formula for the GCC based on the Cox model coefficients was 0.21*CAPRA-S + 5.55*GC, The AUC of integrated model reached 0.80 (95% CI 0.69–0.90). However, after cross-validation optimism adjustment, the AUC was 0.78, which was not significantly different than the individual models, and due to limited number of events in this sample (the optimism adjusted AUC was observed to vary between 0.6 and 1.0 over 10,000 bootstrapped iterations). Decision curve analysis however indicates that modest increases in AUC could be associated with larger gains in utility (Figure 3). Compared to ‘treat none’ and ‘treat all’ scenarios (where no risk prediction model is employed) to make treatment decisions across a range of CSM risk threshold probabilities, GCC had the highest overall net-benefit compared to CAPRA-S or GC alone.
Figure 3. Survival decision curve for predicting 5 years post-RP CSM.
Raw GC, CAPRA-S and GCC scores were converted into 5-year CSM probabilities before estimating net benefit. Genomic-based models demonstrate a higher net benefit.
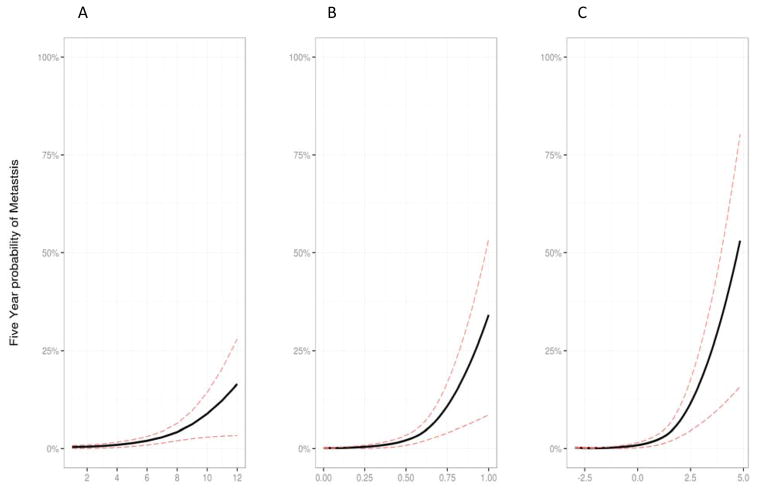
Finally, the 5-year risk probability of CSM as calculated by each of the three risk prediction models for individual patient scores in this study population were evaluated (Figure 4). For all three models, the probability of CSM at 5 years is virtually zero for patients with low risk scores but rises dramatically for higher risk scores. The 5-year probability of CSM ranged between 0.0 – 17% for CAPRA-S (Figure 4A), 0.0 – 34% for GC (Figure 4B) and 0.0 – 53% for the integrated GCC model (Figure 4C). These results further highlight the potential impact of combining genomic and clinical risk prediction models. A wider range of patient scores with the integrated GCC model had a lower predicted probability of CSM and the exponential phase rose more steeply indicating a subset of patients at markedly increased risk of CSM.
Figure 4. Prediction model for likelihood of CSM 5 years post-operatively.
A) CAPRA-S, B) GC, and C) GC + CAPRA-S. The integrated model has a higher risk probability than achieved with either model alone.
Discussion
Integration of tumor genomics into clinical practice for use in individualized patient risk prediction models holds great promise to improve management of high-risk prostate cancer. This investigation follows previous reports on the validation of CAPRA-S15 and GC10, with results here demonstrating that an integrated risk model combining GC and CAPRA-S provides improved risk prediction over either model alone. We have shown in this study both GC and CAPRA-S can accurately predict CSM, and while these risk scores are only modestly correlated, they appear to provide complementary information that can be used for identification of a subset of post-RP patients at extremely high risk for death from prostate cancer. Both CAPRA-S and GC can be determined immediately following RP, in contrast to measures based on PSA kinetics, and therefore these tools can be used to predict clinically significant events before disease progression can be detected clinically, radiographically, or in many cases even biochemically.
The results of this study demonstrate that in an at risk-population (i.e., patients with adverse pathology) individuals with both high GC and CAPRA-S scores represent a group that are at markedly increased risk (>12 fold) of dying of their disease. The patient population sampled in this case-cohort study represents a group of surgical patients that is at considerably higher risk for treatment failure than the average man with prostate cancer treated with RP. At 5 years post-RP the population sampled for this analysis had a 33%, 8% and 2% incidence of biochemical recurrence, metastasis and CSM, respectively.
GC did appear to ‘down-classify’ a significant number of patients classified by CAPRA-S as high risk. In these patients that were classified as high risk by CAPRA-S, but low risk by GC, many experienced biochemical recurrence but not the rapid development of metastasis, and CSM observed in those patients who were classified as high risk by both models. Given that the natural history of BCR is heterogeneous and relatively few men with BCR will experience early metastasis and CSM22, these patients may represent a subset that perhaps could be observed and followed by tracking PSA kinetics.
After adjusting for adjuvant therapy in multivariable analysis, we found that CAPRA-S and GC remained independent and significant predictors of CSM. We did not find an interaction between CAPRA-S and GC with adjuvant therapy. While, the use of adjuvant therapy was not universal and reflected inherent biases among the treating physicians, this does reflect current treatment practices for high-risk prostate cancer. In the study cohort, nearly 12% of patients had positive lymph nodes and 36% received adjuvant therapy; the overall validation of GC in an adjuvant therapy-rich cohort may represent a limitation. It is noteworthy that adjuvant ADT is associated with a negative effect on survival, which is almost certainly due to non-randomized treatment assignments and resulting confounding by indication (i.e., men receiving ADT likely have worse disease beyond what is captured by the variables in the adjusted analyses). This study is limited by its retrospective nature, and without randomization of these treatments these results remain intriguing but hypothesis generating only.
Conclusion
For patients with adverse pathology after RP, outcomes vary greatly. Patients with CAPRA-S>5 and GC>0.6 were associated with a significantly increased risk of CSM in this cohort. As such, these men may benefit from additional secondary therapies, ideally in a clinical trial setting. Conversely, patients with both low CAPRA-S and GC scores had excellent CSM-free survival even after adjusting for use of adjuvant therapy in this cohort. Future studies, ideally randomized-controlled clinical trials, will be required to determine whether use of genomic and clinical risk prediction models actually improve patient cancer-specific and quality of life outcomes.
Supplementary Material
Figure S1: STARD diagram starting from the original cohort, the number selected for the case-cohort study and the final result after samples loss and quality control failure.
Figure S2: Kaplan Meier Plots: Univariable presentation of the probability of CSM for CAPRA-S risk groups (A,C), and GC risk groups (B, D). Plots C and D excluded all patients that received any adjuvant therapy.
Figure S3: Kaplan Meier Plots: Univariable presentation of the probability of CSM for A) CAPRA-S risk groups in GC high-risk group (GC > 0.6), and B) GC risk groups in CAPRA-S high-risk group (CAPRA-S > 5).
Table S1: AUCs curves for predicting CSM five years post-RP for CAPRA-S, GC and clinicopathologic variables alone. The following variables are treated as binary: extracapsular extension, seminal vesicle invasion, positive surgical margins, and positive lymph nodes. Pathologic Gleason score and CAPRA-S are treated as ordinal variables and GC is used a continuous variable.
Table S2: Reclassification of CAPRA-S risk groups by GC high and low risk designations.
Table S3: Univariable and Multivariable Hazard Ratios of CAPRA-S, GC and standard clinicopathologic variables. The analysis reported in Table 2 was repeated using a non-penalized Cox model that was adjusted for the case-cohort design.
Acknowledgments
Funding Support: NIH P50 CA91956 Mayo Prostate Cancer SPORE grant; Richard M. Schulze Family Foundation; National Research Council of Canada Industrial Research Assistance Program, Mayo Foundation For Medical Education and Research and GenomeDx Biosciences Inc.
Dr. Christine Buerki, Kasra Yousefi and Voleak Choeurng for their assistance with the analysis and preparing the manuscript (GenomeDx), Dr. Darby Thompson (EMMES Canada) and Eric Bergstralh (Mayo Depts of Urology and Statistics) for statistical and methodological assistance. The authors appreciate the advice, encouragement and support of Dr. Donald Tindall; PI of the Mayo Prostate Cancer SPORE and Dr. Lesley Esford (National Research Council Canada).
Footnotes
Financial Disclosures: ED, AC and MG are employees of GenomeDx. RJK and MC have previously received unrestricted research grants from GenomeDx. GenomeDx has filed patents on prostate cancer biomarkers including the Decipher panel in which RJB and ED are co-inventors.
References
- 1.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117(22):5039–46. doi: 10.1002/cncr.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Punnen S, Freedland SJ, Presti JC, et al. Multi-institutional Validation of the CAPRA-S Score to Predict Disease Recurrence and Mortality After Radical Prostatectomy. Eur Urol. 2013 doi: 10.1016/j.eururo.2013.03.058. [DOI] [PubMed] [Google Scholar]
- 3.Cary KC, Cooperberg MR. Biomarkers in prostate cancer surveillance and screening: past, present, and future. Ther Adv Urol. 2013;5(6):318–29. doi: 10.1177/1756287213495915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3(5):e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14(1):690. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31(11):1428–34. doi: 10.1200/JCO.2012.46.4396. [DOI] [PubMed] [Google Scholar]
- 7.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–55. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erho N, Crisan A, Vergara Ia, et al. Discovery and Validation of a Prostate Cancer Genomic Classifier that Predicts Early Metastasis Following Radical Prostatectomy. In: Creighton C, editor. PLoS One. 6. Vol. 8. 2013. p. e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100(20):1432–8. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a Genomic Classifier that Predicts Metastasis Following Radical Prostatectomy in an At Risk Patient Population. J Urol. 2013;190(6):2047–53. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross AE, Feng FY, Ghadessi M, et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic Dis. 2013 doi: 10.1038/pcan.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barlow WE, Ichikawa L, Rosner D, Izumi S. Analysis of case-cohort designs. J Clin Epidemiol. 1999;52(12):1165–72. doi: 10.1016/s0895-4356(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 13.Breslow NE, Lumley T, Ballantyne CM, Chambless LE, Kulich M. Using the whole cohort in the analysis of case-cohort data. Am J Epidemiol. 2009;169(11):1398–405. doi: 10.1093/aje/kwp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooperberg MR, Pasta DJ, Elkin EP, et al. The UCSF Cancer of the Prostate Risk Assessment (CAPRA) Score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. 2010;173(6):1938–1942. doi: 10.1097/01.ju.0000158155.33890.e7.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Punnen S, Cooperberg MR, D’Amico AV, et al. Management of Biochemical Recurrence After Primary Treatment of Prostate Cancer: A Systematic Review of the Literature. Eur Urol. 2013:1–11. doi: 10.1016/j.eururo.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 16.Steyerberg EW. Clinical Prediction Models. New York, NY: Springer New York; 2009. [DOI] [Google Scholar]
- 17.Barlow WE. Robust Variance Estimation for the Case-Cohort Design. 2013;50(4):1064–1072. [PubMed] [Google Scholar]
- 18.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Therneau TM, Li H. Computing the Cox model for case cohort designs. Lifetime Data Anal. 1999;5(2):99–112. doi: 10.1023/a:1009691327335. [DOI] [PubMed] [Google Scholar]
- 20.Fine JP, Gray RJ. A Proportional Hazards Model for the Subdistribution of a Competing Risk. J Amrican Stat Assoc. 94(446):496–509. [Google Scholar]
- 21.Korn EL. Censoring distributions as a measure of follow-up in survival analysis. Stat Med. 5(3):255–60. doi: 10.1002/sim.4780050306. [DOI] [PubMed] [Google Scholar]
- 22.Freedland SJ, Humphreys EB, Mangold La, et al. Death in patients with recurrent prostate cancer after radical prostatectomy: prostate-specific antigen doubling time subgroups and their associated contributions to all-cause mortality. J Clin Oncol. 2007;25(13):1765–71. doi: 10.1200/JCO.2006.08.0572. [DOI] [PubMed] [Google Scholar]
- 23.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80(1):27–38. [Google Scholar]
- 24.Heinze G, Schemper M. A solution to the problem of monotone likelihood in Cox regression. Biometrics. 2001;57(1):114–119. doi: 10.1111/j.0006-341x.2001.00114.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: STARD diagram starting from the original cohort, the number selected for the case-cohort study and the final result after samples loss and quality control failure.
Figure S2: Kaplan Meier Plots: Univariable presentation of the probability of CSM for CAPRA-S risk groups (A,C), and GC risk groups (B, D). Plots C and D excluded all patients that received any adjuvant therapy.
Figure S3: Kaplan Meier Plots: Univariable presentation of the probability of CSM for A) CAPRA-S risk groups in GC high-risk group (GC > 0.6), and B) GC risk groups in CAPRA-S high-risk group (CAPRA-S > 5).
Table S1: AUCs curves for predicting CSM five years post-RP for CAPRA-S, GC and clinicopathologic variables alone. The following variables are treated as binary: extracapsular extension, seminal vesicle invasion, positive surgical margins, and positive lymph nodes. Pathologic Gleason score and CAPRA-S are treated as ordinal variables and GC is used a continuous variable.
Table S2: Reclassification of CAPRA-S risk groups by GC high and low risk designations.
Table S3: Univariable and Multivariable Hazard Ratios of CAPRA-S, GC and standard clinicopathologic variables. The analysis reported in Table 2 was repeated using a non-penalized Cox model that was adjusted for the case-cohort design.